The rate and pattern of new mutation is critical to the understanding of human disease and evolution. We used extensive autozygosity in a genealogically well-defined population of Hutterites to estimate the mutation rate over multiple generations. We sequenced whole genomes from five parent-offspring trios and identified 44 segments of autozygosity. We computed the number of meioses separating each pair of autozygous alleles and validated 72 heterozygous single nucleotide variants (SNVs) from 512 Mbp of autozygous DNA providing an SNV mutation rate of 1.20×10−8 (95% confidence interval 0.89–1.43×10−8) mutations per basepair per generation. We observed a 9.5-fold increase for bases within CpG dinucleotides (9.72×10−8) and strong evidence (p = 2.67×10−4) for a paternal bias in the origin of new mutations (85% paternal). We observed a non-uniform distribution of heterozygous SNVs (both novel and known) in the autozygous segments (p = 0.001) suggestive of mutational hotspots or sites of long-range gene conversion.
Various approaches have provided a wide range of SNV mutation rate estimates (1–3×10−8 per basepair per generation). Early studies of mutation rates in humans focused on specific loci or the de novo incidence of disease 1–4. More recent studies have leveraged whole-genome sequencing (WGS) data on a total of three nuclear families to estimate de novo mutation rates for SNVs of about 1×10−8 [Ref. 5–6]. Comparative studies of chimpanzee and human genomes provided higher estimates (i.e. 2.5×10−8) but are highly contingent upon uncertainty in the number of generations since the human-chimpanzee divergence 7.
In contrast to studies focused on identifying new mutations arising in a single generation, populations with a small number of founding individuals provide an ideal resource for estimating mutation rates across a small number of generations. The Hutterites are a population of Anabaptist farmers living in the plains of the United States and Canada who descended from a small group of founders (<90 individuals). The genealogy is completely known and genome-wide single nucleotide polymorphism (SNP) genotype data have been collected from over 1400 individuals who are related to each other in a 13-generation pedigree descended from 64 founders 8–9. Due to increased levels of consanguinity, Hutterite individuals carry large segments of the genome that are autozygous or homozygous by recent decent 10. The alleles in an autozygous segment are descended from a recent common ancestor and have accumulated mutations in the generations since transmission from this individual.
We selected five Hutterite parent-offspring trios for whole-genome sequencing (WGS) where the parents in each trio are related to each other within 6–8 (mean = 6.6) meiotic transmissions (Figure 1). We performed WGS on DNA isolated from whole blood using Illumina paired-end sequencing. We generated 775 gigabases (Gbp) of sequence with an average of 13-fold coverage per individual (Supplementary Table 1). We aligned the sequencing reads for each sample to the human reference genome (NCBI build 36). We identified a total of 5.4 million SNVs based on the intersection of two different algorithms 11–12 (Supplementary Table 2). The SNP genotypes from WGS were highly concordant to SNP microarray (mean genotype concordance = 99.7%) (Supplementary Table 2).
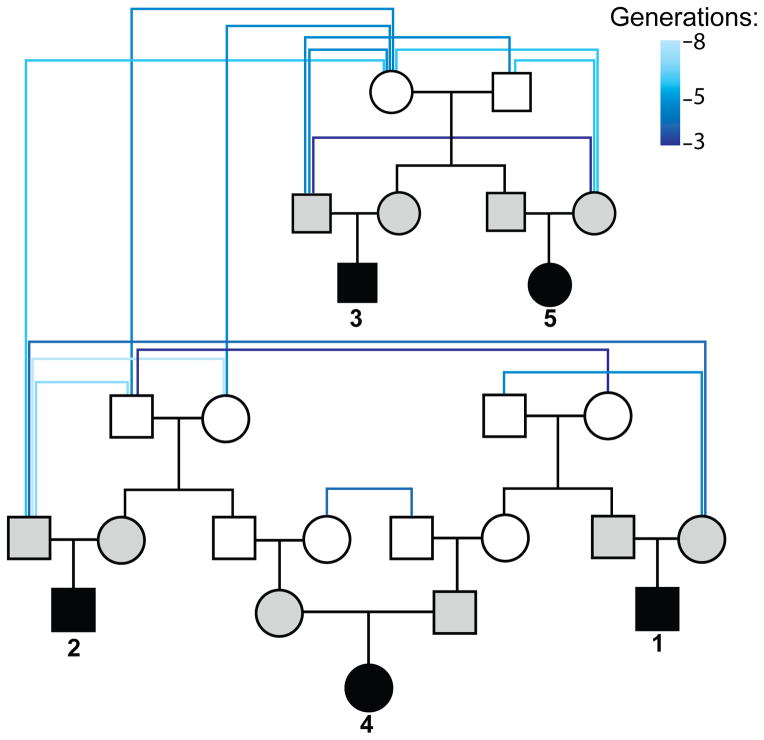
Figure 1. Relationship of sequenced individuals.
