Abstract
Several multiprotein DNA complexes capable of insulator activity have been identified in Drosophila melanogaster, yet only CTCF, a highly conserved zinc finger protein, and the transcription factor TFIIIC have been shown to function in mammals. CTCF is involved in diverse nuclear activities, and recent studies suggest that the proteins with which it associates and the DNA sequences that it targets may underlie these various roles. Here we show that the Drosophila homolog of CTCF (dCTCF) aligns in the genome with other Drosophila insulator proteins such as Suppressor of Hairy wing [SU(HW)] and Boundary Element Associated Factor of 32 kDa (BEAF-32) at the borders of H3K27me3 domains, which are also enriched for associated insulator proteins and additional cofactors. RNAi depletion of dCTCF and combinatorial knockdown of gene expression for other Drosophila insulator proteins leads to a reduction in H3K27me3 levels within repressed domains, suggesting that insulators are important for the maintenance of appropriate repressive chromatin structure in Polycomb (Pc) domains. These results shed new insights into the roles of insulators in chromatin domain organization and support recent models suggesting that insulators underlie interactions important for Pc-mediated repression. We reveal an important relationship between dCTCF and other Drosophila insulator proteins and speculate that vertebrate CTCF may also align with other nuclear proteins to accomplish similar functions.
Insulators were first characterized as regulatory elements that play an important role in establishing proper gene expression in eukaryotic cells. Early studies demonstrated the ability of insulators to act as barriers, preventing the spread of heterochromatin and thereby demarcating chromatin boundaries, as well as enhancer-blockers, preventing enhancers from activating nearby genes in a direction-dependent manner (Gaszner and Felsenfeld 2006; Bushey et al. 2008). Insulators have since been shown to be multiprotein–DNA complexes that can mediate inter- and intrachromosomal interactions important for facilitating proper gene expression at specific loci, and more recently, in genome-wide chromatin organization (Phillips and Corces 2009). Insulator activity in vertebrates requires the essential, highly conserved, CCCTC-binding factor CTCF. Recent genome-wide studies have effectively mapped both the mammalian CTCF binding sites and the chromatin interactions that they facilitate (Kim et al. 2007; Handoko et al. 2011). However, how CTCF mediates these interactions and the nature of the proteins required for functional insulator activity remains poorly understood.
The CTCF insulator protein contains a highly conserved central domain encoding 11 zinc fingers, and is ubiquitously expressed (Klenova et al. 1993). Interestingly, CTCF has been implicated in numerous unique nuclear functions in addition to the classical enhancer-blocking and barrier activities that define insulators. These include X-chromosome inactivation (Chao et al. 2002), nucleolar stability (Guerrero and Maggert 2011), V(D)J recombination (Guo et al. 2011), and global chromatin organization (Kim et al. 2007; Handoko et al. 2011). The combinatorial use of its 11 zinc fingers in binding to discrete DNA target sequences, as well as the diverse, context-dependent protein-interaction networks of CTCF, have been proposed to underlie these numerous roles (Filippova et al. 1996; Zlatanova and Caiafa 2009; Weth and Renkawitz 2011). Meanwhile, recent studies in both D. melanogaster and humans have demonstrated that CTCF appears to demarcate physical chromatin domains (Dixon et al. 2012; Nora et al. 2012; Sexton et al. 2012), including a subset of repressive H3K27me3 domains (Bartkuhn et al. 2009; Cuddapah et al. 2009). However, the proteins with which CTCF associates and the purpose for which CTCF demarcates chromatin boundaries requires further exploration.
In Drosophila, several insulator binding proteins have been identified and characterized, including the Drosophila homolog of CTCF (dCTCF), Boundary element associated factor of 32 kDa (BEAF-32), and Suppressor of Hairy wing [SU(HW)] (Gurudatta and Corces 2009). These DNA-binding proteins require additional proteins for functional insulator activity, including Centrosomal protein 190 (CP190) and Modifier of mdg4 [MOD(MDG4)] (Ghosh et al. 2001; Pai et al. 2004; Gerasimova et al. 2007). We have recently identified the genome-wide binding sites of dCTCF, BEAF-32, SU(HW), and CP190 with high-resolution ChIP-seq and demonstrated that recruitment of these proteins is regulated during the ecdysone response in D. melanogaster (Wood et al. 2011). However, the functional relationship between these different insulator proteins remains unknown.
