Summary
Mutations in the RNA binding protein FUS cause ALS, a fatal adult motor neuron disease. Decreased expression of SMN causes the fatal childhood motor neuron disorder SMA. The SMN complex localizes in both the cytoplasm and nuclear Gems, and loss of Gems is a cellular hallmark of SMA patient fibroblasts. Here, we report that FUS associates with the SMN complex, an interaction mediated by U1 snRNP and by direct interactions between FUS and SMN. Functionally, we show that FUS is required for Gem formation in HeLa cells, and expression of FUS containing a severe ALS-causing mutation (R495X) also results in Gem loss. Strikingly, a reduction in Gems is observed in ALS patient fibroblasts expressing either mutant FUS or TDP-43, another ALS-causing protein that interacts with FUS. The physical and functional interactions between SMN, FUS, TDP-43, and Gems indicate that ALS and SMA share a biochemical pathway, adding strong new support to the view that these motor neuron diseases are related.
Introduction
Mutations in at least 10 genes cause ALS but the disease mechanisms are not yet understood. Approximately 10% of ALS cases are familial while the remainder is sporadic (Boillee et al., 2006; Valdmanis and Rouleau, 2008). Mutations in the RNA binding protein FUS are the cause of a subset of familial and sporadic ALS cases (Kwiatkowski et al., 2009; Vance et al., 2009). FUS has features in common with the RNA binding protein TDP-43, and mutations in TDP43 also cause ALS (Gitcho et al., 2008; Kabashi et al., 2008; Sreedharan et al., 2008). FUS and TDP-43 are nuclear proteins at steady state and shuttle between the nucleus and cytoplasm. Both proteins function in transcription, splicing, mRNP transport and other processes in the nucleus and cytoplasm (Liu-Yesucevitz et al., 2011). These and other observations suggest a relationship between RNA metabolism and motor neuron disease. For example, the childhood motor neuron disease, SMA, is caused by deficiency in the SMN protein (Lefebvre et al., 1995). SMN is a component of the SMN complex, which functions in snRNP biogenesis (Battle et al., 2006), and has been implicated in other RNA-related roles, such as in mRNP transport (Fallini et al., 2012b). RNA metabolism defects may also explain the pathogenicity of C9ORF72, which causes ALS via a repeat expansion in the first intron (DeJesus-Hernandez et al., 2011; Renton et al., 2011). This expansion forms nuclear aggregates that could titrate crucial RNA binding proteins (DeJesus-Hernandez et al., 2011).
In most cell types, including motor neurons, the SMN complex localizes in the cytoplasm and in nuclear Gems (Battle et al., 2006; Gubitz et al., 2004; Liu and Dreyfuss, 1996). Loss of Gems is a cellular hallmark of SMA. Interestingly, Gems are also lost from motor neurons in a TDP-43 knockout mouse (Shan et al., 2010), and mutations in SOD1, which account for a large fraction of familial ALS cases, also lead to Gem deficiency in mouse models (Gertz et al., 2012; Kariya et al., 2012). These mouse model studies suggest that ALS and SMA may be related diseases, but the biochemical pathways involved are not known . These studies have generated considerable interest in understanding how RNA-related proteins cause motor neuron disease. Here, we report that FUS interacts both physically and functionally with SMN. We show that SMN-containing nuclear Gems are lost from FUS knockdown HeLa cells. Strikingly, Gem loss also occurs in HeLa cells transfected with a FUS construct bearing a severe ALS-causing mutation and in ALS patient fibroblasts bearing FUS or TDP-43 mutations. Together, these observations suggest that a common biochemical pathway involving SMN, FUS, TDP-43 and Gems links the motor neuron diseases SMA and ALS.