A simplified pedigree showing the relationship between the 15 sequenced individuals is shown. Children in the five trios are colored in black and their parents in gray. Founders are connected by shades of blue lines denoting the number of generations separating those individuals. For clarity, only the shortest relationships between each individual and the parents of each individual are shown.
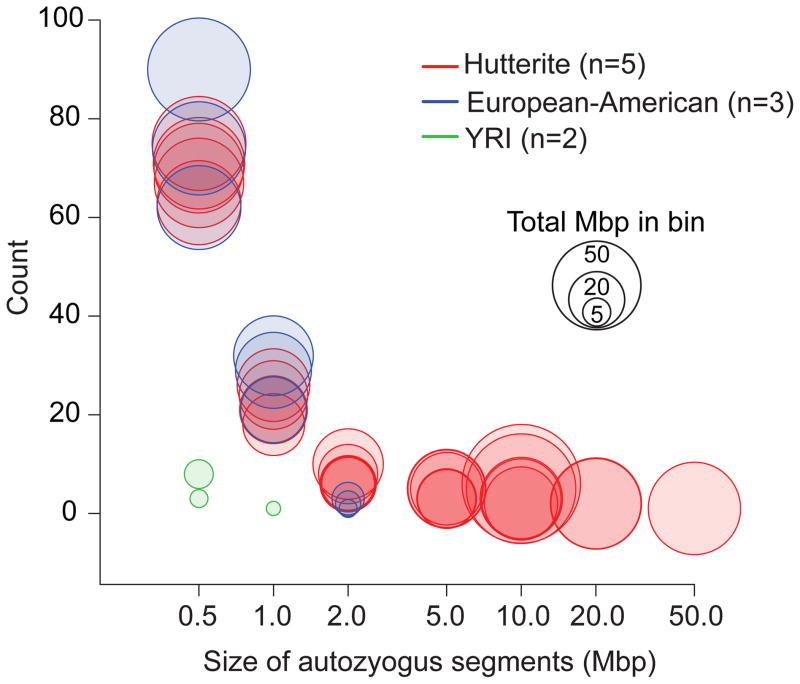
We identified extended regions of homozygosity in the offspring of the five trios and in five previously sequenced genomes (three European Americans and two Yoruba) 13 (Methods). The extent of homozygosity was correlated to the inbreeding coefficients of the Hutterite individuals (Supplementary Table 3; Supplementary Fig. 1; Supplementary Note). As expected, the five Hutterite probands showed significantly greater autozygosity (223 Mbp average per individual) than other European-American individuals (95 Mbp) or the Yoruba individuals (4 Mbp) (Figure 2; Supplementary Table 3). Although the amount of short homozygous segments was similar, we observed a 33-fold increase in autozygous basepairs in segments greater than 2 Mbp in the Hutterite individuals compared to the other European-American individuals (Figure 2; Supplementary Note). We further refined and validated segments that were longer than 5 Mbp by comparing to SNP microarray data for the same samples 10,15 to obtain a final list of 44 regions of autozygosity (6–12 segments per individual; 5–54 Mbp in length). We restricted subsequent analyses to these 512 Mbp of autozygous DNA (Supplementary Fig 2; Supplementary Table 4).
Figure 2. Elevated autozygosity in the Hutterite individuals.
Autozygous segments were binned by size for the five Hutterite individuals, three European-American individuals, and two Yoruba individuals. The x-axis represents bins of autozygous segment sizes and the y-axis is the number of segments in each bin. Each individual is represented by a “bubble” for each bin where the size of the bubble represents the total amount of genomic sequence in that bin.