Here we present a comprehensive map of direct insulator-binding sites throughout the Drosophila genome and show that as many as 40% of dCTCF sites align tightly with the Drosophila specific insulators SU(HW) and BEAF-32. dCTCF sites are enriched for three similar but distinct DNA motifs, potentially representing discrete binding modes throughout the Drosophila genome. Aligned insulators are enriched for additional cofactors and commonly occur at the borders of H3K27me3 domains, where they are essential for maintaining appropriate chromatin structure. Surprisingly, we find that disruption of insulators genome wide by knockdown of insulator components does not significantly affect the expression of genes flanking H3K27me3 domains, nor does H3K27me3 spread beyond domain borders as one might expect based on classical barrier models for insulator function at chromatin boundaries. Instead, H3K27me3 is lost within domains, suggesting that chromatin insulators serve an important role in the maintenance of silenced chromatin in Polycomb (Pc) domains. Our findings support recently proposed models, wherein chromatin insulators are involved in mediating long-range interactions important for Polycomb (Pc)-mediated repression (Pirrotta and Li 2011).
Results
dCTCF sites align with Drosophila-specific insulator proteins SU(HW) and BEAF-32
Two recent studies independently identified the genome-wide binding sites of insulator proteins in Drosophila melanogaster by combining chromatin immunoprecipitation with microarray hybridization (ChIP-chip). Whereas one study demonstrated unique genome-wide distributions and gene ontologies between dCTCF, SU(HW), and BEAF-32 (Bushey et al. 2009), the other observed an enrichment of dCTCF and BEAF-32 as colocalizing, and therefore split insulators into two main classes: dCTCF/BEAF-32/CP190 (Class I) and SU(HW) (Class II) (Negre et al. 2010). However, previous studies have shown that CP190 is an essential component of both the dCTCF and SU(HW) insulators (Pai et al. 2004; Gerasimova et al. 2007). The functional implications of insulator classes and why dCTCF might colocalize with other insulator proteins requires further exploration.
To better determine the genome-wide binding sites of Drosophila insulators with greater accuracy and resolution, we recently re-mapped dCTCF, SU(HW), BEAF-32, and CP190 sites by combining chromatin immunoprecipitation with high-throughput sequencing (ChIP-seq). Here we analyze peaks repeatedly called in three independent ChIP-seq experiments during the ecdysone response in Drosophila Kc cells, which are therefore most likely to represent real, stable insulator binding sites (Wood et al. 2011). We then determined enriched consensus sequence motifs by MEME-ChIP (Machanick and Bailey 2011). Results confirm previously identified position weight matrices for each respective insulator protein (Ramos et al. 2006; Holohan et al. 2007; Negre et al. 2010; Supplemental Fig. S1). Given the ability of distant insulator proteins to interact with each other, it is possible that different insulator proteins bound to these sites may coprecipitate during the ChIP procedure, thus appearing to colocalize, when in fact they are located in different genomic locations. As a consequence, we speculate that genome-wide binding profiles for each insulator protein likely contain many indirect binding sites. Therefore, we excluded sites that do not contain appropriate target sequences for each respective insulator protein (see Methods), thereby providing a stringent list of insulator sites that are highly likely to represent real, direct binding sites for each protein.
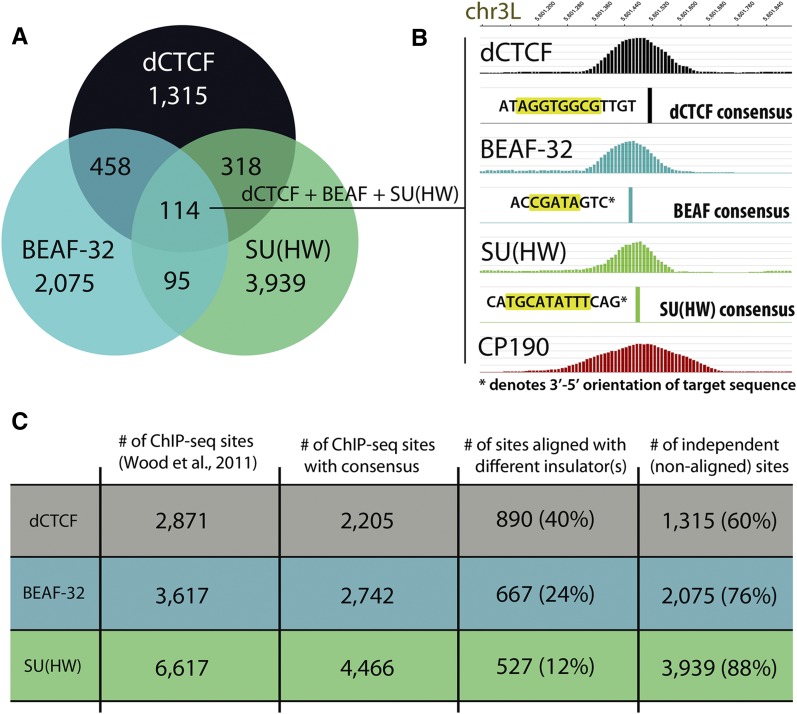