Results
FUS associates with the SMN complex and U1 snRNP
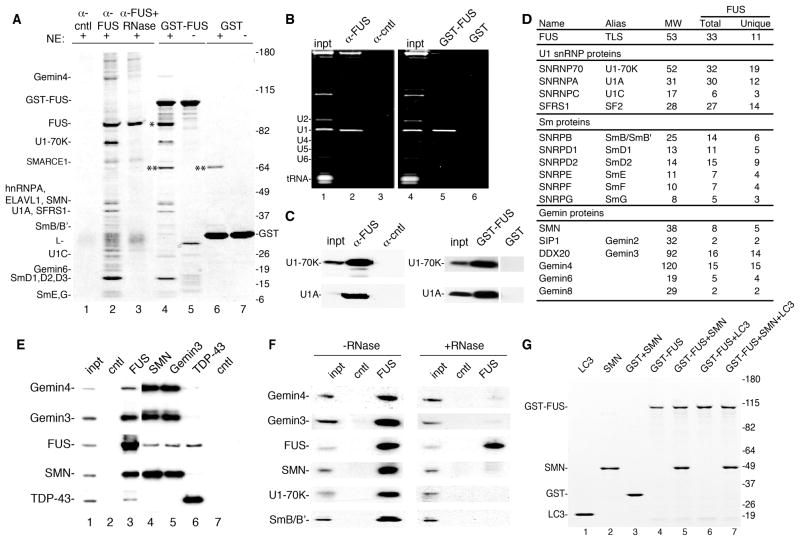
To investigate the mechanisms by which mutations in RNA binding proteins cause ALS, we focused on FUS. Antibodies raised against GST-FUS detect one main band on a Western blot and immunoprecipitate (IP) FUS from HeLa nuclear extracts (Fig. S1). To examine the FUS interactome, the FUS antibody was used for IPs and GST-FUS for pulldowns from nuclear extract, and proteins were analyzed on a Coomassie gel (Fig. 1A). Mass spectrometry of bands excised from the gel showed that U1 snRNP components are among the most abundant proteins associated with FUS (Fig. 1A, lanes 2 and 4). Moreover, U1 snRNA is abundant in the FUS IP and pulldown (Fig. 1B), and Western analysis confirmed the presence of U1 snRNP proteins in the FUS IP and pulldown (Fig. 1C). The interaction between FUS and U1 snRNP is specific, as U1 snRNP components were not immunoprecipitated by the FUS antibody in FUS knockdown nuclear extracts (Fig. S1).
Fig. 1. The SMN complex and U1 snRNP associate with FUS.
(A) IPs were carried out with HeLa nuclear extracts using FUS or negative control antibodies (lanes 1–3). In lane 3, nuclear extract was incubated with RNase A prior to IP. GST-FUS (lanes 4 and 5) or GST (lanes 6 and 7) was used for pulldowns from nuclear extract (lanes 4 and 6) or buffer alone (lanes 5 and 7). Proteins were run on a 4–12% SDS gradient gel and detected by Coomassie. Indicated proteins were identified by mass spectrometry. L: antibody light chain. *: FUS, **: a non-specific band. (B) Total RNA from IP and GST pulldown samples used in panel A. 30% input (inpt) was loaded. RNAs were detected with ethidium bromide. (C). Protein was isolated from samples in panel B followed by Westerns using indicated antibodies. 15% input (inpt) was loaded. (D) Table showing mass spectrometry data for indicated proteins from FUS IP. The number of total peptides (Total) and total unique peptides (Unique) identified by mass spectrometry is shown. (E) IP/Westerns using indicated antibodies. The negative control for the FUS and TDP-43 antibodies was a rabbit polyclonal antibody (lane 2, SAP130), and the negative control for the monoclonal antibodies (SMN and Gemin3) was a monoclonal against HA (lane 7). (F) Same as panel E except nuclear extract was treated or not with RNase prior to the IP. (G) FUS interacts directly with SMN. The indicated purified proteins (2 μg) were mixed in the presence of RNase A followed by GST pulldowns. Proteins were separated on a 4–12% SDS-gradient gel and detected with Coomassie. Markers in kD are indicated.
The FUS IP and pulldown revealed that FUS also associates with components of the SMN complex, including SMN and Gemins 4 and 6 (Fig. 1A). This result is significant as an association between FUS and SMN raises the possibility that ALS and SMA are caused by defects in a shared biochemical pathway. To pursue this possibility, we analyzed the total proteins present in the FUS IP by mass spectrometry. U1 snRNP components and Sm proteins were the most abundant proteins in the data set (Fig. 1D, Table S1). In addition, all of the nuclear components of the SMN complex except for the smallest (Gemin 7, 14 kD) were present (Fig. 1D, Table S1). FUS reciprocally co-immunoprecipitated with SMN and the Gemins, confirming the specificity of the association between the SMN complex and FUS (Fig. 1E). Although previous work reported that FUS associates with TDP-43 (Kim et al., 2010; Ling et al., 2010), we found that the level of TDP43 was not significantly above background in our FUS IP (Table S1). However, IP/Western analysis revealed that TDP-43 and FUS do co-IP in nuclear extract (Fig. 1E). In contrast, TDP-43 does not co-IP with SMN complex components (Fig. 1E). Thus, although FUS and TDP-43 interact, the two proteins have distinct molecular associations. Indeed, TDP-43 associates with components of the miRNA processing machinery (Ling et al., 2010; Sephton et al., 2011).
