Abstract
Variation in the composition of the human oral microbiome in health and disease has been observed. We have characterized inter- and intra-individual variation of microbial communities of 107 individuals in one of the largest cohorts to date (264 saliva samples), using culture-independent 16S rRNA pyrosequencing. We examined the salivary microbiome in up to three time-points during 10 yr spanning adolescence, and determined the influence of human genotype, gender, age, and weight class. Participants, including 27 monozygotic and 18 dizygotic twin pairs, were sampled mainly at ages 12–13, 17–18, and 22–24, with a few sampled as early as 8 yr of age. In contrast to gut or skin microbiomes, there is a core genus-level salivary microbiome. Individuals are more similar to themselves and their co-twins in the 12–17 and in the 17–22 cohorts than to the whole sample population, but not over the 10 yr from 12 to 22; and monozygotic twin pairs are statistically not more similar than dizygotic twin pairs. The data are most consistent with shared environment serving as the main determinant of microbial populations. Twins resemble each other more closely than the whole population at all time-points, but become less similar to each other when they age and no longer cohabit. Several organisms have age-specific abundance profiles, including members of the genera Veillonella, Actinomyces, and Streptococcus. There is no clear effect of weight class and gender. The results of this work will provide a basis to further study oral microbes and human health.
Humans have a lifelong intimate relationship with bacteria (Turnbaugh et al. 2007; Ley et al. 2008), starting with inoculation at birth (Dominguez-Bello et al. 2010). Beneficial microbes perform roles including immune system maturation, pathogen defense, complex polysaccharide digestion, and toxic compound degradation (Turnbaugh et al. 2007). Shifts in microbiota composition may predispose humans to disease (Young 2012), but how the dynamics of naturally occurring variation in microbes affect normal health and development is still largely undetermined.
A major point of bacterial entry into the human body is the mouth. It has both soft-tissue (lips, cheeks, tongue, palate) and hard-tissue surfaces (such as teeth) that support diverse bacteria (Aas et al. 2005). Bacterial communities in the mouth can cause oral (Socransky et al. 1998) and systemic diseases such as bacteremia (Poveda-Roda et al. 2008), endocarditis (Parahitiyawa et al. 2009), and potentially premature birth (Buduneli et al. 2005; Fardini et al. 2010). According to the World Health Organization, “oral health is integral to general health and is essential for well-being” (Petersen 2003). Microbes that colonize oral surfaces, and human oral cells such as epithelial cells and lymphocytes, slough off and accumulate in saliva. Saliva has been used as a readily accessible body fluid for research on oral commensal bacteria (Motisuki et al. 2005) and is routinely used to obtain human DNA for genetic and forensic purposes (Young et al. 2000; Nasidze et al. 2009).
The oral microbiota appears shortly after birth due to vertical transmission of bacteria from family members, especially mother to child (Asikainen et al. 1997; Van Winkelhoff and Boutaga 2005; Li et al. 2007). Despite exposure of the mouth to environmental microbes due to ingestion and oral hygienic practices, oral microbial communities appear especially stable relative to those in other body habitats (Costello et al. 2009). Colonization by horizontal transmission can occur (Asikainen et al. 1997; Van Winkelhoff and Boutaga 2005), but the quantitative importance of this pathway is unclear. The oral microbiome varies most during childhood, when contact with external microbes is highest (Kononen 2000), the population matures, or new habitats arise (Costello et al. 2012), for example when deciduous and later permanent teeth erupt (Crielaard et al. 2011). Some microorganisms may remain in the oral cavity once acquired (Saarela et al. 1999; Lucas et al. 2000) and accumulate with age (Papaioannou et al. 2009).
Although ∼50% of oral bacteria can be cultured (Mullany et al. 2011), culture-based methods still miss much of the community. Studies of variation in oral microbiota in healthy individuals by 16S rDNA sequence analysis have been limited by small sample size (Aas et al. 2005; Costello et al. 2009; Lazarevic et al. 2009, 2010; Zaura et al. 2009; Bik et al. 2010; Caporaso et al. 2011) or number of sequences per subject (Nasidze et al. 2009). To define accurately the oral microbial population, a large-scale study of the population of microbial rDNA in the saliva of hundreds of samples including several hundred individual sequences per sample is needed. Beyond steady-state assessment of the oral microbiome, it is important to understand the long-term stability of the bacterial populations in saliva over time in individuals and in populations. Current data about temporal variation of the oral microbiome do not exceed 15 mo (Costello et al. 2009; Lazarevic et al. 2010; Caporaso et al. 2011), and little is known about changes during adolescence.
