Summary
A large group of E3 ubiquitin ligases is formed by the multisubunit SCF complex, whose core complex (Rbx1/Cul1-Cdc53/Skp1) binds one of many substrate recruiting F-box proteins to form an array of SCF ligases with diverse substrate specificities. It has long been thought that ubiquitylation by SCF ligases is regulated at the level of substrate binding. Here we describe an alternative mechanism of SCF regulation by active dissociation of the F-box subunit. We show that cadmium stress induces selective recruitment of the AAA+ ATPase Cdc48/p97 to catalyze dissociation of the F-box subunit from the yeast SCFMet30 ligase to block substrate ubiquitylation and trigger downstream events. Our results not only provide an additional layer of ubiquitin ligase regulation but also suggest that targeted, signal-dependent dissociation of multisubunit enzyme complexes is an important mechanism in control of enzyme function.
Introduction
Modification of proteins by covalent attachment of ubiquitin regulates many processes in cells. Ubiquitylation is catalyzed by the E1-E2-E3 enzyme cascade and can control abundance, localization, or activity of substrates. E3 ubiquitin ligases have emerged as the central and most diverse components governing these processes because both ubiquitin conjugation and substrate selection depend on the E3 component (Hershko and Ciechanover, 1998; Kerscher et al., 2006; Pickart, 2004; Varshavsky, 2012). One large group of E3 ligases is formed by the multi-subunit cullin-RING ligases (CRLs) (Deshaies and Joazeiro, 2009; Duda et al., 2011; Petroski and Deshaies, 2005; Zimmerman et al., 2010), which include the well-studied ligase subfamily, the SCF (Skp1-cullin-F-box) complex. The SCF core components consist of the RING-finger protein Rbx1, the scaffold Cul1 (yeast Cdc53), and a linker protein Skp1 that associates with different F-box proteins to form a range of SCF ligases (Cardozo and Pagano, 2004; Petroski and Deshaies, 2005; Willems et al., 2004). F-box proteins directly bind substrates and determine substrate selectivity of the various SCF complexes (Jin et al., 2004). Substrate binding to SCF ligases is generally initiated by phosphorylation and other post-translational modifications on target proteins and has been considered the most important regulatory step in ubiquitylation (Petroski and Deshaies, 2005). However, induced disassembly of the multi-subunit SCF-type ubiquitin ligase complexes has also been suggested as a regulatory process to control substrate ubiquitylation (Barbey et al., 2005; Li and Johnston, 1997; Yen et al., 2005; Zhang et al., 2004). The mechanism for SCF complex dissociation is not known.
The most convincing support for SCF complex integrity as a target for regulation stems from studies of the S. cerevisiae ubiquitin ligase SCFMet30, for which dissociation of the F-box subunit Met30 from the core ligase in response to cadmium stress has been demonstrated (Barbey et al., 2005; Yen et al., 2005). SCFMet30 coordinates cell cycle progression with biosynthetic pathways of sulfur-containing metabolites, such as methionine, cysteine, and S-adenosylmethionine. In addition to these metabolic pathways, the cellular response to cadmium and arsenic stress is also largely dependent on the SCFMet30 ubiquitin ligase (Kaiser et al., 2006). Active SCFMet30 ubiquitylates the transcriptional activator Met4 to maintain it in a dormant state during normal growth conditions and ubiquitylates the zinc-finger protein Met32 to induce its degradation by the proteasome (Ouni et al., 2010). Both nutrient and heavy metal stress inactivate SCFMet30, leading to active, non-ubiquitylated Met4 and stabilization of Met32, which together induce cell cycle arrest and activation of a transcriptional program to restore normal levels of sulfur containing metabolites or activate the defense system for protection against heavy metal stress (Kaiser et al., 2006).
The yeast AAA-ATPase Cdc48 was first linked to the UPS many years ago (Ghislain et al., 1996) and has since attracted much attention as an important component of the UPS (Meyer et al., 2012). Cdc48 and the mammalian ortholog p97/VCP associate with a number of cofactors, which typically contain ubiquitin-binding domains and likely specify the diverse biological functions of Cdc48 (Haines, 2010; Schuberth and Buchberger, 2008; Yeung et al., 2008). The biological activity most often associated with Cdc48/p97 is that of a segregase (Jentsch and Rumpf, 2007) because Cdc48/p97 is involved in the separation of macromolecules such as dislocation of unfolded proteins from the ER (Ye et al., 2001), release of membrane-bound transcription factors (Rape et al., 2001; Shcherbik and Haines, 2007), extraction of chromatin-associated proteins (Acs et al., 2011; Franz et al., 2011; Raman et al., 2011; Verma et al., 2011; Wilcox and Laney, 2009), and ribosomal subunit dissociation (Fujii et al., 2012)
Here we report a role for Cdc48 in the ubiquitin system: the signal-induced disassembly of a cullin-RING ubiquitin ligase complex to block ubiquitylation of substrates. Our results highlight the role of Cdc48 in control of E3 ligase activity and suggest active, signal-initiated disassembly of multi-subunit enzymes as a mode of enzyme regulation.
Results
Selective dissociation of Met30 from Skp1 in response to cadmium stress
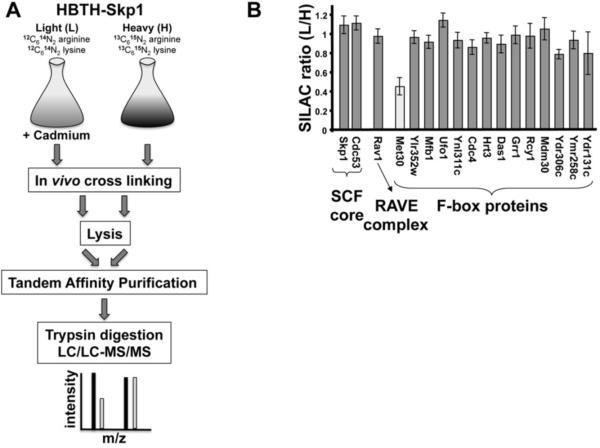
Substrate ubiquitylation by cullin-RING ubiquitin ligases and, in particular, by the SCF subfamily is thought to be primarily regulated at the level of substrate binding. However, direct regulation of SCF complex integrity has been suggested (Barbey et al., 2005; Li and Johnston, 1997; Yen et al., 2005). To test specificity and understand the molecular mechanism of SCF disassembly, we focused on the cadmium-induced disruption of the yeast SCFMet30 ubiquitin ligase (Barbey et al., 2005; Yen et al., 2005). In order to function as a specific regulatory mechanism for substrate ubiquitylation, signal-induced SCF complex disassembly should only affect specific F-box proteins. We therefore asked whether cadmium stress selectively disassembles the SCFMet30 complex or has a more general effect on SCF ligases. Skp1 protein complexes were purified from cells exposed to cadmium stress and compared to Skp1 complexes from unexposed cells by mass spectrometry coupled with stable isotope labeling with amino acids in cell culture (SILAC) (Figure 1A). Quantitative comparison of Skp1-associated F-box proteins showed specific dissociation of Met30, demonstrating selective disruption of the SCFMet30 complex (Figure 1B). Skp1 binding to none of the other detected F-box proteins was sensitive to cadmium stress, and interaction with Cdc53 or the SCF-independent partner Rav1 (Seol et al., 2001) was constant. These data demonstrate selective, signal-induced disruption of an SCF complex as a mechanism for inhibition of substrate ubiquitylation.