Most of the proteins in the FUS IP associate with FUS in an RNA-dependent manner (Fig. 1A, lane 3), including the SMN complex and U1 snRNP (Fig. 1F). Consistent with this observation, analysis of FUS deletion mutants revealed that the RNA-recognition motif is required for the association of FUS with the SMN complex, U1 snRNP proteins, and U1 snRNA (Fig. S1). The SMN complex binds directly to U1 snRNA (Yong et al., 2002). Thus, U1 snRNA may mediate the RNA-dependent binding of the SMN complex to FUS. The GST-pulldown data also revealed that FUS associates with itself, as endogenous FUS in the nuclear extract binds to GST-FUS (indicated by *, Fig. 1A, lane 4). An amino-terminal region on FUS (1–111) is necessary and sufficient for the FUS-FUS interaction (Fig. S1).
To further characterize the association between FUS and SMN, we carried out GST-FUS pulldowns using purified proteins in the presence of RNase. This analysis revealed that GST-FUS interacts efficiently and directly with SMN, but not with negative control proteins (GST and LC3) (Fig. 1G, lanes 1–7). We conclude that the associations between FUS, the SMN complex, and U1 snRNP are mediated by U1 snRNA and also by a direct interaction between FUS and SMN.
FUS is required for Gem formation in HeLa cells
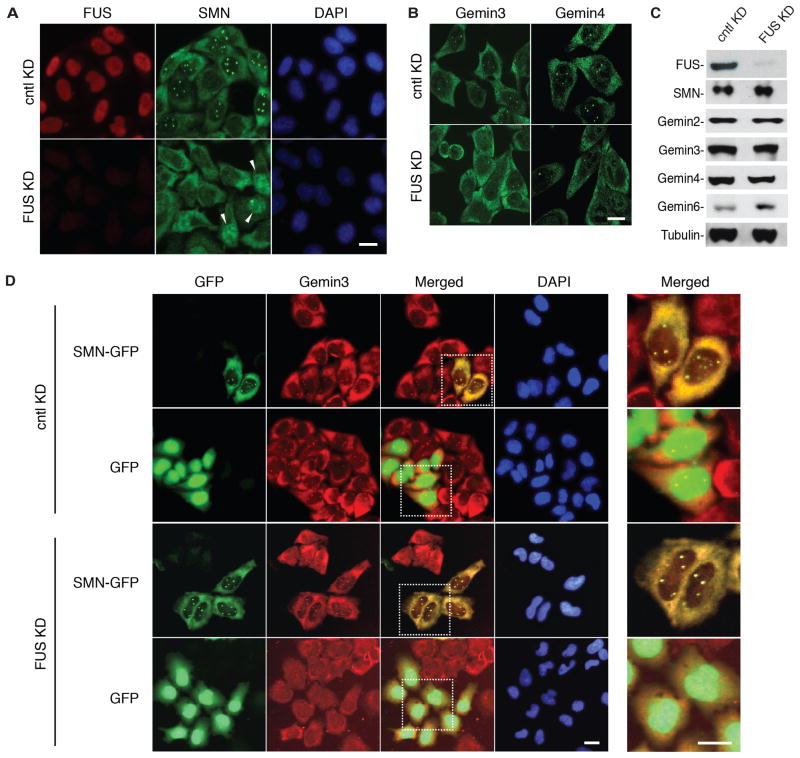
We next asked whether the physical association between FUS and SMN is functionally significant. Accordingly, we targeted FUS with shRNA in HeLa cells and examined the distribution of SMN and FUS by immunofluorescence (IF). IF showed that FUS was efficiently knocked down, and as expected, FUS localized in the nucleus in control knockdown cells (Fig. 2A). Remarkably, the number of SMN-stained nuclear bodies was dramatically reduced in the FUS knockdown cells (Fig. 2A), and diffuse nuclear staining of SMN was observed in a subset of the cells (Fig. 2A, see arrowheads). Loss of Gems was also observed using antibodies to Gemins 3 and 4 (Fig. 2B), indicating that Gems are the nuclear bodies lost in FUS knockdown cells. Consistent with this conclusion, Gems are detected when antibodies to Gemin3 or SMN are used for double IF (Fig. S2). Importantly, however, the levels of SMN and the other Gemin proteins were not affected by the FUS knockdown in total cell lysates (Fig. 2C). We conclude that FUS knockdown results in loss of Gems without affecting the overall levels of SMN/Gemins.