The influence of the human host genome on the composition of the microbiome is still controversial, with limited evidence available (Corby et al. 2007). The gut microbiome is the best studied, but no study with sufficient statistical power to define the effects of human genotype on bacterial composition has been reported (Tims et al. 2011). A study by Benson and coworkers in 2010 identified QTLs that influenced the abundance of several gut bacteria in mice (Benson et al. 2010). The influence of host genetic variation of the human IL-1 gene (Interleukin-1) on periodontitis has been suggested, but is inconclusive (Kornman and di Giovine 1998; Huynh-Ba et al. 2007).
Here we use the oral microbiome of a large human cohort to address three main questions: (1) What is the intra- and inter-individual variation of the oral microbiome in a large, geographically defined cohort (Front Range, Colorado)? (2) Is the composition of the human microbiome heritable? Do monozygotic (MZ) twins differ from dizygotic (DZ) twins? (3) What are the changes of the salivary microbiome during the decade spanning adolescence? We also assessed the influence of weight class and gender on oral microbial community composition.
Results
Study population and data generation
We studied the variability in the microbiome of 264 individual saliva samples derived from 107 individuals between the ages of 8 and 26 (Average age 16.2 ± 4.6 yr standard deviation). Ninety-nine individuals (93.5%) were Non-Hispanic Whites, two individuals (1.9%) Hispanic Whites, four (3.7%) Hispanic of unknown race, one individual (0.9%) American Indian, and one individual (0.9%) multiethnic (all self-report). Saliva samples were obtained from 27 MZ twin pairs, 18 DZ twin pairs, eight unrelated sibling pairs of adopted children, and one unrelated individual from the same cohort. Eighty-two individuals were sampled more than once approximately in 5-yr intervals at up to three time-points (12/13, 17/18, and 22/23/24 yr of age, labeled as 12, 17, and 22 yr of age).
PCR amplification and subsequent 454 pyrosequencing of the 16S rDNA gene hypervariable regions V1 and V2 of 264 samples resulted in 593,220 reads, which were quality-filtered with QIIME (Caporaso et al. 2010b) and with OTUpipe (Robert Edgar, http://drive5.com/otupipe/) to select the most reliable reads. Of the barcoded reads, 427,189 were used for analysis after filtering. Samples with fewer than 698 sequence reads and internal controls were not included in this analysis. The average length of the sequence reads was 367 bp (range 200–513 bp) before denoising. Detailed information about the subjects and sequencing barcodes can be found in Supplemental Table S1.
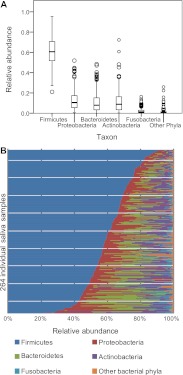
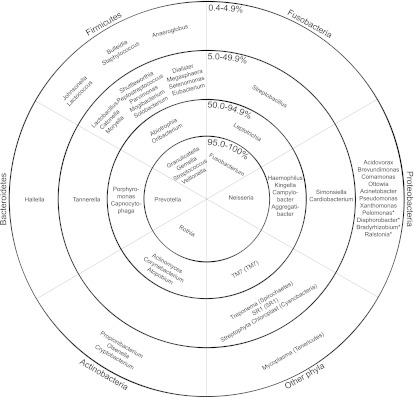
It has been previously described (Bik et al. 2010; Contreras et al. 2010; Lazarevic et al. 2010) that the main bacterial phyla in saliva were Firmicutes (with predominant genera Streptococcus, Veillonella, Gemella, and Granulicatella), Proteobacteria (Neisseria), Bacteroidetes (Prevotella), and Actinobacteria (Rothia). We additionally found Fusobacteria, TM7, Cyanobacteria, SR1, Spirochaetes, and Tenericutes. The relative abundance of each phylum was highly variable between samples (Fig. 1A,B). We defined a core genus-level salivary microbiome based on the percentage of samples in which each genus was found (some rare genera may be missed due to incomplete sampling [Sogin et al. 2006]). In contrast to other human body habitats, such as the gut (Turnbaugh et al. 2009), a core salivary microbiome can be defined at the genus level. Eight genera were observed in >95% of all samples (Streptococcus, Veillonella, Gemella, Granulicatella, Neisseria, Prevotella, Rothia, Fusobacterium); an additional 13 in >50% (Fig. 2; Supplemental Table S2). At a 97% identity level of operational taxonomic units (OTUs), two OTUs (OTU 1 and OTU 578), both of which belong to the genus Streptococcus, were found in all but one sample each (99.6% of samples, 10.5% of all sequence reads). The species of OTU 578 could not be identified with certainty. The closest BLAST match to the Human Oral Microbiome Database (Chen et al. 2010) for OTU 1 was Streptococcus mitis (99.7% identity), the second most common bacterium isolated from the oral cavity by molecular cloning based on the HOMD database (accessed August 31, 2011). S. mitis was also the only oral bacterium found on all oral surfaces from at least four of five individuals examined in Aas et al. (2005). An additional 40 OTUs were shared across >80% of the samples (45.6% of total sequence reads, see Supplemental Tables S3, S4).