Figure 1.
Specific disassembly of SCFMet30 in response to cadmium. (A) Flowchart of QTAX strategy used in panel B. Cells expressing endogenous HBTH-tagged Skp1 were grown in heavy (H: 13C/15N-arginine and lysine) or light (L: 12C/14N-arginine and lysine) media for metabolic proteome labeling. The light culture was exposed to 300μM cadmium chloride for 20 min. Proteins were in vivo cross-linked using 1% formaldehyde to preserve protein complexes and total cell lysates were mixed at equal amounts. HBTHSkp1 complexes were purified under denaturing conditions and tryptic peptides analyzed by LC/LC-MS/MS. (B) Selective disassembly of SCFMet30 during cadmium stress detected by the QTAX strategy. Relative abundance ratios of SCF components are expressed as the average peptide intensity ratios of light (L: cadmium exposed) over heavy (H: unexposed) labeled peptides. Data are represented as mean ± SD. Expression levels of SCF components are relatively unaffected by cadmium stress, with the exception of Met30, which is significantly induced in response to cadmium treatment (Fauchon et al., 2002).
Cdc48 functions in the SCFMet30-mediated cadmium response
Given this specificity of the cadmium signal, we reasoned that we could discover components in the SCFMet30 disassembly pathway by detecting changes in Met30 binding partners. Using a mass spectrometry-based approach, we isolated the homohexameric AAA-ATPase Cdc48/p97 as a cadmium-specific interactor of Met30 (data not shown). Because of its known segregase activity, Cdc48 was a good candidate for factors important in SCFMet30 disassembly. Co-immunopurification experiments confirmed the mass spectrometry results and showed that low amounts of Cdc48 were bound to the F-box protein Met30 under normal growth conditions, but that rapid association of Cdc48 with Met30 was induced under cadmium stress (Figure 2A and B). Importantly, Cdc48 recruitment correlated with dissociation of Met30 from Skp1. Similar disruption of the Met30/Skp1 complex was also detected by immunopurification of Skp1-bound complexes (Figures 1B, 2C, and (Barbey et al., 2005)).
Figure 2.
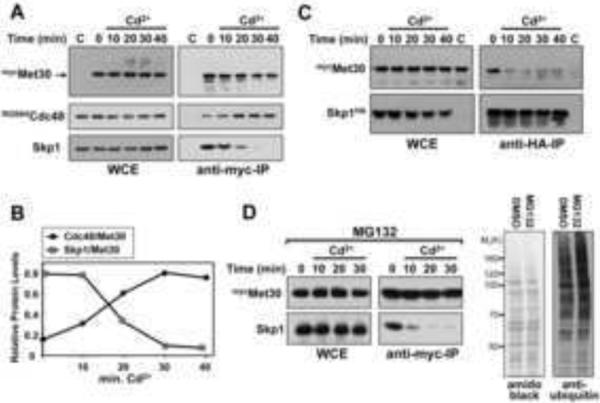
Cdc48 recruitment and disassembly of SCFMet30 during cadmium stress. (A) Cells expressing endogenous Cdc48RGS6His and 12mycMet30 were cultured at 30°C in the presence of 100μM cadmium chloride for the intervals indicated. 12mycMet30 was immunopurified, and co-purified proteins were analyzed by immunoblotting. A yeast strain expressing untagged Met30 was used as a control. WCE: Whole cell extract. (B) Band intensities in the immunocomplexes from panel A were quantified and plotted normalized to Met30. (C) Experiment as in panel A, but cells expressing endogenous Skp1HA and 12mycMet30 were analyzed by immunopurification of Skp1HA. (D) Experiment as in panel A but proteasomes were inhibited with 50μM MG132 for 45 min before cadmium stress. Cells carried a deletion of PDR5 to increase MG132 permeability (Fleming et al., 2002). Proteasome inhibition was confirmed by an increase in total ubiquitin conjugates (right panel).
Cdc48 plays important roles in delivery of ubiquitylated substrates to the proteasome for degradation (Elsasser and Finley, 2005; Richly et al., 2005; Rumpf and Jentsch, 2006). Inhibition of proteasome activity by addition of MG132 did not affect SCFMet30 disassembly demonstrating that the potential role of Cdc48 in separation of Met30 from the SCF core is independent from proteasome activity (Figure 2D).
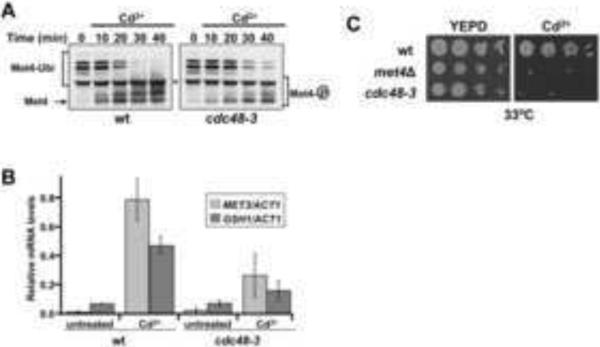
SCFMet30 represses the cadmium stress response during normal growth through ubiquitylation of its substrate Met4 (Barbey et al., 2005; Yen et al., 2005). In response to cadmium exposure, the F-box subunit Met30 dissociates from the SCF ligase core to block ubiquitylation, leading to activation of the stress response program, including induction of cell cycle arrest and expression of genes regulating glutathione and sulfur amino acid biosynthesis (Kaiser et al., 2006). Met4 continued to be ubiquitylated in cdc48 mutants during cadmium stress, indicating that Cdc48 is required for SCFMet30 inactivation (Figure 3A). The small decrease in Met4 ubiquitylation observed in cdc48 mutants is probably due to incomplete inactivation of the temperature sensitive cdc48-3 allele, but could also reflect cadmium-induced deubiquitylation of Met4, which has been reported previously (Barbey et al., 2005). Consistent with the role of Cdc48 in regulation of Met4 ubiquitylation, cadmium-induced expression of Met4 target genes was significantly reduced in cdc48 mutants (Figure 3B). These results predict an important survival function for Cdc48 during cadmium stress because Met4 activation is critical under these conditions (Baudouin-Cornu and Labarre, 2006; Fauchon et al., 2002; Kaiser et al., 2006; Wheeler et al., 2003; Yen et al., 2005). Accordingly, temperature sensitive cdc48 mutants were hypersensitive to cadmium exposure when cultured at permissive temperature (Figure 3C).
