Abstract
In this work, gold nanoparticles perform Boolean logic operations in response to two proangiogenic targets important in cancer diagnosis and treatment: PDGF and VEGF. In the absence of protein target, gold nanoparticles are initially dispersed as a red solution, addition of target proteins causes nanoparticle aggregation, turning the solution blue, as well as the release of dye-labelled aptamer probes which causes an increase of fluorescence. These outputs constitute an AND or OR gate for simultaneous protein detection. We believe this logic-gate-based detection system will become the basis for novel rapid, cheap, and reliable sensors for diagnostic applications.
Keywords: Boolean Logic-based detection, Aptamer, Gold Nanoparticle, Nanoparticle, PDGF, VEGF
1. INTRODUCTION
Gold nanoparticle-based colorimetric detection of nucleic acid or protein targets is fast becoming a cheap, sensitive, and popular signalling method for development of new bioassays. When dispersed in solution, gold nanoparticles are red; however, upon aggregation, their color changes from red to purple. Linking this color change to the presence of target in a solution forms the basis of colorimetric detection using gold nanoparticles.
Consisting of short, single-stranded RNA or DNA, aptamers comprise a class of recognition molecules which selectively bind to their targets. They are selected in vitro from large random libraries by a process called SELEX [1–2], which ensures they will bind with high affinity and selectivity to the target of interest. The large variety of targets includes ions, such as Hg2+, small molecules, such as ATP, and proteins, including vascular endothelial growth factor (VEGF) and platelet-derived growth factor (PDGF). Aptamers are small, chemically synthesized, stable, and easily functionalized. Structurally, they have several advantages over antibodies for biosensor development, especially for surface-based immobilization. For instance, while aptamers selectively bind their protein targets with affinities comparable to antibodies, they can still hybridize with complementary DNA (cDNA), a phenomenon that allows for simple engineering of an antidote or signaling cascade. In addition, upon binding to their targets, aptamers often change their secondary structures. This property has been harnessed to make aptamer molecular beacons [3–5], protein-sensitive DNAzymes [6–8]
Likewise, color-change based detection of targets using aptamer modified AuNPs has been extensively explored [9–13]. Moreover, multivalent binding of oligonucleotides onto the gold nanoparticle (AuNP) surface [14–16] imparts unusual properties which play a significant role in biodetection, diagnostics and nanotherapeutics [17–22].
While these prior applications are useful for detection of a single protein target, there are situations, particularly in cancer medicine, where simultaneous multi-protein detection is needed to improve disease diagnosis. In many cancers, tumors produce their own growth factors, such as, PDGF BB and VEGF A, two potent angiogenic cytokines. VEGF A is a potent inducer of angiogenesis and directs the production of malformed vessels. PDGF BB is often produced later in tumor development and promotes the growth of cells which surround and support newly formed vessels leading to their maturation [23]. While there is evidence that coexpression of VEGF-A and PDGF-BB serves to inhibit angiogenesis [24], many high-grade tumors produce elevated levels of both PDGF-BB and VEGF-A [25–27]. Combination therapies, such as sunitinib malate, target both VEGF and PDGF receptors and have been found to be more successful than therapies that target either receptor individually [28–29].
Designing assays based on Boolean logic, where several inputs produce a single output, can solve this problem. These logic-gate systems have been used for detection of various targets such as small molecules, metal ions, enzymes and proteins, etc [30]. In our test case, the inputs are the proteins PDGF and VEGF, and the gate produces a signal only when a certain pattern of proteins is detected in the sample. This simultaneous detection of two important proangiogenic proteins, may have prognostic importance [31].
2. EXPERIMENTAL SECTION
2.1 Preparation of Citrate-Capped Gold nanoparticles
GNPs were prepared by citrate reduction of tetrachloroauric(III) acid (HAuCl4, 99.999%, Sigma Aldrich) as reported elsewhere [32–34]. In a typical procedure, 38.8 mM trisodium citrate (25 mL) was injected rapidly into 250 mL of 1 mM HAuCl4 aqueous solution under boiling conditions. Within several minutes, the color of the solution changed from pale yellow to deep red. The mixture was allowed to heat under reflux for another 15 min before it was cooled to room temperature with continuous stirring. The concentration of these Au nanoparticles was determined to be 15 nM using UV-Vis spectroscopy, based on an extinction coefficient of 2.7×108 M−1 cm−1 at λ= 520 nm for 13.5 nm GNPs [33].