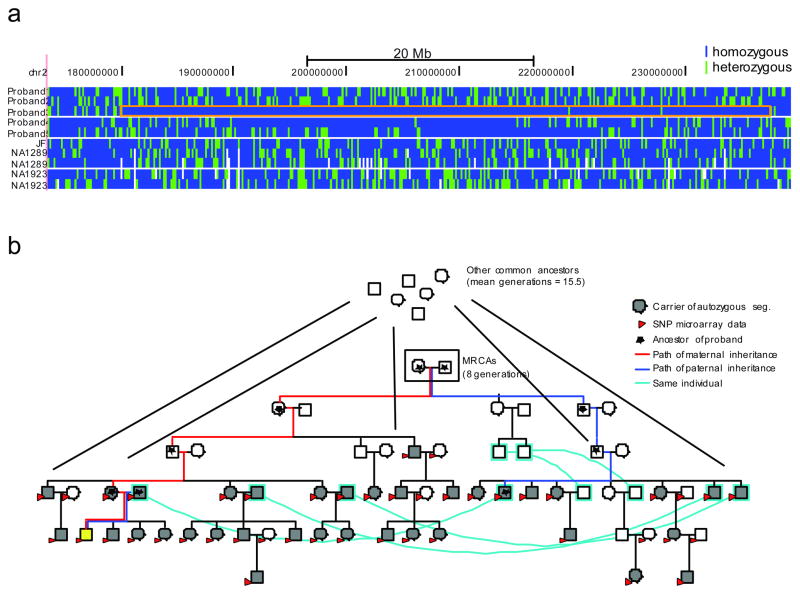
We determined the number of meioses separating each allele within each autozygous segment. The small founding population and complex genealogy (Figure 1) of the Hutterite population made this potentially problematic because of the large number of shared common ancestors (CAs) and multiple paths of descent between any ancestor-descendant pair. To resolve this, we combined both the pedigree structure and genome-wide SNP genotype data9 to identify the most recent common ancestors (MRCAs) based on segregation within the Hutterite genealogy 8 (Figure 3; Supplementary Fig. 3; Supplementary Note). We estimated that the two haplotypes of the 44 autozygous segments are separated by 8–18 meioses based on the MRCAs (Supplementary Table 4).
Figure 3. Determination of the MRCA for an autozygous segment.
A) A 54 Mbp autozygous segment on chromosome 2 in Individual 3. Genomic coordinates (hg18) are represented horizontally, and each individual is represented vertically: the five Hutterite individuals, followed by the three European-American individuals, and then the two Yoruba. Each SNV is represented by a vertical bar colored blue if the variant is homozygous and green if it is heterozygous. The autozygous segment in Individual 3 is boxed in orange. B) Determination of the MRCA for this autozygous segment. The pedigree containing all the haplotype carriers of the autozygous haplotype is shown. Cyan lines connect the same individuals who are represented twice in the pedigree. Individual 3 is shown in yellow. All samples with SNP microarray data are shown with red arrows, and haplotype carriers are shown in gray. These haplotype carriers have two MRCAs (boxed) as well as additional CAs further up the pedigree. The paths from these individuals to the autozygous subject are shown in red for the maternal ancestors and blue for the paternal ancestors; all ancestors of the individual are marked with a star.
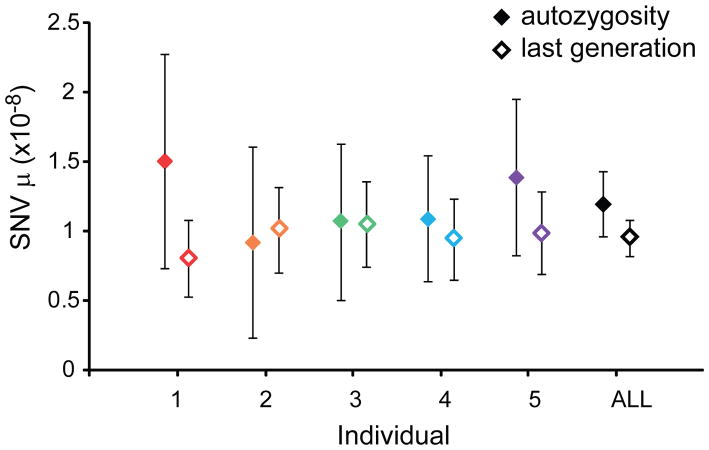
To calculate the SNV mutation rate, we identified heterozygous SNVs within each autozygous segment, excluding regions of common repeats, segmental duplication, and known SNPs (dbSNP132). We validated 72 SNVs as heterozygous by Sanger-based capillary sequencing (Table 1; Supplementary Table 5; Supplementary Note). We calculated an SNV mutation rate (μ) of 1.20×10−8 mutations per basepair per generation (95% confidence intervals (CI): 0.89×10−8 – 1.43×10−8). We observed consistent μ across the five trios with a range between 0.92×10−8 and 1.51×10−8 (Figure 4; Table 1). Among these mutations, we observed an excess of transitions relative to transversions (Ti/Tv = 1.64) that was not significantly different from the genome-wide SNV ratio of 2.17 (two-tailed chi-square p = 0.27). Twelve of the 72 validated heterozygous SNVs (16.7%) mapped to CpG dinucleotides. We calculated μ for CpG sites of 9.72×10−8 per CpG basepair per generation, 9.5X greater than the μ for non-CpG bases (1.02×10−8). We also estimated mutation rate based on de novo mutations in the most recent generation (Supplementary Note; Supplementary Table 6). Based on 176 validated de novo SNVs, we calculated a mutation rate of 0.96×10−8 (95% CI: 0.82×10−8 – 1.09×10−8); although this rate is lower than calculated using autozygosity, the confidence intervals of these rates overlap (Figure 4).
