Abstract
Kidney cancer is not a single disease; it is made up of a number of different types of cancer, including clear cell, type 1 papillary, type 2 papillary, chromophobe, TFE3, TFEB, and oncocytoma. Sporadic, nonfamilial kidney cancer includes clear cell kidney cancer (75%), type 1 papillary kidney cancer (10%), papillary type 2 kidney cancer (including collecting duct and medullary RCC) (5%), the microphalmia-associated transcription (MiT) family translocation kidney cancers (TFE3, TFEB, and MITF), chromophobe kidney cancer (5%), and oncocytoma (5%). Each has a distinct histology, a different clinical course, responds differently to therapy, and is caused by mutation in a different gene. Genomic studies identifying the genes for kidney cancer, including the VHL, MET, FLCN, fumarate hydratase, succinate dehydrogenase, TSC1, TSC2, and TFE3 genes, have significantly altered the ways in which patients with kidney cancer are managed. While seven FDA-approved agents that target the VHL pathway have been approved for the treatment of patients with advanced kidney cancer, further genomic studies, such as whole genome sequencing, gene expression patterns, and gene copy number, will be required to gain a complete understanding of the genetic basis of kidney cancer and of the kidney cancer gene pathways and, most importantly, to provide the foundation for the development of effective forms of therapy for patients with this disease.
Kidney cancer affects nearly 270,000 patients annually worldwide and is responsible for over 115,000 deaths each year (Ferlay et al. 2010). Kidney cancer is not a single disease; rather, it is comprised of a number of different histologically and genetically distinct types of cancer, each with a different histologic type, each of which has a different clinical course and different responses to therapy (Fig. 1; Linehan et al. 2003; Linehan and Zbar 2004). If a patient presents with a small cancer localized to the kidney, this patient can be managed by surgery and can expect a 95% 5- to 10-yr survival. However, if a patient presents with advanced kidney cancer, in which the primary tumor has spread to other organs, nearly 80% will die of this disease within 24 mo (Linehan et al. 2011).
Figure 1.
Kidney cancer is not a single disease; it is made up of a number of cancers that occur in the kidney, each with a distinct histology, each with a different clinical course, each responding differently to therapy, and each caused by a different gene. The VHL gene is mutated in the germline of ∼100% of von Hippel-Lindau families and the majority of sporadic clear cell RCCs. The MET gene is mutated in the germline of ∼100% of hereditary papillary renal carcinoma families and is mutated in a subset of tumors (13%) from patients with sporadic, type 1 papillary kidney cancer. The fumarate hydratase gene (FH) is the gene for the inherited form of type 2 papillary kidney cancer associated with hereditary leiomyomatosis renal cell carcinoma (HLRCC). Sporadic papillary type 2 RCC is composed of a mixture of kidney cancers including collecting duct and medullary RCC. The gene(s) for sporadic type 2 papillary RCC, collecting duct RCC, and medullary RCC is not known. The FLCN is mutated in 96% of families affected with the inherited form of chromophobe RCC and oncocytoma associated with Birt-Hogg-Dubé syndrome. The genes for the sporadic forms of chromophobe RCC and oncocytoma are not known. TFE3, TFEB, and MITF are part of the MiT family of transcription factors. TFE3 and TFEB translocation kidney cancer are sporadic (nonhereditary); germline mutations of MITF have been found in the germline of patients with melanoma and kidney cancer or both. (Fig. 1 adapted from Linehan et al. 2003.)
Studies of the inherited forms of kidney cancer have led to the identification of a number of genes important for this disease. There are a number of different types of familial kidney cancers, including von Hippel-Lindau (VHL), hereditary papillary renal carcinoma (HPRC), Birt-Hogg-Dubé (BHD), hereditary leiomyomatosis renal cell carcinoma (HLRCC), succinate dehydrogenase kidney cancer (SHD-RCC), tuberous sclerosis complex (TSC), and Cowden's disease. Identification of the genes that cause kidney cancer has improved the management of patients with kidney cancer and provided the foundation for the development of therapeutic approaches for patients with advanced forms of this disease.
The conventional approach to therapy for patients with localized clear cell, type 1 papillary, or chromophobe kidney cancer has traditionally been to surgically remove the tumor. For decades, the conventional surgical approach has been to remove the entire kidney (“radical nephrectomy”). Over the past two decades, the surgical approach of partial nephrectomy (removal of the tumor and a small margin of normal renal parenchyma) has been developed and is now the recommended approach for most patients diagnosed with localized kidney cancers that are 7 cm or less in size. The role of adjuvant therapy (such as agents targeting the VHL pathway, discussed below) is currently being evaluated in clinical trials. Systemic therapy is reserved for patients who develop metastatic disease.
Clear cell kidney cancer
Clear cell is the most common histologic type of kidney cancer, occurring in 75% of cases. In the initial studies performed to identify the clear cell kidney cancer gene, investigators showed that loss of heterozygosity on the short arm of chromosome 3 was a consistent feature in tumors from patients with clear cell kidney cancer (Zbar et al. 1987). Subsequent fine-mapping studies pinpointed the location of a kidney cancer gene on chromosome 3 and revealed that the region was too large to search by conventional methods available at the time (Anglard et al. 1991).
Von Hippel-Lindau: Clear cell kidney cancer
In order to identify the clear cell kidney cancer gene, investigators studied families with the hereditary form of clear cell kidney cancer, von Hippel-Lindau. Von Hippel-Lindau is an autosomal dominant, hereditary cancer syndrome in which affected individuals are at risk for the development of tumors in a number of organs, including the kidneys, adrenal glands, retina, central nervous system, pancreas, and epididymis. VHL patients are at risk for the development of early onset, bilateral, multifocal clear cell kidney cancer (Fig. 2). There is a clear relationship between prevalence of kidney cancer and VHL genotype, with, for example, a higher prevalence in patients with partial versus complete deletion of the VHL gene (Maranchie et al. 2004). While there is a wide range of the number of detectable kidney tumors (0–70) in an individual kidney in a patient with VHL (Fadahunsi et al. 2011), it is estimated that VHL patients are at risk for the development of up to 600 microscopic tumors and 1100 cysts per kidney (Walther et al. 1995). Historically, 35%–45% of patients with VHL died of kidney cancer. The growth rate of VHL kidney tumors is known and predictable, and an approach to management has been developed which involves active surveillance of VHL kidney tumors by imaging until the largest tumor reaches the 3-cm threshold, at which time surgical resection is recommended (Herring et al. 2001). When surgical resection is recommended, all visible tumors are removed, while unaffected areas of the kidney are spared. Up to 70 tumors have been removed from a single kidney in a VHL patient. VHL patients are at risk for the development of new tumors and, while surgical treatment does not “cure” the patient of kidney cancer, it is intended to “set back the clock” and to thereby diminish the development of metastases. In a series of VHL patients managed in this fashion, Herring et al. (2001) reported a 10 yr experience in which none of the patients developed metastatic disease when their kidney tumors were removed surgically upon reaching the 3-cm threshold. VHL patients are also at risk for the development of retinal angiomas, spinal and cerebellar hemangioblastomas, inner ear endolymphatic sac tumors, pancreatic cysts, neuroendocrine tumors, pheochromocytomas, and epididymal cystadenomas (Lonser et al. 2003).
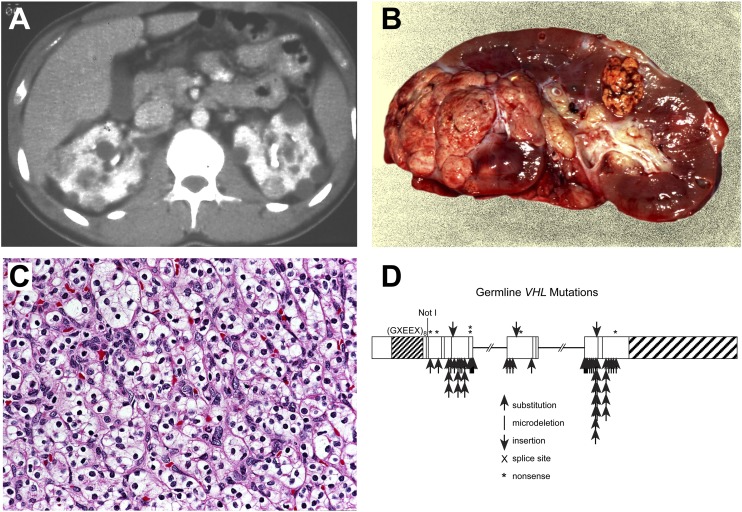
Figure 2.
Clear cell kidney cancer. Von Hippel-Lindau (VHL) is an autosomal hereditary cancer disorder in which affected individuals are at risk for the development of bilateral, multifocal kidney cancer (A,B) of clear cell histologic type (C). VHL is characterized by germline mutation of the VHL gene (D). The VHL gene is mutated in 57%–89% of sporadic, noninherited clear cell kidney cancers (Gnarra et al. 1994; Moore et al. 2011). (Fig. 2 from Linehan et al. 2003.)
