SUMMARY
LIN28 is a conserved RNA binding protein implicated in pluripotency, reprogramming and oncogenesis. Previously shown to act primarily by blocking let-7 microRNA (miRNA) biogenesis, here we elucidate distinct roles of LIN28 regulation via its direct messenger RNA (mRNA) targets. Through cross-linking and immunoprecipitation coupled with high-throughput sequencing (CLIP-seq) in human embryonic stem cells and somatic cells expressing exogenous LIN28, we have defined discrete LIN28 binding sites in a quarter of human transcripts. These sites revealed that LIN28 binds to GGAGA sequences enriched within loop structures in mRNAs, reminiscent of its interaction with let-7 miRNA precursors. Among LIN28 mRNA targets, we found evidence for LIN28 autoregulation and also direct but differing effects on the protein abundance of splicing regulators in somatic and pluripotent stem cells. Splicing-sensitive microarrays demonstrated that exogenous LIN28 expression causes widespread downstream alternative splicing changes. These findings identify important regulatory functions of LIN28 via direct mRNA interactions.
INTRODUCTION
Post-transcriptional regulation of gene expression is fundamentally important to a multitude of cellular processes, including development, homeostasis and differentiation. RNA binding proteins (RBPs) interact directly with RNA transcripts in cells to exert various forms of regulation such as alternative splicing, turnover, localization and translation (Glisovic et al., 2008). Altered expression levels of RBPs often results in genetic diseases and cancer (Lukong et al., 2008). Among these key proteins is LIN28A (herein referred to as LIN28). Conserved across bilaterian animals, LIN28 is highly expressed early in development and is selectively downregulated during differentiation (Moss et al., 1997; Yang and Moss, 2003). Consistent with this pattern of expression, LIN28 has been shown to be important in the maintenance of embryonic stem (ES) cell pluripotency and efficacy of induced pluripotent stem cell (iPSC) derivation (Moss et al., 1997; Newman and Hammond, 2010; Yu et al., 2007). Of the factors used in reprogramming, LIN28 is unique in its classification as an RBP, rather than as a transcription factor. Notably, aberrant upregulation of LIN28 has been found in a range of different cancer cells and primary tumor tissues (Cao et al., 2011; Viswanathan et al., 2009; West et al., 2009).
LIN28 and its only paralog in humans, LIN28B, block the processing of let-7 microRNAs (miRNAs) by binding to the terminal loop of the let-7 precursor (pre-let-7) hairpin via a cold-shock domain (CSD) and two retroviral-like CHCC zinc-finger knuckles (Hagan et al., 2009; Heo et al., 2008; Heo et al., 2009; Nam et al., 2011; Piskounova et al., 2008). Subsequent reports have described several modes of interaction between LIN28 and primary, precursor, and mature forms of let-7 miRNAs (Desjardins et al., 2011; Nam et al., 2011; Rybak et al., 2008; Van Wynsberghe et al., 2011; Viswanathan et al., 2008; Zisoulis et al., 2012). In the context of a negative feedback loop, mature let-7 miRNAs have also been shown to repress LIN28 protein expression (Reinhart et al., 2000; Rybak et al., 2008).
Thus far, the regulation of let-7 miRNAs is the best-studied mechanism by which LIN28 controls gene regulatory networks. Reactivation of LIN28 in cancerous tissues has been proposed to cause downregulation of let-7 and subsequent activation of oncogenes such as K-RAS, C-MYC, and HMGA2 (Bussing et al., 2008). Similarly, LIN28 expression can convey resistance to diet-induced diabetes by releasing let-7 repression of insulin-PI3K-mTOR pathway genes IGF1R, INSR, and IRS2 (Zhu et al., 2011). However, changes in LIN28 expression have also been shown to have phenotypic consequences independent of altered let-7 levels. For example, transgenic mice with muscle-specific deletion of LIN28 exhibited impaired glucose uptake and insulin sensitivity, despite unchanged let-7 levels (Zhu et al., 2011). Other transgenic mice aberrantly expressing LIN28 show phenotypes of greater organ mass even in adult tissues where let-7 was unaffected (Zhu et al., 2010). Furthermore, during neurogliogenesis, constitutive expression of LIN28 has been shown to favor differentiation towards the neural lineage at the expense of glial cell development, prior to any influence on let-7 levels (Balzer et al., 2010). In ES cells, LIN28 has a positive influence on proliferation, in part by binding to and increasing the translation of mRNAs encoding cell-cycle regulators (Peng et al., 2011; Xu et al., 2009). These findings strongly suggest that regulation of other RNA transcripts, beyond let-7 miRNAs, is an equally important function of this protein. Until now, the lack of precise genome-wide LIN28 binding sites in RNA targets has represented a significant hurdle in our understanding of its regulatory network of target genes.
To generate a LIN28 protein-RNA interaction map, we used UV cross-linking and immunoprecipitation followed by high-throughput sequencing (CLIP-seq) (Licatalosi et al., 2008; Sanford et al., 2008; Yeo et al., 2009), which resulted in the discovery of LIN28 binding sites in over 6,000 gene targets. These sites were recapitulated in human ES (hES) cells and in a somatic cell line stably expressing LIN28. The resolution afforded by CLIP-seq enabled us to discover a GGAGA motif enriched in LIN28 binding sites within mRNA sequences. This motif occurs preferentially within predicted hairpins and other unpaired loop structures, similar to its context within pre-let-7. Among its mRNA targets, we find that LIN28 preferentially binds to transcripts encoding RNA processing and splicing factors. In fact, we demonstrate that exogenous expression of LIN28 in somatic cells, independent of altered let-7 miRNA levels, enhances the translation of a subset of RBPs that are known to regulate alternative splicing, namely hnRNP F, TIA-1, FUS/TLS and TDP-43. We showed that binding sites within these mRNAs were sufficient to enhance the activity of reporter constructs. Alternative inclusion of LIN28 binding sites within TDP-43 mRNA also revealed an interesting coupling between alternative splicing and translation control of this transcript. As a consequence of this direct regulation of splicing factors, LIN28 expression in somatic cells results in widespread alteration of splicing patterns. Depletion of LIN28 and LIN28B in hES cells also resulted in protein level changes of splicing factors. Furthermore, LIN28 and LIN28B exerted different effects on their targets in hES cells, hinting at further complexity in target regulation.
RESULTS
LIN28 binding sites found within thousands of human genes
In hES cells where LIN28 is expressed at high levels, we performed CLIP-seq with an antibody that specifically recognizes the endogenous protein (Figure 1A and Figure S1A). To model the reactivation of LIN28 expression observed in many cancer cells, we generated a stable Flp-In HEK293 cell line that constitutively expresses a C-terminal V5-tagged human LIN28 protein at physiological levels, but 5-6 fold below that of endogenous LIN28 in hES cells (LIN28-V5 293 cells; Figure S1B). We performed CLIP-seq on these cells, in this case with a V5 antibody (Figure 1A and Figure S1C). LIN28-bound RNA fragments from transcripts expressed in hES and LIN28-V5 293 cells were represented by 4.8 and 2.8 million sequenced reads that mapped to non-repetitive regions of the human genome, respectively (Table S1), comparable to previously published CLIP-seq experiments performed with hES cells (Yeo et al., 2009).
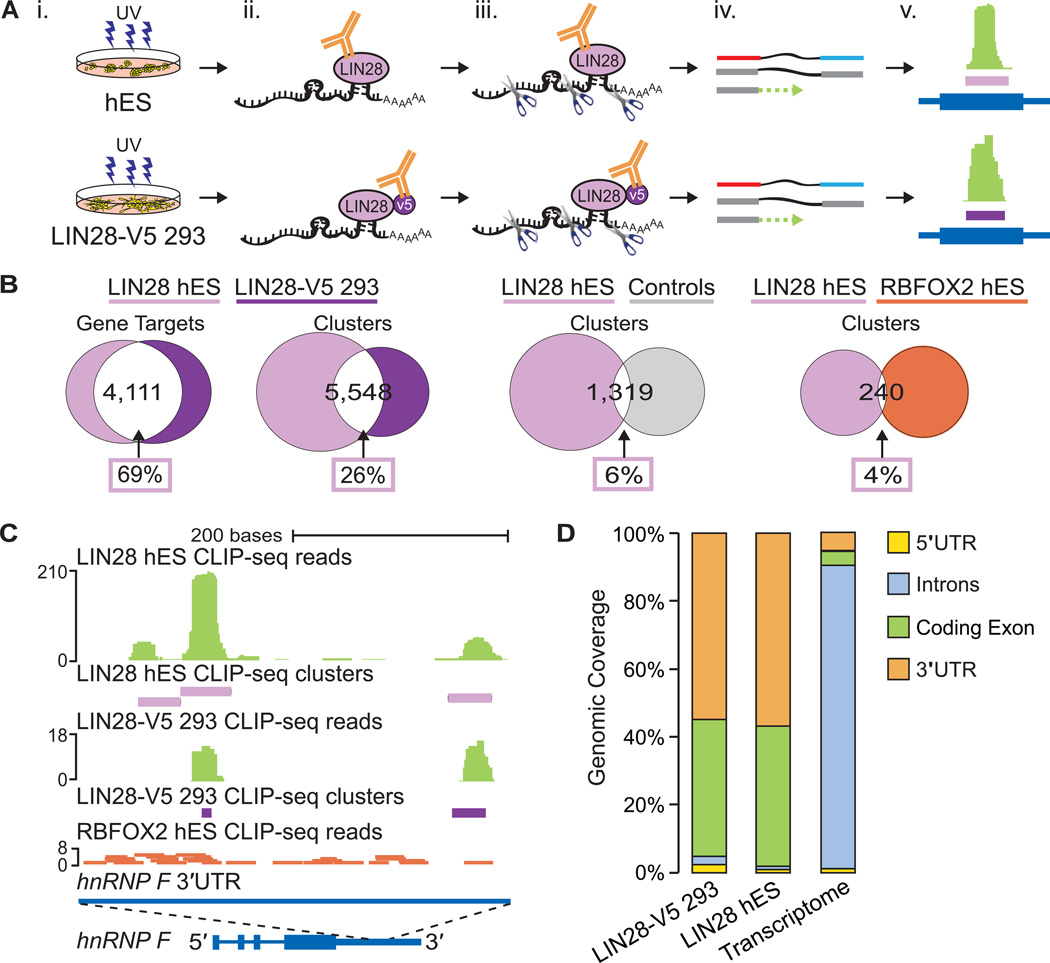
Figure 1. CLIP-seq identifies LIN28 binding sites in thousands of human genes.
