Abstract
Aging is characterized by clonal expansion of myeloid-biased hematopoietic stem cells and by an increased risk of myeloid malignancies. Exome sequencing of 3 elderly females with clonal hematopoiesis demonstrated by X-inactivation analysis identified somatic TET2 mutations. Recurrence testing found TET2 mutations in 10 out of 182 individuals with X-inactivation skewing. TET2 mutations were specific to individuals with clonal hematopoiesis without hematologic malignancies and were associated with alterations in DNA methylation.
Keywords: TET2, X-inactivation, clonality, skewing, aging, clonal hematopoiesis, methylation, hydroxymethylation, HUMARA
The aging hematopoietic system is characterized by an altered hematopoietic stem cell (HSC) compartment notable for myeloid lineage bias and for an increased incidence of myeloid malignancies(1, 2). Previous studies measuring X-inactivation ratios in females identified age-associated skewing (AAS) in blood cells, particularly within the myeloid compartment(3, 4). One of several postulated causes of AAS is the acquisition of somatic mutation(s) conferring a growth advantage and resulting in clonal hematopoiesis(5). Recent studies in mice have led investigators to hypothesize that age-related myeloid lineage bias can be explained by clonal expansion of intrinsically myeloid-biased HSCs(1, 2). However, a genetic basis for this phenomenon has not been elucidated, nor have recurrent somatic mutations been observed in patients with clonal hematopoiesis without clinically apparent hematopoietic malignancies.
We hypothesized that somatic genetic abnormalities occur in the normal aging population and are a significant cause of AAS in the myeloid compartment. We performed exome sequencing of neutrophils (PMNs) and buccal epithelial cells from three elderly individuals with AAS (Supplementary Figure 1A). In one subject we identified somatic mutations in TET2, DNMT3A, SLC39A12, ERCC6, and KIAA1919 (Supplementary Table 1). We did not identify validated somatic mutations in the other two individuals with median 110x exome sequencing coverage. Importantly, the TET2 mutation was a nonsense mutation, analogous to the inactivating TET2 mutations observed in patients with myeloid malignancies (Figure 1A and Supplementary Figure 1B)(6).
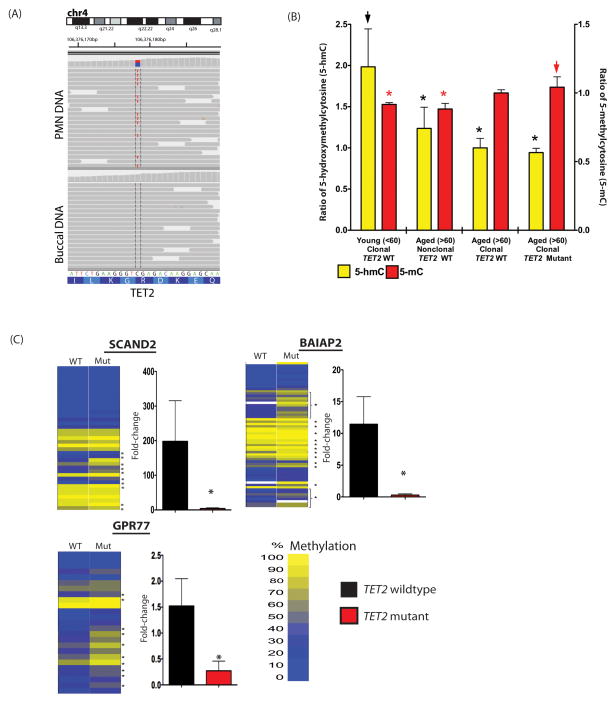
Figure 1. Somatic TET2 mutations are present in normal elderly individuals with myeloid skewing and associated with epigenetic alterations.
Exome sequencing identifies somatic TET2 mutation in myeloid cells of an elderly normal subject with non-random X-inactivation (A). LC/MS analysis reveals a decrease in 5-hmC with age and increased 5-mC in PMNs of elderly individuals with TET2 mutations (asterisk indicates p<0.05; arrowheads indicate comparator) (B). This is associated with site-specific hypermethylation and transcriptional silencing (C) in normal individuals with TET2 mutations as revealed by MassArray Epityper and qRT-PCR analysis in the PMNs of 6 TET2 wildtype and 6 mutant individuals. Error bars represent standard deviation relative to mean in panels B and C.