Identification of the VHL gene
In order to identify the gene for von Hippel-Lindau, genetic linkage analysis in VHL families was performed (Hosoe et al. 1990). In a study that took nearly 10 yr to complete, investigators reported the identification of the VHL gene in the spring of 1993 (Latif et al. 1993). Mutations of the VHL gene have been found in nearly 100% of VHL families (Stolle et al. 1998), and a simple blood test for germline mutation of the VHL gene is now used to make the diagnosis. VHL germline mutation testing makes it possible to make the correct diagnosis of von Hippel-Lindau and to determine which at-risk family members are affected. Development of carcinomas in patients with mutation in the VHL gene fits the Knudson model of a classic two-hit tumor suppressor gene, i.e., affected individuals in VHL families inherited a mutated copy of the VHL gene and the second allele is generally deleted in the VHL-associated tumors (Knudson 1995).
Chromosome 3 translocation kidney cancer
In 1979, Cohen et al. reported a family in which multiple individuals were found to have bilateral, multifocal clear cell kidney cancer. Whereas bilateral, multifocal clear cell kidney cancer can occur in VHL patients in their second decade, the kidney tumors in the chromosome 3 translocation families tended to occur in older individuals in the fourth or fifth decades of life. Affected individuals were found to have a balanced germline 3;8 translocation t(3;8)(p14.2;q24.1) between the short arm of chromosome 3 and the long arm of chromosome 8 (Cohen et al. 1979). Subsequent studies revealed that the chromosome 3 to 8 translocation, which included the VHL gene, was deleted in the clear cell tumors and that the remaining somatic VHL allele was mutated in the tumors (Schmidt et al. 1995). Other kidney cancer families have been identified with chromosome 3 translocations involving chromosome 6, t(3:6)(q12;q15), and chromosome 2, t(2:3)(q12;q15) (Eleveld et al. 2001; Podolski et al. 2001; Van Erp et al. 2003). The finding that three successive events (“hits”), including germline translocation as the first event, followed by deletion of the long arm of chromosome 8 containing the translocated VHL gene, and finally, by mutation of the remaining somatic VHL allele, could explain why the tumor formation in chromosome 3 translocation kidney cancers is characterized by later onset and fewer kidney tumors than, for example, VHL.
Sporadic, noninherited clear cell kidney cancer: The VHL gene
In order to determine if the VHL gene was associated with the common form of noninherited, sporadic clear cell kidney cancer, VHL gene mutation and loss of heterozygosity analyses were performed in patients with kidney cancer. In patients with sporadic kidney cancer, VHL gene mutation or methylation has been found in a high percentage (nearly 90%) of tumors from patients with clear cell kidney cancer (Gnarra et al. 1994; Herman et al. 1994; Nickerson et al. 2008; Moore et al. 2011). VHL gene mutation is not found in tumors from patients with papillary, chromophobe, collecting duct, or medullary kidney cancer. Thus, the VHL gene is the predominant cancer gene in sporadic clear cell kidney cancers (Knudson 1995).
VHL gene pathway: Oxygen sensing
When the VHL gene was identified, it was a novel gene with no known function. The initial key to elucidation of the function the VHL gene was the identification of the VHL protein binding partners, elongin C, elongin B, CUL2, and Rbx1. The VHL complex targets the hypoxia-inducible factors, HIF1A and EPAS1 (also known as HIF2A), for ubiquitin-mediated degradation in an oxygen-dependent fashion (Duan et al. 1995; Kibel et al. 1995; Kamura et al. 1999; Maxwell et al. 1999; Ohh et al. 2000; Jaakkola et al. 2001). When there are normal levels of oxygen in the cell (normoxia), two proline residues on the hypoxia-inducible factors (HIF1A and HIF2A) are hydroxylated through the action of prolyl hydroxylase enzymes, and this oxygen-dependent hydroxylation enables the VHL complex to bind and ubiquitinate the hypoxia-inducible factors, which targets them for subsequent degradation by the proteosome. When the cellular oxygen concentrations decrease, i.e., there is hypoxia, the VHL complex cannot target and degrade the hypoxia-inducible factors, and the nonhydroxylated hypoxia-inducible factors accumulate. The HIFs are transcription factors. Under hypoxic conditions, when they accumulate, the transcription of factors such as vascular endothelial growth factor (VEGFA), platelet derived growth factor (PDGF), the glucose transporter, SLC2A1 (also known as GLUT1), and the epidermal growth factor receptor (EGFR) is increased. When there is mutation of the VHL gene, even in normoxia, HIFs accumulate, and the transcription of a large number of downstream genes, such as KDR (also known as VEGFR), PDGFRB, EGFR, and SLC2A1, is increased (for review, see Kaelin 2008).
Therapeutic approaches targeting the VHL pathway
Understanding the VHL pathway provided the foundation for the development of therapeutic approaches for patients with advanced clear cell kidney cancer. The first study targeting the VHL pathway in patients with advanced clear cell kidney cancer involved the use of an antibody targeting vascular endothelial growth factor, bevacizumab. Yang et al. (2003) conducted a randomized trial evaluating the effect of bevacizumab on patients with advanced clear cell kidney cancer. While there was no difference in overall survival between the groups, there was a statistically significant increase in time to progression in the high-dose-antibody group (4.8 mo) as compared to the placebo group (2.5 mo, P < 0.001) (Yang et al. 2003; Yang 2004). Subsequent studies have identified agents targeting the VHL pathway such as sunitinib maleate, sorafinib, temsirolimus, everolimus, pazopanib, and axitinib as having significant anti-tumor activity in patients with advanced clear cell kidney cancer (Escudier et al. 2007; Hudes et al. 2007; Motzer et al. 2007, 2008; Sternberg et al. 2010; Rini et al. 2011). While with some of these agents there are responses in nearly 40% of patients and increased survival, most patients eventually progress, and many eventually die of this disease.
In a very elegant recent study, Gerlinger et al. (2012) reported exome sequencing, chromosome aberration analysis, and ploidy profiling on multiple tumor biopsies from primary clear cell kidney cancers and associated metastatic sites. While they found common “truncal” mutations (in genes such as VHL), they also found remarkable somatic intratumoral mutational heterogeneity. When they evaluated the metastatic sites in one patient, there was remarkable similarity among the metastatic sites that correlated with a single site in the primary tumor (suggesting that the metastases had originated from that single site within the tumor). These findings suggest that genomic analysis from a single site may significantly underestimate the genetic heterogeneity of the tumor and that identification of common truncal mutations may provide critical information for the development of effective forms of therapy for an individual cancer (Gerlinger et al. 2012).
Sporadic, noninherited clear cell kidney cancer: The PBRM1 gene
In 2010, Dalgliesh et al. established the importance of the chromatin modification machinery in clear cell kidney cancer when they reported sequencing 101 tumors through 3544 protein-coding genes. They identified inactivating mutations of genes involved in histone modification, including SETD2, a histone H3 lysine 36 methyltransferase, JARID1C, a histone H3 lysine 4 demethylase, as well as mutations in the histone H3 lysine 27 demethylase, UTX (Dalgliesh et al. 2010). Varela et al. (2011) recently reported sequencing of the protein coding exome in clear cell renal cancers and identified truncating mutations of the SWI/SNF chromatin remodeling complex gene, PBRM1, in 41% of tumors. These two seminal studies opened chromatin biology as an entirely new area of significant importance in understanding the genetic basis of kidney cancer. Detailed expression and mechanistic analysis will be required to determine the important functional consequences of mutations of the histone modifying genes in clear cell kidney cancer, as reported by Dalgliesh et al. (2010) and Varela et al. (2011).
Genomics approaches in clear cell kidney cancer
Although seven FDA-approved agents that target the VHL pathway have been developed, new agents need to be developed which will be effective in all patients with this disease. Understanding gene copy number variations as well as expression profile changes in the various types of kidney cancer mentioned above will provide potential improvement in disease classification and stratification and could provide novel significant insight into potential therapeutic approaches to therapy. Beroukhim et al. (2009) performed an integrated, genome-wide analysis of copy-number changes and gene expression profiles in 90 sporadic and hereditary clear cell tumors and identified 14 regions of nonrandom copy number change, including seven regions of amplification (1q, 2q, 5q, 7q, 8q, 12q, and 20q) and seven regions of deletion (1q, 3p, 4q, 6q, 8p, 9p, and 14q). VHL was identified as a deleted gene on 3p, as were CDKN2A and CDKN2B from 9p. MYC was found to be amplified in the 8q amplification peak (Beroukhim et al. 2009). Gene expression profiles using DNA microarrays in clear cell kidney cancer have been performed which predict survival, and individual transcripts whose expression was associated with survival have been identified (Vasselli et al. 2003; Zhao et al. 2006). Gene expression profiles have been performed to define a molecular subclassification of clear cell kidney cancer as well as to identify novel biomarkers (Takahashi et al. 2003a,b). Although significant progress has been made in many aspects of kidney cancer, a number of critical questions remain; in-depth genomics approaches will be essential in answering these.