(A) CLIPseq experimental approach performed using H9 human ES (hES) and LIN28-V5 293 cells. (i) UV cross-linking; (ii) immunoprecipitation of LIN28 protein-RNA complexes; (iii) micrococcal nuclease treatment, SDS PAGE gel size selection, and protease digestion; (iv) cDNA library preparation and high-throughput sequencing; and (v) cluster identification (in purple) based on the density of reads (in green) mapped to genes (in dark blue). (B) Venn diagrams illustrating the number of LIN28 target genes and clusters in common between hES and LIN28-V5 293 cells. For comparison, randomly located clusters in the same genes and genic regions as LIN28-V5 293 clusters and clusters from RBFOX2 in hES cells were used. The percentage of LIN28 hES gene targets or clusters in common with each comparison dataset are indicated within boxes. (C) LIN28 binding sites identified within the 3′UTR of the hnRNP F gene in both hES and LIN28-V5 293 cells. Clusters are depicted by purple rectangles representing the highest density of CLIP-seq reads (graphed as continuous densities in green). Individual RBFOX2 hES CLIP-seq reads are shown in red for comparison. The scale to the left indicates the height of aligned reads. (D) LIN28 binding enrichment in coding exons and 3′UTR sequences in both hES and LIN28-V5 293 cells, as compared to the observed percentage of nucleotides in the annotated transcriptome.
High-confidence LIN28 clusters (binding sites) were defined using a published computational procedure (Polymenidou et al., 2011; Yeo et al., 2009; Zisoulis et al., 2010). We found that 5,969 and 6,061 protein-coding genes in hES and LIN28-V5 293 cells contained at least one LIN28 cluster (Table S1). Despite differences in the variety and copy number of transcripts expressed between these two cell types, we found that over half (4,111) of the genes with at least one cluster in hES cells (69%) were also targets in the LIN28-V5 293 cells (68%) (Figure 1B). Thus, when expressed in somatic cells, LIN28 binds a significant portion of its mRNA targets that are naturally found in hES cells. In comparison with the 1,259 mRNA transcripts previously identified as LIN28 targets in hES cells (Peng et al., 2011) using a RNA immunoprecipitation (RIP) technique (which suffers from the caveat that the absence of cross-linking allows re-association of RNAs and RBPs after cell lysis (Mili and Steitz, 2004)), an average of 67% of the previously identified targets were detected in our CLIP-seq experiments (Figure S1D and S1E). While 82% of the 273 highest ranked RIP targets (Peng et al., 2011) were identified in our CLIP-seq datasets, more than 85% of the transcripts we have identified are not previously described as LIN28 targets (Figure S1F and S1G).
LIN28 binding sites are enriched within exons and 3′ untranslated regions of mRNAs
LIN28 was observed to bind in multiple locations within mRNA transcripts in hES and LIN28-V5 293 cells. Each target gene had ~3.5 significant clusters, approximately 35 nucleotides or less in length, totaling 26,279 hES and 15,028 LIN28-V5 293 binding sites. Within mRNAs that were expressed in both cell types, 26% of LIN28 hES clusters overlapped with a cluster identified in LIN28-V5 293 cells by at least 1 nucleotide (Figure 1B), comprising 47% of LIN28-V5 clusters. This was 4.3-fold higher than expected (6%) when LIN28 hES clusters were compared to randomly located clusters within the same genic regions (Figure 1B; p < 10−4, hypergeometric test). To illustrate the concordance of LIN28 binding sites in hES and LIN28-V5 293 cells, clusters from both CLIP-seq experiments were found in overlapping positions within the 3′ untranslated region (3′UTR) of the gene encoding the heterogeneous nuclear ribonucleoparticle protein F (hnRNP F) (Figure 1C). As a testament to the specificity of LIN28 binding, reads from a CLIP-seq experiment for the splicing factor RBFOX2 in hES cells (Yeo et al., 2009) were sparse in this region (Figure 1C). Indeed, only 4% of all LIN28 and RBFOX2 clusters in hES gene targets overlapped (Figure 1B).
We observed significant enrichment of LIN28 binding within coding exons and 3′UTRs, compared to the expected percentage of these regions in the transcriptome (Figure 1D). Less than 7% of LIN28 CLIP-seq clusters were found within intronic regions, indicating that LIN28 largely interacts with sequences within mature mRNA transcripts, consistent with the dominant localization of LIN28 protein in the cytoplasm (Balzer and Moss, 2007). In addition, LIN28 binding sites were found uniformly distributed across exons and 3′UTRs (Figure S1H). The concordance between our hES and LIN28-V5 293 datasets suggests that when aberrantly expressed, LIN28 interacts with similar loci within mRNAs as it does in transcripts expressed in ES cells.
CLIP-seq confirms binding of LIN28 to pre-miRNAs
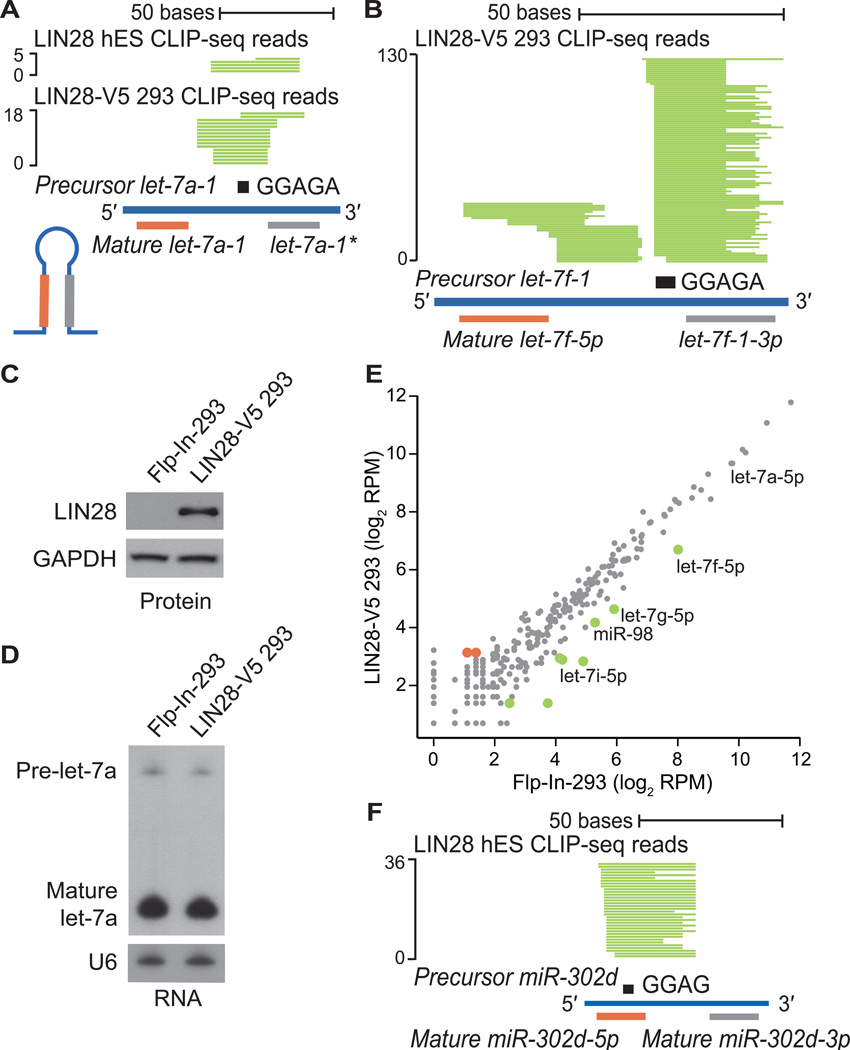
We identified 32 and 56 pre-miRNAs in hES and LIN28-V5 293 cells that featured LIN28 CLIP-seq reads, 15 of which were common between the two cell types (Table S2). Of the 17 pre-miRNA targets unique to hES cells, the majority were miRNAs that are more abundant in hES relative to LIN28-V5 293 cells. Similarly, more than half of the pre-miRNA targets in LIN28-V5 293 cells were more highly expressed in these cells, as compared to hES cells (Table S2). This suggests that LIN28 target specificity depends in part upon differences in cell type specific expression levels of miRNAs. Consistent with previous publications, we found evidence of LIN28 binding within all let-7 family members, such as let-7a-1, let-7f, let-7g, let-7i and miR-98 pre-miRNAs (Figure 2A, 2B and Table S2)(Hagan et al., 2009; Heo et al., 2008; Heo et al., 2009; Piskounova et al., 2008). CLIP-seq reads centered on the let-7 precursor loop fall precisely within the reported LIN28 interaction site at a GGAGA motif (Figure 2A and 2B) (Heo et al., 2009; Nam et al., 2011). To minimize the contribution of let-7 regulation in our study, we have selected a LIN28-V5 293 cell line with expression of LIN28 that did not alter the levels of highly abundant mature let-7a (Figure 2C, S1B and S1I), as confirmed both by Northern blot analysis (Figure 2D) and by deep sequencing the small RNA fraction of these cells (Figure 2E). Nevertheless, LIN28 targets let-7f, let-7g, let-7i and miR-98, which are expressed ~10–100 fold lower than let-7a, were reduced in the presence of LIN28-V5 expression (Figure 2E). CLIP-seq also identified other LIN28-interacting miRNAs, such as miR-302 family members (Figure 2F and Table S2), consistent with a previous report (Balzer et al., 2010). Of these many LIN28-interacting miRNAs, only the levels of let-7 family members appear to be directly affected by LIN28 binding in this system.