2.2 Aptamer Synthesis
All DNA oligomers were synthesized on an ABI3400 DNA/RNA synthesizer (Applied Biosystems, Foster City, CA) in our laboratory. All DNA bases and 5′-modifiers were purchased from Glen Research, Sterling, VA. The PDGF and VEGF aptamers were labeled with 5′-TAMRA and 5′-FITC, respectively. Similar dye-modifications were followed for PDGF and VEGF mutant sequences. Thiol-Modified CpG beads (Glen Research, USA) were used for the 3′-SH modification of the carrier strands. For the deprotection procedures, sequences labeled with 5′-TAMRA-Modifier were deprotected in TAMRA deprotection solution (0.05M potassium carbonate in methanol) at 65°C for 3–4 hours. All other sequences were deprotected in AMA (ammonium hydroxide/40% aqueous methylamine 1:1) at 65°C for 20 minutes. After deprotection, all sequences were further purified by reversed phase HPLC (ProStar, Varian, Walnut Creek, CA) on a C-18 column using acetonitrile/aqueous triethylammonium acetate as the mobile phase. A Cary Bio-300 UV spectrometer (Varian, Walnut Creek, CA) was used to measure absorbance to quantify the fabricated sequences. (See Table S1, Supporting Info.)
2.3 Preparation of DNA-Modified GNPs
The protocol for functionalizing gold nanoparticles with thiol-modified oligonucleotides followed a published procedure [35]. Before DNA loading, the thiol functionality on the oligonucleotides was deprotected. The 3′-SH carrier strands (P-SH (PDGF system) and V-SH (VEGF system)) (1 mM) were deprotected by 1 M Dithiothreitol (DTT, Sigma Aldrich, USA) in PBS buffer (pH 8.3 – 8.5) for 30 minutes at room temperature; then the excess DTT was removed by extraction four times with 400 μL of ethylacetate solution [36]. Freshly cleaved thiolated carrier strands (P-SH and V-SH) were added to 1 mL GNP solutions for surface modification. After 12 h, 10× Phosphate Buffered Saline (PBS) (pH 7.4, Fisher Scientific, Pittsburgh, PA) was added to the reaction mixture followed by another addition after 12 h to make a final concentration of 1× PBS solution (pH 7.4 ± 0.1) with 11.9 mM PO4 3−, 137mM NaCl and 2.7 mM KCl. The reaction mixture was then allowed to react for 48 h. Unbound oligonucleotides were subsequently removed by centrifugation and resuspension of the pellet (14,000 rpm, twice). DNA-coated gold NPs were resuspended, and the concentrations were determined by measuring the absorptions using a Cary Bio-300 UV spectrometer (Varian, Walnut Creek, CA). Condensed to a concentration of 10 nM, the DNA-gold NP conjugates were stored in a 1× PBS solution (11.9 mM PO4 3−, 137 mM NaCl and 2.7 mM KCl, pH 7.4 ± 0.1).
The target binding aptamers, 5′-FITC-extension-VEGF and 5′-TAMRA-extension-PDGF (50 μL of 100 μM stock solution) were hybridized to 1.0 mL of their corresponding carrier strand-coated gold nanoparticles (suspended in 1× PBS) by initially heating the solution at 70 °C for 5 min followed by incubation at room temperature for 30 min. The unreacted target aptamers were removed from the aptamer-carrier-gold conjugates by centrifugation (2×) at 12000 g for 30 min. The amount of target aptamers (5′-TAMRA-extension-PDGF and 5′-FITC-extension-VEGF) hybridized with respective carrier strands (P-SH and V-SH) was estimated around 62 ±7 and 73±9 per Au nanoparticle, resectively. . The number was calculated indirectly by measuring the amount of unhybridized dye-tagged target aptamers remaining after purification of the hybridized conjugate by centrifugation.