Fig. 2. FUS is required for Gem formation in HeLa cells.
(A) FUS or control knockdown (KD) HeLa cells were used to detect Gems (green in nucleus) and FUS (red) by IF with SMN and FUS antibodies, respectively. FUS was knocked down using an shRNA against FUS. Scrambled shRNA was used as a negative control. DAPI shows the nucleus. Scale bar, 20 μm. (B). IF staining of FUS or control knockdown HeLa cells was carried out with the indicated antibodies. Scale bar, 20 μm. (C) Western analysis of FUS and control knockdowns in HeLa cells using the indicated antibodies. Tubulin was used as loading control. (D) SMN-GFP or GFP was expressed in control or FUS knockdown HeLa cells. Gems were detected with SMN-GFP and co-staining with the Gemin3 antibody. DAPI shows the nucleus. Scale bar, 20 μm. Right panels (Merged) show high magnification of the dashed squares indicated in left panels.
To further examine the role of FUS in Gem formation, we asked whether the loss of Gems in FUS knockdown cells could be rescued by expression of exogenous SMN. FUS knockdown or control cells were transfected with SMN-GFP and then analyzed for SMN-GFP expression and by IF for Gemin3. Their co-localization was used to identify Gems. As shown in Fig. 2D, Gems were lacking in FUS knockdown cells that did not express SMN-GFP or that express GFP alone. In contrast, Gems were efficiently restored in FUS knockdown cells expressing SMN-GFP. In control knockdown cells, the Gem levels were similar in non-transfected cells and in cells transfected with SMN-GFP or GFP alone (Fig. 2D). We conclude that increased levels of SMN can bypass the requirement for FUS in Gem formation, suggesting that SMN acts downstream of FUS in a shared pathway required for Gem formation.
Expression of ALS-causing FUS mutation causes loss of Gems
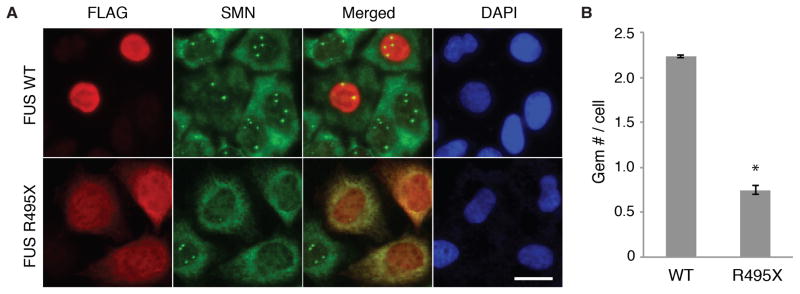
Many of the mutations in FUS that cause ALS are found in the nuclear localization sequence (NLS) and result in varying degrees of mis-localization of FUS to the cytoplasm (Dormann et al., 2010). As these ALS mutations are dominant, we next asked whether transfection of a construct bearing the FUS R495X ALS mutation affected Gem levels in HeLa cells. This mutation, in which the NLS is lacking, causes a severe form of ALS (Bosco et al., 2010; Waibel et al., 2010). When wild type FUS was expressed in HeLa cells, FUS properly localized to the nucleus (Fig. 3A). In contrast, high levels of FUS R495X were detected in the cytoplasm (Fig. 3A). Strikingly, Gem levels were dramatically reduced in the cells containing FUS R495X compared to cells transfected with WT FUS (Fig. 3A, B). We conclude that normal Gem levels require nuclear localization of FUS. The NLS of FUS is not required for SMN binding (Fig. S3). Thus, it is possible that FUS R495X sequesters SMN in the cytoplasm to an extent that results in loss of Gems. Alternatively, FUS R495X may inhibit the normal function of FUS in Gem formation by acting as a dominant negative via the FUS-FUS interaction.
Figure 3. Gems are lost in HeLa cells transfected with ALS-causing R495X FUS mutation.
(A) Representative images showing the expression of FLAG tagged FUS or FUS R495X in HeLa cells (red). Gems were detected using SMN antibodies (green in nucleus). Scale bar, 20 μm. (B) Quantitation of Gem levels in HeLa cells expressing the indicated proteins. The mean and standard deviation of Gem number per cell were calculated from three independent experiments. At least 100 cells were observed in each experiment. P-values were calculated by comparison with three controls. * indicates p < 0.01, Student’s t test.