Table 1.
SNV mutation rates determined from segments of autozygosity
| individual | Segments (>5 Mbp) | Total callable (Mbp)* | Mean meioses (MRCA)† | SNVs‡ | SNV μ | 95% CI§ |
|---|---|---|---|---|---|---|
| 1 | 7 | 63.4 | 13.8 | 13 | 1.51×10−8 | 0.62–2.28×10−8 |
| 2 | 6 | 55.9 | 13.8 | 7 | 0.92×10−8 | 0.17–1.72×10−8 |
| 3 | 9 | 124.8 | 9.9 | 13 | 1.07×10−8 | 0.45–1.63×10−8 |
| 4 | 10 | 147.6 | 12.0 | 19 | 1.09×10−8 | 0.56–1.55×10−8 |
| 5 | 12 | 120.8 | 12.0 | 20 | 1.40×10−8 | 0.73–1.96×10−8 |
| ALL | 44 | 512.4 | 11.9 | 72 | 1.20×10−8 | 0.89–1.43×10−8 |
Non-segmental duplication, non-simple repeat and non-dbSNP132 with at least six mapped reads.
Weighted by length of segment.
Validated novel heterozygous.
Based on a Poisson distribution.
Figure 4. SNV mutation rate estimates.
The SNV mutation rate point estimates are shown for each individual and all five individuals combined with the error bars representing the 95% confidence intervals based on a Poisson distribution. μ – SNV mutations per generation per basepair. Filled diamonds are estimates from autozygous segments and open diamonds are estimates from the most recent generation.
We identified and validated one potential gene conversion event involving paralogs of segmental duplications containing the genes C4A and C4B—a region where lower copy number has been associated with lupus 16. Although individual 4 has a total diploid copy number of six for this CNV, we determined that the sequence content of the two alleles differ (Supplementary Fig. 4) likely as a result of gene conversion between paralogous copies of, at a minimum, the gene TNX (6 kbp).
Both theoretical and experimental analyses have predicted that the male germline contributes disproportionately to de novo mutations as compared to the female germline 7,17–18. However, a recent analysis on two parent-offspring trios reported a paternal bias in mutation in one trio and a maternal bias in the other 5. Given the complexity of the Hutterite pedigree and transmissions through multiple female and male ancestors, we focused on the putative genome-wide de novo mutations in the most recent generation. We used molecular phasing 5,12,18 to determine the parental origin of 26 of the 176 validated de novo SNVs, and we found that 84.6% (22/26) of de novo SNPs originated on the paternal haplotype confirming a male bias for new SNVs (two-tailed binominal p = 2.67×10−4; 95% CI – 70.8–98.5%).
One advantage of using autozygosity is the ability to identify potential gene conversion events between homologous chromosomes. Such events could lead to clusters of heterozygous SNVs (including known SNPs) within regions of autozygosity, and we identified four clusters (two or more SNVs mapping within 10 kbp of each other) (Table 2). One of these clusters is 309 kbp suggesting that this is most likely a product of crossover events 19. Excluding this large cluster, the average distance between heterozygous SNPs in the remaining three clusters is 2723 bp (range: 7–7839 bp). We tested this distribution by simulation (n = 10,000 replicates) and determined a significant excess of “clustered” SNVs (empirical p = 0.001).
Table 2.
Clusters of heterozygous SNVs
| Individual | chr | start | end | length | heterozygous SNVs (novel)* | CpG SNVs† | GC%‡ | Meioses§ |
|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 4403120 | 4414313 | 11193 | 3 (1) | 0 | 0.46 | 15.5 |
| 1 | 1 | 74906779 | 74909810 | 3031 | 2 (2) | 0 | 0.36 | 14.0 |
| 5 | 1 | 10788610 | 10793450 | 4840 | 5 (0) | 1 | 0.50 | 9.0 |
| 5 | 2 | 211397873 | 211706890 | 309017 | 66 (1) | 8 | 0.35 | 9.0 |
| 1 | 7 | 45928832 | 45930883 | 2051 | 2 (2) | 0 | 0.47 | 2.0 |
| 1 | 16 | 77252309 | 77256230 | 3921 | 2 (2) | 0 | 0.43 | 2.0 |
| 2 | 1 | 189717619 | 189717626 | 7 | 2 (2) | 0 | 0.31 | 2.0 |
Total number of SNVs in the cluster with the number of novel (non-dbSNP132) SNVs in parentheses.