Figure 1.
(A) Box-and-whiskers plot of the five major bacterial phyla of 264 human saliva samples. The top of the box represents the 75th percentile, the bottom of the box points to the 25th percentile, and the black line in the middle shows the median. The whiskers represent the highest and lowest values up to 1.5 times the interquartile range; extreme values and outliers are represented by empty circles. (B) Relative abundance of the five major bacterial phyla of all individual saliva samples, sorted by decreasing Firmicutes content.
Figure 2.
Presence of bacterial genera based on occurrence in samples. Taxonomic identity is based on RDP classification (Cole et al. 2007). The rings represent the percentage of samples where a given genus was observed (0.4%–4.9% means that genus was found in 1–12 samples, 5.0%–49.9% means that genus was found in 13–131 samples, 50.0%–94.9% means that genus was found in 132–250 samples, and 95.0%–100% means that genus was found in 251–264 samples). The pie slices subdivide the chart into the various bacterial phyla. (*) Genera that have only been found in one sample.
Is the composition of the oral microbiome heritable?
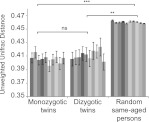
Comparison of the sharing of microbiomes of MZ and DZ twins permits a powerful assessment of heritability (i.e., the influence of the human genotype on phenotype). MZ twin pairs, who share 100% of their alleles, are expected to have oral microbiomes that are more similar to each other than do DZ twin pairs, who share ∼50% of their alleles. We compared 59 MZ and 39 DZ same-aged twin saliva sample pairs obtained between the ages of 12, 17, and 22. The metric used for comparison was the unweighted UniFrac distance (Lozupone and Knight 2005; Lozupone et al. 2006), a widely used qualitative (presence/absence) community comparison measure based on phylogenetic information. UniFrac values range from 0 (identical communities) to 1 (maximum difference). We observed a slight trend toward more similarity among cohabiting MZ pairs than DZ pairs (Fig. 3), but as previously shown in the gut (Turnbaugh et al. 2009), this difference was not statistically significant. This observation could be due either to a small genetic influence relative to overall variation or to other cofounding effects. However, if we compare both MZ and DZ pairs to unrelated individuals who live in different homes at the same age, the difference becomes highly significant. Because the MZ–DZ comparison was nonsignificant, we pooled MZ and DZ twins together for all following analysis.
Figure 3.
Genetic effect on the salivary microbiome at the ages of 12, 17, and 22 yr. Averaged pairwise unweighted UniFrac distances of same aged MZ (n = 59) and DZ (n = 39) twin pairs and same aged sample population (n = 7882 pairs) at ages 12–13, 17–18, and 22–24 (±SEM). The data set was randomly subsampled 10 times at a sequencing depth of 800 sequences/sample, and each subsampling is shown as a separate bar. The statistical analysis was a Mann-Whitney U-test. The P-value outcomes are denoted as follows: (ns) nonsignificant, (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. The P-value of each permutation was recorded and the lowest significance level that has occurred in at least nine out of 10 rarefactions is presented.