Figure 3.
Cdc48 is required for the cellular response to cadmium stress. (A) Cdc48 is required for Met4 activation during cadmium stress. Wild type cells and temperature sensitive cdc48-3 mutants were shifted to 37°C for 90 minutes and treated with 100μM cadmium chloride for the time interval indicated. Whole cell lysates were analyzed by immunoblotting with anti-Met4 antibodies to follow the ubiquitylation status of Met4. Asterisks label a protein cross-reacting with the antibody. (B) Cadmium-induced gene expression requires Cdc48. Wild type cells and cdc48-3 mutants were shifted to 37°C for 90 min and exposed to 100μM cadmium chloride for 40 min. Expression of the Met4 target genes MET3 and GSH1 was analyzed by rt-qPCR (n=3). Data are represented as mean ± SD. (C) Cdc48 is important for resistance to cadmium stress. Wild type cells, met4Δ mutants, and temperature sensitive cdc48-3 mutants were grown at 25°C to midlog phase and serial dilutions of cells were spotted on YEPD plates with or without 50μM cadmium chloride. Plates were incubated at 33°C for 3 days.
Cdc48 is required for SCFMet30 disassembly
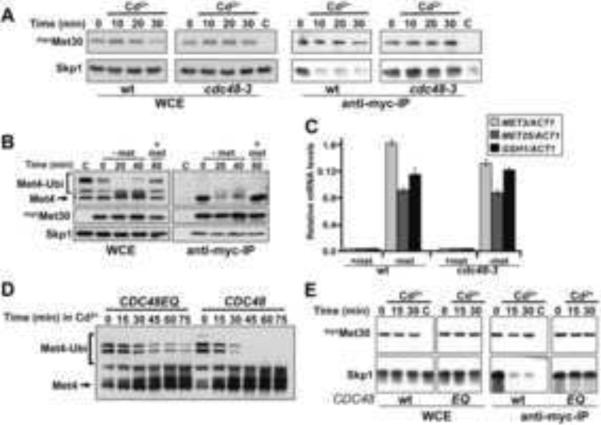
To directly test the role of Cdc48 in SCFMet30 disassembly, we analyzed Met30 immunocomplexes from temperature sensitive cdc48 mutants during cadmium stress. When temperature sensitive cdc48-3 mutants were shifted to the restrictive temperature to inactivate Cdc48, dissociation of Met30 from the SCF core was prevented (Figure 4A). Blocked SCFMet30 disassembly was not due to mitotic arrest of cdc48 mutants because Met30 readily dissociated from the SCF core in wild type cells arrested in metaphase (Figure S1A). Taken together, these results suggest a role for Cdc48 in SCF regulation.
Figure 4.
SCFMet30 disassembly depends on Cdc48 and is cadmium specific. (A) SCFMet30 disassembly depends on Cdc48. Wild type cells and cdc48-3 mutants expressing endogenous 12mycMet30 were grown at 37°C for 90 minutes, exposed to 100μM cadmium chloride and 12mycMet30 immunocomplexes were analyzed by immunoblotting. Cells expressing untagged Met30 were used as control. (B) Nutrient stress regulates SCFMet30 at the level of substrate binding. Met4 activation during methionine limitation was analyzed in cells shifted to methionine-depleted growth medium for the time intervals indicated. Met4 was re-repressed by addition of 1mM methionine to the medium. 12mycMet30 was immunopurified from cells and co-purified proteins were analyzed by immunoblotting. (C) Met4 target gene expression during methionine stress is independent of Cdc48 activity. Wild type cells and cdc48-3 mutants were shifted to 37°C for 90 minutes and then transferred to methionine-depleted growth medium for 30 minutes to activate Met4. Expression levels of the Met4-target genes MET3, GSH1, and MET25 were analyzed by rt-qPCR (n=3). Data are represented as mean ± SD. (D) ATPase activity of Cdc48 is required for its function in the cadmium stress response. Cells expressing either wild type Cdc48 or the ATPase activity deficient Cdc48EQ mutant (Ye et al., 2003) under control of the GAL1 promoter were grown in sucrose medium to early-log phase and expression of Cdc48 or Cdc48EQ was induced by addition of galactose at 30°C. Cells were exposed to 100μM cadmium chloride for the time intervals indicated and samples were analyzed by immunoblotting to monitor the Met4 ubiquitylation status. (E) Cdc48 ATPase activity is required for SCFMet30 disassembly. Immunopurification of 12mycMet30 as described in figure 2A, but cells were expressing Cdc48 (wt) or Cdc48EQ from the GAL1 promoter for 4 hrs before addition of cadmium.
Signal-specific and ATP-hydrolysis dependent SCFMet30 regulation by Cdc48
Two different signals are known to impinge on SCFMet30, namely limitation of sulfur-containing metabolites, most notably methionine, and heavy metal stress (Kaiser et al., 2006). Gene expression programs responding to sulfur-metabolite and heavy metal stress are connected because of greater demand for the cysteine-containing tripeptide glutathione for heavy metal detoxification. Increased glutathione synthesis is thus accompanied by up-regulation of sulfur amino acid production, and both metabolic pathways are coordinated by their dependence on the transcriptional activator Met4. Both methionine limitation and cadmium stress block ubiquitylation of Met4 to activate down-stream events, therefore we asked whether methionine starvation would also induce disruption of the SCFMet30 complex. In stark contrast to cadmium stress, depletion of methionine in the growth medium prevented binding of Met4 to the F-box protein Met30 but did not affect the integrity of the SCFMet30 complex as indicated by constant interaction between Skp1 and Met30 (Figure 4B). Therefore, two different molecular mechanisms account for SCFMet30 inactivation; sulfur amino acid starvation acts through the canonical pathway by blocking the substrate (Met4) F-box interaction, and cadmium exposure induces SCFMet30 disassembly. Only the latter was dependent on Cdc48 (Figure 4A), which predicted that cdc48 mutants should be selectively deficient for activation of Met4 by cadmium stress but not by methionine starvation. Accordingly, expression of Met4-dependent genes in response to methionine depletion was largely unaffected in cdc48 mutants (Figure 4C) but severely reduced in response to cadmium exposure (Figure 3B). These results indicate a direct role of the AAA+ ATPase Cdc48 in SCFMet30 disassembly and excluded a role in modulating Met4 activity downstream of its ubiquitylation.