Gems are deficient in ALS patient fibroblasts bearing FUS or TDP-43 mutations
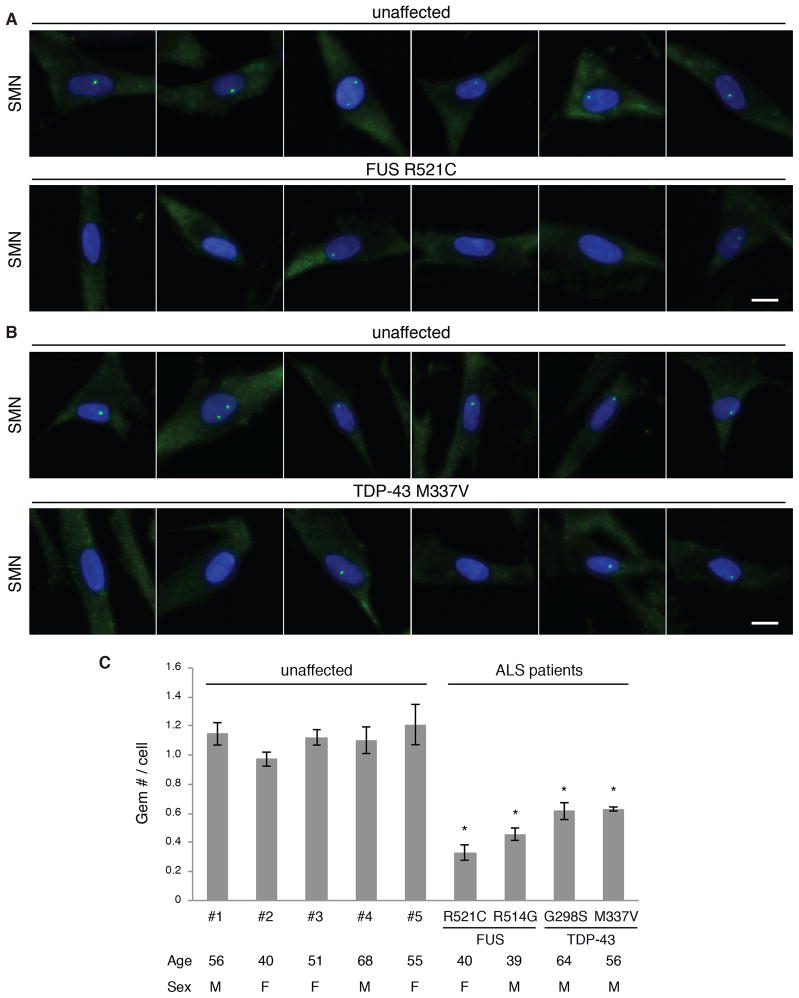
Previous work showed that Gems are lost from SMA patient fibroblasts (Coovert et al., 1997). In addition, Gems are deficient in both TDP-43 and SOD1 ALS mouse models (Gertz et al., 2012; Kariya et al., 2012; Shan et al., 2010). We therefore asked whether Gem levels are affected in ALS patient fibroblasts. Co-staining with SMN and Gemin antibodies was used to verify the detection of Gems in the fibroblasts (Fig. S4). We first examined two ALS patient fibroblast lines, one bearing a FUS R521C mutation and another bearing a TDP-43 M337V mutation. Strikingly, Gem deficiency was observed in both FUS and TDP-43 patient fibroblasts compared to age- and sex-matched fibroblasts from unaffected individuals (Fig. 4A and B). To extend these results, we used an automated system to collect images from three biological replicates of each fibroblast line and then counted Gems in a total of more than 800 cells for each. We also used this system to examine fibroblasts from an additional FUS (R514G) and TDP43 (G298S) patient and 5 unaffected individuals. These data revealed that the average Gem number was 2–3 fold lower in the FUS fibroblasts and 1.8 fold lower in the TDP-43 fibroblasts relative to controls (Fig. 4C). We conclude that a single dominant amino acid substitution in FUS or TDP-43 results in Gem deficiency in these ALS patient fibroblasts. Both of the FUS patient mutations that we examined are in the NLS, and we observe significant mislocalization of FUS to the cytoplasm in these fibroblast lines relative to the controls (Fig. S4). Thus, the decreased level of Gems may be explained by the decrease in levels of nuclear FUS.
Figure 4. Gems are deficient in FUS and TDP-43 ALS patient fibroblasts.