Figure 2. CLIP-seq defines LIN28 binding sites within miRNA precursors.
(A, B) Individual LIN28 CLIP-seq reads (in green) aligned to (A) precursor miRNA let-7a-1 and (B) precursor miRNA let-7f-1, with the mature miRNA boundaries depicted below. The sequence GGAGA in the hairpin loop is depicted as a black rectangle. The scale to the left indicates the number of aligned reads. (C) Western blot analysis of LIN28 protein levels in control Flp-In-293 and LIN28-V5 293 cells. GAPDH serves as a loading control. (D) Northern blot analysis of the human let-7a miRNA in control Flp-In-293 and LIN28-V5 293 cells. The U6 snRNA serves as a loading control. (E) Scatter plot comparing the log2 RPM (reads per million mapped) for expressed mature miRNAs in control Flp-In-293 and LIN28-V5 293 cells (gray), showing significantly upregulated (red) and downregulated (green) miRNAs. (F) LIN28 CLIP-seq reads (in green) aligned to precursor mir-302d, centered on the motif GGAG (black rectangle).
LIN28 binds mRNA sequences at GGAGA(U) motifs
The resolution of binding sites identified by CLIP-seq was exploited to identify motifs that characterize the interaction of LIN28 with mRNA sequences. The pentamer with the strongest statistical enrichment in LIN28 binding sites from both hES and LIN28-V5 293 cells was GGAGA (p < 10−4, Z-score analysis) (Figure 3A). Despite occurring two-fold higher than control clusters, this exact pentamer was neither necessary nor sufficient for LIN28 interaction, as only 13% (or 8%) of LIN28 hES (or LIN28-V5 293) clusters contained the sequence GGAGA. Nevertheless, this sequence element is enriched even in binding sites within lowly expressed transcripts, showing that we have captured LIN28 interaction with genes expressed across a wide spectrum of levels (Figure S2A). HOMER, a de novo differential motif discovery algorithm (Heinz et al., 2010) confirmed statistically significant enrichment for degenerate GGAGA (LIN28 hES) and GGAGAU (LIN28-V5) motifs (Figure 3B; P < 10−46). These motifs were prominently located at the center of LIN28 clusters in hES and LIN28-V5 293 cells in both coding exons (Figure 3C and S2B) and also within 3′UTRs (Figure 3D and Figure S2C), confirming that this signal is not attributed to nucleotide biases within coding regions.
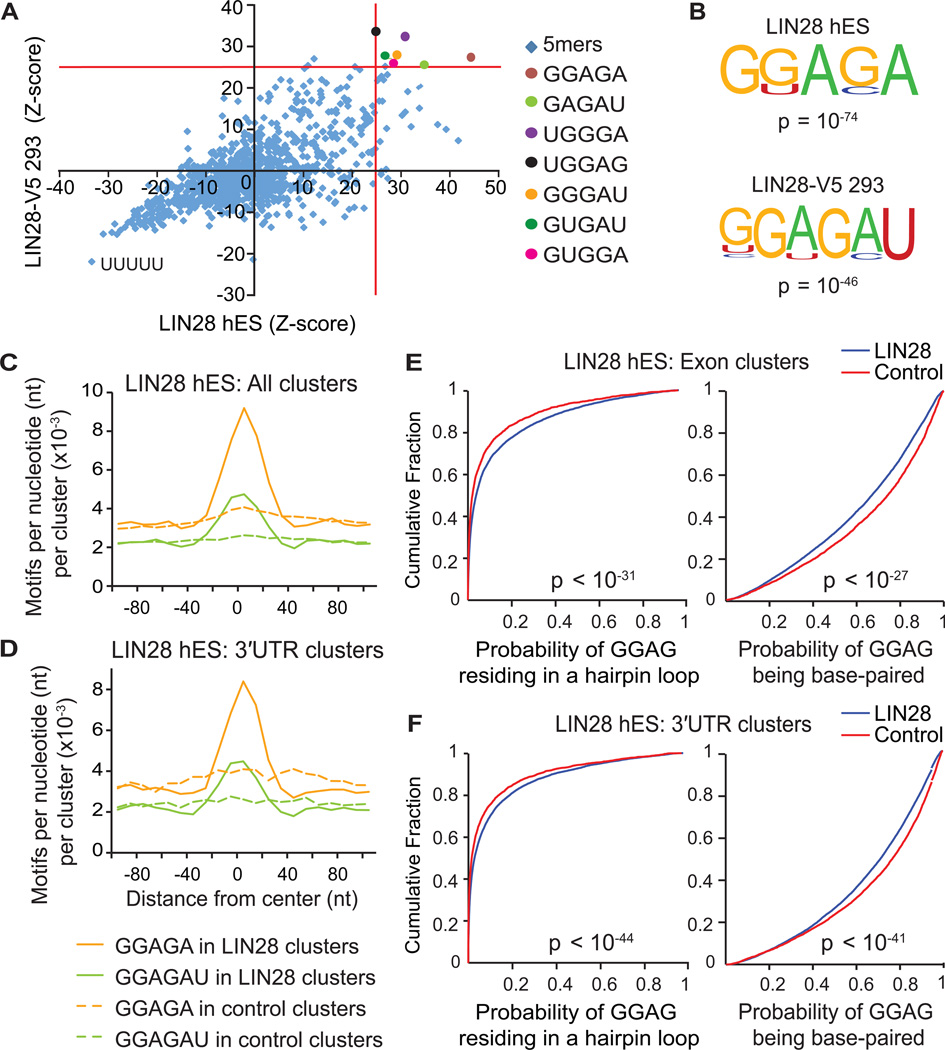
Figure 3. LIN28 binds GGAGA(U) motifs in mRNA sequences within hairpin loop structures.
(A) Scatter plot comparing pentamer Z-scores in hES and LIN28-V5 293 cells. Pentamers overrepresented (p < 10−4) in both cell-types are highlighted by colored circles, and defined on the right. (B) Consensus motifs within LIN28 clusters identified by the HOMER algorithm (Heinz et al., 2010) in hES and LIN28-V5 293 cells with corresponding p-values shown below the motif. (C and D) The positional frequency of consensus motifs GGAGA and GGAGAU relative to the center of (C) all LIN28 hES clusters and (D) clusters only in 3′UTRs. Dashed lines correspond to the positional frequency of these motifs within randomly distributed control clusters from the same type of genic region. (E and F) Cumulative distribution plots display the probability that each nucleotide of a GGAG sequence found within LIN28 hES clusters (blue) or control clusters (red) resides in a predicted hairpin loop (left panel) or base-paired region (right panels) of mRNA; (E) considering clusters only in exons, or (F) clusters only in 3′UTRs (p-values calculated by two-sample Kolgomorov-Smirnov test).
Although the sequence GGAG has been reported as the functional binding site of LIN28 in the terminal loop of let-7 miRNAs (Heo et al., 2009), we observed that the full sequence GGAGAU is conserved across let-7 pre-miRNA family members at this location. Crystal structures of mouse Lin28 in complex with let-7 pre-miRNAs confirmed that the zinc-finger knuckles of LIN28 interact with this GGAG motif (Nam et al., 2011), and also provided evidence that the CSD binds another discrete structural element within the precursor terminal loop containing the consensus motif NGNGAYNN (Y = pyrimidine; N = any base), which constitutes the expanded sequence GGAGAU that we have identified. Thus, we conclude that LIN28 interacts with a consensus GGAGA(U) motif within miRNA, as well as mRNA, sequences.