Hereditary papillary renal cell carcinoma: Type 1 papillary kidney cancer
Hereditary papillary renal carcinoma is a highly penetrant, autosomal dominant hereditary cancer syndrome in which affected individuals are at risk for the development of bilateral, multifocal, type 1 papillary kidney cancer (Fig. 3; Zbar et al. 1994). While there is a wide range of the number of detectable kidney tumors (0–51) in an individual kidney in a patient with HPRC (Herring et al. 2001), it is estimated that HPRC patients are at risk for the development of 1100 to 3400 microscopic tumors per kidney (Ornstein et al. 2000). By carefully monitoring the growth parameters and metastatic potential of HPRC renal tumors over an 18-yr time period, a clinical management approach has been developed involving active surveillance until the largest kidney tumor reaches 3 cm, at which time surgical intervention is recommended ( Ornstein et al. 2000; Chernoff et al. 2001; Herring et al. 2001).
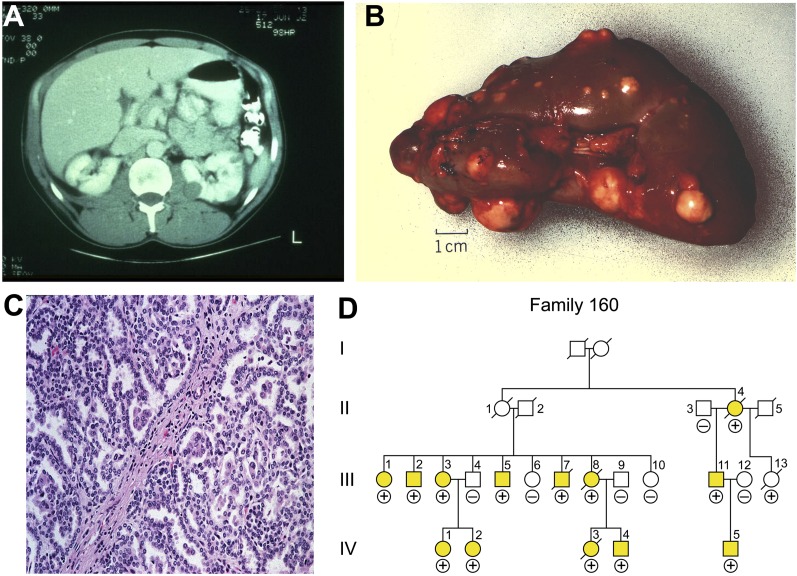
Figure 3.
Type 1 papillary kidney cancer. Hereditary papillary renal carcinoma (HPRC) is an autosomal dominant hereditary cancer syndrome (D) in which affected individuals are at risk for the development of bilateral kidney cancer (A,B) that is type 1 papillary histologic type (C). Patients affected with HPRC are characterized by germline mutation of the MET gene. MET mutations are also found in a subset of tumors (13%) from patients with sporadic, nonhereditary papillary kidney cancer. (Fig. 3 from Linehan et al. 2003.)
The HPRC gene: MET
Genetic linkage analysis was performed in the HPRC families, and the proto-oncogene, MET, was identified as the HPRC gene (Schmidt et al. 1997). MET mutations have also been found in a subset of tumors (13%) from patients with sporadic, nonhereditary papillary kidney cancer (Schmidt et al. 1997). The finding that the MET mutations in the HPRC families are in the tyrosine kinase domain of the gene has led to the evaluation of a targeted approach to therapy with an agent with dual kinase activity against the MET and VEGF receptors in patients with HPRC-associated as well as sporadic papillary kidney cancer. Effectiveness of this agent against tumors with and without MET mutation or amplification is being evaluated (Choueiri et al. 2012).
Birt-Hogg-Dubé: The hereditary form of chromophobe kidney cancer
Birt-Hogg-Dubé is an autosomal dominant, hereditary cancer syndrome in which affected individuals are at risk for the development of bilateral, multifocal kidney tumors as well as pulmonary cysts and cutaneous fibrofolliculomas (Fig. 4; Zbar et al. 2002). Whereas patients affected with von Hippel-Lindau develop clear cell kidney cancer and patients with HPRC develop type 1 papillary kidney cancer, patients affected with BHD are at risk for the development of a number of different histologic types of kidney cancer, including chromophobe, hybrid oncocytic, clear cell, and oncocytoma (Pavlovich et al. 2002). BHD kidney cancer is managed in the same fashion as is kidney cancer in patients with VHL and HPRC; patients with BHD-associated kidney cancer are managed with active surveillance until the largest tumor reaches the 3-cm threshold. At this point, surgical intervention is recommended (Pavlovich et al. 2005).
Figure 4.
Chromophobe kidney cancer. Birt-Hogg-Dubé (BHD) is an autosomal dominant hereditary cancer syndrome in which affected individuals are at risk for the development of cutaneous fibrofolliculomas, pulmonary cysts, and bilateral, multifocal chromophobe and hybrid oncocytic kidney cancer and oncocytoma. BHD is characterized by germline mutation of the FLCN gene. (Fig. 4 from Linehan et al. 2003.)
Identification of the BHD gene: FLCN
Genetic linkage analysis performed in BHD families led to the identification of FLCN as the BHD gene (Nickerson et al. 2002). FLCN has the characteristics of a tumor suppressor gene; mutation or loss of heterozygosity of the wild-type allele of the FLCN gene was identified in 70% of tumors (Vocke et al. 2005). Germline testing has identified mutations of the FLCN gene in a high percentage of BHD families (Toro et al. 2008).
Function of FLCN: Nutrient sensing
FLCN is in the LKB1/AMPK pathway. The FLCN protein binds two proteins, FNIP1 and FNIP2, which bind the γ-subunit of AMPK (also known as PRKAG2) (Baba et al. 2006; Hasumi et al. 2008). In preclinical and clinical models, when FLCN is deficient, mTORC1 and mTORC2 are activated (Baba et al. 2008; Hasumi et al. 2009). These findings have provided a potential approach to therapy for FLCN-deficient kidney cancer involving targeting the mTOR pathway. Promising results have been seen targeting the mTORC1 pathway in a kidney-specific FLCN knockout model (Baba et al. 2008), and clinical trials in BHD patients targeting this pathway are planned.
Hereditary leiomyomatosis renal cell carcinoma: Type II papillary kidney cancer
Hereditary leiomyomatosis renal cell carcinoma (HLRCC) is an autosomal dominant, hereditary cancer syndrome in which affected individuals are at risk for the development of cutaneous and uterine leiomyomas and kidney cancer (Fig. 5; Zbar et al. 1995; Launonen et al. 2001). HLRCC-associated kidney cancer is a very aggressive form of kidney cancer in which very small tumors have the potential to metastasize (Grubb et al. 2007; Merino et al. 2007). In some patients, advanced disease can be fatal within months. In other patients, the disease can be stable for a longer period of time. These tumors should be managed very differently from VHL-, HPRC-, or BHD-associated kidney cancer. When a localized HLRCC kidney cancer is detected, it should be surgically removed. Active surveillance is not recommended for patients who are found to have localized HLRCC-associated kidney cancer. The surgical procedure for removal of an HLRCC kidney cancer should be much more extensive than that performed in other types of kidney cancer. Due to the aggressive, invasive nature of HLRCC-associated kidney cancer, a larger margin of normal renal tissue should be removed than is required, for example, in a patient with a VHL-, HPRC-, or BHD-associated clear cell kidney cancer. Moreover, it is very important to avoid releasing resected tumor cells into the peritoneum or retroperitoneum during surgery, as these aggressive tumor cells may form multiple metastatic foci post-operatively. Very careful annual imaging with abdominal magnetic resonance imaging and/or abdominal computed tomography scans to evaluate for the presence of kidney tumors in individuals affected with hereditary leiomyomatosis renal cell carcinoma should be performed.
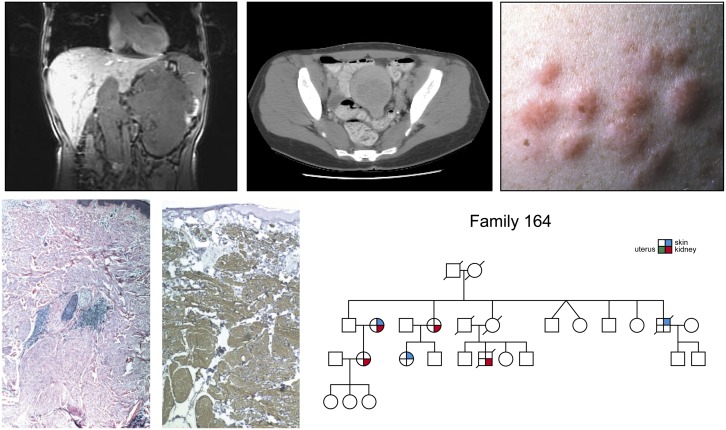
Figure 5.
Type 2 papillary kidney cancer. Hereditary leiomyomatosis renal cell carcinoma (HLRCC) is an autosomal dominant hereditary cancer syndrome (right lower panel) in which affected individuals are at risk for the development of type 2 kidney cancer (upper left and upper middle panels), cutaneous leiomyomas (right upper, left lower, and middle lower panels) and early onset uterine leiomyomas (fibroids). HLRCC is characterized by germline mutation of the gene for the Krebs cycle enzyme, fumarate hydratase. (Fig. 5 from Linehan et al. 2003.)