Results from this analysis suggest that insulator proteins indeed cluster together in the genome often as previously reported (Negre et al. 2010) and do so while binding their own discrete target sequence. As many as 40% of all dCTCF sites align with SU(HW) or BEAF-32, and as many as 5% of all dCTCF sites tightly align with both SU(HW) and BEAF-32 (Fig. 1A,B). Though previous studies broke insulators into two or three classes, we find that dCTCF aligns with SU(HW) (432; 49% of aligned sites) and/or BEAF-32 (572; 64% of aligned sites) at many sites. Given the number of SU(HW) binding sites throughout the genome [4466 sites with SU(HW) consensus, Fig. 1C], earlier correlation analyses of colocalization were likely biased by thousands of independent SU(HW) sites. The high resolution obtained by ChIP-seq demonstrates that these insulators align tightly, within only 200–300 bp (Fig. 1B), and sequential ChIP for insulator proteins dCTCF, BEAF-32, and SU(HW) suggests that these proteins colocalize at these sites in individual cells (Supplemental Fig. S2). In addition, by removing insulator sites lacking known target sequences, we demonstrate that each insulator protein binds to its own target sequence (notice DNA sequence) (Fig. 1B), and overlap is not a consequence of indirect binding. The alignment of dCTCF with both SU(HW) and BEAF-32 suggests the possibility of synergistic cooperation in insulator function.
Figure 1.
dCTCF aligns with Drosophila insulator-binding proteins SU(HW) and BEAF-32. (A) Venn diagram depicting overlap of dCTCF binding sites with those of BEAF-32 and SU(HW). Overlap represented as number of sites (summits ±150 bp) in which dCTCF intersects BEAF-32 and/or SU(HW) and target sequences are present for each insulator protein, suggesting close alignment (within 150 bp). (B) Example of ChIP-seq profile for dCTCF, BEAF-32, SU(HW), and CP190 on chromosome 3L; in which case dCTCF aligns with both BEAF-32 and SU(HW), where each cognate target is present. (C) Number of sites in which dCTCF, BEAF-32, and SU(HW) contain appropriate target sequences. Percentages of sites in which dCTCF, BEAF-32, and SU(HW) align closely with other DNA-binding insulator proteins. Most (90%) alignments include dCTCF, and as many as 40% of dCTCF sites align with either BEAF-32 and/or SU(HW).
dCTCF sites are enriched for multiple DNA motifs
In addition to its ability to interact with numerous nuclear proteins, the versatility of CTCF in genome biology may also be attributable to its wide range of potential target sequences. However, genome-wide analyses of CTCF binding sites have revealed a primarily enriched core target sequence that is strikingly similar between invertebrates and vertebrates despite millions of years of evolution (Holohan et al. 2007; Supplemental Fig. S1). This is not entirely surprising given that CTCF encodes 11 highly conserved zinc fingers that confer target specificity. However, early characterization of CTCF identified its ability to bind to a wide range of sequences dependent on its combinatorial use of zinc fingers (Filippova et al. 1996; Ohlsson et al. 2001), and recent work has identified similar regulatory elements bound by CTCF in the human genome (Xie et al. 2007). These data suggest that CTCF may bind to unique DNA target sequences not represented in the conserved target sequence.
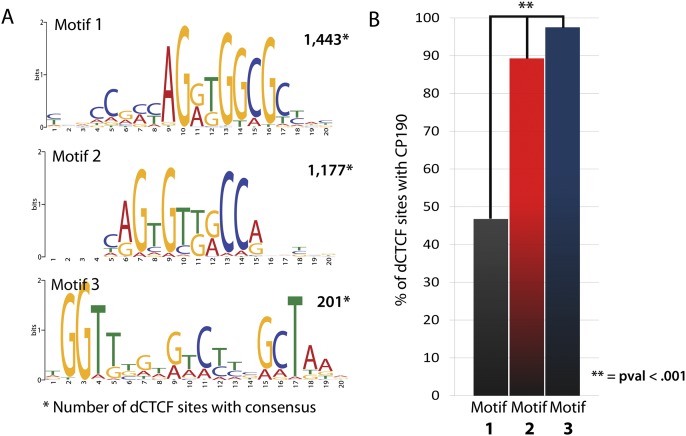
Motif analysis of dCTCF ChIP-seq data by MEME-ChIP (Machanick and Bailey 2011) indeed identifies the primary consensus sequence of dCTCF as previously reported (Fig. 2; Supplemental Fig. S1). However, the results also indicate strong enrichments for a strikingly similar but novel secondary consensus sequence (Fig. 2), also independently obtained using Weeder 1.3 (Pavesi et al. 2004), suggesting that the variability in target sequence specificity holds true for Drosophila. There is also enrichment for an additional motif accounting for <10% of dCTCF sites (Fig. 2). Comparison of dCTCF read intensities at these three motifs suggests that the highly conserved core consensus (motif 1) recruits higher occupancy levels of dCTCF, whereas motifs 2 and 3 recruit lower but similar occupancy levels (Supplemental Fig. S3). This finding is similar to previous reports suggesting that CTCF targets different occupancy-based motif classes in vertebrates (Essien et al. 2009), and suggests that these unique target sites may underlie distinct roles.