LIN28 shows a preference for unpaired mRNA regions of secondary structure
Since LIN28-miRNA interactions occur in the context of RNA secondary structures, we hypothesized that LIN28 might also interact with its motifs within a structural context in mRNA transcripts. As previously performed for a range of RBPs (Kazan et al., 2010; Li et al., 2010; Zisoulis et al., 2010), we applied the algorithm RNAplfold (Bernhart et al., 2006) to analyze LIN28-bound mRNA regions for structural features. Using RNA folding simulations, we calculated the likelihood for each position in two variants of our consensus motif, GGAG or GNGAY, to base-pair within stretches of ~200 nucleotides. These calculations enabled us to assign a probability that the motif frequently occurs in a hairpin, external, internal, or multi-loop, or is base-paired. Our results indicate a significant preference for GGAG and GNGAY motifs within LIN28 clusters to reside in hairpin and other loop structures (Figures 3E, 3F, S2D, S2E, left panels, and S2F), both in exons (Figures 3E and S2D) and in 3′UTRs (Figures 3F and S2E), relative to instances of these motifs in control clusters. We also concluded that GGAG and GNGAY motifs within LIN28 binding sites are less frequently base-paired (Figures 3E, 3F, S2D, S2E, right panels and S2F). While complex structures with an ‘A’ bulge in a handful of genes have been suggested to interact with LIN28 (Lei et al., 2011), our results demonstrate that LIN28 preferentially interacts directly with mRNA transcripts at GGAGA(U) sequence motifs within regions of unpaired secondary structure.
LIN28 binds to its own mRNA as a mode of autoregulation
CLIP-seq in hES cells provided evidence that LIN28 binds within its own mRNA, primarily in its 3′UTR where there were 13 significant clusters, the majority of which harbored GGAGA motifs (Figure 4A). Previous studies have suggested that LIN28 may bind to its own mRNA; however, experimental support was not presented (Polesskaya et al., 2007). We confirmed this interaction by RIP analysis of LIN28 in the HUES6 hES cell line (to complement our independent CLIP-seq experiments using the H9 line) (Figure 4B). Quantitative RT-PCR using primers recognizing the 3′UTR of the endogenous LIN28 mRNA showed a three-fold increase in steady state mRNA in LIN28-V5 293 cells compared to control Flp-In-293 cells (Figure 4C).
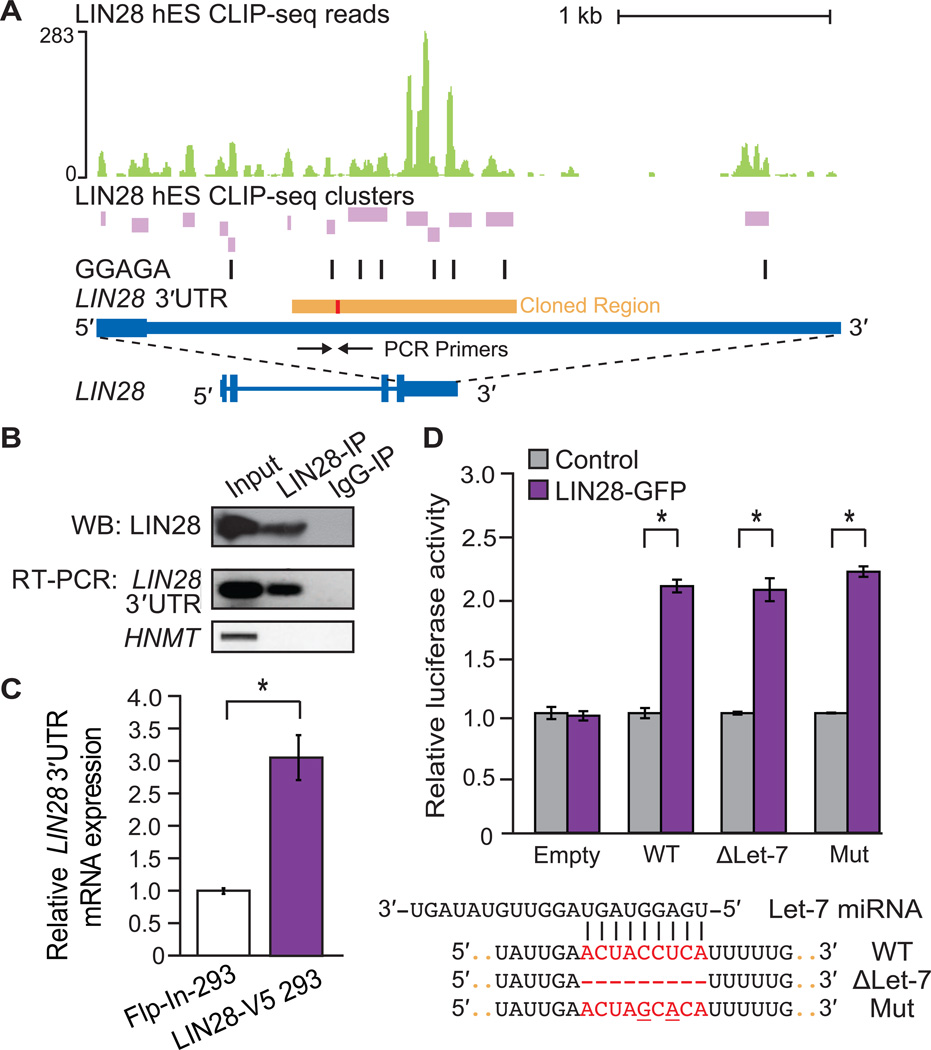
Figure 4. LIN28 binds to its own 3′UTR to positively autoregulate.
(A) LIN28 H9 hES CLIP-seq reads (graphed as continuous densities in green) and clusters (in purple) falling within the LIN28 3′UTR. Instances of GGAGA motifs within clusters are shown (black boxes). The scale to the left indicates the height of aligned reads. A portion of the LIN28 3′UTR (orange) containing a let-7 binding site (red) was cloned downstream of a luciferase open-reading frame (ORF) reporter (see (D)). (B) Western blot (WB) analysis using an antibody recognizing endogenous LIN28 in lysates after immunoprecipitation (IP) of LIN28 and bound RNA transcripts in HUES6 hES cells. IgG was used as an IP control. RNA isolated from the IP was also used for RT-PCR experiments to confirm IP of the endogenous LIN28 3′UTR (primers shown as arrows in (A)), and a negative control, HMNT, that is not bound by LIN28. (C) Quantitative RT-PCR analysis showing increased mRNA levels of endogenous LIN28 in LIN28-V5 293 relative to control Flp-In-293 cells, and normalized to GAPDH levels (*p < 0.05, Student’s t-test, error bars ± s.d.). (D) Relative luciferase activity of reporters containing a portion of the wild-type (WT) LIN28 3’UTR (as depicted in A), or deletion (ΔLet-7) of or mutations (Mut) within a sequence complementary to the let-7f miRNA, when co-transfected in Flp-In-293 cells with a LIN28-GFP expression vector (purple) or with an unrelated control vector (grey) (*p < 0.001, Student’s t-test, error bars ±s.d.). A control luciferase reporter lacking the partial LIN28 3′UTR (Empty) was unaffected by LIN28-GFP.
To evaluate if the LIN28-bound sequence within the LIN28 3′UTR is sufficient to enhance expression levels in a heterologous context, the region containing the highest density of LIN28 clusters (Figure 1A, “Cloned Region”) was inserted downstream of a luciferase reporter. Co-transfection of the reporter with a plasmid expressing LIN28-GFP demonstrated that this region of the LIN28 3′UTR is sufficient to enhance luciferase activity, whereas transfection of a control plasmid had no effect (Figure 4D). As it is thought that LIN28 can be regulated by let-7, we noted that the increased luciferase activity might be due to a relief from repression by let-7. However, neither deletion nor mutation of the let-7 complementary site (as performed by Mayr and colleagues (Mayr et al., 2007)) within the LIN28 3′UTR reporter construct increased luciferase levels beyond those observed from LIN28-GFP overexpression. Therefore, we conclude that LIN28 directly enhances its own expression level by binding to sites within its 3′UTR, revealing a mechanism of positive feed-forward regulation by LIN28. The transcription factors OCT4, SOX2 and NANOG, which are required for propagation of undifferentiated ES cells and are important for reprogramming, also collaborate to autoregulate themselves in feed-forward loops (Boyer et al., 2005). Our results suggest that LIN28 exhibits the same ability to affect its own protein levels.