(A) IF using the SMN antibody was used to detect Gems in fibroblasts from an unaffected individual or an ALS patient carrying a FUS R521C mutation. DAPI shows the nucleus. Scale bar, 20 μm. (B) Same as A except using fibroblasts bearing a TDP-43 M337V mutation. (C) Graph showing Gem levels in ALS patients and unaffected individuals. The mean and standard deviation of Gem numbers per cell were calculated from three independent experiments. At least 150 cells were analyzed in each experiment. P-values were calculated by comparison with three controls. * indicates p < 0.01, Student’s t test. The age (years) and sex (M: Male or F: Female) of the individuals are indicated.
Previous work showed that Gems are decreased in a TDP-43 knockout mouse model (Shan et al., 2010). Consistent with this conclusion and with our patient fibroblast data, Gems are significantly decreased when TDP-43 is knocked down in HeLa cells (Fig. S4). We found that TDP-43 was properly localized in the nucleus in both of the ALS patient fibroblast lines containing TDP-43 mutations (Fig. S4). We conclude that these mutations in TDP-43 or knockdown of TDP-43 affect the normal levels of Gems.
Discussion
Here we report several independent lines of evidence that ALS and SMA are motor neuron diseases linked by a common molecular pathway. Specifically, we show that FUS, which is mutated in ALS, interacts with SMN, the protein deficient in SMA. SMN is a component of the SMN complex (Battle et al., 2006), and we show that FUS associates with this complex. One of the functions of the SMN complex is in snRNP biogenesis and we found that U1 snRNP is abundantly associated with FUS. In the nucleus, the SMN complex is present in Gems and we found that FUS is required for Gem formation. We furthermore found that Gems are lost from HeLa cells transfected with an ALS-causing FUS mutation. Previous work showed that Gems are lacking in SMA patient fibroblasts (Coovert et al., 1997). Strikingly, we found that single dominant mutations in FUS result in decreased Gem levels in ALS patient fibroblasts. FUS is known to interact directly with TDP-43 (Ling et al., 2010) and FUS acts downstream of TDP-43 in shared genetic pathways required for normal survival and motor function in Drosophila and zebrafish (Kabashi et al., 2011; Wang et al., 2011). As observed for SMN and FUS, TDP-43 is required for normal Gem levels (Shan et al., 2010), and we found that Gems are significantly decreased in ALS patient fibroblasts bearing TDP-43 point mutations. Taken together, these observations suggest a model in which TDP-43 functions upstream of FUS, which in turn is required for assembly of SMN into Gems. Thus, SMA and ALS share a common pathway involving TDP-43, FUS, SMN, and Gems. Interestingly, Gems were recently shown to be deficient in SOD1 mouse models of ALS (Gertz et al., 2012; Kariya et al., 2012). Thus, disruption of the Gem pathway may be a common feature of SMA and multiple types of familial ALS. Our data showing that FUS associates with itself may explain why FUS mutations are dominant. By interacting with wild type FUS, mutant FUS may inhibit the normal function(s) of FUS or sequester normal FUS, forming aggregates that are toxic to motor neurons. Similar self-interactions have been observed for TDP-43, which may also explain the dominance of these mutations (Da Cruz and Cleveland, 2011).
Our studies also led to important new insights into Gem formation. Specifically, we found that Gems are lost in FUS knockdown cells despite containing normal levels of SMN and the Gemin proteins. These data indicate that the requirement for FUS in Gem formation is not due to an effect on SMN/Gemin levels. However, our data revealed that the FUS requirement for Gem formation could be bypassed by overexpression of SMN, suggesting that the role of FUS in Gem formation may be to associate with SMN and increase its effective concentration and/or its association with other Gem components. Our observation that Gems are deficient in HeLa cells transfected with FUS R495X, which lacks the NLS, indicates that the nuclear localization of FUS is required for normal Gem levels. Many of the known patient mutations in FUS that cause ALS are located in the NLS (Dormann et al., 2010), including both of the mutations we analyzed in FUS patient fibroblast lines. Our data show that these lines display significant mislocalization of FUS to the cytoplasm and Gem deficiency. Together, these data raise the possibility that other patient mutations in the FUS NLS might have the Gem deficiency phenotype in ALS patient fibroblasts. When we analyzed Gem levels in a few examples of ALS patient fibroblasts with unknown mutations, no obvious Gem phenotype was detected (TY and RR, unpublished). Thus, a large number of ALS patient fibroblast lines must be examined to determine the generality of the phenotype. TDP-43, FUS and SMN also have other functions. For example, both TDP-43 and FUS are nucleocytoplasmic shuttling proteins that are present in cytoplasmic axonal mRNP transport granules together with the SMN complex (Fallini et al., 2012a, b; Liu-Yesucevitz et al., 2011). Thus, mutant FUS, TDP-43, and SMN may cause motor neuron disease by disrupting axonal transport of mRNAs encoding proteins essential for motor neuron function. At present, it remains to be determined which function(s) of FUS, TDP-43 and SMN are mechanistically involved in SMA/ALS and which may be a signature of these diseases.