Figure 2.
dCTCF sites are enriched for three distinct DNA motifs, including a similar but novel secondary motif enriched for insulator protein CP190. (A) Position weight matrices for primary target sequence and secondary target sequences obtained by MEME-ChIP, and confirmed with Weeder 1.3. Number of sites provided represents sites in which dCTCF summits are ±150 bp from the DNA motif. (B) Percentage of dCTCF sites in which CP190 is present when containing each DNA motif.
Studies in CP190 mutants demonstrated a dependence of dCTCF on CP190 for binding to a subset of its DNA-binding sites on polytene chromosomes (Gerasimova et al. 2007; Mohan et al. 2007). Earlier studies have shown that although CP190 physically associates with insulator proteins SU(HW) and MOD(MDG4)2.2 [also referred to as MOD(MDG4)67.2] and is essential for functional insulator activity, it does not directly bind to insulator sequences present on the gypsy retrotransposon (Pai et al. 2004), and therefore likely relies on dCTCF and SU(HW) to associate with insulator sites. In support of this notion, recent biochemical studies demonstrated that CP190 function at dCTCF, SU(HW), and BEAF-32 sites requires its BTB/POZ (protein interaction) domains, whereas its zinc fingers were dispensable (Oliver et al. 2010). Interestingly, we find significant enrichments for CP190 at dCTCF sites containing motifs 2 or 3 when compared with the primary conserved consensus (Fig. 2B). Given the dependence of dCTCF on CP190 for binding to a subset of its sites, we speculate that interactions between dCTCF and CP190 facilitate its interaction with these low-occupancy and presumably lower-affinity target sequences. Furthermore, we find enrichments for the novel secondary sequence at sites where dCTCF aligns with BEAF-32 and where dCTCF aligns with both BEAF-32 and SU(HW) (Supplemental Fig. S4), suggesting that these DNA target sequences exhibit distinct features with respect to insulator recruitment and alignment.
dCTCF recruits unique MOD(MDG4) isoform(s)
The recruitment of CP190 is of particular interest given its ability to form stable homodimers and homotetramers in vitro, supporting the notion that active insulators function through loop formation via interactions with other insulators. CP190 contains a unique BTB domain that excludes it from the ttk (tramtrack) group of BTB/POZ proteins and inhibits it from interacting with ttk members (Bonchuk et al. 2011). However, SU(HW) was first characterized as recruiting an additional BTB/POZ protein, MOD(MDG4), also essential for insulator activity (Gerasimova et al. 1995; Ghosh et al. 2001). MOD(MDG4), in fact, belongs to the ttk group of BTB/POZ containing proteins, and has been shown to form higher-order homo-oligomers (Bonchuk et al. 2011). Meanwhile, the observation that in diploid cell nuclei insulators form large structures called insulator bodies suggests that many insulators associate together within the nucleus, and the presence of CP190 and MOD(MDG4) supports this possibility (Ghosh et al. 2001; Gerasimova et al. 2007).