Next we asked whether Cdc48 actively disassembles SCFMet30. We tested requirement of the Cdc48 ATPase activity for SCFMet30 regulation during cadmium stress by utilizing the Cdc48EQ mutant, which carries a glutamate to glutamine substitution in the Walker B motif of the second (D2) ATPase domain (Ye et al., 2003). This substitution has been shown to block ATP hydrolysis of Cdc48/p97 while having no effect on cofactor binding, indicating that structural integrity is not affected (Ye et al., 2003). We induced expression of either wild type Cdc48 or Cdc48EQ and monitored their effect on the cadmium stress response. Expression of Cdc48EQ significantly blocked SCFMet30 inactivation, as evidenced by a failure to block Met4 ubiquitylation (Figure 4D.) Because structural integrity is not affected significantly in Cdc48EQ (Ye et al., 2003) these results argue against an important role of Cdc48-associated deubiquitylation activity in Met4 activation. Furthermore, no single deubiquitylating enzyme had major effects on Met4 activation during cadmium stress (Figure S1B), indicating that Met4 deubiquitylation depends on redundant deubiquitylation activities, which are most likely recruited to Met4 independent of Cdc48. Most importantly, overexpression of Cdc48EQ in otherwise wild type cells was sufficient to completely block dissociation of Met30 from the SCF core (Figure 4E). Both ectopically expressed Cdc48EQ and endogenous Cdc48 were recruited to Met30 (Figure S1C), demonstrating that Cdc48EQ did not prevent binding of Cdc48 to SCFMet30 and is likely more directly blocking the dissociation step. Although we cannot definitely exclude that Cdc48EQ sequesters unknown factors required for SCFMet30 disassembly, these data strongly suggest that Cdc48 mediated disassembly of SCFMet30 is an active, ATP-driven process.
Cdc48 is recruited to the ubiquitylated F-box subunit Met30
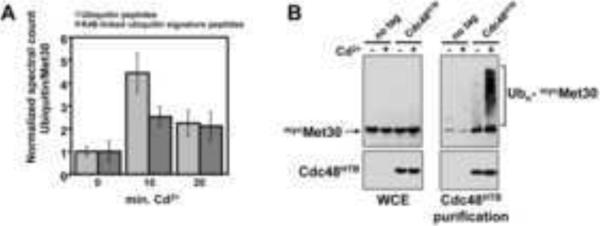
Ubiquitylation is thought to be an important part of the recognition signal for Cdc48/p97 complexes (Jentsch and Rumpf, 2007; Meyer et al., 2012; Schuberth and Buchberger, 2008; Ye et al., 2001). Mass spectrometric analysis of Met30 complexes showed a significant increase of ubiquitin when complexes were purified from cadmium stressed cells (data not sown). This was most likely due to cadmium-stimulated ubiquitylation of Met30 itself, because we detected a similar increase in ubiquitin abundance after tandem purification of Met30 under fully denaturing conditions in 8M urea buffers and washes with 2%SDS (Figure 5A). This increase in ubiquitylated Met30 was not due to a global increase in ubiquitylated proteins under these conditions (Figure S2), and likely reflects increased modification with lysine-48 linked ubiquitin chains because the abundance of signature peptides (Peng et al., 2003) characteristic for this chain topology also increased after cadmium stress (Figure 5A). No other ubiquitin-chain linkages were detected. Consistent with induction of Met30 ubiquitylation by cadmium stress, we detected multiple ubiquitylated lysine residues in Met30 specifically after cadmium treatment (Figure S3). Several ubiquitin attachment sites in Met30 might indicate modification by multiubiquitylation. However, the observed increase in chain-specific ubiquitin peptides is consistent with polyubiquitylation at flexible lysine residues and suggests that the precise site of modification is probably not critical. Indeed, mutation of all six identified ubiquitin acceptor lysines to arginine neither prevented cadmium-induced Met30 ubiquitylation nor was Cdc48 recruitment or Met30/Skp1 dissociation affected (data not shown), suggesting that these lysine residues are preferred, but not necessary, ubiquitylation sites.
Figure 5.
Cdc48 is recruited to ubiquitylated Met30. (A) Transient increase in Met30 ubiquitylation during cadmium stress. HBTH-tagged Met30 was purified in the presence of 8M urea by sequential binding to Ni2+-sepharose and streptavidin agarose from cells untreated or treated with 200μM cadmium chloride. Samples were analyzed by LC-MS/MS. Ubiquitin spectral counts and spectral counts for lysine48-linked ubiquitin chain signature peptides (Peng et al., 2003) normalized to total Met30 spectral counts are shown. Data are represented as mean ± SD. (B) Cdc48 preferentially binds ubiquitylated Met30. Cells expressing endogenous 12mycMet30 and Cdc48HTB were either treated with 100μM cadmium chloride for 30 minutes or untreated. Cdc48HTB was sequentially purified on Ni2+-sepharose and streptavidin agarose under native conditions and co-purified Met30 species were analyzed by immunoblotting with antibodies against the myc-epitope. A yeast strains expressing untagged Cdc48 was used as control.
The potential role of Met30 ubiquitylation in SCFMet30 disassembly was likely attributed to a signaling function and not to degradation because inhibition of proteasome activity with MG132 did not prevent SCFMet30 inactivation during cadmium stress (Flick et al., 2006), nor did it prevent Met30 dissociation from the core SCF (Figure 2D). We therefore asked whether Cdc48 binds preferentially to ubiquitylated forms of Met30. Cdc48 was sequentially purified by Ni2+ chelate chromatography and binding to streptavidin resins from cells carrying an insertion of the HTB tag (Tagwerker et al., 2006) at the C-terminus of the CDC48 locus. The Cdc48-bound Met30 fraction was dramatically enriched for ubiquitylated species, indicating specific recruitment of Cdc48 to ubiquitylated Met30 (Figure 5B). Some binding of deubiquitylated Met30 was also observed and could be a result of loss of the modification during the purification procedure, or reflect asymmetric Met30 ubiquitylation in the context of the SCFMet30 dimer (Tang et al., 2007). These results suggest Met30 ubiquitylation is the signal for recruitment of Cdc48, which induces dissociation of Met30 from the core SCF.