HLRCC gene: Fumarate hydratase—the Warburg effect
The gene for HLRCC encodes the Krebs cycle enzyme, fumarate hydratase (FH) (Tomlinson et al. 2002). Germline testing revealed mutations of the FH gene in a high percentage of HLRCC families (Toro et al. 2003; Wei et al. 2006). Fumarate hydratase-deficient kidney cancer models are characterized by aerobic glycolysis and are very dependent on glucose transport and glycolysis for energy production (Isaacs et al. 2005; Yang et al. 2010; Tong et al. 2011). Fumarate hydratase-deficient kidney cancer conforms to the Warburg model of cancer. In the 1920s, Otto Warburg suggested that aerobic glycolysis was a fundamental aspect of cancer (Warburg et al. 1924; Warburg 1956). In tumors that manifest the “Warburg effect,” the cancer depends on aerobic glycolysis instead of oxidative phosphorylation for energy production. In fumarate hydratase-deficient kidney cancer, mitochondrial respiration is severely impaired, and these cancers are very dependent on glycolysis for ATP production (Yang et al. 2010; Tong et al. 2011). In FH-deficient kidney cancer, the Krebs cycle pathway is remodeled, and a reductive glutamine-dependent pathway predominates to support fatty acid synthesis and tumor growth (Mullen et al. 2011). Understanding the fumarate hydratase pathway in FH-deficient kidney cancer has provided the foundation for novel approaches to therapy targeting the tumor vasculature and glucose transport. At the National Cancer Institute, a clinical trial is currently in progress evaluating the role of bevacizumab and erlotinib in patients with advanced forms of HLRCC-associated as well as sporadic, non-HLRCC type 2 papillary kidney cancer to evaluate the effectiveness of this regimen in patients with FH −/− as well as FH +/+ papillary kidney cancer.
Succinate dehydrogenase kidney cancer
Succinate dehydrogenase kidney cancer is a hereditary cancer syndrome in which affected individuals are at risk for the development of pheochromocytomas and paragangliomas and kidney cancer (Vanharanta et al. 2004). SDH-RCC can also be a very aggressive form of kidney cancer, and surgical removal should be performed as soon as a kidney tumor is identified in an affected individual. SDH-RCC has been found in association with germline mutations of the SDHB and SDHD genes, and these cancers represent additional examples of the Warburg effect in cancer (Neumann et al. 2004; Ricketts et al. 2008; Gill et al. 2011).
Tuberous sclerosis complex: TSC1/2
Tuberous sclerosis complex (TSC) is a familial hamartoma syndrome in which affected individuals are at risk for the development of tumorous growths in a number of organs, including facial angiofibroma, cutaneous shagreen patches, CNS lesions including cortical tubers and subependymal giant-cell tumors, cardiac rhabdomyomas, pulmonary lymphangiomatosis, and renal tumors (for a comprehensive review, see Crino et al. 2006). The renal tumors are most often benign angiomyolipomas; however, TSC patients are also at risk for the development of renal cell carcinomas. Genetic linkage analysis was used to identify two genes for tuberous sclerosis complex, TSC1, which is located on chromosome 9q34, and TSC2, on chromosome 16p13. The production of TSC1 (hamartin) and TSC2 (tuberin) form a heterodimer which has been shown to inhibit the mammalian target of rapamycin (mTOR) pathway (Crino et al. 2006). Mutation/inactivation of both copies of either TSC1 or TSC2 activates the mTOR pathway and results in increased cellular growth seen in TSC-associated hamartomas/cancers. A clinical trial was recently reported evaluating the role of sirolimus, an agent which suppresses mTOR signaling, in TSC patients. In this trial, Bissler et al. (2008) reported regression of angiomyolipomas and improvement in pulmonary parameters of TSC patients treated with sirolimus. Although the angiomyolipomas tended to increase in volume after the therapy was discontinued, this important study showed that targeting the TSC1/2-mTOR pathway can have a definite phenotypic effect in these patients and might be a useful approach to ameliorate the manifestations of this disorder (Bissler et al. 2008).
MiT family kidney cancer: TFE3, TFEB, and MITF
TFE3, TFEB, and MITF, members of the MiT family of transcription factors, have also been implicated as causes of kidney cancer. TFE3 has been described as involved in a translocation involving chromosome 1 and the X chromosome, t(X:1)(p11.2;p34), primarily in children and young adults (Clark et al. 1997; Argani et al. 2010). TFEB has been described as involved in a translocation involving chromosome 6 and chromosome 11, t(6;11)(p21;q13), which is also seen primarily in the pediatric population (Argani et al. 2001; Davis et al. 2003). The microphthalmia-associated transcription factor, MITF, has been implicated in melanoma and kidney cancer. Bertolotto et al. (2011) recently reported the detection of germline missense MITF mutations in patients with kidney cancer, melanoma, or both.
Kidney cancer is a metabolic disease
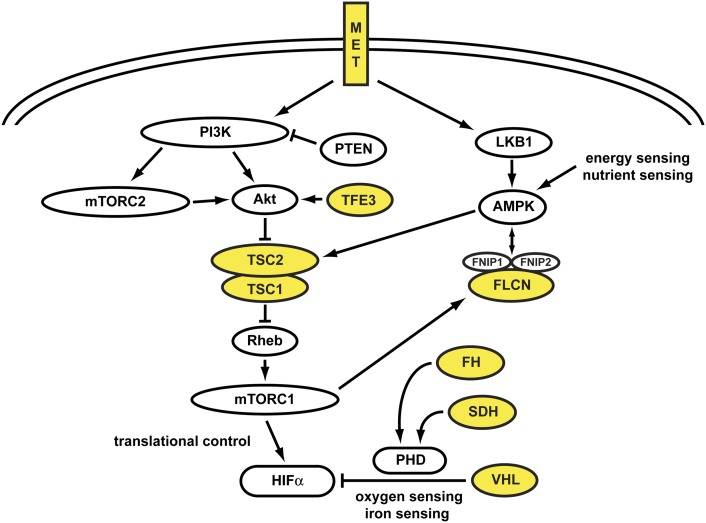
Each of the kidney cancer genes VHL, MET, FLCN, fumarate hydratase, succinate dehydrogenase, TSC1, TSC2, and TFE3 affects the cell's ability to sense iron, oxygen, nutrients, or energy. Kidney cancer is fundamentally a metabolic disease. Understanding the fundamental metabolic aspects of kidney cancer will hopefully provide the foundation for the development of novel approaches to therapy for this disease (Fig. 6).
Figure 6.
Kidney cancer gene pathways. Kidney cancer is fundamentally a metabolic disease. Each of the kidney cancer genes, VHL, MET, FLCN, FH, SDH, TSC1, TSC2, and TFE3 affects the cell's ability to sense oxygen, iron, nutrients, and most particularly in the Krebs cycle enzymes, FH and SDH, and energy. Further genomics studies elucidating these kidney cancer gene pathways will provide the foundation for the development of targeted approaches for therapy for these diseases. (Fig. 6 adapted from Linehan et al. 2010.)
Sporadic (nonfamilial) kidney cancer
What is the genetic basis of clear cell kidney cancer?
The Cancer Genome Atlas (TCGA) is conducting an exhaustingly comprehensive study of 500 clear cell kidney cancers, including complete genome sequencing, gene expression and microRNA analysis, analysis of methylation, copy number analysis, and fusion protein analysis. These analyses will be correlated with clinical parameters, including stage, response to therapy, and survival. A number of important questions are being addressed, such as: Are there other mutated genes which contribute to the development and/or progression of clear cell kidney cancer? Systematic gene expression studies will assess novel pathways that may be activated in the initiation or progression phase of this disease. Systematic gene copy number assessment is being performed to determine if there are gains or losses of genes involved in initiation or progression. One of the major accomplishments of TCGA is likely to be the integration of these complex/disparate data types to provide novel insights about pathways and gene sets. The initial findings are being carefully catalogued and will be made available as an invaluable resource for investigators studying this disease.
Clear cell kidney cancer that does not have VHL gene mutation is associated with a more aggressive phenotype and diminished survival (Patard et al. 2008). This is most likely because VHL +/+ clear cell kidney cancer is caused by a different gene(s) than VHL −/− clear cell kidney cancer; it appears that it is simply a different disease (with a more aggressive clinical phenotype).
In two studies in which the pathologic phenotype of clear cell kidney cancer was determined in 470 cases by a single, experienced kidney cancer pathologist and where the tumor material was carefully dissected to eliminate noncancerous material, VHL mutation or methylation was detected in nearly 90% of cases (Nickerson et al. 2008; Moore et al. 2011). Other studies have detected a lower VHL mutation rate in clear cell RCC (Young et al. 2009), which could be secondary to variation in histologic evaluation, sample preparation, or ethnic origin of the patient material. Two important unanswered questions that genomic approaches (i.e., whole genome sequencing) will answer are: (1) What are the other genes besides VHL that are involved in the genesis of clear cell kidney cancer? and (2) what is the genetic cause(s) of the non-VHL-deficient (VHL +/+) clear cell kidney cancers?
What is the genetic basis of non-clear cell kidney cancer?