In order to determine if mutations in these genes recur in subjects with AAS, we performed resequencing of all five genes mutated in the index subject in 284 healthy, elderly women aged >65 years old (179 with X-inactivation skewing, 105 without skewing) and 96 younger women <60 years-old (all with X-inactivation skewing). We identified missense, nonsense, and frameshift somatic TET2 mutations, present in granulocyte DNA but not in lymphocyte or buccal DNA, in 10/182 (5.6%) of elderly subjects with AAS, in 0/105 elderly subjects without AAS, and in 0/96 younger subjects with AAS, suggesting that TET2 mutations are highly enriched in elderly subjects with AAS (p<0.007; linear logistic) (Table 1). Additional somatic mutations in DNMT3A, SLC39A12, ERCC6, and KIAA1919 were not observed (Supplementary Table 2). The TET2 mutation present in each subject was easily detectable as a dominant mutant clone (Supplementary Figure 1B and 1C), and in each case X-inactivation patterns at the HUMARA locus in PMNs further confirmed that TET2 mutant individuals had fully skewed X-inactivation consistent with clonally derived hematopoiesis (Supplementary Methods). Quantitative Ion Torrent sequencing to measure TET2 mutant allele burden in patients with TET2 mutations revealed close concordance with the degree of skewing identified by the HUMARA assay in the majority of subjects studied (Supplementary Table 3). These data are consistent with recent murine data demonstrating that Tet2 deletion or haploinsufficiency results in increased HSC self-renewal, competitive growth advantage, and bias towards the myeloid lineage(7–10)
Table 1.
Details of TET2 somatic mutations found in normal elderly individuals (n=10)1.
| Nucleotide substitution | Amino-acid substitution | Chromosome | Position |
|---|---|---|---|
| c.286_298delCGCACAGTTAGTG | p.Arg96Asnfs*12 | 4 | 106,155,385 |
| c.1330delA | p.Thr444Hisfs*6 | 4 | 106,156,429 |
| c.1348delA | p.Lys450Lysfs*2 | 4 | 106,156,447 |
| c.1547delC | p.Pro516Hisfs*16 | 4 | 106,156,646 |
| c.1630C>T | p.Arg544* | 4 | 106,156,729 |
| c.3311_3312insAT | p.Phe1104Leufs*3 | 4 | 106,158,411 |
| c.3991A>C | p.Thr1331Pro | 4 | 106,182,952 |
| c.5200delA | p.Met1734Leufs*11 | 4 | 106,196,867 |
| c.5575insT | p.Ile1859tyrfs*16 | 4 | 106,197,239 |
| c.5725G>T | p.Glu1909* | 4 | 106,197,392 |
Reference sequence used to annotate TET2 mutations was NM_001127208.
Although TET2 mutant elderly individuals had X-inactivation ratios compatible with clonal hematopoiesis, hematologic parameters in TET2 mutant elderly individuals did not differ from age-matched counterparts consistent with the absence of an overt hematological phenotype (Supplementary Figure 2 and Supplementary Table 4). Notably, we obtained clinical follow-up for 7 of the TET2 mutant subjects for 5 years or longer after mutational analysis. Although 6 patients remained without clinical evidence of a hematological malignancy, one TET2 mutant subject subsequently presented with JAK2V617F-positive essential thrombocytosis. The increased prevalence of TET2 mutations in older patients with hematological cancers(11), and the age dependence of TET2 mutations in our cohort of elderly subjects further support an initiating role for TET2 mutation in the pathogenesis of age-associated hematological cancers.