Changes of the salivary microbiome over a decade
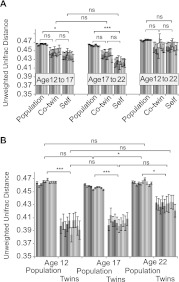
To detect patterns of dynamic, temporal changes in the microbiome during adolescence, we analyzed the salivary microbiome of 82 individuals over time (198 saliva samples). We compared each individual to itself and to its twin sibling at a later time-point. We also compared the salivary microbiome of the cohort population of the same age from age 12 to age 17, from age 17 to age 22, and from age 12 to 22, spanning a period of 5 and 10 yr. After both 5-yr spans, the oral microbiome of an individual resembles itself more closely than that of the population, based on unweighted UniFrac distances (Fig. 4A). After 10 yr (12–22), the oral microbiome has a trend toward self-similarity, but this trend is not statistically significant. Within the twin sample, we compared the oral microbiome of one individual twin at a younger age with the oral microbiome of the co-twin at an older age (e.g., twin A at age 12 to his co-twin B at age 17). We found that comparing microbiomes at age 12 to age 17 between the twins is statistically no different than comparing the microbiome in the same individual going from age 12 to age 17. The similarity across the twin pairs is reduced between the ages of 17 and 22, when at least 21 out of 25 twin pairs (≥84%) no longer cohabitate (Supplemental Text 2). Figure 4B shows the same trend, where twins sampled at ages 12 and 17 are more similar to each other. At age 22, the differences between the pairs increase. At all ages examined, co-twins are statistically more similar to each other than to the whole population. Therefore, even in the human oral microbiome, where one may anticipate frequent environmental perturbations, there is remarkable stability over long time periods during development up to 5 yr. It should be noted that changes in individuals occurring between age 17 and 22 tend to be less extreme than changes seen between age 12 and 17 (Fig. 4A). In this time interval, there are significant developmental changes that occur (e.g., puberty [Guncu et al. 2005] or behavioral changes) that could be the contributing factors.
Figure 4.
(A) Time progression of individuals, their twins, and the sample population at ages 12, 17, and 22. MZ and DZ pairs were pooled. Age 12–17: Population (n = 6165 pairs), Co-twin (n = 58 pairs), Self (n = 64 pairs); age 17 to 22: Population (n = 5060 pairs), Co-twin (n = 50 pairs), Self (n = 52 pairs); Age 12 to 22: Population (n = 3685 pairs), Co-twin (n = 28 pairs), Self (n = 34 pairs). (B) Similarities based on age of sampling (ages 12, 17, and 22 yr) of twins and the sample population. MZ and DZ pairs were pooled. Pairwise unweighted UniFrac distances of the same person, the person and his/her twin, and the sample population at different ages were calculated (±SEM). The data set was randomly subsampled 10 times at a sequencing depth of 800 sequences/sample. The statistical analysis was a Mann-Whitney U-test. The P-value outcomes are denoted as follows: (ns) nonsignificant, (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. The P-value of each permutation was recorded, and the lowest significance level that has occurred in at least nine out of 10 rarefactions is presented.
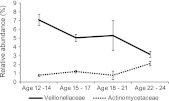
Besides exploring how whole bacterial communities change over time using the UniFrac distance metric, it is important to test for changes in taxa at different levels. To account for differences in sample numbers at each age and to aid in visualization, we grouped the individual samples into four age groups (ages 12–14, ages 15–17, ages 18–21, and ages 22–24). On a bacterial family level, we observed a negative correlation of Veillonellaceae (Phylum: Firmicutes, Class: Clostridia, Order: Clostridiales, Pearson product-moment correlation coefficient [Pearson's r] = −0.28700, Bonferroni corrected p = 3.98 × 10−4) and a positive correlation of Actinomycetaceae (Phylum: Actinobacteria, Class: Actinobacteria, Order: Actinomycetales, Pearson's r = 0.32188, Bonferroni corrected p = 3.02 × 10−5, Fig. 5) with age. There is a significant and substantial increase in the proportion of Actinomycetaceae with age, especially in young adults at age 22–24. Actinomyces species have been found preferentially in early-stage caries in children and young adults (Aas et al. 2008), and so this may reflect a general decline in dental health with age. Even though Actinomyces and Veillonella have been shown to coaggregate (Shen et al. 2005), their abundance changes do not follow the same pattern in our study. There are OTUs at a 97% identity level that are positively and negatively correlated with age in the genera Actinomyces, Veillonella, and Streptococcus (Supplemental Figs. S1–S3; Supplemental Table S5; Supplemental Text 1). All of the reported significant Pearson product-moment correlation coefficients were at a P-value of <0.05 with Bonferroni correction. They were also recovered from an ANOVA with a P-value <0.05 with Bonferroni correction.
Figure 5.