Met30 autoubiquitylation signals Cdc48 recruitment and SCFMet30 disassembly
We next asked whether cadmium stress triggers autoubiquitylation of Met30, which initiates disassembly through Cdc48 recruitment. Consistent with this idea, inactivation of the cognate SCFMet30 ubiquitin conjugating enzyme Cdc34 blocked cadmium-induced Met30 ubiquitylation (Figure 6A). Importantly, Met30 was ubiquitylated in cdc48 mutants, excluding a role for Cdc48 in the ubiquitylation process (Figures 6A and B). These results were further confirmed by detection of ubiquitin after purification of Met30 under denaturing conditions. The ubiquitin signal detected under these experimental conditions reflects ubiquitylated Met30 because control cells expressing untagged Met30 showed no detectable signal (Figure 6B). Significantly, wild type cells and cdc48 mutants showed cadmium-induced Met30 ubiquitylation while ubiquitylation of Met30 was blocked in cdc34 and cdc53 mutants (Figure 6B). Consistent with our previous data showing that SCFMet30 dissociation is a specific response to cadmium stress and not a general mechanism for Met4 activation (Figures 4B and 4C), no increase in ubiquitylated Met30 species was observed during activation of the Met4 pathway through methionine depletion (Figure 6C). These findings support a model where the cadmium signal induces autoubiquitylation of the F-box component Met30, which leads to Cdc48 recruitment and disassembly of SCFMet30. This model predicts that blocking cadmium-induced autoubiquitylation should prevent Cdc48 recruitment to Met30 and subsequent SCFMet30 disassembly. We first analyzed the behavior of a mutant version of Met30 where the interaction with the core SCF was prevented by mutating the F-box motif (Figure 6D), and autoubiquitylation is thus suppressed. Cdc48 was no longer recruited to Met30ΔF-box in response to cadmium stress (Figure 6D). Similarly, Cdc48 was no longer recruited to Met30 in response to cadmium stress when Met30 autoubiquitylation was inhibited by inactivation of the ubiquitin conjugating enzyme Cdc34 (Figure 6E). Most importantly, cadmium-induced disassembly of SCFMet30 was also blocked in cdc34 mutants (Figure 6E). Furthermore, deletion of Met4, which itself is a substrate and a substrate receptor for SCFMet30 (Ouni et al., 2010) did not prevent Cdc48 recruitment (Figure 6F) excluding Met4 ubiquitylation or ubiquitylation of any other known SCFMet30 substrate as the recruitment signal for Cdc48. These results strongly suggest that Met30 autoubiquitylation presents the signal for Cdc48 recruitment and the subsequent dissociation of Met30.
Figure 6.
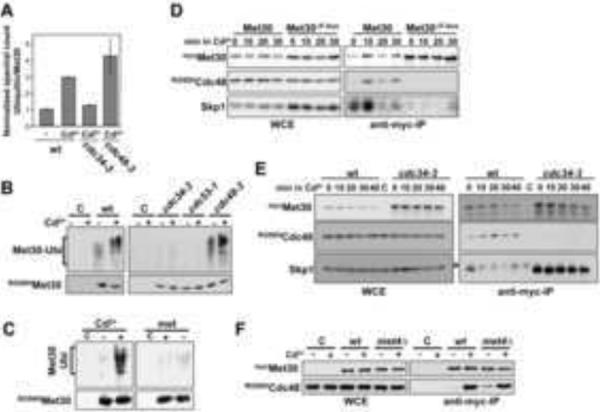
SCFMet30 autoubiquitylation signals Cdc48 recruitment. (A) Cdc34-dependent ubiquitylation of Met30 during cadmium stress. Experimental conditions as in figure 5A, but cells were shifted to 37°C for 90 min before cadmium exposure to inactivate temperature sensitive alleles. Data are represented as mean ± SD. (B) SCFMet30 autoubiquitylation in response to cadmium stress. Cells expressing RGS6HMet30 were shifted to 37°C for 90 min to inactivate temperature sensitive alleles and treated with 200μM cadmium chloride for 10 min. RGS6HMet30 was purified on Ni2+-sepharose under denaturing conditions and analyzed by immunoblotting using antibodies directed against ubiquitin (upper panel) or the RGS6H epitope (lower panel). Cells expressing untagged Met30 were processed as control. (C) SCFMet30 ubiquitylation is induced in response to cadmium but not methionine stress. Cells were treated with 100μM cadmium chloride or shifted to methionine free growth medium for 30 min. Met30 ubiquitylation was analyzed as in panel B. (D) The F-box motif in Met30 is required for Cdc48 recruitment. Cells expressing endogenous Cdc48RGS6H and 12mycMet30 or 12mycMet30ΔF-box lacking the F-box motif (residues 187–227 deleted) were cultured at 30°C in the presence of 100μM cadmium for the time intervals indicated. Met30 immunocomplexes were purified and analyzed by immunoblotting. (E) Met30 autoubiquitylation is required for Cdc48 recruitment and Skp1 dissociation. Wild type cells and cdc34-3 mutants were shifted to 37°C for 90 min and exposed to 100μM cadmium chloride for the time intervals indicated. Met30 immunocomplexes were analyzed as above. (F) Cdc48 recruitment is independent of Met4. Cdc48 recruitment to Met30 was analyzed as above in wild type cells and met4Δ mutants expressing Cdc48RGS6H and 12mycMet30.
Discussion
Our results describe a regulatory mechanism of substrate ubiquitylation that differs fundamentally from the extensively studied substrate recruitment step, which is generally initiated by substrate modification (Petroski and Deshaies, 2005). We demonstrate that selective, signal-induced disassembly of the SCF ubiquitin ligase complex is used to block ubiquitylation (Figure 7). The ubiquitylated F-box component serves as a recruitment platform for Cdc48, which separates the substrate adapter from the core ligase. Indications for signal-dependent regulation of subunit composition for other SCF-type ligases have been described previously in yeast and mammals (Li and Johnston, 1997; Zhang et al., 2004). It is thus likely that the association and dissociation of substrate adapters within cullin-RING ubiquitin ligases is highly regulated and used to modulate the activity of individual CRLs in response to signals.
Figure 7.
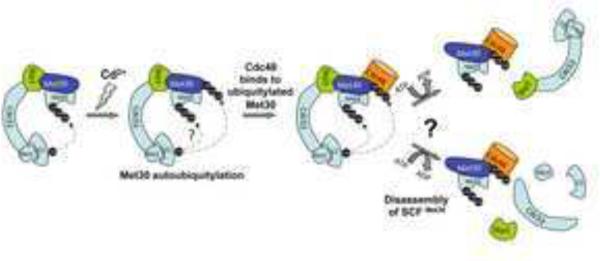
Model of signal-induced disassembly of SCFMet30. Cadmium stress induces Met30 autoubiquitylation through an unknown mechanism. Whether substrate ubiquitylation continues during this phase or the ubiquitin transfer is completely shifted to Met30 is currently unknown, as indicated by the question mark. Autoubiquitylated Met30 recruits Cdc48, which actively dissociates Met30 from the SCF core to prevent substrate ubiquitylation. Deubiquitylation of Met4 is required as a final step for Met4 activation (not shown).