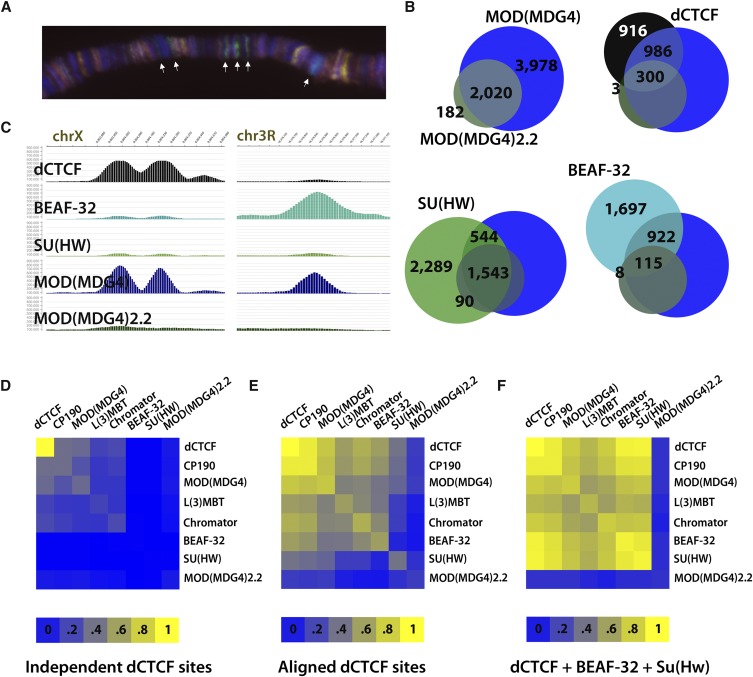
Whereas CP190 has been shown to associate with dCTCF, SU(HW), and BEAF-32 (Bushey et al. 2009), currently only the SU(HW) insulator has been shown to recruit MOD(MDG4). Although the mod(mdg4) gene encodes for at least 26 alternatively spliced variants, each containing a common N-terminal region encoding the ttk-family BTB/POZ domain (Dorn and Krauss 2003; Labrador and Corces 2003), SU(HW) insulator activity requires one specific isoform, MOD(MDG4)2.2 (Gerasimova et al. 1995). Staining of Drosophila polytene chromosomes reveals MOD(MDG4)-specific bands, unaccounted for with MOD(MDG4)2.2 staining alone (Fig. 3A), suggesting that additional isoforms are recruited to DNA. Whether dCTCF and BEAF-32 recruit unique MOD(MDG4) isoforms is currently unknown. We therefore carried out ChIP-seq analyses in Drosophila Kc cells using two different antibodies that recognize either all MOD(MDG4) isoforms or MOD(MDG4)2.2.
Figure 3.
dCTCF and BEAF-32 recruit isoform(s) of MOD(MDG4) different from MOD(MDG4)2.2. Aligned dCTCF sites are enriched for MOD(MDG4) and additional cofactors. (A) Immunofluorescence microscopy of MOD(MDG4) (green) and MOD(MDG4)2.2 (red) on Drosophila polytene chromosomes. MOD(MDG4) staining includes many discrete bands not accounted for by MOD(MDG4)2.2 specific antibodies, depicted with white arrows. (B) Genome-wide overlap of dCTCF, BEAF-32, and SU(HW) with MOD(MDG4) and MOD(MDG4)2.2. Many dCTCF (45%) and BEAF-32 (34%) sites overlap MOD(MDG4) isoform(s) not represented by MOD(MDG4)2.2. Meanwhile, many SU(HW) (37%) sites overlap MOD(MDG4)2.2 sites, as expected. (C) ChIP-seq profile for MOD(MDG4) and MOD(MDG4)2.2 reveals many unique peaks specifically in the MOD(MDG4) profile, accounted for at BEAF-32 and dCTCF sites. (D–F) Heatmap representation of percentages of dCTCF sites in which CP190, MOD(MDG4), MOD(MDG4)2.2, BEAF-32, SU(HW), L(3)MBT, and/or Chromator co-occur at independent dCTCF sites, aligned dCTCF sites, and sites where dCTCF aligns with both BEAF-32 and SU(HW).
The binding profile for MOD(MDG4)2.2 is significantly accounted for at SU(HW) sites (Fig. 3B). Given that MOD(MDG4)2.2 is required for SU(HW) insulator activity, the MOD(MDG4)2.2 map may reveal a subset of active SU(HW) sites throughout the Drosophila genome. Here, we find that the ChIP-seq profile of MOD(MDG4), which includes significantly more binding sites than MOD(MDG4)2.2 alone (Fig. 3B), reveals unique peaks at discrete dCTCF and BEAF-32 sites, suggesting that additional isoforms of MOD(MDG4) recruited by dCTCF and BEAF-32 must exist (Fig. 3C). Whereas average read intensities for MOD(MDG4)2.2 are strongest at SU(HW) sites, dCTCF and BEAF-32 sites show an opposite trend, with stronger read intensities for MOD(MDG4) (Supplemental Fig. S5). Finally, as many as 64% of dCTCF sites and 38% of BEAF-32 sites colocalize with an isoform of MOD(MDG4). These data suggest that dCTCF and BEAF-32 indeed recruit unique isoforms of MOD(MDG4), and that all three Drosophila insulators function similarly through the recruitment of BTB domain-containing proteins CP190 and MOD(MDG4).