SNVs in CpG dinucleotides.
Percent of G and C bases in the heterozygous cluster.
Number of meioses in which event(s) occurred based on the MRCA.
We also tested whether the de novo SNVs in the most recent generation were uniformly distributed in the genome. Interestingly, we observed three clusters of validated de novo variants (Table 2; Supplementary Table 5) and a significant excess (empirical p = 6×10−6) of de novo SNVs in close proximity (<10 kbp) based on simulation (n = 1,000,000 replicates).
There has recently been much interest in using massively parallel sequence data to obtain an accurate estimate of mutation rate using nuclear families 5–6. We developed an approach using extended regions of autozygosity to discover new mutations that have emerged within a few generations. Compared to analyses focused on de novo mutations in a single generation, we significantly reduced the number of false positives and somatic mutations, since most mutations in autozygous segments are transmitted from one of the parents. In addition, given the relationship between paternal age and number of de novo mutations 18,20, our approach reduces this confounding effect by yielding an average mutation rate over 8–18 meioses. Disadvantages include uncertainties in ancestry of the autozygous segments (Supplementary Note), the smaller genomic “search space” (512 Mbp), the potential to confound new mutation and gene conversion events, and an increased potential for purifying selection to eliminate a small fraction of new mutations, although fraction of such events should be negligible 21. We have tried to reduce the confounding effect of gene conversion by limiting our analysis to novel SNVs. We estimated an SNV mutation rate of 1.20×10−8 using autozygous segments, which is higher than the rate of 0.96×10−8 that we estimated for the most recent generation and the rate 1.1×10−8 previously published for the whole genome 5–6 yet lower than the rate estimated in a recent resequencing study 22. While this manuscript was under review, two additional studies of mutation rate were published. First, a mutation rate of 1.2×10−8 was calculated from an analysis of WGS in over 70 trios20, which is equal to that obtained in our analysis suggesting the accuracy of using autozygosity to estimate mutation rate. Interestingly, the quantification of the correlation between number of mutations and paternal age reported by Kong et al.20 suggests that the relatively young age of the father of the trios analyzed here (21–30 years old at the time of the child’s birth) may provide an explanation for the lower mutation rate we observed in the most recent generation. In a second publication, an inferred mutation rate of 1.82×10−8 was calculated by modeling population genetic parameters based on the mutational properties of microsatellites23; the difference between this estimate and our estimates are likely due to differences in methodology.
We observed a non-uniform distribution of a small fraction of mutations within autozygous segment that appear to be evidence of recent allelic gene conversion. We observed three clusters that were unlikely generated by crossover mechanisms and might represent potential allelic gene conversion events in the autozygous segments, although one cluster was larger (11 kbp) than expected for typical gene conversion events 24. Only one of the ten SNVs in these clusters was in a CpG, and the GC content (0.36–0.50) of these three regions was not consistent with a model of recurrent mutation due to CpG methylation and demethylation. The average distance between heterozygous SNPs in these clusters was 2723 bp, ruling out compound mutation 25 as a likely mechanism. Intriguingly, one of these clusters of heterozygous SNVs is comprised of two novel SNVs (i.e. not in dbSNP) and could be further evidence of a non-uniform distribution of new mutations similar to what we observed with the de novo mutations. In addition, we observed an excess of heterozygous bases at dbSNP positions in autozygous segments (N = 22), most of which were not clustered with other heterozygous variants (16 out of 22) but may also be evidence of recent gene conversion.
Notably, we observed a non-uniform distribution (three clusters with two de novo SNVs within 10 kbp; range 7–3921 bp) (empirical p = 6×10−6) among the validated de novo events. One of these clusters contained SNVs 7 bp apart suggesting a compound mutational event 25; based on this event, 0.97% of de novo mutations were part of multi-nucleotide mutations (95% CI (Wilson method) – 0.27–3.5%). While this is somewhat lower than the estimate of 2–3% of de novo mutations in compound mutational events based on WGS of two trios 25, the confidence intervals do overlap. The remaining two clusters contained SNVs greater than 2 kbp apart suggesting that, even at greater distances, mutational mechanisms do not produce uniform distributions of new mutations.