Papillary kidney cancer
Papillary kidney cancer is traditionally subclassified by histologic (microscopic) morphology as type 1 and type 2. Type 1 papillary kidney cancer is characterized by tightly formed, cribiform papillae covered by small cuboidal cells with a single line of uniform nuclei and small nucleoli (Delahunt and Eble 1997; Leroy et al. 2002). Type I papillary kidney cancer is much more common than type 2 and tends to have a significantly better prognosis. Sporadic type 1 papillary kidney cancer is most often multifocal, it can be bilateral, and its growth rate often may be very slow. Type 2 papillary kidney cancer is characterized by large and less well-organized papillae with pleomorphic nuclei, prominent nucleoli, and nuclear pseudostratification (Delahunt and Eble 1997; Leroy et al. 2002). Type 2 papillary kidney cancer tends to be solitary and has a tendency to metastasize early. There is no effective form of therapy for patients affected with either type 1 or type 2 kidney cancer.
Type I papillary kidney cancer
HPRC-associated type 1 kidney cancer is associated with germline mutation of the MET gene on chromosome 7. However, MET gene mutations are found in only a subset of sporadic type 1 kidney cancers, and the genetic basis of sporadic type 1 papillary kidney cancer is not known (Schmidt et al. 1999). Both HPRC-associated as well as sporadic type 1 papillary kidney cancer are characterized by trisomy of chromosome 7; however, the role of trisomy 7 in papillary kidney cancer and whether or not amplification of MET on chromosome 7 will be a marker of response to therapy by agents which target MET has yet to be determined.
Non-type I papillary renal cell carcinoma
Non-type I papillary renal cell carcinoma comprises a mixed group of histologies, including type II papillary renal carcinoma, collecting duct carcinoma, medullary renal cell carcinoma, the MiTF kidney cancers (TFE3, TFEB), and HLRCC-associated kidney cancer. Other than the MiTF and HLRCC cancers, little is known about their origin or genetic basis, and there is no effective form of therapy for patients with advanced type II papillary kidney cancer. These tumors are frequently confused with each other, and it is often difficult to make an accurate diagnosis or classification. Even the term “type II papillary kidney cancer” is poorly defined, which is why these cancers are sometimes grouped together as “non-type I kidney cancer.”
The Cancer Genome Atlas is also currently conducting an extensive project to perform complete genome sequencing, copy number analysis, and gene expression analysis in a large number of papillary kidney cancers. It is hoped that this project will identify other genes and other pathways that are involved with the initiation and progression of papillary kidney cancer which will hopefully provide the foundation for the development of effective forms of therapy for this disease.
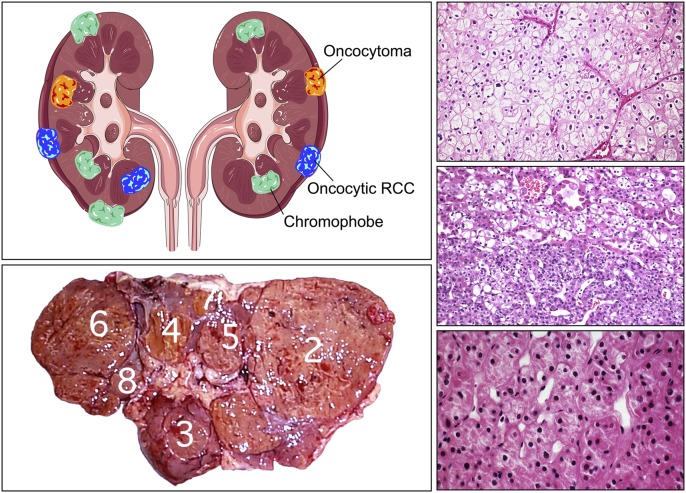
Chromophobe kidney cancer and oncocytoma
BHD-associated chromophobe kidney cancers and oncocytomas are characterized by germline mutation and somatic inactivation of the somatic FLCN allele (Vocke et al. 2005). However, FLCN mutations are not a consistent feature of either sporadic chromophobe RCC or oncocytoma, and the causal genes are not known. Gad et al. (2007) detected FLCN mutations in five of 46 (10.9%) chromophobe renal carcinomas, and TP53 mutations were detected in 11 of 46 (23.9%). The Cancer Genome Atlas has recently instituted a special study to evaluate the genetic basis of chromophobe kidney cancer. Oncocytomas are benign kidney tumors that do not metastasize. A critical biologic question is: What is the genetic basis of oncocytoma and why are oncocytomas benign and nonmetastastic, whereas nearly all other kidney tumors are malignant?
Discussion
Is there an inherited predisposition to all kidney cancers? A central unanswered question that needs to be addressed is what role inheritance plays in the development of kidney cancer. Currently, 5%–8% of kidney cancer is considered to have an inherited basis. However, while the autosomal dominant kidney cancer syndromes (VHL, HPRC, BHD, HLRCC, etc.) may comprise 5% of the cases of kidney cancer, there is a much higher incidence of families in which multiple individuals develop kidney cancer with no known genetic basis. Gudbjartsson et al. (2002) conducted a population-based study that included all 1078 patients diagnosed with kidney cancer in Iceland between 1955 and 1999. In this study, 58% of all cases of kidney cancer were found in families with two or more affected members. These authors concluded that “germline mutations are significantly involved in what has been defined as sporadic RCC” (Gudbjartsson et al. 2002). Genome-wide association studies have identified susceptibility loci in patients with “sporadic” kidney cancer (Purdue et al. 2011). The use of complete germline genomic sequencing will likely identify additional genes that predispose to the development of kidney cancer and will hopefully determine the role and extent of genes involved in susceptibility to kidney cancer.
Are mutations in one gene sufficient, or must mutations accrue in seven genes in order to create a kidney cancer? A fundamentally important question is how many mutations it takes to cause kidney cancer and which genetic pathway represents the most plausible target for a targeted therapeutic approach. It is a widely held view, based on many elegant models and studies by accomplished and thoughtful leaders in the field, that development of a cancer is a multiple step process and that it may take up to seven gene mutations to cause a cancer (Vogelstein et al. 1988). However, is that the case in kidney cancer? Is it possible that kidney cancers, unlike colon or other cancers, can be caused by loss of function of a single gene? Patients affected with VHL are at risk for the development of up to 600 tumors per kidney (Walther et al. 1995). These patients inherit a mutation of the VHL gene, and the wild-type (somatic) allele is deleted in the vast majority of the tumors. Do all 600 of these tumors have mutations in VHL and an additional six genes? Based on precise VHL-associated kidney tumor measurements generated over years, it is estimated that 25 years elapse from the time of initiation of the tumor until the tumor reaches a size of 2 cm. It has long been known that cells circulate for years before metastases develop. Is it possible that the process of metastasis in VHL- (or HPRC- or BHD-) associated kidney cancer is simply a function of time and not necessarily dependent on alteration of additional genes, i.e., that as tumors grow over the years and kidney cancer cells continue to circulate until the conditions are right for implantation and growth and the development of clinically detectable metastases? If seven genes are required for tumor development, which gene pathway should be targeted for therapy? When a VHL-deficient kidney cancer cell line is implanted in an athymic nude mouse, it forms a tumor. When the VHL gene is re-expressed in the cell line, either no tumor or a very small tumor appears. Is it possible that mutation of this single gene is sufficient to cause VHL-deficient kidney cancer? Patients affected with hereditary papillary renal carcinoma (HPRC), which is caused by activating mutation of the proto-oncogene, MET, are at risk for the development of up to 1900 tumors per kidney (Ornstein et al. 2000). These tumors are characterized by trisomy of chromosome 7 (where the MET gene is located) and nonrandom duplication of the mutant MET allele (Zhuang et al. 1998). Do each of these 1900 tumors have seven mutations or is duplication of the mutant MET allele sufficient for tumorigenesis? It is estimated that patients affected with Birt-Hogg-Dubé (BHD) are at risk for the development of up to 3000 tumors per kidney. When a FLCN-deficient human kidney cancer cell line is implanted in an athymic nude mouse, it forms a tumor. When the FLCN-restored cell line is implanted in the mouse, either no tumor or a small tumor appears. Similarly, when a fumarate hydratase-deficient cell line (derived from an HLRCC patient) is implanted into an athymic nude mouse, it rapidly forms a tumor, whereas when the enzyme fumarate hydratase is restored in the FH-deficient kidney cancer cell line, it is no longer tumorigenic. It is certainly possible that overexpression of a tumor suppressor gene (such as VHL or FLCN) would inhibit tumorigenesis of a cancer cell line. However, it is quite remarkable that restoration of fumarate hydratase activity is sufficient to reverse the tumorigenic phenotype.
What is the genetic basis of the metabolic shift to aerobic glycolysis? We have shown that fumarate hydratase-deficient kidney cancer is characterized by aerobic glycolysis; mitochondrial respiration is significantly impaired, and the cells depend on increased glucose transport and glycolysis for ATP production. The metabolic glycolytic shift in FH-deficient kidney cancer results in increased energy production, decreased AMPK, and increased mTOR activity and fatty acid biosynthesis, which are required for the rapid anabolic growth that characterizes this form of kidney cancer (Tong et al. 2011). The FH-deficient kidney cancer cells preferentially use glutamine over glucose as a carbon source for lipid biosynthesis, utilizing citrate derived from reductively carboxylated glutamine for fatty acid production (Mullen et al. 2011). Metallo et al. (2012) recently demonstrated that a similar reductive carboxylation pathway for glutamine metabolism by IDH1 mediates de novo lipogenesis in VHL-deficient kidney cancer.