Aligned dCTCF sites are enriched for CP190, MOD(MDG4) isoforms, and additional cofactors
The tight alignment of dCTCF with BEAF-32 and SU(HW), combined with their common insulator function, suggests that insulators synergize, perhaps to create a more active insulator complex. Given that CP190 and MOD(MDG4) form homo-oligomers, it is also intuitive to imagine closely aligned insulators cooperatively recruiting CP190 and MOD(MDG4), increasing the likelihood that each member of the insulator cluster is functionally active. In support of this hypothesis, we find enrichment for aligned dCTCF sites containing CP190 and MOD(MDG4) when compared with independent dCTCF insulators (Fig. 3D–F). This suggests that by associating with BEAF-32 and/or SU(HW), dCTCF might ensure that it will become a functionally active insulator complex by recruiting essential cofactors.
Many additional proteins have been functionally associated with insulators in D. melanogaster, suggesting that these insulator clusters may represent hubs for recruiting other cofactors. For example, Lethal (3) malignant brain tumor [L(3)MBT] has been recently shown to colocalize with the Drosophila chromatin insulators dCTCF, BEAF-32, SU(HW), and CP190 (Richter et al. 2011), and other studies found direct interactions between L(3)MBT and the SU(HW) insulator protein (Guruharsha et al. 2011). L(3)MBT imparts transcriptional regulation of the Salvador-Warts-Hippo (SWH) pathway, likely repressing SWH target genes important for cell proliferation and organ size control. Whereas recently published L(3)MBT sites (Richter et al. 2011) are enriched primarily at independent dCTCF sites over the BEAF-32 and SU(HW) insulators, we find enrichment of L(3)MBT sites, identified independently by ChIP-seq in Drosophila Kc cells, at aligned dCTCF sites when compared with independent dCTCF sites (Fig. 3D–F). In addition to SU(HW), L(3)MBT interacts with a chromodomain protein, Chromator, that has also recently been shown to colocalize and cooperate with the BEAF-32 insulator (Giot et al. 2003; Gan et al. 2011). Indeed, using publicly available ChIP-chip data for Chromator in Drosophila Kc cells (Celniker et al. 2009), aligned dCTCF sites also show an apparent enrichment for Chromator (Fig. 3D–F). Interestingly, Chromator and zinc finger protein Z4 are important for maintaining polytene chromosome structure (Eggert et al. 2004), suggesting a functional relationship between insulators and Chromator in chromatin domain organization.
Together, these data suggest that dCTCF may team up with Drosophila-specific insulator proteins in order to more efficiently recruit cofactors essential for insulator activity. These insulator sites are enriched for additional insulator-related proteins L(3)MBT and Chromator, suggesting that these sites are different from independent insulator sites and appear to represent large complexes of proteins associated with insulator activity.
Aligned dCTCF sites commonly flank the borders of H3K27me3 domains
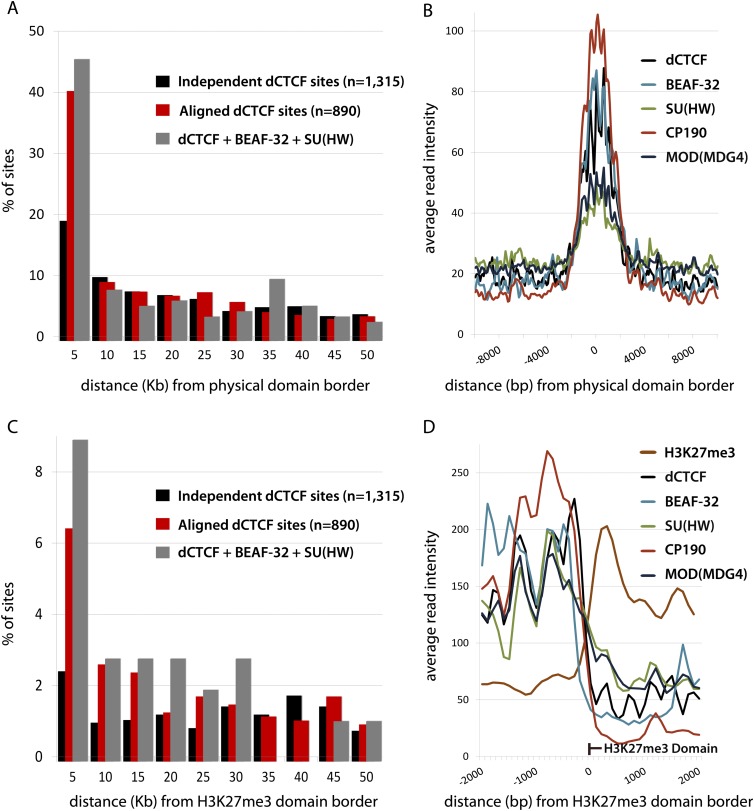
The correlation of dCTCF with SU(HW) and BEAF-32 is striking, but why dCTCF clusters with other insulator proteins requires further exploration. Recent interrogation of chromosome architecture in D. melanogaster revealed recurrent combinations of insulators and active histone marks at the borders of physical domains, including enrichment for Chromator (Sexton et al. 2012). Comparison with physical domains analyzed by Sexton et al. (2012) reveals enrichment for aligned dCTCF sites within 5 kb of domain borders, suggesting that these tandemly aligned insulators are involved in demarcating chromatin domains (Fig. 4A,B). Recent studies in both Drosophila and humans have also demonstrated an enrichment of CTCF and other insulators at the borders of H3K27me3 domains (Bartkuhn et al. 2009; Cuddapah et al. 2009; Negre et al. 2010).