2.4 Protein Incubation
The protein stock solutions were made following the supplier’s protocol (PDGF-BB, Human recombinant, R&D Systems, USA and VEGF, Human recombinant, Sigma Aldrich, USA). All protein incubation experiments were carried out in 0.2 M PBS (200 mM NaCl, 5 mM sodium phosphate, pH 7.4) for 1 h at RT. For the fluorescence spectroscopy measurements, the protein nanoparticle mixture was centrifuged 1 h post-incubation, and the supernatant was measured for fluorescence signals (FITC and TAMRA in response to VEGF and PDGF, respectively). The nanoparticle pellet obtained post-centrifugation was redispersed in 0.01 M PBS (phosphate buffer, 300 mM NaCl) and checked for UV/Vis absorption measurements and colorimetric detection. BSA (Bovine Serum Albumin, New England BioLabs Inc., USA) and lysozyme (Sigma Aldrich, USA) were used as control proteins with similar concentrations and experimental conditions as described above.
3. RESULTS AND DISCUSSION
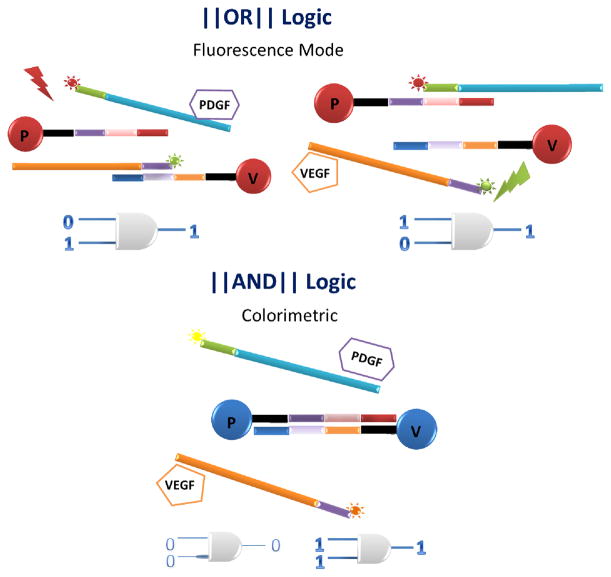
As shown in Scheme 1, our aptamer-based Boolean logic detection system consists of two gated conditions that work simultaneously: a colorimetric AND system and a fluorescent OR system. The AND system exploits the ability of an aptamer either to hybridize with its complementary strand or to bind with its target and uses AuNPs to realize visual detection. More specifically, two aptamer-modified AuNPs are functionalized with complementary strands of DNA. Each aptamer recognizes only one of the two input proteins, either PDGF or VEGF. Color change from red (dispersed AuNP’s) to purple (aggregated AuNP’s) occurs only when both protein targets are recognized.
Scheme 1.
(Top) Two types of nanoparticles that make up the nanoparticle logic gate with no target in the system and each aptamer (purple and orange) protecting its complementary DNA. (Bottom) Components used in the system. Note: Each particle holds more than one carrier/aptamer extension pair.
In the absence of target, the carrier strands are hybridized to their respective fluorescently labeled aptamers, which cover a portion of the carrier strand preventing any hybridization between the gold nanoparticles, leaving the solution red. Under these conditions, when only one type of input protein is added, it displaces the corresponding aptamer from the gold nanoparticle and makes the complementary strand on the gold nanoparticle accessible for hybridization with its complementary DNA on the other type of gold nanoparticle. However, if the other target protein is not present, the gold nanoparticle carrying the corresponding aptamer is still protected and there is no aggregation of nanoparticles and hence, no color change. It is the presence of both target proteins in the AND gate system that causes the aptamer strands to bind their respective target proteins and release their corresponding carrier strands, as shown in Scheme 2. The exposed carrier strands on both types of nanoparticles are now free to hybridize with each other, inducing Au nanoparticle aggregation and causing a corresponding color change from red to purple. As shown below, this system can detect equimolar concentrations of both targets in the nanomolar range, as well as targets in other concentration ratios.
Scheme 2.
Schematic of Au nanoparticle-based OR (Top) and AND (Bottom) logic gate designs for fluorescence detection and colorimetric output mode, respectively
In addition to the colorimetric AND gate described above, the system also supports an OR gate in the fluorescence mode. Here the quenching properties of gold nanoparticles are used in a manner similar to that in the research on nanoflares [37]. Basically, fluorophores are quenched when close to a gold surface. Once released from the surface, they are no longer quenched. Each aptamer extension is labeled with a unique fluorophore; when they are released after binding, the fluorescent signal specific for that protein increases. Thus, this system is capable of simultaneous AND/OR logic, depending on the desired detection modality.