We have presented a novel approach for estimating mutation rate in humans. We based our analysis on autozygosity and WGS from whole blood DNA to remove the effects somatic mutations and cell line artifacts. In addition, we were able to detect other recent changes in the genome including gene conversion events. Furthermore, we believe that the application of this approach to additional families has the potential to elucidate the dynamics of other forms of mutation including CNVs and indels.
METHODS
DNA samples and whole-genome sequencing
We selected five parent-offspring trios for WGS from a 13-generation pedigree of Hutterites from South Dakota (Figure 1). Detailed phenotyping data are available for all individuals in this study design. None of the individuals studied have a Mendelian disorder. All individuals consented, and the project was approved by the institutional review boards (IRBs) at The University of Chicago and the University of Washington.
One library for each individual was constructed from DNA isolated from whole blood using Illumina recommended protocols. Briefly, 1 to 3 ug of DNA were fragmented using sonication. The resulting fragments were end-repaired, adenosine overhangs added, and adaptors ligated. Size selection was performed by running the library on an agarose gel and cutting a band around 400 bp. The size-selected libraries were amplified using quantitative PCR. The resulting libraries were sequenced using an Illumina HiSeq 2000 to generate 51 and 101 bp paired-end reads. We generated 36–68 Gbp of sequencing data for each of the 15 individuals (10–17X coverage of the human genome) (Supplementary Table 1). Sequence data have been deposited into dbGAP; accession numbers pending.
Sequence data analysis and SNV identification
We aligned the paired-end reads to the NCBI build 36 reference of the human genome using BWA (version 0.5.9) with standard parameters 26. The quality scores for the mapped reads were recalibrated and PCR duplicates were removed using Picard (version 1.43). Then, we realigned the reads around potential indels to reduce spurious SNV calls due to misalignment in these regions using the Genome Analysis Toolkit (GATK) software (version 1.0.5777) 12. We used both GATK and SAMtools (version 0.1.8) 11 to identify SNVs. After generating an initial list of SNVs, we used the following approaches to generate a high-confidence variant list. First, we applied variant recalibration to the variants identified with GATK and used a variant quality score recalibration (VQSR) threshold of 2.30 (99% of known high-quality SNPs identified) to generate a final list of GATK variants 27. In addition, we applied the standard recommended GATK filters and filtered out SNVs located near indels. For the SAMtools call set, we filtered calls with a quality score less than 10. Then, we used the intersection of the GATK and SAMtools call sets for further analysis.
Concordance with SNP microarray genotypes and false negative rate estimation
To assess the quality of our SNV genotypes from WGS, we compared these data to genotype data generated for these same individuals on Affymetrix SNP microarrays (versions 500K and 6.0) 9. We assessed the genotype concordance for the 211,438–468,681 SNPs that passed quality control metrics on the microarray for each individual (Supplementary Table 1).
We estimated the false negative rate for heterozygous variants, since our calculations of mutation rate are based on heterozygous SNVs and these variants are more likely to be missed in moderate coverage WGS data. For each individual, we determined the number of heterozygous SNPs genotyped by SNP microarray and then determined how many of these variants were missed in the WGS (i.e. called as homozygous). We calculated false negative rate as the number of missed heterozygous SNPs divided by the total number of heterozygous SNPs (Supplementary Table 1).
Identification and definition of autozygous segments
We identified long stretches of homozygosity in the genomes of the five children of the Hutterite trios, two European Americans from the Centre d’Etude du Polymorphisme Humain collection (CEU) 13, two Yoruba from Ibadan, Nigeria (YRI) 13, and one additional European-American male. We ran PLINK 14 on all filtered SNV genotypes with variants in segmental duplications removed to avoid artifacts due to paralogous sequence variation. We required a minimum homozygosity length of 600 kbp, a maximum gap in homozygosity of 100 kbp, and a maximum of three heterozygous SNPs per 5 Mbp. We merged all segments within 50 kbp, since it appeared that some regions, especially in the Hutterites, were erroneously split. We did not consider regions with >20% gaps or segmental duplications in the reference assembly.
We compared the resulting list of large autozygous segments (>5 Mbp) to regions of likely autozygosity determined with SNP microarray genotypes and the 3555 member pedigree using IBDLD 15. The intersection of regions determined by genome sequence data and those determined from SNP microarrays gave us a final list of autozygous regions greater than 5 Mbp. We trimmed all 44 segments by 100 kbp because we observed an excess of heterozygous variants at the edges (Supplementary Fig. 5).