Figure 4.
Aligned dCTCF sites are enriched at the borders of H3K27me3 and physical domains. (A) Percentage of independent and aligned dCTCF sites within 5 kb from recently mapped physical domain boundaries (Sexton et al. 2012). (B) Average read intensity for insulator proteins at physical domain boundaries, ±10 kb. Comparison of insulator profiles normalized by total read numbers. (C) Percentage of independent and aligned dCTCF sites within 5 kb from H3K27me3 domain borders. (D) Average read intensity for H3K27me3 and insulator proteins at H3K27me3 domain borders, ±2 kb. Comparison of insulator profiles normalized by total read numbers.
In order to determine whether aligned dCTCF insulator sites occur at H3K27me3 domain borders in Drosophila Kc cells, we independently mapped repressive chromatin domains by ChIP-seq against H3K27me3. We find an enrichment of insulator proteins immediately outside of H3K27me3 domain borders, and significant enrichment of aligned dCTCF sites within 5 kb of H3K27me3 domains (Fig. 4C,D). Interestingly, read intensities for each insulator protein flanking domain borders (Fig. 4D) suggest a periodicity of insulator presence beginning with dCTCF, consistent with the observation that insulators tandemly align. There is no significant enrichment for dCTCF aligned with BEAF-32 vs. dCTCF aligned with SU(HW) at domain borders (Supplemental Fig. S6), suggesting that dCTCF aligns with either BEAF-32 and/or SU(HW) at the borders of repressed chromatin domains. However, the functional significance of insulators at chromatin domain borders and dCTCF alignment remains poorly characterized
RNAi depletion of insulator proteins results in H3K27me3 loss within repressed domains
Previous analyses of H3K27me3 levels immediately flanking domain borders in dCTCF and CP190 mutants suggest that these sites functionally maintain chromatin architecture at these domains by preventing the spread of heterochromatin (Bartkuhn et al. 2009). Given recent data that CTCF associates with various nuclear proteins in a context-dependent fashion, it is conceivable that dCTCF tightly aligns with other insulators to establish a robust barrier insulator at the borders of repressive domains to effectively prevent the spread of heterochromatin. Therefore, we next sought to determine whether insulators are important for maintaining appropriate chromatin architecture and gene expression at these H3K27me3 domains by combinatorial knockdown of insulator proteins in Drosophila Kc cells (Supplemental Fig. S7).
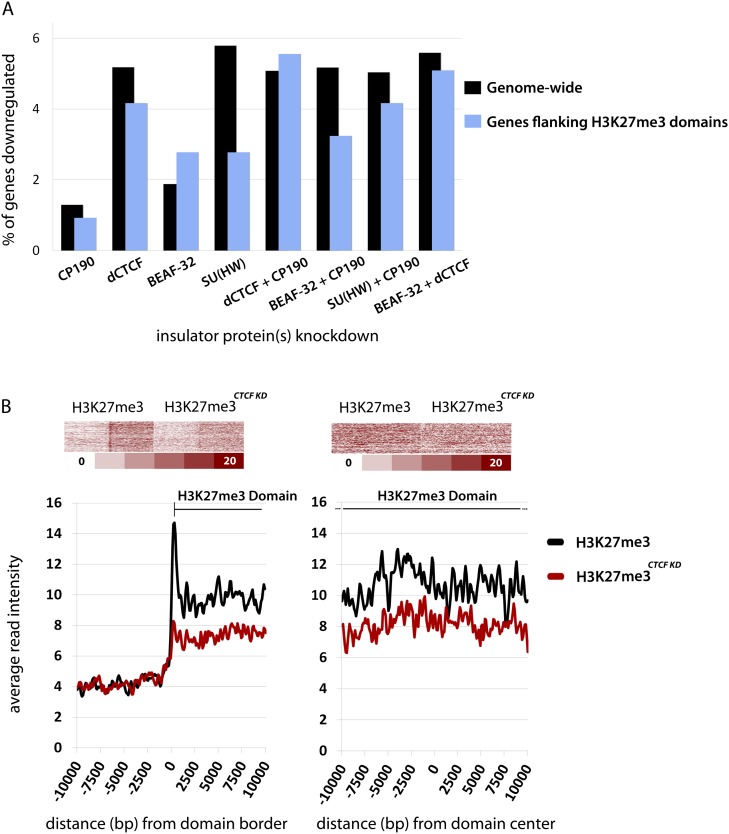