TET2 catalyzes the hydroxylation of 5-methylcytosines (5-hmC) throughout the mammalian genome, resulting in loss of DNA methylation (5-mC)(12). Consonant with these functional data we have previously shown TET2-mutant acute myeloid leukemia (AML) patients are characterized by a hypermethylation phenotype with site-specific cytosine methylation of target genes. Quantitative mass spectrometric measurement of 5-hydroxymethylcytosine (5-hmC) and 5-mC levels revealed that TET2 mutant elderly individuals with acquired clonal hematopoiesis (n=6) had reduced 5-hmC compared with younger individuals with skewed X-inactivation ratios (n=10), and increased 5-mC compared with younger clonal individuals or age-matched polyclonal controls (n=15) (Figure 1B). We also observed a decrease in global 5-hmC levels in myeloid cells with age suggesting that decreases in 5-hmC may be an important feature in the aging hematopoietic system, of which acquired TET2 mutations are one mechanism. In order to determine if the global changes in 5-mC/5-hmC were associated with site-specific changes in promoter methylation, we performed quantitative DNA methylation analysis of loci known to be hypermethylated in AML patients with TET2 mutations(13). This revealed significant, focal increases in site-specific promoter methylation in elderly subjects with TET2 mutations (7 individuals with TET2 mutations and 7 TET2 wildtype age-matched controls), which were associated with transcriptional silencing of these target genes (n=3 genes) (Figure 1C).
Here we report somatic, recurrent TET2 mutations in normal elderly individuals with acquired clonal hematopoiesis. These data, in combination with recent studies in murine systems, support a model where TET2 mutation in a HSC confers enhanced self-renewal and clonal expansion resulting in age-related myeloid lineage bias. More broadly, we report the existence of clonal, somatic mutations in the hematopoietic compartment of normal individuals without overt hematopoietic malignancies. The potential risk of transformation of such mutations must be determined prospectively in larger cohorts of aging individuals, and additional studies will likely identify additional somatic mutations which cause clonal hematopoiesis and which predispose to the development of hematologic malignancies.
Supplementary Material
Acknowledgments
We thank members of the Levine and Busque laboratories for helpful comments and discussion. This work was supported by a grant from the National Cancer Institute Physical Sciences Oncology Center (U54CA143798-01) to RLL and AM, by grant 1R01CA138234-01 to RLL, and by grants CA129831 and CA129831-03S1 to LAG. AV is supported by an NIH F32 award. MEF is supported by a Leukemia and Lymphoma Society Special Fellow award and a Doris Duke Charitable Foundation Clinical Scientist Development Award. OAW is an American Society of Hematology Basic Research Fellow. AM is a Burroughs Wellcome Clinical Translational Scholar and is also supported by the Sackler Center for Biomedical and Physical Sciences. RLL is an Early Career Award Recipient of the Howard Hughes Medical Institute. RLL and AM are funded by Leukemia and Lymphoma Society Scholar Awards.
Footnotes
AUTHOR CONTRIBUTIONS
L.B., J.P.P, O.A-W., and R.L.L. designed the study. L.B., S.P., Z.H., and L.M. collected samples and provided genetic and clinical annotation. J.P.P, J.L., A.V., A.H., M.H., N.S., P.K.B., C.E.M., and C.B., performed sequence analysis and validated mutations. M.F., A.V., A.M., and L.E.G. performed methylation analysis. M.G. performed statistical analysis. L.B., J.P.P, O.A-W., and R.L.L. wrote the manuscript.
References
- 1.Beerman I, Maloney WJ, Weissmann IL, Rossi DJ. Curr Opin Immunol. 2010;22:500. doi: 10.1016/j.coi.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beerman I, et al. Proc Natl Acad Sci U S A. 2010;107:5465. doi: 10.1073/pnas.1000834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busque L, et al. Blood. 1996;88:59. [PubMed] [Google Scholar]
- 4.Gale RE, Fielding AK, Harrison CN, Linch DC. Br J Haematol. 1997;98:512. doi: 10.1046/j.1365-2141.1997.2573078.x. [DOI] [PubMed] [Google Scholar]
- 5.Busque L, Gilliland DG. Leukemia. 1998;12:128. doi: 10.1038/sj.leu.2400936. [DOI] [PubMed] [Google Scholar]
- 6.Delhommeau F, et al. N Engl J Med. 2009;360:2289. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 7.Moran-Crusio K, et al. Cancer cell. 2011;20:11. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quivoron C, et al. Cancer cell. 2011;20:25. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, et al. Blood. 2011 [Google Scholar]
- 10.Ko M, et al. Proc Natl Acad Sci U S A. 2011;108:14566. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tefferi A, et al. Leukemia. 2009;23:905. [Google Scholar]
- 12.Ko M, et al. Nature. 2010;468:839. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figueroa ME, et al. Cancer Cell. 2010;18:553. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.