Relative abundance of bacterial families Veillonellaceae and Actinomycetaceae and their negative and positive correlation with age from adolescence to early adulthood. Veillonellaceae: P-value (Bonferroni corrected) 3.98 × 10−4; Pearson's r = −0.28700; Actinomycetaceae: P-value (Bonferroni corrected) 3.02 × 10−5, Pearson's r = 0.32188. Sample sizes: Age 12–14: n = 72; age 15–17: n = 88; age 18–21: n = 19; age 22–24: n = 55. The average relative abundance and ±SEM are plotted.
Corelation between weight, gender, and the oral microbiome
Besides the age of the subject, other metrics were examined, namely, weight and gender. The interplay between human weight (lean vs. obese) and the gut microbiome is under intense investigation (Ley et al. 2005, 2006; Turnbaugh et al. 2006, 2009; Spor et al. 2011). However, very little is known about the oral microbiome in this context. There is evidence of an increase in a particular oral bacterium (Tannerella forsythia) with BMI (Haffajee and Socransky 2009), although there has been no broad 16S rDNA sequencing approach applied to compare phylum level correlations. To test for associations between saliva microbiome and BMI, we divided subjects into underweight, normal, overweight, and obese weight classes for boys and girls based on the age-appropriate target BMI (Kuczmarski et al. 2000). In contrast to the gut microbiome (Ley et al. 2006), we found no significant correlation between any OTUs at a 97% or higher identity taxonomic level and overall weight class in human saliva.
As reported in other microbiomes (skin [Fierer et al. 2008], gut [Costello et al. 2009]), there was no significant difference between male and female individuals.
Discussion
The analysis of salivary microbial DNA from banked, frozen samples that were obtained from a relatively large population of individuals of known genetic relationships and well-characterized environmental factors has been presented. Exploiting the availability of samples from preexisting genetic studies vastly reduces costs and allowed us to select the most informative collections of individuals retrospectively. From the 264 saliva samples studied, we have been able to define a core oral microbiome on the genus level. This is the first such definition of a common set of organisms in the human mouth in an appropriately sized sample. Using a very conservative removal of sequencing “noise” with OTUpipe (Robert Edgar, http://drive5.com/otupipe/), it could be argued that this is an underestimate since we did not include rare phylotypes in this sample definition. However, rare phylotypes cannot be reliably detected in principle due to sampling considerations, so it is appropriate to use such a conservative approach.
We analyzed microbial communities of saliva derived from monozygotic and dizygotic twins. Although there was a trend toward a higher similarity among monozygotic twin pairs, this similarity was not statistically significant. This suggests that the overall genetic make-up of the host has little or no apparent role at ages 12–24 in explaining salivary phylogenetic composition measured by unweighted UniFrac distance. The environment has a higher impact on the overall composition of the oral microbiome. This finding does not rule out the possibility that individual gene variations may still have an effect on the overall composition or influence individual bacterial organisms or groups. Perhaps of greater interest, individuals and their co-twins are not significantly distinguishable during their progression from age 12 to age 17 and progression from age 17 to age 22. However, the same individuals differ significantly from unrelated individuals at ages 12–17, and even more at ages 17–22. The majority of twins changed their environment (i.e., move to a new home) between the ages of 17 and 22, which may increase differences. However, changes in individuals between ages 17 and 22 are still less than changes occurring between ages 12 and 17. The greater changes occurring in early adolescence could potentially be explained by puberty (Guncu et al. 2005), or behavioral changes that are prominent in this period.
The long-term stability of the oral microbiome over many years is remarkable. One expects changes in diet, oral hygiene, or romantic partners to occur in these years, and yet stability remains very high. These findings are similar to those for the gut, where the microbial community is in flux only in the first few months of life, mainly determined by environmental events (Koenig et al. 2011). Later on, the microbiome stabilizes and becomes less susceptible to disruptions (Spor et al. 2011). We would expect that the oral microbiome becomes increasingly stable after the teenage and preadolescent years, when hygiene and eating routines develop. Even though there is no systematic change in beta diversity, based on the unweighted UniFrac metric, during adolescence and young adulthood, we did observe systematic patterns of change in the genera Veillonella (decrease with age), Actinomyces (increase with age), and Streptococcus (increase with age).
Data collected from our sample included many personal preferences and characteristics that could influence the oral microbiome including gender, obesity, and food preferences. Interestingly, although we conclude that environmental factors provide the greatest influence on oral microbial composition, we could find no clear effects of weight class, gender, or food preferences in the analyses.