LIN28 directly regulates the protein levels of RNA binding proteins
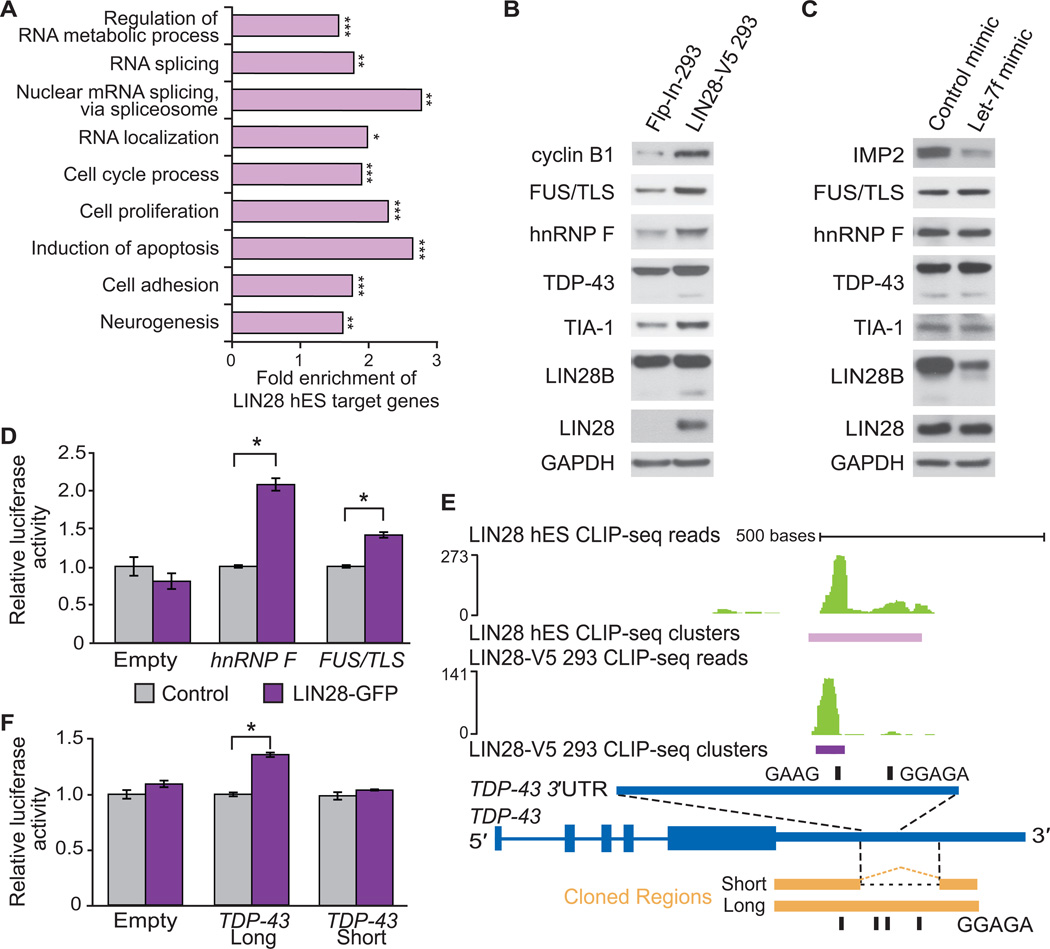
To explore potential pathways affected by LIN28, Gene Ontology (GO) analysis identified “regulation of RNA metabolic processes” (1386 target genes), “RNA splicing” (234), and “RNA localization” (87) as statistically significant RNA-related categories enriched among LIN28 target genes, as well as categories consistent with its known roles in cellular proliferation and neurogenesis (Figure 5A). To specifically address if RBPs were enriched as LIN28 targets, we analyzed a compiled set of 443 RBPs for the presence of LIN28 clusters (Huelga et al., 2012). Out of these RBPs, 248 (56%) and 236 (53%) were found to be direct targets of LIN28 in hES and LIN28-V5 293 cells, respectively, (p < 10−4, hypergeometric test).
Figure 5. LIN28 binds and regulates splicing factors.
(A) Enriched Gene Ontology (GO) terms for hES LIN28 target genes were identified using the DAVID algorithm (Huang et al., 2009). Statistical comparisons to all genes with transcripts expressed in H9 hES cells were made (*p < 10−10, **p < 10−15, ***p < 10−40). (B) Western blot analysis of splicing factors in control Flp-In-293 and LIN28-V5 293 cells. Cyclin B1 is shown as a positive control (Xu et al., 2009). All membranes were probed for LIN28 and GAPDH (used as a loading control). (C) Western blot analysis of splicing factors in LIN28-V5 293 cells transfected with a let-7f miRNA mimic or control mimic. The let-7 target IMP2 was used as a positive control. (D) Relative luciferase activity of reporters containing cloned portions of the hnRNP F or FUS/TLS LIN28-bound RNA regions co-transfected into Flp-In-293 cells with a LIN28-GFP expression vector (purple) or with a control vector (grey) (*p < 0.001, Student’s t-test, error bars ±s.d.). A control luciferase reporter lacking a LIN28-bound region (Empty) was unchanged by LIN28-GFP. (E) LIN28 CLIP-seq reads (in green) and clusters (in purple) mapped to an intronic region within the 3′UTR of the human TDP-43 gene (in blue). The scale to the left indicates the height of aligned reads. Portions of the homologous mouse TDP-43 3’UTR that contain (long) or lack (short) the intronic region that harbors the majority of LIN28 binding sites are shown aligned (in orange). These regions were inserted downstream of a luciferase reporter as previously described (Polymenidou et al., 2011). Instances of GGAGA and GAAG motifs in the respective organisms are shown (black rectangles). (F) Relative luciferase activity of reporters containing the TDP-43 3’UTR with LIN28 binding sites (long), and the TDP-43 3’UTR without LIN28 binding sites (short) co-transfected into Flp-In-293 cells with a LIN28-GFP expression vector (purple) or a control vector (grey) (*p < 0.001, Student’s t-test, error bars ±s.d.). A control luciferase reporter lacking any LIN28-bound region (Empty) was unchanged by LIN28-GFP.
To establish if direct LIN28 targets, such as genes encoding RBPs, were regulated by LIN28 at the RNA level, we conducted triplicate microarray gene expression analysis of LIN28-V5 293 and control Flp-In-293 cells (Figure S3A). Our results indicated that genes with altered expression levels were not enriched for binding relative to unchanged genes (at a p < 0.01 cutoff, chi-square test; Figure S3B), suggesting that direct targets of LIN28 are neither frequently nor significantly affected at the steady-state mRNA level when LIN28 is expressed. This result was also recapitulated with deep sequencing of cDNAs (RNA-seq) from hES cells transduced with lentivirus encoding an shRNA targeting LIN28 (Figures S3B, S3C, S3D and S3E).
To determine if LIN28 targets were instead controlled at the level of translation, we first evaluated the protein level of cyclin B1. We observed higher levels of cyclin B1 in the LIN28-V5 293 compared to control Flp-In-293 cells (Figure 5B and S3F), consistent with published results indicating that murine cyclin B1 decreases upon LIN28 depletion in mouse ES cells (Xue et al., 2009). Next we selected a number of LIN28 targets, focusing on RBPs which have published roles in regulating splicing, including FUS/TLS, hnRNP F, TDP-43 and TIA-1. These genes all increased by at least two-fold at the protein level in LIN28-V5 cells compared to control cells, but were unaltered at the mRNA level (as measured by the microarrays) (Figure 5B).
Since higher levels of LIN28 reduced let-7f expression (Figure 2E), we introduced let-7f mimics (artificial mature miRNA duplexes) that were insensitive to LIN28 regulation into LIN28-V5 293 cells to determine if the levels of these RBPs were higher due to lack of let-7f. Compared to a control mimic, the protein levels of IMP2, a known let-7 target (Yun et al., 2011) was effectively downregulated in the presence of the let-7f mimic (Figure 5C). We also noted that the paralog LIN28B protein was downregulated upon increased let-7f expression, suggesting that LIN28B is likely regulated by let-7f (Guo et al., 2006). Importantly, FUS/TLS, hnRNP F, TDP-43, and TIA-1 were unaffected in their protein levels by let-7f expression (Figure 5C and S3G), supporting the conclusion that these RBPs are directly regulated by LIN28-mRNA interactions, and not through let-7f.
Next we set out to determine whether specific LIN28-bound regions of target genes were sufficient to convey LIN28-dependent translational regulation. We cloned mRNA regions from hnRNP F (coding region; Figure S4A) and FUS/TLS (coding region and 3′UTR; Figure S4B) that contained LIN28 binding sites in both hES and LIN28-V5 293 cells downstream of a luciferase reporter. Consistent with our western blot results (Figure 5B), co-expression of LIN28-GFP, but not a control plasmid, significantly enhanced luciferase activity (p < 0.001, Figure 5D), confirming that LIN28 binding sites are sufficient to increase translational regulation of hnRNP F and FUS/TLS.