A central unanswered question is whether the metabolic remodeling that occurs in fumarate hydratase- and VHL-deficient kidney cancers is attributable to a coordinated process mediated by methylation or acetylation of metabolic enzymes, to enable coordination of different carbon source utilization to provide building blocks, cofactors, and energy to fuel cell growth (Wang et al. 2010), or whether additional mutations are required to facilitate this process. On one hand, Wang et al. (2010) identified a process of differential and reversible acetylation of central metabolic enzymes in response to different carbon sources, which is conserved in both human and prokaryotes. This reversible acetylation of metabolic enzymes could be the type of mechanism that ensures that cells are able to rapidly respond to changing energy status, such as a metabolic shift to aerobic glycolysis in FH-deficient kidney cancer. An alternate possibility is that mutations of additional genes are required to effect this type of metabolic shift. Insight into a potential mutation-based mechanism for adaptation to, for example, the cell's need for increased glucose uptake and remodeling of the Krebs cycle to adapt to increased bioenergetic needs of a rapidly growing cancer cell comes from the elegant study of Meyer et al. (2012) who studied the adaptive development of mutations that allowed a virus, bacteriophage λ, to evolve to infect its host, Escherichia coli, through a novel pathway. They discovered that four precise mutations were required to evolve a new site of attachment, and these mutations developed repeatedly within weeks under the appropriate selective pressure (Meyer et al. 2012). A similar type of biological adaptability, evidenced by a stochastic selection of multiple precise additional mutations, could also underlie the transition to a fully transformed phenotype in human cancers. The answer to the question of whether it takes many mutations, or additional gene methylations, or histone acetylations, or copy number changes, or altered miRNAs, versus whether mutation of a single gene can cause cancer over time by remodeling metabolism in each of the various forms of renal cancer will most likely be discovered in comprehensive genomic analyses.
Summary
Genomics approaches have changed the way we think about kidney cancer and provided the foundation for the development of novel approaches to therapy for these diseases.
Historically, kidney cancer was thought of as a single disease. Now we know it is made up of a number of genetically and histologically different types of cancer that occur in the kidney. The identification of the genes for kidney cancers allows clinicians to manage patients according to the genotype. The kidney cancers in patients with germline mutations of the VHL, MET, and FLCN gene are each managed very differently from those, for example, in patients with germline mutations of the FH and SDH genes. Importantly, it has become clear through gene identifications that metabolic remodeling is an important part of renal cancer that could be exploited in therapies. For instance, the AMPK activator, metformin, may prove to be a candidate for treatment of FH-associated kidney cancers. There is still much to be done in the genomics of kidney cancers. Deep sequencing of clear cell kidney cancer, type 1 papillary kidney cancer, chromophobe kidney cancer, type II papillary kidney cancer, collecting duct cancer, and medullary kidney cancer will be needed to determine which gene or genes are critical for initiation of these diseases and for progression and metastasis. While a significant effort has been put into targeting the VHL pathway in clear cell kidney cancer, we currently have no targeted therapies which result in complete response for the majority of patients with this disease. Recent findings implicating inactivation of histone modifying genes opens new avenues for prevention as well as therapy of this malignancy. Genomic studies evaluating copy number as well as gene expression patterns in these cancers will also be needed to uncover other cancer gene pathways that will be critical for the development of effective forms of therapy for patients with these diseases.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The author thanks Tracey A. Rouault, M.D. for the manuscript review and Georgia Shaw for editorial and graphics support.
Footnotes
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.131110.111.
References
- Anglard P, Brauch TH, Weiss GH, Latif F, Merino MJ, Lerman MI, Zbar B, Linehan WM 1991. Molecular analysis of genetic changes in the origin and development of renal cell carcinoma. Cancer Res 51: 1071–1077 [PubMed] [Google Scholar]
- Argani P, Hawkins A, Griffin CA, Goldstein JD, Haas M, Beckwith JB, Mankinen CB, Perlman EJ 2001. A distinctive pediatric renal neoplasm characterized by epithelioid morphology, basement membrane production, focal HMB45 immunoreactivity, and t(6;11)(p21.1;q12) chromosome translocation. Am J Pathol 158: 2089–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argani P, Hicks J, De Marzo AM, Albadine R, Illei PB, Ladanyi M, Reuter VE, Netto GJ 2010. Xp11 translocation renal cell carcinoma (RCC): Extended immunohistochemical profile emphasizing novel RCC markers. Am J Surg Pathol 34: 1295–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Hong SB, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF III, Hartley JL, et al. 2006. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci 103: 15552–15557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Furihata M, Hong SB, Tessarollo L, Haines DC, Southon E, Patel V, Igarashi P, Alvord WG, Leighty R, et al. 2008. Kidney-targeted Birt-Hogg-Dubé gene inactivation in a mouse model: Erk1/2 and Akt-mTOR activation, cell hyperproliferation, and polycystic kidneys. J Natl Cancer Inst 100: 140–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Brunet JP, Di NA, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, et al. 2009. Patterns of gene expression and copy-number alterations in von-Hippel Lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res 69: 4674–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, Dessen P, d'Hayer B, Mohamdi H, Remenieras A, et al. 2011. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 480: 94–98 [DOI] [PubMed] [Google Scholar]
- Bissler JJ, McCormack FX, Young LR, Elwing JM, Chuck G, Leonard JM, Schmithorst VJ, Laor T, Brody AS, Bean J, et al. 2008. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N Engl J Med 358: 140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff A, Choyke PL, Linehan WM, Walther MM 2001. Parenchymal sparing surgery in a patient with multiple bilateral papillary renal cancer. J Urol 165: 1623–1624 [PubMed] [Google Scholar]
- Choueiri TK, Vaishampayan U, Rosenberg JE, Logan TF, Harzstark AL, Bukowski RM, Rini BI, Srinivas S, Stein MN, Adams LM, et al. 2012. A phase II and biomarker study of the dual MET/VEGFR-2 inhibitor foretinib in patients with papillary renal-cell carcinoma. J Clin Oncol (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, Lu YJ, Sidhar SK, Parker C, Gill S, Smedley D, Hamoudi R, Linehan WM, Shipley J, Cooper CS 1997. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene 15: 2233–2239 [DOI] [PubMed] [Google Scholar]
- Cohen AJ, Li FP, Berg S, Marchetto DJ, Tsai S, Jacobs SC, Brown RS 1979. Hereditary renal-cell carcinoma associated with a chromosomal translocation. N Engl J Med 301: 592–595 [DOI] [PubMed] [Google Scholar]
- Crino PB, Nathanson KL, Henske EP 2006. The tuberous sclerosis complex. N Engl J Med 355: 1345–1356 [DOI] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. 2010. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463: 360–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IJ, Hsi BL, Arroyo JD, Vargas SO, Yeh YA, Motyckova G, Valencia P, Perez-Atayde AR, Argani P, Ladanyi M, et al. 2003. Cloning of an α-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci 100: 6051–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahunt B, Eble JN 1997. Papillary renal cell carcinoma: A clinicopathologic and immunohistochemical study of 105 tumors. Mod Pathol 10: 537–544 [PubMed] [Google Scholar]
- Duan DR, Pause A, Burgess WH, Aso T, Chen DY, Garrett KP, Conaway RC, Conaway JW, Linehan WM, Klausner RD 1995. Inhibition of transcription elongation by the VHL tumor suppressor protein. Science 269: 1402–1406 [DOI] [PubMed] [Google Scholar]
- Eleveld MJ, Bodmer D, Merkx G, Siepman A, Sprenger SH, Weterman MA, Ligtenberg MJ, Kamp J, Stapper W, Jeuken JW, et al. 2001. Molecular analysis of a familial case of renal cell cancer and a t(3;6)(q12;q15). Genes Chromosomes Cancer 31: 23–32 [DOI] [PubMed] [Google Scholar]
- Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, Negrier S, Chevreau C, Solska E, Desai AA, et al. 2007. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med 356: 125–134 [DOI] [PubMed] [Google Scholar]