Ubiquitin ligases typically target several different proteins for ubiquitylation. Dissociation of substrate adapters such as F-box proteins will inactivate ubiquitin ligase activity towards all its substrates. In contrast, the well-described mechanism of posttranslational modifications on substrates as initiators for substrate binding and ubiquitylation allows for much more directed, less global control. What then is the benefit of inhibiting all substrates of a particular ligase in response to one signal? We speculate that substrates of individual ligases are not completely unrelated and control different pathways that require co-regulation in response to certain signals. For example, SCFMet30 substrates control both glutathione synthesis and cell cycle control (Kaiser et al., 2006). Cadmium stress simultaneously blocks ubiquitylation of all SCFMet30 substrates and so coordinates heavy metal defense, detoxification, and cell cycle arrest. It is noteworthy that the substrate repertoire of the two other well-studied yeast SCF ligases, SCFGrr1 and SCFCdc4, also includes both metabolic pathway and cell cycle regulators (Skaar et al., 2009). It is thus reasonable to hypothesize that conditions that require coordinated regulation of metabolic pathways and the cell cycle may act through modulation of the Grr1-Skp1 or Cdc4-Skp1 interaction. Evidence for such a regulation has not been reported for SCFCdc4, but has been shown for the Grr1-Skp1 (Li and Johnston, 1997). Individual CRLs may therefore target a group of substrates, which under specific stress signals require coordinated regulation. For such instances, modulating posttranslational modifications for each individual substrate to synchronously block ubiquitin ligase binding seems less effective than direct inactivation of the CRL complex.
Our results strongly support cadmium-induced autoubiquitylation as the initial step for Cdc48 recruitment and subsequent SCFMet30 disassembly. How cadmium stress enhances Met30 autoubiquitylation is currently unclear. Addition of cadmium to in vitro reconstituted SCFMet30 reactions did not trigger Met30 modification, however, we cannot exclude that a specific redox environment may be required to mimic in vivo conditions. Alternatively, cadmium may activate a signaling cascade that leads to posttranslational modifications on Met30 to stimulate autoubiquitylation. Mechanistically we envision direct or indirect effects of cadmium acting on Met30 to induce subtle conformational changes or to slightly alter positioning within the SCF complex to enhance automodification, which ultimately triggers Met30 dissociation.
Cdc48/p97 is one of the most abundant proteins in the cell and uses ATP hydrolysis to separate macromolecules. Substrate selection and processing by Cdc48/p97 is thought to rely on cofactors (Alexandru et al., 2008; Haines, 2010; Halawani and Latterich, 2006; Schuberth and Buchberger, 2008; Yeung et al., 2008). The identity of cofactors required for ubiquitin-mediated recruitment of Cdc48 to SCFMet30 remains unclear. Similar to our observation that the hypomorph cdc48-3 mutant is sensitive to cadmium stress (Figure 3C), we expected Cdc48 cofactors essential for cadmium-induced SCFMet30 disassembly to be required for cellular cadmium resistance. We tested the known yeast Cdc48 cofactors and found that ufd3 and shp1 mutants showed significant sensitivity to cadmium (Figure S4A). Hypersensitivity of ufd3 mutants to cadmium exposure was unrelated to the SCFMet30/Met4 pathway because induction of Met4-dependent gene expression was unaffected in ufd3 mutants (data not shown). Shp1 may be involved in SCFMet30 dissociation as suggested by delayed dissociation of Met30 from the core SCF during cadmium stress (Figure S4B). However, Shp1 is not essential for Cdc48 recruitment (data not shown), but could be required for Cdc48 activation or correct topological arrangement for SCFMet30 disassembly. Even so there is some concern about interpretation of the results obtained with shp1 mutants because of their very slow growth rate. More detailed analyses of Met4 activation (Figure S4C) and SCF Met30 disassembly (Figure S4D) in temperature sensitive ufd1-2 and npl4-1 mutants excluded the heterodimeric, essential Cdc48 cofactor Npl4/Ufd1 as an important component in this pathway. Finally, recently human Ubxd7 and its yeast ortholog Ubx5 have been shown to link p97/Cdc48 to neddylated cullins (Besten et al., 2012). Cadmium-induced recruitment of Cdc48 to SCFMet30 likely occurs through a different binding mechanism because ubx5 mutants are not cadmium sensitive (Figure S4A). We expect that Cdc48 recruitment to ubiquitylated Met30 is either independent of known Cdc48 cofactors or several cofactors function redundantly. Alternatively, Cdc48 my directly recognize ubiquitylated Met30 because both yeast Cdc48 and its mammalian ortholog p97 have been shown to contain intrinsic ubiquitin binding activity (Dai and Li, 2001; Rape et al., 2001).
Cdc48/p97 has long been functionally connected to the ubiquitin proteasome system (Dai et al., 1998; Ghislain et al., 1996). More recently, physical association of p97 with several CRLs has been reported and shown to be required for turnover of the CUL2/VHL substrate HIF1alpha (Alexandru et al., 2008). Molecular mechanisms for Cdc48/p97 in CRL substrate turnover are unclear but it is generally expected that Cdc48/p97 is involved in proteasome targeting and/or processing of ubiquitylated substrates (Elsasser and Finley, 2005). Our results suggest that the AAA-ATPase Cdc48 actively removes the substrate adapter Met30 from the SCF core to inhibit substrate ubiquitylation in response to environmental signals. It is tempting to speculate that the segregase role of Cdc48/p97 plays additional roles in CRL regulation. Cdc48/p97 may be important for global CRL dynamics. General assembly and disassembly of SCF complexes and likely other CRLs is thought to be driven by degradation of adapter subunits through an autoubiquitylation mechanism (Galan and Peter, 1999; Zhou and Howley, 1998). This process is controlled at several levels including F-box deubiquitylation (Zhou et al., 2003), binding of the ligase inhibitor Glomulin (Duda et al., 2012; Tron et al., 2012), and SCF dimerization (Min et al., 2012). Whether Cdc48/p97 contributes to control of adaptor abundance by catalyzing post(auto)ubiquitylation steps in F-box degradation, or helps sculpting the global CRL landscape by dynamic disassembly of E3 complexes needs to be addressed in future studies.
Our study describes a critical role for Cdc48/p97 in signal-induced disassembly of the SCF ubiquitin ligase complex for coordinated inhibition of substrate ubiquitylation, and suggests a potentially general role for AAA+ ATPases such as Cdc48/p97 in inhibition of multisubunit enzymes by active complex disassembly. In addition, our results expand the repertoire of regulatory mechanisms for ubiquitin ligases and describe yet another link between Cdc48/p97 and the ubiquitin-proteasome system.