Within the 3′UTR of TDP-43, we observed LIN28 binding sites overlapping with purine-rich (GGAGA) motifs in a retained intronic region (Figure 5E). This region was previously reported to be bound and spliced by TDP-43 itself, thereby eliciting nonsense-mediated decay (NMD) to reduce its mRNA levels (Polymenidou et al., 2011). We hypothesized that when this 3′UTR-embedded intron remains unspliced and the TDP-43 mRNA is exported to the cytoplasm, the LIN28 protein could interact with binding sites in the 3′UTR to enhance translation of the mRNA. However, a spliced TDP-43 3′UTR would not contain LIN28 binding sites, and thus would not be affected by LIN28 expression. To test this hypothesis, we utilized two reporter constructs containing different arrangements of the homologous mouse TDP-43 3′UTR downstream of a luciferase open reading frame (Polymenidou et al., 2011) (Figure 5E). The first reporter, referred to as “short,” contained the spliced 3′UTR, which removed the majority of LIN28 binding sites. The second reporter, referred to as “long”, harbors an unspliced region of the TDP-43 3′UTR homologous to the human region containing LIN28 binding sites. Co-transfection of these reporter constructs demonstrated that the reporter containing the LIN28 binding sites was significantly enhanced at the translational level when LIN28-GFP was overexpressed; however, the spliced “short” construct was not (Figure 5F). Deletion of one of the four LIN28 GGAGA binding motifs within the “long” reporter reduced its translational output by ~15% in the presence of LIN28-GFP expression, suggesting that site-specific interactions of LIN28 contributes to its ability to enhance translation (Figure S4C). We conclude that LIN28 regulates TDP-43 protein levels by interacting with specific binding sites within a retained intron in the TDP-43 3′UTR. Importantly, if this intron is spliced these binding sites are not available for control of protein levels, offering an interesting example of a coupling between the regulation of splicing and translation.
Increased levels of LIN28 in somatic cells causes widespread changes in alternative splicing
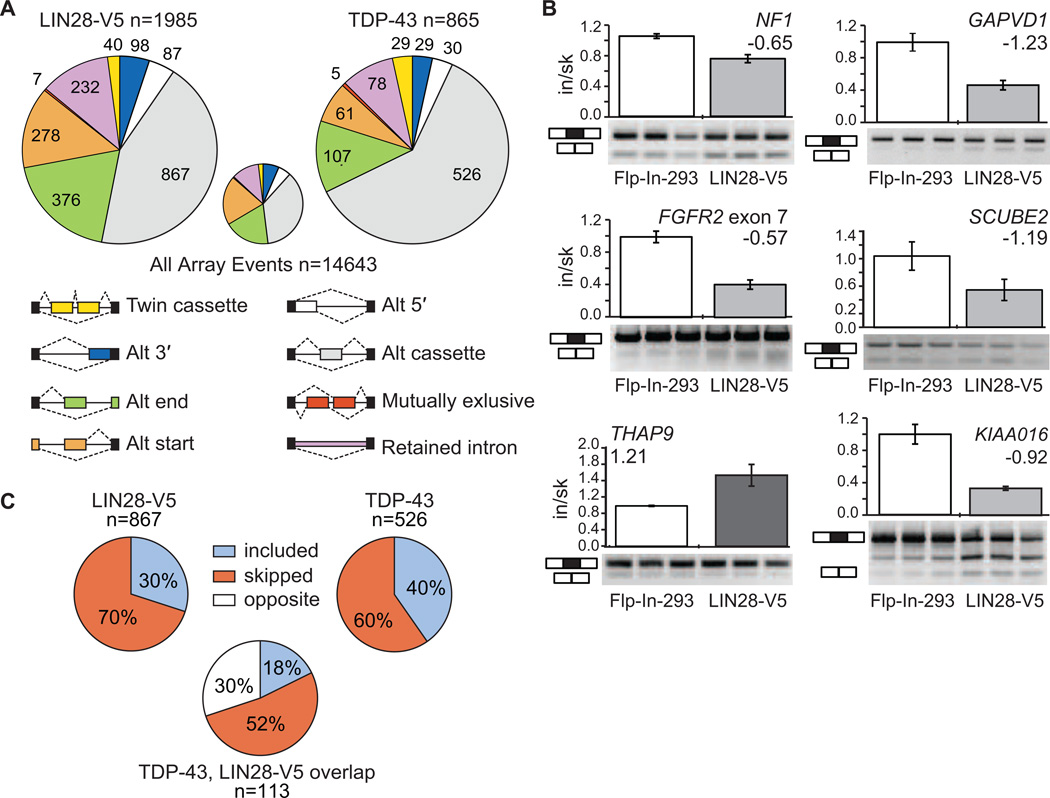
If LIN28 regulates the translation of many splicing factors, we expect that LIN28 misregulation will result in changes in alternative splicing (AS). To test this, we subjected total RNA from LIN28-V5 293 cells and control Flp-In-293 cells to splicing-sensitive microarray (HJAY) analysis. We identified 1,985 differentially regulated AS events in the presence of LIN28 expression, out of 14,643 events detected on the array (Figure 6A). These events are comprised of isoform changes in approximately 1,965 genes. This number of AS events is comparable to the numbers regulated by well-studied splicing factors such as hnRNP proteins, RBFOX2 and HuR (Huelga et al., 2012; Mukherjee et al., 2011; Venables et al., 2009). Since we found little evidence of LIN28 binding to intronic regions (Figure 1D), we reasoned that LIN28 likely interacts with cytoplasmic, mature mRNA transcripts, which suggests that the observed AS events are most likely the downstream result of LIN28 regulation of splicing factors. We successfully validated a number of these AS changes by semi-quantitative RT-PCR with an 85% validation rate (Figure 6B and S5A). As an interesting example, we validated the alternative splicing of a 63 nucleotide (nt) cassette exon 23a in the neurofibrimin 1 (NF1) gene, which is skipped upon expression of LIN28-V5 (Figure 6B). As a known negative regulator of the Ras signaling pathway, accurate control of NF1 isoforms are important in cancer and neuronal differentiation (Patrakitkomjorn et al., 2008), thereby providing a glimpse into signaling pathways that LIN28 may affect through regulation of AS.
Figure 6. LIN28 expression in somatic cells results in thousands of alternative splicing events, in part through regulation of TDP-43 levels.
(A) The pie charts display the number of each type of alternative splicing event changed upon overexpression of LIN28-V5 (left) or TDP-43 (right) in Flp-In-293 cells, as detected by splicing-sensitive microarray analyses. The small pie chart (center) represents the distribution of alternative splicing event types detected on the microarray. (B) RT-PCR validations of alternative cassette events detected by microarray analysis. All plots show significant differences between control Flp-In-293 and LIN28-V5 293 cells (p < 0.05, Students t-test). Bars represent an average and error bars represent the standard deviation across biological triplicates. (C) The percent of included versus skipped alternative cassette exons upon overexpression of LIN28-V5 (left) or TDP-43 (right) in Flp-In-293 cells. For the alternative cassette exons that changed in both conditions (n = 113), the percent of exons affected in the same (where the exon is included or skipped in both conditions) or opposite direction are shown below.
To analyze the extent of alternative splicing events affected due to the regulation of a single splicing factor by LIN28, we overexpressed a plasmid harboring the open reading frame of TDP-43 fused to a C-terminal GFP in Flp-In-293 cells, reproducing the upregulation of TDP-43 upon LIN28 expression observed in LIN28-V5 293 cells. We subjected total RNA to splicing-sensitive microarray analysis (Figure S5B), identifying a total of 865 AS events that changed, including 526 differentially spliced cassette exons (Figure 6A). Of the cassette exons affected by stable LIN28-V5 expression in our cell line, we identified a significantly overlapping subset of 113 cassettes (13%) that were also affected upon upregulation of TDP-43 (p < 10−5, hypergeometric test), with 70% of the cassette events changing in the same direction (Figure 6C). Of the hundreds of splicing factors that LIN28 is predicted to regulate, LIN28 affects a statistically significant overlapping set of alternative splicing events with at least one splicing factor, TDP-43.