For each protein (PDGF and VEGF), 13 nm gold nanoparticles were used. They were first functionalized via thiol (SH) interactions with (i) 3′-SH short carrier strands (20mer). The complementary portion of these carrier strand- functionalized nanoparticles was next protected by addition of (ii) 5′-Dye -modified target-specific aptamer strands which are partially complementary to the carrier strands conjugated with the nanoparticles (Au PDGF aptamer/P-SH and Au VEGF aptamer/V-SH ) (Scheme 1). An extension of five bases was also introduced at the 5′ end of the aptamers to aid hybridization with the carrier strand (Scheme 1). A list of DNA sequences is supplied in the supplementary information (Table S1). In the absence of targets, the carrier strands are protected by their hybridization to the aptamer strands, and the AuNPs remain dispersed and the solution red. At the same time, the 5′-Dye is quenched by its close proximity to the Au nanoparticle surface. Our system is also designed to generate target-specific signal for either one or both (OR logic) targets using fluorescence mode, whereas a visual color change from red to purple (colorimetric) occurs only in the presence of both targets (AND logic) (Scheme 2).
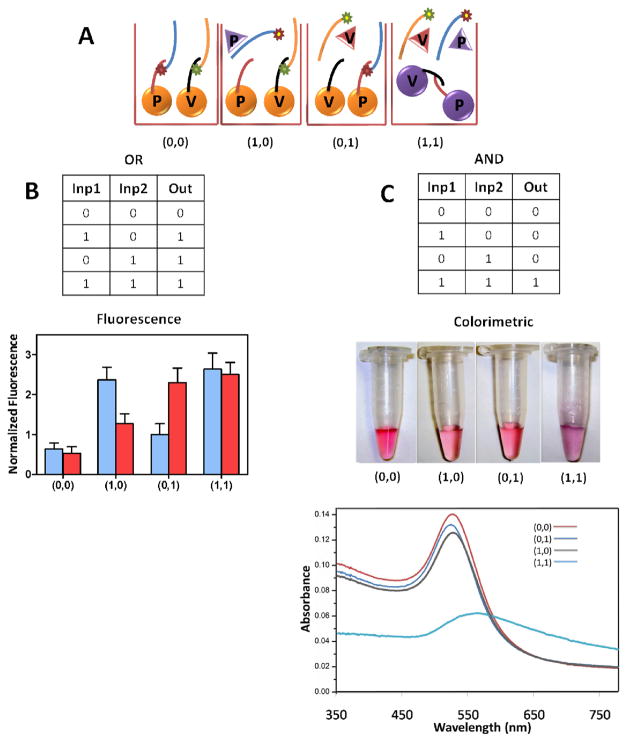
Figure 1A shows the various input conditions that are applicable to both logic systems. A single fluorescence signal, either FITC (blue bar) or TAMRA (red bar), is visualized in the presence of equimolar concentrations of 200 nM for both VEGF and PDGF proteins, respectively, whereas a positive readout fluorescence for both dyes is visualized when the two target proteins coexist. These signals arise because target binding results in the displacement of 5′-Dye-modified aptamers away from the nanoparticle surface (dequenching) (Scheme 2). Figure 1B shows the experimental results, which correspond to ideal OR gate responses. Following the same system and target concentrations, but using the colorimetric detection mode, the AND gate is also visualized. Only when both input proteins are added (and both types of aptamers are competed off their respective gold nanoparticles), will the complementary strands on each type of gold nanoparticle become unprotected and able to hybridize, thus causing nanoparticle aggregation and a corresponding color change from red to purple, as shown in Figure 1C.
Figure 1.
(A) Various input conditions followed during the experiments applicable to both logic systems. (B) Results for each path of the OR gate (see OR truth table) characterized by fluorescence spectroscopy (blue bars: FITC signal and red bars: TAMRA signal). (C) Results of the AND gate (see AND truth table) visualized in the photograph of the Au nanoparticle solutions and their respective UV-Vis extinction spectra. (Target proteins; PDGF=VEGF=200 nM)
In addition to visual inspection, the system was also investigated by using UV/Vis spectroscopy (Fig. 1C). The absorption maximum for the Au nanoparticle solution in the presence of either of the target proteins (0, 1) (blue curve) or (1, 0) (grey curve) is located at 525 nm, which is similar to the absorption of the system in the absence of any target protein (0, 0) (red curve). Only when the system is subject to both protein inputs (1, 1) does the peak shift to approximately 570 nm by the aggregation of Au nanoparticles (Fig. 1C). See Supporting Information (Fig. S2 and S3) for additional information on these AuNP’s UV/Vis spectra.