We observed about a 2–3 fold decrease in Gem levels in ALS patient fibroblasts whereas Gem levels in SMA patient fibroblasts with severe type I disease are reduced by ~20 fold and by ~3–4 fold for the less severe SMA types II and III (Coovert et al., 1997). The observation that Gem levels are deficient in ALS patient fibroblasts raises the interesting possibility that Gem levels could be used as a rapid diagnostic marker. For example, Gem levels may be potentially useful for sub-typing FUS and TDP-43 forms of the disease. However, further ALS patient fibroblasts containing FUS, TDP-43, or other mutations must be analyzed in future work to determine the generality of the Gem phenotype. The observation that both SMA and ALS (at least some sub-types) have a Gem phenotype also raises the possibility that drug candidates identified for SMA may be efficacious for ALS. Our observation that overexpression of SMN rescues Gem levels in FUS knockdown cells and recent work showing that overexpression of SMN delays disease onset in an SOD1 mouse ALS model (Kariya et al., 2012), provide a rationale for testing SMA therapeutics that both increase Gem levels and rescue motor neuron defects.
The multiple links identified among FUS, TDP-43, SOD1, the SMN complex, U1 snRNP and Gems add strong new support to the view that defects in RNA metabolism are involved in the pathogenesis of motor neuron disease. In further work, it is important to assess components of these RNA complexes for mutations that may be candidates for ALS or SMA susceptibility genes or risk factors.
Experimental Procedures
Plasmids, proteins, antibodies
His-SMN and His-LC3 proteins were obtained from Enzo Life Sciences. The SMN-GFP expression plasmid was from Origene. Rabbit polyclonal antibodies were raised against GST-FUS (Covance). Antibodies to SMN (2B1), Sm (Y12), Gemin3 (12H12) were from Abcam, U1-70K (9C4.1) and Gemin2 (2E17) from Millipore, TDP43 from Proteintech, and U1A (BJ-7), HA, Tubulin, Gemin4 (E-8) and Gemin6 (20H8) from Santa Cruz, and FLAG from Sigma. SAP130 and HA were used as negative controls for polyclonal and monoclonal antibodies, respectively.
IP, GST pulldown and mass spectrometry
IPs and GST pulldowns were performed as described (Das et al., 2007). Gel samples were trypsin digested and peptides analyzed by LC-MS/MS. FUS and control IPs were TCA-precipitated and analyzed by LC-MS/MS. Keratin and likely contaminants, such as, desmoplakin, actin, tubulin, myosin, and translation proteins were not included in Table S1. Proteins found in the negative control IP were not included in Table S1 if the total peptides were less than 3 fold lower than in the FUS IP. Proteins greater than 300 amino acids for which only 7 peptides or less were identified by mass spectrometry were not included in Table S1. Proteins with less than 2 unique peptides were also omitted.
RNAi
Lentiviruses were made by transfecting 293FT cells with 3 packaging plasmids (pLP1, pLP2, and pLP/VSVG) using the method provided (ViraPower™ Lentiviral Expression System, Invitrogen). Infected cells were selected with 3 μg/ml puromycin (Sigma). For siRNAs, cells were treated with Lipofectamine 2000 (Invitrogen) for 48 hrs according to manufacturer’s protocol.
Immunofluorescence (IF) and Gem imaging
IF was carried out using FUS (1:1000), TDP-43 (1:1000), SMN (1:400), Gemin3 (1:400), Gemin4 (1:400) and FLAG (1:1000) antibodies. HeLa cells were fixed with 4% paraformaldehyde in PBS for 15 min, and fibroblasts were fixed with methanol and acetone (1:1) for 15 min. Cells were permeabilized with 0.1% TritonX-100 in PBS for 15 min. For IF, cells were incubated in 10 antibody overnight at 4°C. After 3 washes in PBS, 20 antibody was added for 1 hr at RT, followed by 3 washes in PBS. 10 antibodies were diluted in 10% calf serum in PBS. 20 antibodies were mouse Alexa-488 and rabbit Alexa-647 diluted 1:1000 in 10% calf serum in PBS. HeLa cells images were captured with a Nikon TE2000U inverted microscope with a PerkinElmer ultraview spinning disk confocal and a 20X objective using Metamorph software (Molecular Devices, Sunnyvale, CA). See Extended Experimental Procedures for details of all methods.