Decreased levels of LIN28 and LIN28B in embryonic stem cells modulates translation of RNA binding proteins
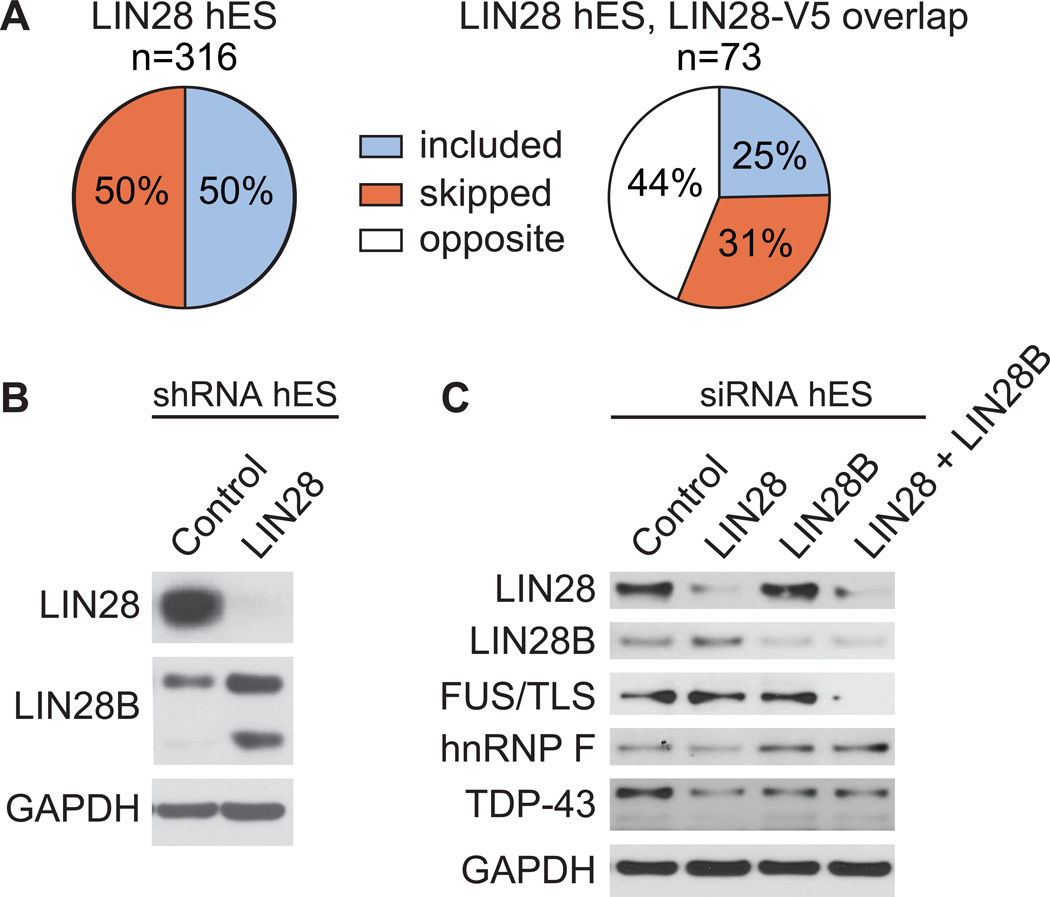
We were surprised to find that depletion of LIN28 in hES cells resulted in less than half of the number of AS events as in LIN28-V5 293 cells, and that few of these events were reciprocal (Figure S5C and 7A). In addition, despite the high concordance between hES and LIN28-V5 293 cells of the location of LIN28 binding sites on target mRNAs, its splicing factor targets did not display a decrease in protein levels expected upon knockdown of LIN28 (Figure 7C). Given that LIN28B, the paralog of LIN28, was significantly enhanced when LIN28 was depleted in hES cells (Figure 7B) and that LIN28 and LIN28B interact with a common set of mRNAs encoding splicing factors (Figure S5D), we hypothesized the LIN28B may compensate for loss of LIN28. To address this relation between LIN28 and LIN28B, we electroporated hES cells with siRNAs that individually depleted LIN28 and LIN28B, as well as both proteins simultaneously (Figure 7C). Interestingly, we observed that hnRNP F increases at the protein level with depletion of LIN28B, TDP-43 is downregulated when either LIN28 or LIN28B was depleted but not further downregulated by depletion of both, and FUS/TLS was reduced only when both LIN28 and LIN28B were concurrently depleted. Therefore, LIN28 and LIN28B may exhibit synergistic (FUS/TLS), and both repressive (hnRNP F) and enhancing (TDP-43, FUS/TLS) effects on translation of their mRNA targets in stem cells. Our observations that LIN28 and LIN28B have differing effects on their targets, and that LIN28 levels affect LIN28B expression (Figure 7B), reveal another layer of complexity ripe for future investigation. These studies will be important to address the extent of this functional overlap between LIN28 and LIN28B, and to identify co-factor complexes that underlie differences in cell type and gene-specific regulation by these proteins.
Figure 7. LIN28 and LIN28B affect splicing factors differently in human ES cells.
(A) The percent of included versus skipped alternative cassette exons upon depletion of LIN28 in hES cells is shown. Of the alternative cassette events changed in both the LIN28 hES and LIN28-V5 293 experiments (n = 73), the percent of exons affected in the same (where the exon is included or skipped in both conditions) or opposite direction are shown below. The direction of cassette exon splicing changes due to LIN28 depletion in hES cells is flipped to correspond to LIN28 overexpression. (B) Western blot analysis of LIN28B levels upon shRNA-mediated depletion of LIN28 in hES cells. GAPDH serves as a loading control. (C) Western blot analysis of LIN28, LIN28B, and splicing factors upon siRNA-mediated depletion of LIN28, LIN28B, or both in hES cells.
DISCUSSION
Systematic, genome-wide identification of thousands of LIN28 binding sites revealed that more than 6,000 genes are targets of LIN28 in hES cells and in somatic cells where LIN28 was exogenously introduced. We report the identification of a GGAGA(U) motif within LIN28 mRNA binding sites which resembles the sequence and structural context of the interaction with let-7 miRNA precursors. We also provide evidence of LIN28 autoregulation by direct binding to its own mRNA. Independent of prerequisite alteration of let-7 levels, we find that LIN28 binds to mRNA regions within transcripts that code for splicing factors, including TDP-43, FUS/TLS, TIA-1, and hnRNP F and controls their protein abundance. Upregulation of protein levels of these targets in response to an increase in LIN28 in somatic cells leads to widespread changes in alternative splicing patterns. Surprisingly, downregulation of LIN28 in hES cells does not always result in reciprocal changes for these RBPs. Furthermore, LIN28B does not in general compensate for lack of LIN28 function, despite also interacting with mRNAs encoding these RBPs, and has different, or sometimes synergistic, effects on these targets. This cell type specific control of gene regulatory targets by LIN28 presents an alternative mechanism through which LIN28 and LIN28B expression can shape cell fate and homeostasis.
Aside from alternative splicing, the RBP targets of LIN28 are also involved in other RNA processing steps, expanding the breadth of known effects of LIN28 on gene regulation. Both TDP-43 and FUS/TLS regulate mRNA transport, translation, turnover and miRNA processing, and disruption of either protein leads to amyotrophic lateral sclerosis (Lagier-Tourenne et al., 2010). TIA-1 is a central player in the formation of stress granules, which safeguards selected mRNAs by controlling their translation and stability during cellular stress (Kedersha and Anderson, 2002). Our finding that LIN28 regulates TIA-1 expression provides another link between LIN28 and RNA regulation through control of stress granule formation (Balzer and Moss, 2007). HnRNP F protein, as well as the structurally similar hnRNP H1 protein, has been observed to co-immunoprecipitate with LIN28 (Polesskaya et al., 2007). Of note, hnRNP F and H1 (Caputi and Zahler, 2001) are known to recognize GGGA sequences in RNA. With our finding that LIN28 also binds GNGAY motifs, it is possible that these hnRNP proteins and LIN28 regulate a common set of targets. To summarize, our genome-wide study reveals avenues by which LIN28 impacts gene regulatory networks through direct regulation of its mRNA targets, and provides a valuable framework for future characterization of the molecular roles of LIN28 and LIN28B in biological pathways.
EXPERIMENTAL PROCEDURES
Cell culture and stable cell line generation
The LIN28 open reading frame (Homo sapiens, GenBank: DQ896719) was cloned from a Gateway pENTR221 vector (Open Biosystems) into the Gateway pEF5/FRT/V5 destination vector (Life Technologies) to generate the V5-tagged LIN28. To generate LIN28-V5 293 stable cell lines, pEF5/FRT/LIN28-V5 plasmid was co-transfected along with the FLP Recombinase expressing plasmid pOG44 into Flp-In-293 cells. Stably transected clones were propagated in media supplemented with 75–100 µg/ml hygromycin B (Life Technologies) and several independent clonal cell lines were established. Human ES cell lines H9 and HUES6 were grown in feeder-free conditions with mTeSR media (STEMCELL Technologies) and on Matrigel (BD Biosciences).
RNA IP experiments
RNA immunoprecipitation (RIP) experiments were performed as described (Van Wynsberghe et al., 2011) with lysates from HUES6 or Flp-In-293 cells using antibodies against LIN28 (Abcam ab46020), LIN28B (Cell Signaling 4196), or IgG (Caltag Laboratories 10500C) with beads pre-bound with either antibody.
Western blot analysis
Membrane incubations with anti-GAPDH (Abcam ab8245), anti-LIN28 (Abcam ab46020), anti-LIN28B (Cell Signaling 4196), anti-FUS/TLS (Santa Cruz Biotechnologies SC-47711), anti-TDP43 (Aviva ARP35837_P050), anti-Tia1 (Santa Cruz Biotechnologies SC-1751), anti-cyclin B1 (Abcam ab72), anti-hnRNP F (Santa Cruz Biotechnologies SC-10045), and IMP2 (MBL RN008P) were performed overnight. Secondary antibodies were used at 1:10,000 (anti-rabbit Calbiochem 401393 or Cell Signaling 7074, anti-mouse Cell Signaling 7076, anti-Goat Promega V-4771) and chemiluminescence reagents (Thermo Pierce) according to manufacturers’ recommendations.