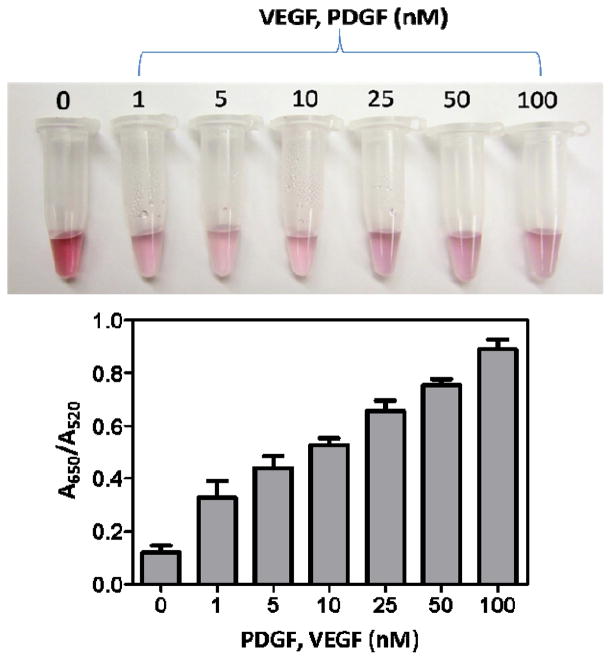
Once the design was established, concentration-dependent studies (Fig. 2) were performed on equal concentrations of both target proteins, PDGF and VEGF (AND system), and were monitored for the extent of agglomeration, both visually (colorimetric) (Fig. 2A) and spectroscopically (UV/Vis) (Fig. 2B). The system was designed to generate a quick visual response to the presence of both protein targets (AND logic) in a qualitative manner, as seen in Figure 2A. As mentioned elsewhere [38], in addition to the inter-particle distance, the size of the aggregate also affects the optical properties. At high target concentrations (25, 50, and 100 nM), the color changes to purple, but at lower concentrations (1, 5 and 10 nM) the change is to light pink or colorless. The effect of target concentration on the extent of agglomeration was quantified using UV/Vis spectroscopy. The ratio of the extinction at 650 nm, which corresponds to aggregated particles, to that at 520 nm, which corresponds to dispersed particles, was used to quantify the aggregation process or color change. As shown in Figure 2B, with the increase of target concentration, the extinction ratio (A650/A520) increases by the formation of large-sized 3D Au nanoparticle aggregates.
Figure 2.
(A) Colorimetric and (B) UV/Vis detection of the extent of agglomeration in the presence of different concentrations of an equimolar target mixture.
Simultaneous detection of two or more targets or proteins is required for the development combinatorial therapies [28, 29]. One major concern in real samples is that one target may exist at a higher or lower concentration than the other. To address this challenge, our AND-gated gold nanoparticle-based detection system is able to signal a color change (red-to-colorless or to pale pink) in the mere presence of both targets, irrespective of their individual concentrations. Figure 3 shows a visual color change in Au nanoparticle solutions with several VEGF:PDGF concentration ratios. This is likely due to the polyvalent nature of DNA-conjugated Au nanoparticles. This favors interaction in three dimensions, all oriented perpendicular to the nanoparticle surface, allowing efficient base-pairing once the carrier strands are freely exposed as a result of aptamer-target binding [39]. We confirmed that the aggregation and fluorescence changes observed were a result of aptamer-specific interactions and not nonspecific interactions with the added proteins in an extensive series of control experiments (Table S4, Fig. S5–S7, Supporting Info).
Figure 3.