Further studies are needed to relate the variation in the oral microbiome to specific environmental factors. These may include dietary behaviors and oral hygiene, as well as more pathological factors such as smoking, alcohol, and consumption of illicit drugs such as methamphetamines that are well known to have severe negative effects on dental health (Shetty et al. 2010). Crucially, cases and controls must be age-matched for correct conclusions to be drawn, given changes in the oral microbiome over adolescence. This study thus highlights the requirement for a broad sampling of humans of different ages and lifestyles for microbiome studies, especially those targeted at understanding the effects of specific diseases.
Methods
Sample selection and DNA extraction
Genomic DNA was isolated from the saliva of participants in the Longitudinal Twin Study and Colorado Adoption Project, Institute for Behavioral Genetics, University of Colorado from 1997 to present (see also Supplemental Methods). Informed consent was obtained from all individuals, and the anonymity of all subjects is ensured by separation of all personal information from the sample and replacing it by a numerical identifier. Written informed consent was obtained and approved by the University of Colorado Human Research Committee (protocol 0399.11). Samples were collected after 2 h of abstinence from eating. Ten milliliters of Scope mouthwash was vigorously swished in the mouth for a minimum of 30 sec and released into a 50-mL Falcon tube and stored at 4°C until extraction. The total DNA of the sample was isolated using the Puregene (QIAGEN) extraction kit, dissolved in TE buffer and stored at −80°C until needed. Although no mechanical disruption method was used in the extraction of the DNA from saliva, we readily detected known “hard to lyse” species including Actinomyces and Streptococcus in comparable abundances to previously published studies that used zirconia beads to disrupt bacterial cell walls (Keijser et al. 2008). However, we do not exclude a systematic bias of our DNA extraction method compared with other studies.
16S ribosomal specific PCR and 454 sequencing
Fragments of bacterial 16S ribosomal RNA genes were amplified from DNA samples by polymerase chain reaction using primers that are specific to bacterial variable region 1 and 2 (V1–V2). The composite primers include 454 specific sequencing regions as well as a unique error-correcting 12-nt barcode for each sample as previously described (Liu et al. 2007; Fierer et al. 2008; Hamady et al. 2008; Costello et al. 2009). Bacterial specific primers were chosen to avoid amplification of the human 18S rRNA gene and because the majority of organisms in the oral cavity is assigned to the domain of life bacteria. PCR conditions and reagents were similar to those of Costello et al. (2009), specifics can be found in the Supplemental Methods section.
Sequence analysis
The data set was denoised to remove sequences with potential sequencing errors and chimeras with OTUpipe (Robert Edgar, http://drive5.com/otupipe/). The software package QIIME (versions 1.2.0 and 1.3.0 [Caporaso et al. 2010b]) was used for downstream data analysis (see also Supplemental Methods). Similar sequences were clustered into operational taxonomic units (OTUs) with a sequence identity of at least 97% using uclust. A representative sequence of each cluster was chosen and aligned with PyNAST (Caporaso et al. 2010a) to the Greengenes database (DeSantis et al. 2006). To assign taxonomy, the RDP database was used to assign the genera for Figure 2 with a confidence of 0.8. For all other taxonomic assignments, a BLAST against the Greengenes data set (version: 4feb2011 [DeSantis et al. 2006]) was used. This had the advantage of using a manually curated data set but lacks an assignment below the taxonomic family level. The alignment parameters were a minimum of 150 bp length and a minimum identity of 75%. The alignment was masked according to the QIIME tutorial instructions (Caporaso et al. 2010b). The phylogenetic tree was built with FastTree (Price et al. 2009) and subsequently used to calculate the unweighted UniFrac distances between pairs of communities (Lozupone et al. 2006). To obtain Figures 3 and 4, each sample was randomly subsampled 10 times at a sequencing depth of 800 sequences/sample to account for variations in sequencing efforts. The average UniFrac distance and the SEM of all pairwise comparisons in each group and each of the 10 rarefactions were calculated. Statistical testing was done using the Mann-Whitney U-test in R (R programming environment, http://www.R-project.org) for nonparametrical hypothesis testing for each of the rarefactions. We scored each P-value as one of the four significance categories: (ns) >0.05, (*) <0.05, (**) <0.01, (***) <0.001. We then reported the significance category in which at least nine out of 10 rarefactions fell in, which is a very conservative approach. Correlations of bacterial taxa were done on the actual ages of subjects with a Pearson correlation. All P-values are Bonferroni-corrected for multiple testing.
Data access
The sequence data from this study have been submitted to the EBI-SRA-database under study number ERP001346.