Luciferase assays
The spliced (long) and unspliced retained intron (short) forms of the mouse TDP-43 3′UTR were cloned into the psiCHECK-2 vector (Promega) (Polymenidou et al., 2011). A portion of the human 3′UTR of LIN28 (‘Cloned Region’ Figure 4A), and mRNA sequences of FUS/TLS and hnRNP F (Figure S4A and B) were amplified using cDNA derived from LIN28-V5 293 cells. To disrupt let-7 binding to the 3’UTR of LIN28, the let-7 seed region was removed (ΔLet-7) or mutated (Mut) within a psiCHECK-2 construct containing the cloned portion of the LIN28 3’UTR. Transfection of Flp-In-293 cells was performed using Fugene 6 (Roche Applied Science) with reporter plasmid and pcDNA3.1 (Life Technologies) or LIN28-GFP (Balzer and Moss, 2007). Luciferase activity was determined using the Dual-Luciferase Reporter system (Promega). Renilla activity was normalized to firefly activity, which is used as the internal control.
Lentiviral shRNA-mediated and siRNA-mediated knockdown of LIN28 and LIN28B
To achieve knockdown of LIN28, we utilized an shRNA construct targeting human LIN28 in the pLKO.1 vector (TRCN0000102579; Open Biosystems). As a control, a pLKO.1 vector containing an shRNA toward GFP was used (Open Biosystems). To deplete LIN28 and LIN28B, we utilized On-TARGETplus SMARTpool siRNAs from Dharmacon (LIN28A: L-018411-01-0005, LIN28B: L-028584-01-0005, On-TARGETplus Non-Targeting Pool: D-001810-10-05).
TDP-43 overexpression
Flp-In-293 cells were grown to ~70% confluency and transfected with TDP-43-GFP (Liu-Yesucevitz et al., 2010) or control pEGFP-C2 (Clontech) plasmid using Lipofectamine-2000 (Life Technologies) according to manufacturer’s instructions.
Let-7f expression
Rescue of let-7f expression levels in LIN28-V5 293 cells was achieved via replicate transfections of cells with a final concentration of 5 nM human let-7f mimic (miScript syn-hsa-let-7f; Qiagen MSY0000067) or a control miRNA mimic (AllStars Negative Control; Qiagen 1027280) using Lipofectamine RNAiMax (Life Technologies).
Small RNA-Seq
Small RNA libraries were generated from total RNA isolated from H9, untreated Flp-In-293, and LIN28-V5 293 cell line using the Illumina's Small RNA Digital Gene Expression v1.5 protocol and sequenced on the Illumina GAII for 36 cycles.
CLIP-seq experiments and data analysis
Confluent human H9 or LIN28-V5 293 cells were subjected to UV cross-linking on ice. CLIP-seq libraries were constructed for LIN28 as previously described (Yeo et al., 2009) using an antibody against endogenous LIN28 (Abcam ab46020) in H9 cells, or an antibody to the V5 epitope (Sigma V8137) in LIN28-V5 293 cells. Read mapping from CLIP-seq experiments and data processing was performed as published (Polymenidou et al., 2011).
RNA-seq data processing and gene expression analysis
Strand-specific RNA-seq reads were mapped to our annotated gene structure database (Bowtie version 0.12.2, with parameters –q -e 70 –y –l 25 –n 2 –m 5 –k 5 --best --strata). Gene expression was measured as the number of reads uniquely mapped to exons of a gene, per kilobase of exon sequence for that gene, normalized by the total number of million mapped reads to genes (RPKM). Differentially expressed genes were identified using a Z-score analysis as previously described with a cutoff of Z < -2 (downregulated) or Z > 2 (upregulated) (Polymenidou et al., 2011).
Small RNA-seq data processing and mature miRNA expression analysis
Small RNA reads were mapped to the human (hg19) genome using Bowtie short read aligner (Langmead et al., 2009) and associated with coordinates of known miRNAs from mirBase18 (Griffiths-Jones, 2004; Kozomara and Griffiths-Jones, 2011). Changes in miRNA expression were calculated by Z-score analyses of the log2 fold change (RPM LIN28-V5 over RPM Flp-In-293 cells) for all miRNAs with an RPM >= 1. Mature miRNAs with an absolute Z-score >= 2 and an RPM > 1 in both cell types were considered significantly changed.
Splicing array analysis for splicing and RNA expression changes
Microarray data analysis for LIN28-V5 293 cells, untreated Flp-In-293 cells, TDP-43 overexpression in Flp-In-293 cells and H9 hES cells with control or LIN28 knockdown conditions were performed using a previously described method (Sugnet et al., 2006), with cutoff of q-value < 0.05 and an absolute separation score > 0.5 to identify alternative splicing events.
Motif analysis
Motif analysis was performed as previously described (Yeo et al., 2009) using LIN28 clusters and the randomly distributed set of control clusters counting all possible pentamers. De novo motif finding was also applied using the HOMER v3.4 differential motif discovery algorithm (Heinz et al., 2010).
Supplementary Material
HIGHLIGHTS.
LIN28 interacts with thousands of exonic and 3’UTR binding sites in human genes
LIN28 binds mRNA sequences at GGAGA(U) motifs within unpaired, secondary structures
LIN28 controls splicing factor abundance, independent of altered let-7 levels
Regulation of splicing factors by LIN28 leads to widespread splicing changes
ACKNOWLEDGEMENTS
The authors would like to thank Jonathan Scolnick for critical reading of the manuscript, D. Cleveland for the TDP-43 luciferase constructs, E. Moss for the LIN28-GFP construct, and B. Wolozin for the TDP-43-GFP construct. We thank L. Shiu, J.P.D and M. Ares for assistance with splicing array analysis. M.L.W. and T.J.S. were supported in part by the UCSD Genetics Training Program through an institutional training grant from the National Institute of General Medical Sciences, T32 GM008666. S.C.H. was funded by a National Science Foundation Graduate Research Fellowship. M.T.L. was supported by a fellowship from Genentech. This work was supported by grants to G.W.Y. from the US National Institutes of Health (HG004659, GM084317 and NS075449) and the California Institute for Regenerative Medicine (RB1-01413 and RB3-05009). G.W.Y. is an Alfred P. Sloan Research Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
All Illumina sequencing and splicing array data is accessible through the Gene Expression Omnibus (GEO) accession number XXXXX.
REFERENCES
- Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development. 2010;137:891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- Bernhart SH, Hofacker IL, Stadler PF. Local RNA base pairing probabilities in large sequences. Bioinformatics. 2006;22:614–615. doi: 10.1093/bioinformatics/btk014. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Cao D, Allan RW, Cheng L, Peng Y, Guo CC, Dahiya N, Akhi S, Li J. RNA-binding protein LIN28 is a marker for testicular germ cell tumors. Hum Pathol. 2011;42:710–718. doi: 10.1016/j.humpath.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Caputi M, Zahler AM. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H'/F/2H9 family. J Biol Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- Desjardins A, Yang A, Bouvette J, Omichinski JG, Legault P. Importance of the NCp7-like domain in the recognition of pre-let-7g by the pluripotency factor Lin28. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and posttranscriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T, Aburatani H. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. Integrative Genome-wide Analysis reveals cooperative regulation of Alternative Splicing by hnRNP Proteins. Cell Reports. 2012;1:83–184. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan H, Ray D, Chan ET, Hughes TR, Morris Q. RNAcontext: a new method for learning the sequence and structure binding preferences of RNA-binding proteins. PLoS computational biology. 2010;6:e1000832. doi: 10.1371/journal.pcbi.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic acids research. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei XX, Xu J, Ma W, Qiao C, Newman MA, Hammond SM, Huang Y. Determinants of mRNA recognition and translation regulation by Lin28. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Quon G, Lipshitz HD, Morris Q. Predicting in vivo binding sites of RNAbinding proteins using mRNA secondary structure. RNA. 2010;16:1096–1107. doi: 10.1261/rna.2017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderweyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PloS one. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular Basis for Interaction of let-7 MicroRNAs with Lin28. Cell. 2011 doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Hammond SM. Lin-28: an early embryonic sentinel that blocks Let-7 biogenesis. Int J Biochem Cell Biol. 2010;42:1330–1333. doi: 10.1016/j.biocel.2009.02.023. [DOI] [PubMed] [Google Scholar]
- Patrakitkomjorn S, Kobayashi D, Morikawa T, Wilson MM, Tsubota N, Irie A, Ozawa T, Aoki M, Arimura N, Kaibuchi K, et al. Neurofibromatosis type 1 (NF1) tumor suppressor, neurofibromin, regulates the neuronal differentiation of PC12 cells via its associating protein, CRMP-2. J Biol Chem. 2008;283:9399–9413. doi: 10.1074/jbc.M708206200. [DOI] [PubMed] [Google Scholar]
- Peng S, Chen LL, Lei XX, Yang L, Lin H, Carmichael GG, Huang Y. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 2011;29:496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Sanford JR, Coutinho P, Hackett JA, Wang X, Ranahan W, Caceres JF. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS One. 2008;3:e3369. doi: 10.1371/journal.pone.0003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet CW, Srinivasan K, Clark TA, O'Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, Pasquinelli AE. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol. 2011;18:302–308. doi: 10.1038/nsmb.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E, et al. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol. 2009;16:670–676. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. Rna. 2009;15:357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DH, Moss EG. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr Patterns. 2003;3:719–726. doi: 10.1016/s1567-133x(03)00140-6. [DOI] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet. 2010;42:626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature. 2012;486:541–544. doi: 10.1038/nature11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.