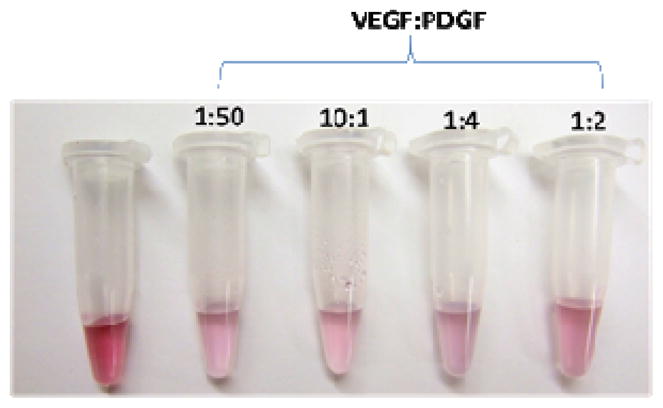
Colorimetric detection for the presence of various VEGF:PDGF concentration ratios
In summary, we have developed a simple system which can perform logic-gate-based detection in fluorescence and colorimetric modes for OR and AND gates, respectively. The dye-modified aptamer-conjugated Au nanoparticle system aids dual detection using only one system regardless of protein concentration ratio. The simultaneous detection of minute traces of oncoproteins, such as PDGF BB and VEGF-A present in both equal and unequal concentrations, will establish a rational scientific platform for the further development of combinatorial therapy based on logical detection.
Supplementary Material
Acknowledgments
This research was supported by the grant from National Science Foundation (NSF). We are thankful to the interdisciplinary Center for Biotechnology Research (ICBR) at University of Florida for TEM technical support.
Footnotes
Aptamer sequences with modifications used in experiments; the UV/Vis spectra of AuP-SH and AuV-SH DNA modified and a hybridization mixture of both types of Au nanoparticles. Control experiments, colorimetric and fluorescence detection of samples and TEM images of control experiments/samples. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ellington AD, Szostak JW. Nature. 1990;346:818. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Science. 1990;249:505. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Tyagi S, Kramer FR. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 4.Venkatesan N, Seo Y, Kim BH. Chem Soc Rev. 2008;37:648–663. doi: 10.1039/b705468h. [DOI] [PubMed] [Google Scholar]
- 5.Wang K, Tang Z, Yang CJ, Kim Y, Fang X, Li W, Wu Z, Medley CD, Cao Z, Li J, Colon P, Lin H, Tan W. Angew Chem, Int Ed. 2009;48:856–870. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Kim Y, Liu H, Zhu Z, Bamrungsap S, Tan W. J Am Chem Soc. 2009;131:8221–8226. doi: 10.1021/ja901132y. [DOI] [PubMed] [Google Scholar]
- 7.Moshe M, Elbaz J, Willner I. Nano Lett. 2009;9:1196–1200. doi: 10.1021/nl803887y. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Brown AK, Meng X, Cropek DK, Istok JK, Watson DB, Lu Y. Proc Natl Acad Sci USA. 2007;104:2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao W, Chiuman W, Brook MA, Li Y. ChemBioChem. 2007;8:727–731. doi: 10.1002/cbic.200700014. [DOI] [PubMed] [Google Scholar]
- 10.Medley CD, Smith JE, Tang Z, Wu Y, Bamrungsap S, Tan W. Anal Chem. 2008;80:1067–1072. doi: 10.1021/ac702037y. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Lu Y. Angew Chem Int Ed. 2006;45:90–94. [Google Scholar]
- 12.Liu j, Lu Y. Adv Mat. 2006;18:1667–1671. [Google Scholar]
- 13.Liu J, Lu Y. Biosens Bioelectron. 2001;16:799–809. doi: 10.1016/s0956-5663(01)00223-8. [DOI] [PubMed] [Google Scholar]
- 14.Hurst SJ, Lytton-Jean AKR, Mirkin CA. Anal Chem. 2006;78:8313–8318. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Seferos DS, Giljohann DA, Mirkin CA. J Am Chem Soc. 2008;130:5430–5431. doi: 10.1021/ja800797h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. J Am Chem Soc. 1998;120:1959–1964. [Google Scholar]
- 17.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Lu Y. J Am Chem Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 19.Medley CD, Smith JE, Tang Z, Wu Y, Bamrungsap S, Tan W. Anal Chem. 2008;80:1067–1072. doi: 10.1021/ac702037y. [DOI] [PubMed] [Google Scholar]
- 20.He L, Musick MD, Nicewarner SR, Salinas FG, Benkovic SJ, Natan MJ, Keating CD. J Am Chem Soc. 2000;122:9071–9077. [Google Scholar]