Determination of common ancestors for autozygous segments
The 15 sequenced individuals are part of a larger 13-generation pedigree of Hutterites, and we made use of this pedigree information to trace each autozygous segment to the MRCA of all carriers as previously described 8. We used genome-wide SNP genotypes of 1415 individuals from this large pedigree 8–9 to identify all individuals who carried at least one allele identical by state (IBS) to sequenced individuals with the autozygous segment across the majority of the SNPs in the autozygous region. We considered individuals to be a carrier of the autozygous haplotype if he/she was IBS >= 1 with the sequenced individual at >99% of the genotyped SNPs in the autozygous segment; the median number of SNPs used was 1204 (range: 84–5493). After we identified all haplotype carriers for an autozygous segment, we examined the pedigree to identify all CAs of these individuals. We defined the MRCA as the individual with the smallest mean number of meioses to all haplotype carriers. To estimate mutation rate, we used the mean number of meioses in all paths from the MRCA(s) to the autozygous individual (Supplementary Note).
Identification of heterozygous SNVs in regions of autozygosity
We intersected our list of autozygous segments in each sample with the list of quality-filtered SNVs identified in that sample. Then, for each heterozygous SNV identified in the autozygous sample, we applied an additional read-depth filter requiring a minimum of six sequencing reads at that location in the heterozygous individual. We did not consider SNVs in simple-repeats (based on the Tandem Repeats Finder track for hg18 in the UCSC Genome Browser), segmental duplications, or in dbSNP132. We identified a total of 85 novel, heterozygous SNVs in the refined autozygous regions (Supplementary Table 5), and attempted to validate these variants with Sanger sequencing (Supplementary Note).
Determination of SNV mutation rate using autozygosity
For our mutation rate calculations, we determined the number of basepairs in each autozygous segment that we were able to test for heterozygous variants. We counted the number of unique (i.e. not in segmental duplications), non-simple repeat, non-dbSNP132 bases with a read depth of at least six that were callable by GATK; these “callable” bases served as the denominator in our mutation rate calculation. To calculate mutation rate for each individual and across all five individuals, we applied the following formula:
where N is the number of validated heterozygous SNVs in the autozygous regions for that individual, FNR is the false negative rate for heterozygous SNVs, G is the weighted average of the number of meioses separating the alleles of autozygous segments (weighted by length of segment) for that individual, and L is the sum of callable basepairs in autozygous segments in that individual. The same formula was applied to calculate the mutation rate across all five individuals where N is the 72 validated heterozygous SNVs, G is 11.9 — the weighted mean (by segment length) number of meioses separating the two alleles of all autozygous segments, and L is the 512.4 Mbp of total callable basepairs in autozygous segments in the five individuals. To calculate the mutation rate at CpG basepairs, we considered only the estimated number of true heterozygous SNVs at CpG bases divided by “callable” CpG basepairs and the number of generations to the MRCA.
Determination of mutation rate using de novo mutations in the most recent generation
We identified putative de novo SNVs across the whole genome as those variants observed only in a single individual (i.e. not observed in the parents or any of the other Hutterite genomes). We did not consider variants in segmental duplications, simple repeats, or known SNPs (dbSNP132), and we required a read depth of at least six for all individuals in the trio. After applying these filters, we obtained a list of 632 putative de novo variants (Supplementary Table 7). Using molecular inversion probes (MIPs) 28–29, we attempted to capture and resequence these putative de novo variants (Supplementary Note). We calculated the mutation rate using the following equation:
where N is the total number of putative de novo mutations, FDR is the false discovery rate for putative de novo SNVs (Supplementary Note; Supplementary Table 6), FNR is the false negative rate, and L is the total callable basepairs with read depth of at least six in all members of the trio in the genome.
Simulations of mutation clusters
To determine whether there was a non-random distribution of heterozygous SNVs in autozygous segments, we performed the following simulations. We randomly permuted the positions of heterozygous SNVs in autozygous blocks that contained greater than one heterozygous SNV and determined the number of clusters of SNVs with less than 10 kbp. We repeated this process 10,000 times and determined an empirical p-value based on the number of simulations with at least three heterozygous SNV clusters (i.e. heterozygous SNVs within 10 kbp). We performed a similar analysis for the de novo SNVs by permuting 1,000,000 times the locations of the estimated number of true de novo SNVs in each sample within the callable regions of the genome.