Experimental Procedures
Yeast Strains and Growth Conditions
Yeast strains used in this study are isogenic to 15DaubΔ, a bar1Δ ura3Δns, a derivative of BF264-15D (Reed et al., 1985). Specific strains are listed in supplementary table S1. All strains were cultured in standard media, and standard yeast genetic techniques were used unless otherwise indicated (Guthrie and Fink, 1991). References to the use of cadmium (Cd2+) are specifically to cadmium chloride (CdCl2). For cell spotting assays, strains were cultured to logarithmic growth phase, sonicated, and then counted. Serial dilutions were made and spotted onto YEPD plates supplemented with or without indicated amounts of cadmium via a pin replicator (V&P Scientific, San Diego, CA).
Protein Analysis
For immunoblot analysis, yeast whole cell lysates were prepared under denaturing conditions in urea buffer (8M urea, 200mM NaCl, 100mM Tris-HCl pH7.5, 0.2% SDS, 10mM Na-pyrophosphate, 5mM EDTA, 5mM EGTA, 50mM NaF, 0.1mM orthovanadate, 1mM phenylmethylsulfonyl fluoride [PMSF]) as described (Flick et al., 2006). For immunopurification, cells were lysed with glass beads in Triton X-100 buffer (50mM HEPES, pH 7.5, 0.2% Triton X-100, 200mM NaCl, 10% glycerol, 1mM dithiothreitol, 10mM Na-pyrophosphate, 5mM EDTA, 5mM EGTA, 50mM NaF, 0.1mM orthovanadate, 1mM phenylmethylsulfonyl floride [PMSF], and 1μg/ml each aprotinin, leupeptin, and pepstatin) as described (Flick et al., 2006). 2mg of total protein lysates were used for immunopurification with anti-myc antibodies (SC-789-G; Santa Cruz Biotechnology, Santa Cruz, CA). For purification of HTB-tagged Cdc48 cells were lysed as described for immunoprecipitations but 5mM N-ethylmaleimide and 40mM imidazole were included in the lysis buffer. 4mg of total protein lysates were used for binding to Ni2+-sepharose (GE Healthcare). Beads were washed 3 times in binding buffer and eluted in 60μl binding buffer containing 350mM imidazole and 1mM EDTA. 400μl lysis buffer were added to the eluate before binding to streptavidin agarose (Thermo Scientific, Rockford, IL) for 90 minutes. Samples were eluted by boiling in 2× SDS loading buffer for 10 minutes.
For immunoblot analyses proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. Proteins were detected with the following primary antibodies: anti-Met4 (1:10000; a gift from M. Tyers), anti-Skp1 (1:5000; a gift from R. Deshaies), anti-Cdc48 (1:10000; a gift from E. Jarosch), anti-Myc (1:2000; Covance, Princeton, NJ), anti-RGS4H (1:2000; Qiagen, Germantown, MD), anti-ubiquitin antibodies (1:500; P4G7, #sc-53509, Santa Cruz). Band intensities of immunoblots were quantified using a Fuji LAS-4000 imaging system and analyzed with the Multi Gauge v3 software.
RNA Analyses and PCR
RNA samples were isolated, and analyzed by real-time Reverse Transcriptase (RT)-PCR as described (Flick et al., 2006). A minimum of three biological replicates were analyzed.
Mass spectrometry
Cells expressing HBTH-tagged Skp1 (Figure 1A) or Met30 (Figures 5A and 6A) were used. The HBTH-tag is a combination of a 6xHis sequence (H), a biotinylation signal (B), a TEV protease site (T), and an RGS6xHis sequence (H). The nucleotide sequence of the tag can be downloaded at (https://webfiles.uci.edu:443/pkaiser/www/pk-webpage%20folder/pk-webpage/HB-plasmids/HB-plasmids.html). Three sets of experiments were conducted to investigate Met30 or Skp1 protein-protein interactions and posttranslational modifications. Experiments are described in detail in Supplemental Experimental Procedures.
Supplementary Material
Highlights
-
-
Stress induced SCFMet30 autoubiquitylation is recruitment signal for Cdc48
-
-
Active disassembly of SCFMet30 by Cdc48 to inhibit ubiquitin ligase activity
-
-
ATPase activity is required for F-box dissociation from the core SCF
-
-
SCFMet30 disassembly is stress signal specific
Acknowledgments
We thank S. Jentsch, T. Rapoport, P. Silver, and R. Hampton for yeast strains; R. Deshaies, W. Harper, M. Tyers, T. Sommer, and E. Jarosch for antibodies; C. Guerrero for help with initial mass spectrometric analysis; and H. Zhang for technical support. We are grateful for support from the National Institute of Health (GM66164 and GM66164AS1 to P.K.; GM74830 and S10RR023552 to L.H.; an NIH-NRSA postdoctoral fellowship GM082125 to C.V.P.; a Ruth Kirschstein Postdoctoral Fellowship T32 CA-113265 to I.O.; and a NCI Institutional Training Grant Award 5 T32 CA009054-32 to A.T.). We would like to dedicate this work to our late colleague Masayasu Nomura.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acs K, Luijsterburg MS, Ackermann L, Salomons FA, Hoppe T, Dantuma NP. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat Struct Mol Biol. 2011;18:1345–1350. doi: 10.1038/nsmb.2188. [DOI] [PubMed] [Google Scholar]
- Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbey R, Baudouin-Cornu P, Lee TA, Rouillon A, Zarzov P, Tyers M, Thomas D. Inducible dissociation of SCF(Met30) ubiquitin ligase mediates a rapid transcriptional response to cadmium. Embo J. 2005;24:521–532. doi: 10.1038/sj.emboj.7600556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudouin-Cornu P, Labarre J. Regulation of the cadmium stress response through SCF-like ubiquitin ligases: comparison between Saccharomyces cerevisiae, Schizosaccharomyces pombe and mammalian cells. Biochimie. 2006;88:1673–1685. doi: 10.1016/j.biochi.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Besten W, Verma R, Kleiger G, Oania RS, Deshaies RJ. NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway. Nat Struct Mol Biol. 2012;19:511–516. S511. doi: 10.1038/nsmb.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Dai RM, Chen E, Longo DL, Gorbea CM, Li CC. Involvement of valosin-containing protein, an ATPase Co-purified with IkappaBalpha and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IkappaBalpha. J Biol Chem. 1998;273:3562–3573. doi: 10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- Dai RM, Li CC. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat Cell Biol. 2001;3:740–744. doi: 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Duda DM, Olszewski JL, Tron AE, Hammel M, Lambert LJ, Waddell MB, Mittag T, Decaprio JA, Schulman BA. Structure of a Glomulin-RBX1-CUL1 Complex: Inhibition of a RING E3 Ligase through Masking of Its E2-Binding Surface. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Scott DC, Calabrese MF, Zimmerman ES, Zheng N, Schulman BA. Structural regulation of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2011;21:257–264. doi: 10.1016/j.sbi.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- Fauchon M, Lagniel G, Aude JC, Lombardia L, Soularue P, Petat C, Marguerie G, Sentenac A, Werner M, Labarre J. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol Cell. 2002;9:713–723. doi: 10.1016/s1097-2765(02)00500-2. [DOI] [PubMed] [Google Scholar]