Supplementary Material
Highlights.
Shared molecular pathway altered in ALS and SMA motor neuron diseases
ALS and SMA proteins, FUS and SMN, physically and functionally interact
Gems are deficient in human ALS patient fibroblasts
Acknowledgments
We are grateful to Dr. M. Winkelbauer-Hurt and the Nikon Imaging Center at HMS for assistance with microscopy. HeLa cells were from the National Cell Culture Center. TDP-43 (G298S) and control (#3, 4, 5) fibroblasts were from Coriell Institute. This work was supported by NIH grant GM043375 to RR, grant-in-aid from Toyobo Biotechnology Foundation to TY, and by ALS Therapy Alliance to JCT. MAC was funded by the ALS Association and TM by NIH grant DP1OD003930.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. The SMN complex: an assembly machine for RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:313–320. doi: 10.1101/sqb.2006.71.001. [DOI] [PubMed] [Google Scholar]
- Boillee S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ, Jr, Sapp P, McKenna-Yasek D, Brown RH, Jr, Hayward LJ. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet. 2010;19:4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011;21:904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, Than ME, Mackenzie IR, Capell A, Schmid B, et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C, Bassell GJ, Rossoll W. The ALS disease protein TDP-43 is actively transported in motor neuron axons and regulates axon outgrowth. Hum Mol Genet. 2012a;21:3703–3718. doi: 10.1093/hmg/dds205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C, Bassell GJ, Rossoll W. Spinal muscular atrophy: The role of SMN in axonal mRNA regulation. Brain Res. 2012b;1462:81–92. doi: 10.1016/j.brainres.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz B, Wong M, Martin LJ. Nuclear Localization of Human SOD1 and Mutant SOD1-Specific Disruption of Survival Motor Neuron Protein Complex in Transgenic Amyotrophic Lateral Sclerosis Mice. Journal of neuropathology and experimental neurology. 2012;71:162–177. doi: 10.1097/NEN.0b013e318244b635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, Levitch D, Hatanpaa KJ, White CL, 3rd, Bigio EH, Caselli R, et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubitz AK, Feng W, Dreyfuss G. The SMN complex. Exp Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- Kabashi E, Bercier V, Lissouba A, Liao M, Brustein E, Rouleau GA, Drapeau P. FUS and TARDBP but not SOD1 interact in genetic models of amyotrophic lateral sclerosis. PLoS Genet. 2011;7:e1002214. doi: 10.1371/journal.pgen.1002214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nature genetics. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- Kariya S, Re DB, Jacquier A, Nelson K, Przedborski S, Monani UR. Mutant superoxide dismutase 1 (SOD1), a cause of amyotrophic lateral sclerosis, disrupts the recruitment of SMN, the spinal muscular atrophy protein to nuclear Cajal bodies. Hum Mol Genet. 2012;21:3421–3234. doi: 10.1093/hmg/dds174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Shanware NP, Bowler MJ, Tibbetts RS. Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA. J Biol Chem. 2010;285:34097–34105. doi: 10.1074/jbc.M110.154831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Ling SC, Albuquerque CP, Han JS, Lagier-Tourenne C, Tokunaga S, Zhou H, Cleveland DW. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci U S A. 2010;107:13318–13323. doi: 10.1073/pnas.1008227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem. 2011;286:1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Chiang PM, Price DL, Wong PC. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc Natl Acad Sci U S A. 2010;107:16325–16330. doi: 10.1073/pnas.1003459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdmanis PN, Rouleau GA. Genetics of familial amyotrophic lateral sclerosis. Neurology. 2008;70:144–152. doi: 10.1212/01.wnl.0000296811.19811.db. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waibel S, Neumann M, Rabe M, Meyer T, Ludolph AC. Novel missense and truncating mutations in FUS/TLS in familial ALS. Neurology. 2010;75:815–817. doi: 10.1212/WNL.0b013e3181f07e26. [DOI] [PubMed] [Google Scholar]
- Wang JW, Brent JR, Tomlinson A, Shneider NA, McCabe BD. The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span. J Clin Invest. 2011;121:4118–4126. doi: 10.1172/JCI57883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Pellizzoni L, Dreyfuss G. Sequence-specific interaction of U1 snRNA with the SMN complex. EMBO J. 2002;21:1188–1196. doi: 10.1093/emboj/21.5.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.