Supplementary Material
Acknowledgments
We are grateful to M. Przeworski for thoughtful comments on the manuscript. We thank C. Lee, B. Paeper, J. Smith, and M. Rieder for assistance with sequence data generation, and J. Huddleston for technical advice. We are grateful to T. Brown for assistance with manuscript preparation. C.D.C. was supported by a Ruth L. Kirschstein National Research Service Award (NRSA) (F32HG006070). This work was supported by an American Asthma Foundation Senior Investigator Award to E.E.E, and R01 HD21244 and R01 HL085197 to C.O. E.E.E. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
URLs
Picard -http://picard.sourceforge.netSequence data for additional European American - http://aws.amazon.com/datasets/3357
AUTHOR CONTRIBUTIONS
C.D.C, J.X.C, C.O. and E.E.E designed the study. C.D.C. performed the genome sequencing analysis, MIP targeted resequencing analysis, and mutation rate calculations. J.X.C. performed analyses to determine the ancestry of the autozygous segments. M.M. performed and analyzed validation experiments including Sanger sequencing, microarray hybridizations, and MIP capture. A.K. and P.H.S. performed the read-depth copy number analysis. B.L.D. identified and analyzed the clusters of heterozygous SNVs in the autozygous segments. L.H. and M.A. performed the autozygosity analysis with SNP microarray data. L.V. and B.J.O. created the sequencing libraries. B.J.O. designed the MIP oligos. L.V. along with M.M. performed the MIP capture. E.E.E, C.O., M.A., and J.S. supervised the project. C.D.C and E.E.E wrote the manuscript with input and approval from all co-authors.
COMPETING FINANCIAL INTEREST
E.E.E. is on the scientific advisory boards for Pacific Biosciences, Inc., SynapDx Corp., and DNAnexus, Inc.
References
- 1.Haldane JBS. The rate of spontaneous muation of a human gene. Journal of Genetics. 1935;31:317–326. doi: 10.1007/BF02717892. [DOI] [PubMed] [Google Scholar]
- 2.Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat. 2003;21:12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- 3.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–86. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci U S A. 2010;107:961–8. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conrad DF, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43:712–4. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–9. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chong JX, et al. A common spinal muscular atrophy deletion mutation is present on a single founder haplotype in the US Hutterites. Eur J Hum Genet. 2011;19:1045–51. doi: 10.1038/ejhg.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cusanovich DA, et al. The combination of a genome-wide association study of lymphocyte count and analysis of gene expression data reveals novel asthma candidate genes. Hum Mol Genet. 2012;21:2111–23. doi: 10.1093/hmg/dds021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abney M, Ober C, McPeek MS. Quantitative-trait homozygosity and association mapping and empirical genomewide significance in large, complex pedigrees: fasting serum-insulin level in the Hutterites. Am J Hum Genet. 2002;70:920–34. doi: 10.1086/339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han L, Abney M. Identity by descent estimation with dense genome-wide genotype data. Genet Epidemiol. 2011;35:557–67. doi: 10.1002/gepi.20606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet. 2007;80:1037–54. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haldane JB. The mutation rate of the gene for haemophilia, and its segregation ratios in males and females. Ann Eugen. 1947;13:262–71. doi: 10.1111/j.1469-1809.1946.tb02367.x. [DOI] [PubMed] [Google Scholar]
- 18.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fledel-Alon A, et al. Broad-scale recombination patterns underlying proper disjunction in humans. PLoS Genet. 2009;5:e1000658. doi: 10.1371/journal.pgen.1000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong A, et al. Rate of de novo mutations and the importance of father/’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khalak HG, et al. Autozygome maps dispensable DNA and reveals potential selective bias against nullizygosity. Genet Med. 2012;14:515–9. doi: 10.1038/gim.2011.28. [DOI] [PubMed] [Google Scholar]
- 22.Awadalla P, et al. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. Am J Hum Genet. 2010;87:316–24. doi: 10.1016/j.ajhg.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun JX, et al. A direct characterization of human mutation based on microsatellites. Nat Genet advance online publication. 2012 doi: 10.1038/ng.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JM, Cooper DN, Chuzhanova N, Ferec C, Patrinos GP. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 2007;8:762–75. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 25.Schrider DR, Hourmozdi JN, Hahn MW. Pervasive multinucleotide mutational events in eukaryotes. Curr Biol. 2011;21:1051–4. doi: 10.1016/j.cub.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porreca GJ, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–6. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 29.Turner EH, Lee C, Ng SB, Nickerson DA, Shendure J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat Methods. 2009;6:315–6. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.