Acknowledgments
We thank the participating subjects, the Institute for Behavioral Genetics staff for recruitment, biological samples, and detailed phenotypes, and specifically Sally-Ann Rhea for her assistance with the selection of the twin samples. We also acknowledge Elizabeth Costello, Douglas Wendel, and Gail Ackermann for data analyses and submission help. Funding was provided by NIH grants HD-010333 and DA-011015.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.140608.112.
References
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE 2005. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 43: 5721–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, Leys EJ, Paster BJ 2008. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol 46: 1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asikainen S, Chen C, Alaluusua S, Slots J 1997. Can one acquire periodontal bacteria and periodontitis from a family member? J Am Dent Assoc 128: 1263–1271 [DOI] [PubMed] [Google Scholar]
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, et al. 2010. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci 107: 18933–18938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, Nelson KE, Gill SR, Fraser-Liggett CM, Relman DA 2010. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J 4: 962–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buduneli N, Baylas H, Buduneli E, Turkoglu O, Kose T, Dahlen G 2005. Periodontal infections and pre-term low birth weight: A case-control study. J Clin Periodontol 32: 174–181 [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R 2010a. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. 2010b. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, et al. 2011. Moving pictures of the human microbiome. Genome Biol 12: R50 doi: 10.1186/gb-2011-12-5-r50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE 2010. The Human Oral Microbiome Database: A web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010: baq013 doi: 10.1093/database/baq013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM 2007. The ribosomal database project (RDP-II): Introducing myRDP space and quality controlled public data. Nucleic Acids Res 35: D169–D172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras M, Costello EK, Hidalgo G, Magris M, Knight R, Dominguez-Bello MG 2010. The bacterial microbiota in the oral mucosa of rural Amerindians. Microbiology 156: 3282–3287 [DOI] [PubMed] [Google Scholar]
- Corby PM, Bretz WA, Hart TC, Schork NJ, Wessel J, Lyons-Weiler J, Paster BJ 2007. Heritability of oral microbial species in caries-active and caries-free twins. Twin Res Hum Genet 10: 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R 2009. Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336: 1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJ 2011. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics 4: 22 doi: 10.1186/1755-8794-4-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci 107: 11971–11975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardini Y, Chung P, Dumm R, Joshi N, Han YW 2010. Transmission of diverse oral bacteria to murine placenta: Evidence for the oral microbiome as a potential source of intrauterine infection. Infect Immun 78: 1789–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R 2008. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci 105: 17994–17999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guncu GN, Tozum TF, Caglayan F 2005. Effects of endogenous sex hormones on the periodontium review of literature. Aust Dent J 50: 138–145 [DOI] [PubMed] [Google Scholar]
- Haffajee AD, Socransky SS 2009. Relation of body mass index, periodontitis and Tannerella forsythia. J Clin Periodontol 36: 89–99 [DOI] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5: 235–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh-Ba G, Lang NP, Tonetti MS, Salvi GE 2007. The association of the composite IL-1 genotype with periodontitis progression and/or treatment outcomes: A systematic review. J Clin Periodontol 34: 305–317 [DOI] [PubMed] [Google Scholar]
- Keijser BJ, Zaura E, Huse SM, van der Vossen JM, Schuren FH, Montijn RC, ten Cate JM, Crielaard W 2008. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res 87: 1016–1020 [DOI] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci (Suppl 1) 108: 4578–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononen E 2000. Development of oral bacterial flora in young children. Ann Med 32: 107–112 [DOI] [PubMed] [Google Scholar]
- Kornman KS, di Giovine FS 1998. Genetic variations in cytokine expression: A risk factor for severity of adult periodontitis. Ann Periodontol 3: 327–338 [DOI] [PubMed] [Google Scholar]
- Kuczmarski RJ, Ogden CL, Grummer-Strawn LM, Flegal KM, Guo SS, Wei R, Mei Z, Curtin LR, Roche AF, Johnson CL 2000. CDC growth charts: United States. Adv Data 2000: 1–27 [PubMed] [Google Scholar]