- Fadahunsi AT, Sanford T, Linehan WM, Pinto PA, Bratslavsky G 2011. Feasibility and outcomes of partial nephrectomy for resection of at least 20 tumors in a single renal unit. J Urol 185: 49–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM 2010. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127: 2893–2917 [DOI] [PubMed] [Google Scholar]
- Gad S, Lefevre SH, Khoo SK, Giraud S, Vieillefond A, Vasiliu V, Ferlicot S, Molinie V, Denoux Y, Thiounn N, et al. 2007. Mutations in BHD and TP53 genes, but not in HNF1β gene, in a large series of sporadic chromophobe renal cell carcinoma. Br J Cancer 96: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill AJ, Pachter NS, Clarkson A, Tucker KM, Winship IM, Benn DE, Robinson BG, Clifton-Bligh RJ 2011. Renal tumors and hereditary pheochromocytoma-paraganglioma syndrome type 4. N Engl J Med 364: 885–886 [DOI] [PubMed] [Google Scholar]
- Gnarra JR, Tory K, Weng Y, Schmidt LS, Wei MH, Li H, Latif F, Liu S, Chen F, Duh FM, et al. 1994. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet 7: 85–90 [DOI] [PubMed] [Google Scholar]
- Grubb RL III, Franks ME, Toro J, Middelton L, Choyke L, Fowler S, Torres-Cabala C, Glenn GM, Choyke P, Merino MJ, et al. 2007. Hereditary leiomyomatosis and renal cell cancer: A syndrome associated with an aggressive form of inherited renal cancer. J Urol 177: 2074–2080 [DOI] [PubMed] [Google Scholar]
- Gudbjartsson T, Jonasdottir TJ, Thoroddsen A, Einarsson GV, Jonsdottir GM, Kristjansson K, Hardarson S, Magnusson K, Gulcher J, Stefansson K, et al. 2002. A population-based familial aggregation analysis indicates genetic contribution in a majority of renal cell carcinomas. Int J Cancer 100: 476–479 [DOI] [PubMed] [Google Scholar]
- Hasumi H, Baba M, Hong SB, Hasumi Y, Huang Y, Yao M, Valera VA, Linehan WM, Schmidt LS 2008. Identification and characterization of a novel folliculin-interacting protein FNIP2. Gene 415: 60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasumi Y, Baba M, Ajima R, Hasumi H, Valera VA, Klein ME, Haines DC, Merino MJ, Hong SB, Yamaguchi TP, et al. 2009. Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc Natl Acad Sci 106: 18722–18727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, Samid D, Duan DS, Gnarra JR, Linehan WM, et al. 1994. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci 91: 9700–9704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring JC, Enquist EG, Chernoff A, Linehan WM, Choyke PL, Walther MM 2001. Parenchymal sparing surgery in patients with hereditary renal cell carcinoma: 10-year experience. J Urol 165: 777–781 [PubMed] [Google Scholar]
- Hosoe S, Brauch H, Latif F, Glenn GM, Daniel L, Bale S, Choyke P, Gorin M, Oldfield E, Berman A, et al. 1990. Localization of the von Hippel-Lindau disease gene to a small region of chromosome 3. Genomics 8: 634–640 [DOI] [PubMed] [Google Scholar]
- Hudes G, Carducci M, Tomczak P, Dutcher J, Figlin R, Kapoor A, Staroslawska E, Sosman J, McDermott D, Bodrogi I, et al. 2007. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N Engl J Med 356: 2271–2281 [DOI] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, Merino MJ, Trepel J, Zbar B, Toro J, et al. 2005. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: Novel role of fumarate in regulation of HIF stability. Cancer Cell 8: 143–153 [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. 2001. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292: 468–472 [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr 2008. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer 8: 865–873 [DOI] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG Jr, Elledge SJ, Conaway RC, et al. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284: 657–661 [DOI] [PubMed] [Google Scholar]
- Kibel A, Iliopoulos O, DeCaprio JA, Kaelin WG Jr 1995. Binding of the von Hippel-Lindau tumor suppressor protein to elongin B and C. Science 269: 1444–1446 [DOI] [PubMed] [Google Scholar]
- Knudson AG 1995. VHL gene mutation and clear-cell renal carcinomas. Cancer J 1: 180–181 [PubMed] [Google Scholar]
- Latif F, Tory K, Gnarra JR, Yao M, Duh F-M, Orcutt ML, Stackhouse T, Kuzmin I, Modi W, Geil L, et al. 1993. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science 260: 1317–1320 [DOI] [PubMed] [Google Scholar]
- Launonen V, Vierimaa O, Kiuru M, Isola J, Roth S, Pukkala E, Sistonen P, Herva R, Aaltonen LA 2001. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc Natl Acad Sci 98: 3387–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy X, Zini L, Leteurtre E, Zerimech F, Porchet N, Aubert JP, Gosselin B, Copin MC 2002. Morphologic subtyping of papillary renal cell carcinoma: Correlation with prognosis and differential expression of MUC1 between the two subtypes. Mod Pathol 15: 1126–1130 [DOI] [PubMed] [Google Scholar]
- Linehan WM, Zbar B 2004. Focus on kidney cancer. Cancer Cell 6: 223–228 [DOI] [PubMed] [Google Scholar]
- Linehan WM, Walther MM, Zbar B 2003. The genetic basis of cancer of the kidney. J Urol 170: 2163–2172 [DOI] [PubMed] [Google Scholar]
- Linehan WM, Srinivasan R, Schmidt LS 2010. The genetic basis of kidney cancer: A metabolic disease. Nature Reviews Urology 7: 277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan WM, Yang JC, Rini B 2011. Cancer of the kidney. In Cancer: Principles and practice of oncology (ed. VT DeVita et al.), pp. 1161–1182. Lippincott Williams & Wilkins, New Haven, CT [Google Scholar]
- Lonser RR, Glenn GM, Walther MM, Chew EY, Libutti SK, Linehan WM, Oldfield EH 2003. von Hippel-Lindau disease. Lancet 361: 2059–2067 [DOI] [PubMed] [Google Scholar]
- Maranchie JK, Afonso A, Albert PS, Kalyandrug S, Phillips JL, Zhou S, Peterson J, Ghadimi BM, Hurley K, Riss J, et al. 2004. Solid renal tumor severity in von Hippel Lindau disease is related to germline deletion length and location. Hum Mutat 23: 40–46 [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399: 271–275 [DOI] [PubMed] [Google Scholar]
- Merino MJ, Torres-Cabala C, Pinto PA, Linehan WM 2007. The morphologic spectrum of kidney tumors in hereditary leiomyomatosis and renal cell carcinoma (HLRCC) syndrome. Am J Surg Pathol 31: 1578–1585 [DOI] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. 2012. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481: 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JR, Dobias DT, Weitz JS, Barrick JE, Quick RT, Lenski RE 2012. Repeatability and contingency in the evolution of a key innovation in phage λ. Science 335: 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LE, Nickerson ML, Brennan P, Toro JR, Jaeger E, Rinsky J, Han SS, Zaridze D, Matveev V, Janout V, et al. 2011. Von Hippel-Lindau (VHL) inactivation in sporadic clear cell renal cancer: Associations with germline VHL polymorphisms and etiologic risk factors. PLoS Genet 7: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, Oudard S, Negrier S, Szczylik C, Kim ST, et al. 2007. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med 356: 115–124 [DOI] [PubMed] [Google Scholar]
- Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, Grunwald V, Thompson JA, Figlin RA, Hollaender N, et al. 2008. Efficacy of everolimus in advanced renal cell carcinoma: A double-blind, randomised, placebo-controlled phase III trial. Lancet 372: 1432–1439 [DOI] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ 2011. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481: 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann HP, Pawlu C, Peczkowska M, Bausch B, McWhinney SR, Muresan M, Buchta M, Franke G, Klisch J, Bley TA, et al. 2004. Distinct clinical features of paraganglioma syndromes associated with SDHB and SDHD gene mutations. JAMA 292: 943–951 [DOI] [PubMed] [Google Scholar]
- Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn GM, Turner ML, Duray P, Merino MJ, Choyke P, Pavlovich CP, et al. 2002. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell 2: 157–164 [DOI] [PubMed] [Google Scholar]
- Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, Matveev V, Janout V, Kollarova H, Bencko V, et al. 2008. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res 14: 4726–4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, Pavletich N, Chau V, Kaelin WG 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the β-domain of the von Hippel-Lindau protein. Nat Cell Biol 2: 423–427 [DOI] [PubMed] [Google Scholar]
- Ornstein DK, Lubensky IA, Venzon D, Zbar B, Linehan WM, Walther MM 2000. Prevalence of microscopic tumors in normal appearing renal parenchyma of patients with hereditary papillary renal cancer. J Urol 163: 431–433 [PubMed] [Google Scholar]
- Patard JJ, Fergelot P, Karakiewicz PI, Klatte T, Trinh QD, Rioux-Leclercq N, Said JW, Belldegrun AS, Pantuck AJ 2008. Low CAIX expression and absence of VHL gene mutation are associated with tumor aggressiveness and poor survival of clear cell renal cell carcinoma. Int J Cancer 123: 395–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovich CP, Walther MM, Eyler RA, Hewitt SM, Zbar B, Linehan WM, Merino MJ 2002. Renal tumors in the Birt-Hogg-Dubé syndrome. Am J Surg Pathol 26: 1542–1552 [DOI] [PubMed] [Google Scholar]