- Fleming JA, Lightcap ES, Sadis S, Thoroddsen V, Bulawa CE, Blackman RK. Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341. Proc Natl Acad Sci U S A. 2002;99:1461–1466. doi: 10.1073/pnas.032516399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick K, Raasi S, Zhang H, Yen JL, Kaiser P. A ubiquitin-interacting motif protects polyubiquitinated Met4 from degradation by the 26S proteasome. Nat Cell Biol. 2006;8:509–515. doi: 10.1038/ncb1402. [DOI] [PubMed] [Google Scholar]
- Franz A, Orth M, Pirson PA, Sonneville R, Blow JJ, Gartner A, Stemmann O, Hoppe T. CDC-48/p97 coordinates CDT-1 degradation with GINS chromatin dissociation to ensure faithful DNA replication. Mol Cell. 2011;44:85–96. doi: 10.1016/j.molcel.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Kitabatake M, Sakata T, Ohno M. 40S subunit dissociation and proteasome-dependent RNA degradation in nonfunctional 25S rRNA decay. Embo J. 2012;31:2579–2589. doi: 10.1038/emboj.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JM, Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc Natl Acad Sci U S A. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Dohmen RJ, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. Embo J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. Vol 194. Academic Press, Inc.; San Diego: 1991. [Google Scholar]
- Haines DS. p97-containing complexes in proliferation control and cancer: emerging culprits or guilt by association? Genes Cancer. 2010;1:753–763. doi: 10.1177/1947601910381381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halawani D, Latterich M. p97: The cell's molecular purgatory? Mol Cell. 2006;22:713–717. doi: 10.1016/j.molcel.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Rumpf S. Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P, Su NY, Yen JL, Ouni I, Flick K. The yeast ubiquitin ligase SCF-Met30: connecting environmental and intracellular conditions to cell division. Cell Div. 2006;1:16. doi: 10.1186/1747-1028-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Li FN, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. Embo J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- Min SH, Lau AW, Lee TH, Inuzuka H, Wei S, Huang P, Shaik S, Lee DY, Finn G, Balastik M, et al. Negative regulation of the stability and tumor suppressor function of fbw7 by the pin1 prolyl isomerase. Mol Cell. 2012;46:771–783. doi: 10.1016/j.molcel.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouni I, Flick K, Kaiser P. A transcriptional activator is part of an SCF ubiquitin ligase to control degradation of its cofactors. Mol Cell. 2010;40:954–964. doi: 10.1016/j.molcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- Raman M, Havens CG, Walter JC, Harper JW. A genome-wide screen identifies p97 as an essential regulator of DNA damage-dependent CDT1 destruction. Mol Cell. 2011;44:72–84. doi: 10.1016/j.molcel.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- Reed SI, Hadwiger JA, Lorincz AT. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc Natl Acad Sci U S A. 1985;82:4055–4059. doi: 10.1073/pnas.82.12.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Rumpf S, Jentsch S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell. 2006;21:261–269. doi: 10.1016/j.molcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Shevchenko A, Deshaies RJ. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat Cell Biol. 2001;3:384–391. doi: 10.1038/35070067. [DOI] [PubMed] [Google Scholar]
- Shcherbik N, Haines DS. Cdc48p(Npl4p/Ufd1p) binds and segregates membrane-anchored/tethered complexes via a polyubiquitin signal present on the anchors. Mol Cell. 2007;25:385–397. doi: 10.1016/j.molcel.2007.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Pagan JK, Pagano M. SnapShot: F box proteins I. Cell. 2009;137:1160–1160. e1161. doi: 10.1016/j.cell.2009.05.039. [DOI] [PubMed] [Google Scholar]
- Tagwerker C, Zhang H, Wang X, Larsen LS, Lathrop RH, Hatfield GW, Auer B, Huang L, Kaiser P. HB tag modules for PCR-based gene tagging and tandem affinity purification in Saccharomyces cerevisiae. Yeast. 2006;23:623–632. doi: 10.1002/yea.1380. [DOI] [PubMed] [Google Scholar]
- Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Tron AE, Arai T, Duda DM, Kuwabara H, Olszewski JL, Fujiwara Y, Bahamon BN, Signoretti S, Schulman BA, DeCaprio JA. The glomuvenous malformation protein Glomulin binds Rbx1 and regulates cullin RING ligase-mediated turnover of Fbw7. Mol Cell. 2012;46:67–78. doi: 10.1016/j.molcel.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. Cdc48/p97 Mediates UV-Dependent Turnover of RNA Pol II. Mol Cell. 2011;41:82–92. doi: 10.1016/j.molcel.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler GL, Trotter EW, Dawes IW, Grant CM. Coupling of the transcriptional regulation of glutathione biosynthesis to the availability of glutathione and methionine via the Met4 and Yap1 transcription factors. J Biol Chem. 2003;278:49920–49928. doi: 10.1074/jbc.M310156200. [DOI] [PubMed] [Google Scholar]
- Wilcox AJ, Laney JD. A ubiquitin-selective AAA-ATPase mediates transcriptional switching by remodelling a repressor-promoter DNA complex. Nat Cell Biol. 2009;11:1481–1486. doi: 10.1038/ncb1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems AR, Schwab M, Tyers M. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen JL, Su NY, Kaiser P. The Yeast Ubiquitin Ligase SCFMet30 Regulates Heavy Metal Response. Mol Biol Cell. 2005;16:1872–1882. doi: 10.1091/mbc.E04-12-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung HO, Kloppsteck P, Niwa H, Isaacson RL, Matthews S, Zhang X, Freemont PS. Insights into adaptor binding to the AAA protein p97. Biochem Soc Trans. 2008;36:62–67. doi: 10.1042/BST0360062. [DOI] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Wee S, Rhee E, Naumann M, Dubiel W, Wolf DA. Fission yeast COP9/signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme Ubp12p. Mol Cell. 2003;11:927–938. doi: 10.1016/s1097-2765(03)00136-9. [DOI] [PubMed] [Google Scholar]
- Zhou P, Howley PM. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol Cell. 1998;2:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.