- 21.Thompson DJ, Enright A, Faulds K, Smith WE, Graham D. Anal Chem. 2008;80:2805–2810. doi: 10.1021/ac702403w. [DOI] [PubMed] [Google Scholar]
- 22.Nam J, Thaxton CM, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 23.Lederle W, Linde N, Heusel J, Bzyl J, Woenne EC, Zwick S, Skobe M, Kiessling F, Fusenig NE, Mueller M. Am J Pathol. 2010;176:981–994. doi: 10.2353/ajpath.2010.080998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenberg JL, Shields DJ, Barillas SG, Acevedo LM, Murphy E, Huang J, Scheppke L, Stockmann C, Johnson RS, Angle N, Cheresh DA. Nature. 2008;456:809–813. doi: 10.1038/nature07424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumoto S, Yamada Y, Narikiyo M, Ueno M, Tamaki H, Miki K, Wakatsuki K, Enomoto K, Yokotani T, Nakajima Y. Anticancer Research. 2007;27:2409–2414. [PubMed] [Google Scholar]
- 26.Abdeen A, Chou AJ, Healey JH, Khanna C, Osborne TS, Hewitt SM, Kim M, Wang D, Moody K, Gorlick R. Cancer. 2009;115:5243–5250. doi: 10.1002/cncr.24562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ilhan A, Gartener W, Neziri D, Czech T, Base W, Hörl WH, Wagner L. Anticancer Research. 2009;29:731–736. [PubMed] [Google Scholar]
- 28.Erber R, Thurnher A, Katsen AD, Groth G, Kerger H, Hammes H, Menger MD, Ullrich A, Vajkoczy P. FASEB J. 2004;18:338–340. doi: 10.1096/fj.03-0271fje. [DOI] [PubMed] [Google Scholar]
- 29.Socinski MA, Novello S, Brahmer JR, Rosell R, Sanchez JM, Belani CP, Govindan R, Atkins JN, Gillenwater HH, Pallares C, Tye L, Selaru P, Chao RC, Scagliotti GVJ. Clin Oncol. 2008;26:650–656. doi: 10.1200/JCO.2007.13.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.(a) Xu X, Zhang J, Yang F, Yang X. Chem Commun. 2011;47:9435–9437. doi: 10.1039/c1cc13459k. [DOI] [PubMed] [Google Scholar]; (b) Liu JW, Lu Y. J Am Chem Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]; (c) Liu JW, Lu Y. Anal Chem. 2004;76:1627–1632. doi: 10.1021/ac0351769. [DOI] [PubMed] [Google Scholar]; (d) Lee JS, Han MS, Mirkin CA. Angew Chem Int Ed. 2007;46:4093–4096. doi: 10.1002/anie.200700269. [DOI] [PubMed] [Google Scholar]; (e) Ogawa A, Maeda M. Chem Commun. 2009:4666–4668. doi: 10.1039/b910288d. [DOI] [PubMed] [Google Scholar]; (f) Bi S, Yan YM, Hao SY, Zhang SS. Angew Chem Int Ed. 2010;49:4438–4442. doi: 10.1002/anie.201000840. [DOI] [PubMed] [Google Scholar]
- 31.Loven D, Be’ery E, Yerushalmi R, Koren C, Sulkes A, Lavi I, Shaked Y, Fenig E. Acta Oncologica. 2008;47:104–109. doi: 10.1080/02841860701472470. [DOI] [PubMed] [Google Scholar]
- 32.Turkevitch J, Stevenson PC, Hillier J. Discuss Faraday Soc. 1951 [Google Scholar]
- 33.Hill HD, Mirkin CA. Nat Protoc. 2006;1:324–336. doi: 10.1038/nprot.2006.51. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Lu Y. Nat Protoc. 2006;1:246–252. doi: 10.1038/nprot.2006.38. [DOI] [PubMed] [Google Scholar]
- 35.Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. J Am Chem Soc. 1998;120:1959–1964. [Google Scholar]
- 36.Xu DK, Ma LR, Liu YQ, Jiang ZH, Liu ZH. Analyst. 1999;124:533–536. [Google Scholar]
- 37.Zheng D, Seferos DS, Giljohann DA, Patel PC, Mirkin CA. Nano Lett. 2009;9:3258–3261. doi: 10.1021/nl901517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Storhoff JJ, Lazarides AA, Mucic RC, Mirkin CA, Letsinger RL, Schatz GC. J Am Chem Soc. 2000;122:4640–4650. [Google Scholar]
- 39.Hurst SJ, Hill HD, Mirkin CA. J Am Chem Soc. 2008;130:12192–12200. doi: 10.1021/ja804266j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.