- Pavlovich CP, Grubb RL, Hurley K, Glenn GM, Toro J, Schmidt LS, Torres-Cabala C, Merino MJ, Zbar B, Choyke P, et al. 2005. Evaluation and management of renal tumors in the Birt-Hogg-Dube syndrome. J Urol 173: 1482–1486 [DOI] [PubMed] [Google Scholar]
- Podolski J, Byrski T, Zajaczek S, Druck T, Zimonjic DB, Popescu NC, Kata G, Borowka A, Gronwald J, Lubinski J, et al. 2001. Characterization of a familial RCC-associated t(2;3)(q33;q21) chromosome translocation. J Hum Genet 46: 685–693 [DOI] [PubMed] [Google Scholar]
- Purdue MP, Johansson M, Zelenika D, Toro JR, Scelo G, Moore LE, Prokhortchouk E, Wu X, Kiemeney LA, Gaborieau V, et al. 2011. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat Genet 43: 60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricketts C, Woodward ER, Killick P, Morris MR, Astuti D, Latif F, Maher ER 2008. Germline SDHB mutations and familial renal cell carcinoma. J Natl Cancer Inst 100: 1260–1262 [DOI] [PubMed] [Google Scholar]
- Rini BI, Escudier B, Tomczak P, Kaprin A, Szczylik C, Hutson TE, Michaelson MD, Gorbunova VA, Gore ME, Rusakov IG, et al. 2011. Comparative effectiveness of axitinib versus sorafenib in advanced renal cell carcinoma (AXIS): A randomised phase 3 trial. Lancet 378: 1931–1939 [DOI] [PubMed] [Google Scholar]
- Schmidt LS, Li F, Brown RS, Berg S, Chen F, Wei MH, Tory K, Lerman I, Zbar B 1995. Mechanism of tumorigenesis of renal carcinomas associated with the constitutional chromosome 3;8 translocation. Cancer J Sci Am 1: 191–195 [PubMed] [Google Scholar]
- Schmidt LS, Duh FM, Chen F, Kishida T, Glenn GM, Choyke P, Scherer SW, Zhuang Z, Lubensky I, Dean M, et al. 1997. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet 16: 68–73 [DOI] [PubMed] [Google Scholar]
- Schmidt LS, Junker K, Nakaigawa N, Kinjerski T, Weirich G, Miller M, Lubensky I, Neumann HP, Brauch H, Decker J, et al. 1999. Novel mutations of the MET proto-oncogene in papillary renal carcinomas. Oncogene 18: 2343–2350 [DOI] [PubMed] [Google Scholar]
- Sternberg CN, Davis ID, Mardiak J, Szczylik C, Lee E, Wagstaff J, Barrios CH, Salman P, Gladkov OA, Kavina A, et al. 2010. Pazopanib in locally advanced or metastatic renal cell carcinoma: Results of a randomized phase III trial. J Clin Oncol 28: 1061–1068 [DOI] [PubMed] [Google Scholar]
- Stolle C, Glenn GM, Zbar B, Humphrey JS, Choyke P, Walther MM, Pack S, Hurley K, Andrey C, Klausner R, et al. 1998. Improved detection of germline mutations in the von Hippel-Lindau disease tumor suppressor gene. Hum Mutat 12: 417–423 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Sugimura J, Yang X, Vogelzang N, Teh BS, Furge K, Teh BT 2003a. Gene expression profiling of renal cell carcinoma and its implications in diagnosis, prognosis, and therapeutics. Adv Cancer Res 89: 157–181 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Yang XJ, Sugimura J, Backdahl J, Tretiakova M, Qian CN, Gray SG, Knapp R, Anema J, Kahnoski R, et al. 2003b. Molecular subclassification of kidney tumors and the discovery of new diagnostic markers. Oncogene 22: 6810–6818 [DOI] [PubMed] [Google Scholar]
- Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, et al. 2002. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet 30: 406–410 [DOI] [PubMed] [Google Scholar]
- Tong WH, Sourbier C, Kovtunovych G, Jeong SY, Vira M, Ghosh M, Romero VV, Sougrat R, Vaulont S, Viollet B, et al. 2011. The glycolytic shift in fumarate-hydratase-deficient kidney cancer lowers AMPK levels, increases anabolic propensities and lowers cellular iron levels. Cancer Cell 20: 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro JR, Nickerson ML, Wei MH, Warren MB, Glenn GM, Turner ML, Stewart L, Duray P, Tourre O, Sharma N, et al. 2003. Mutations in the fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North America. Am J Hum Genet 73: 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro JR, Wei MH, Glenn GM, Weinreich M, Toure O, Vocke CD, Turner M, Choyke P, Merino MJ, Pinto PA, et al. 2008. BHD mutations, clinical and molecular genetic investigations of Birt-Hogg-Dube syndrome: A new series of 50 families and a review of published reports. J Med Genet 45: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Erp F, Van Ravenswaaij C, Bodmer D, Eleveld M, Hoogerbrugge N, Mulders P, Geurts K 2003. Chromosome 3 translocations and the risk to develop renal cell cancer: A Dutch intergroup study. Genet Couns 14: 149–154 [PubMed] [Google Scholar]
- Vanharanta S, Buchta M, McWhinney SR, Virta SK, Peczkowska M, Morrison CD, Lehtonen R, Januszewicz A, Jarvinen H, Juhola M, et al. 2004. Early-onset renal cell carcinoma as a novel extraparaganglial component of SDHB-associated heritable paraganglioma. Am J Hum Genet 74: 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. 2011. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasselli JR, Shih JH, Iyengar SR, Maranchie J, Riss J, Worrell R, Torres-Cabala C, Tabios R, Mariotti A, Stearman R, et al. 2003. Predicting survival in patients with metastatic kidney cancer by gene-expression profiling in the primary tumor. Proc Natl Acad Sci 100: 6958–6963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocke CD, Yang Y, Pavlovich CP, Schmidt LS, Nickerson ML, Torres-Cabala CA, Merino MJ, Walther MM, Zbar B, Linehan WM 2005. High frequency of somatic frameshift BHD gene mutations in Birt-Hogg-Dube-associated renal tumors. J Natl Cancer Inst 97: 931–935 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL 1988. Genetic alterations during colorectal-tumor development. N Engl J Med 319: 525–532 [DOI] [PubMed] [Google Scholar]
- Walther MM, Lubensky IA, Venzon D, Zbar B, Linehan WM 1995. Prevalence of microscopic lesions in grossly normal renal parenchyma from patients with von Hippel-Lindau disease, sporadic renal cell carcinoma and no renal disease: Clinical implications. J Urol 154: 2010–2014 [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. 2010. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327: 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O 1956. On the origin of cancer cells. Science 123: 309–314 [DOI] [PubMed] [Google Scholar]
- Warburg O, Posener K, Negelein E 1924. Metabolism of the carcinoma cell. Biochem Z 152: 309–344 [Google Scholar]
- Wei MH, Toure O, Glenn GM, Pithukpakorn M, Neckers L, Stolle C, Choyke P, Grubb R, Middelton L, Turner ML, et al. 2006. Novel mutations in FH and expansion of the spectrum of phenotypes expressed in families with hereditary leiomyomatosis and renal cell cancer. J Med Genet 43: 18–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JC 2004. Bevacizumab for patients with metastatic renal cancer: An update. Clin Cancer Res 10: 6367S–6370S [DOI] [PubMed] [Google Scholar]
- Yang JC, Haworth L, Sherry RM, Hwu P, Schwartzentruber DJ, Topalian SL, Steinberg SM, Chen HX, Rosenberg SA 2003. A randomized trial of bevacizumab, an anti-vascular endothelial growth factor antibody, for metastatic renal cancer. N Engl J Med 349: 427–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Valera VA, Padilla-Nash HM, Sourbier C, Vocke CD, Vira MA, Abu-Asab MS, Bratslavsky G, Tsokos M, Merino MJ, et al. 2010. UOK 262 cell line, fumarate hydratase deficient (FH−/FH−) hereditary leiomyomatosis renal cell carcinoma: In vitro and in vivo model of an aberrant energy metabolic pathway in human cancer. Cancer Genet Cytogenet 196: 45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AC, Craven RA, Cohen D, Taylor C, Booth C, Harnden P, Cairns DA, Astuti D, Gregory W, Maher ER, et al. 2009. Analysis of VHL gene alterations and their relationship to clinical parameters in sporadic conventional renal cell carcinoma. Clin Cancer Res 15: 7582–7592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbar B, Brauch H, Talmadge C, Linehan WM 1987. Loss of alleles of loci on the short arm of chromosome 3 in renal cell carcinoma. Nature 327: 721–724 [DOI] [PubMed] [Google Scholar]
- Zbar B, Tory K, Merino MJ, Schmidt LS, Glenn GM, Choyke P, Walther MM, Lerman M, Linehan WM 1994. Hereditary papillary renal cell carcinoma. J Urol 151: 561–566 [DOI] [PubMed] [Google Scholar]
- Zbar B, Glenn GM, Lubensky IA, Choyke P, Magnusson G, Bergerheim U, Pettersson S, Amin M, Hurley K, Linehan WM, et al. 1995. Hereditary papillary renal cell carcinoma: Clinical studies in 10 families. J Urol 153: 907–912 [PubMed] [Google Scholar]
- Zbar B, Alvord WG, Glenn GM, Turner M, Pavlovich CP, Schmidt LS, Walther MM, Choyke P, Weirich G, Hewitt SM, et al. 2002. Risk of renal and colonic neoplasms and spontaneous pneumothorax in the Birt-Hogg-Dube syndrome. Cancer Epidemiol Biomarkers Prev 11: 393–400 [PubMed] [Google Scholar]
- Zhao H, Ljungberg B, Grankvist K, Rasmuson T, Tibshirani R, Brooks JD 2006. Gene expression profiling predicts survival in conventional renal cell carcinoma. PLoS Med 3: e13 doi: 10.1371/journal.pmed.0030013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z, Park WS, Pack S, Schmidt LS, Pak E, Pham T, Weil RJ, Candidus S, Lubensky IA, Linehan WM, et al. 1998. Trisomy 7-harbouring non-random duplication of the mutant MET allele in hereditary papillary renal carcinomas. Nat Genet 20: 66–69 [DOI] [PubMed] [Google Scholar]