- Lazarevic V, Whiteson K, Huse S, Hernandez D, Farinelli L, Osteras M, Schrenzel J, Francois P 2009. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Methods 79: 266–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V, Whiteson K, Hernandez D, Francois P, Schrenzel J 2010. Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics 11: 523 doi: 10.1186/1471-2164-11-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI 2005. Obesity alters gut microbial ecology. Proc Natl Acad Sci 102: 11070–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI 2006. Microbial ecology: Human gut microbes associated with obesity. Nature 444: 1022–1023 [DOI] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI 2008. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6: 776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ismail AI, Ge Y, Tellez M, Sohn W 2007. Similarity of bacterial populations in saliva from African-American mother–child dyads. J Clin Microbiol 45: 3082–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R 2007. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res 35: e120 doi: 10.1093/nar/gkm541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R 2005. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7: 371 doi: 10.1186/1471-2105-7-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas VS, Beighton D, Roberts GJ 2000. Composition of the oral streptococcal flora in healthy children. J Dent 28: 45–50 [DOI] [PubMed] [Google Scholar]
- Motisuki C, Lima LM, Spolidorio DM, Santos-Pinto L 2005. Influence of sample type and collection method on Streptococcus mutans and Lactobacillus spp. counts in the oral cavity. Arch Oral Biol 50: 341–345 [DOI] [PubMed] [Google Scholar]
- Mullany P, Warburton P, Allan E 2011. The human oral metagenome. In Metagenomics of the human body (ed. KE Nelson), Chapter 9, pp. 165–173. Springer, New York [DOI] [PubMed] [Google Scholar]
- Nasidze I, Li J, Quinque D, Tang K, Stoneking M 2009. Global diversity in the human salivary microbiome. Genome Res 19: 636–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou W, Gizani S, Haffajee AD, Quirynen M, Mamai-Homata E, Papagiannoulis L 2009. The microbiota on different oral surfaces in healthy children. Oral Microbiol Immunol 24: 183–189 [DOI] [PubMed] [Google Scholar]
- Parahitiyawa NB, Jin LJ, Leung WK, Yam WC, Samaranayake LP 2009. Microbiology of odontogenic bacteremia: Beyond endocarditis. Clin Microbiol Rev 22: 46–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen PE 2003. The World Oral Health Report 2003: Continuous improvement of oral health in the 21st century—The approach of the WHO Global Oral Health Programme. Community Dent Oral Epidemiol (Suppl 1) 31: 3–23 [DOI] [PubMed] [Google Scholar]
- Poveda-Roda R, Jimenez Y, Carbonell E, Gavalda C, Margaix-Munoz MM, Sarrion-Perez G 2008. Bacteremia originating in the oral cavity. A review. Med Oral Patol Oral Cir Bucal 13: E355–E362 [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP 2009. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarela MH, Dogan B, Alaluusua S, Asikainen S 1999. Persistence of oral colonization by the same Actinobacillus actinomycetemcomitans strain(s). J Periodontol 70: 504–509 [DOI] [PubMed] [Google Scholar]
- Shen S, Samaranayake LP, Yip HK 2005. Coaggregation profiles of the microflora from root surface caries lesions. Arch Oral Biol 50: 23–32 [DOI] [PubMed] [Google Scholar]
- Shetty V, Mooney LJ, Zigler CM, Belin TR, Murphy D, Rawson R 2010. The relationship between methamphetamine use and increased dental disease. J Am Dent Assoc 141: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr 1998. Microbial complexes in subgingival plaque. J Clin Periodontol 25: 134–144 [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc Natl Acad Sci 103: 12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A, Koren O, Ley R 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9: 279–290 [DOI] [PubMed] [Google Scholar]
- Tims S, Zoetendal EG, de Vos WM, Kleerebezem M 2011. Host genotype and the effect on microbial communities. In Metagenomics of the human body (ed. KE Nelson), Chap. 2, pp. 15–41. Springer, New York [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031 [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI 2007. The human microbiome project. Nature 449: 804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. 2009. A core gut microbiome in obese and lean twins. Nature 457: 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Winkelhoff AJ, Boutaga K 2005. Transmission of periodontal bacteria and models of infection. J Clin Periodontol (Suppl 6) 32: 16–27 [DOI] [PubMed] [Google Scholar]
- Young VB 2012. The intestinal microbiota in health and disease. Curr Opin Gastroenterol 28: 63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SE, Stallings MC, Corley RP, Krauter KS, Hewitt JK 2000. Genetic and environmental influences on behavioral disinhibition. Am J Med Genet 96: 684–695 [PubMed] [Google Scholar]
- Zaura E, Keijser BJ, Huse SM, Crielaard W 2009. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9: 259 doi: 10.1186/1471-2180-9-259 [DOI] [PMC free article] [PubMed] [Google Scholar]