Surprisingly, we find no evidence for down-regulation of domain-flanking genes compared with genome-wide averages when insulators are disrupted genome wide (Fig. 5A), as one might expect if heterochromatin spreads beyond domain boundaries. We therefore carried out ChIP-seq for H3K27me3 in Drosophila Kc cells after dCTCF knockdown. Results revealed decreased levels of H3K27me3 immediately within domain borders as well as throughout H3K27me3 domains, but not an increase outside of domain boundaries (Fig. 5B). H3K27me3 levels were specifically reduced in Polycomb (Pc) domains containing dCTCF, indicating that loss of H3K27me3 is a direct effect of dCTCF knockdown rather than a general consequence of disrupted chromatin architecture (Supplemental Fig. S8). ChIP–PCR against H3K27me3 levels at several loci confirms significant loss of H3K27me3 in response to RNAi depletion of dCTCF, as well as under various conditions of gene expression knockdown for insulator proteins, including MOD(MDG4), suggesting the recruitment of MOD(MDG4) to dCTCF sites and enrichment at aligned insulators is functionally significant (Fig. 6). Importantly, the expression of the Enhancer of zeste [E(z)] gene, which encodes the methyltransferase responsible for H3K27me3, is unaffected by any combination of insulator knockdown, and nuclear levels of H3K27me3 remain unchanged (Supplemental Fig. S7), indicating that this is not an indirect consequence of disruption in methyltransferase activity. The reduction in H3K27me3 levels suggests insulators actively play a role important for the maintenance of H3K27me3 levels within Pc domains. Despite reduced H3K27me3 levels within repressive domains, gene expression is relatively unaffected for genes within these domains after knockdown of insulator proteins (Supplemental Fig. S8), meaning that the mechanisms underlying gene repression in Pc domains have not been entirely compromised, or that additional steps are necessary for the activation of the Pc domain containing genes.
Figure 5.
RNAi depletion of insulator proteins causes H3K27me3 depletion within domains but has no effect on H3K27me3 domain flanking genes. (A) Percentage of genes down-regulated (more than twofold) in Drosophila Kc cells after knockdown of insulator protein expression. Comparison of genes flanking H3K27me3 domains with genome-wide averages. (B) Average read intensity for H3K27me3 in Drosophila Kc cells at domain borders and surrounding domain centers. H3K27me3 levels were determined by ChIP-seq before and after dCTCF knockdown. For appropriate comparison, ChIP-seq data for H3K27me3 was rank normalized as previously described (Whyte et al. 2012) and represented as average read intensity.
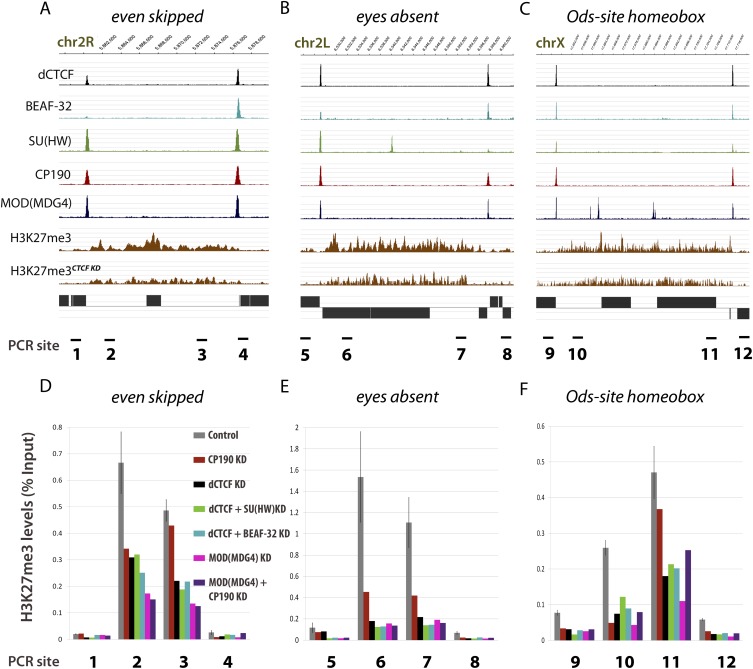
Figure 6.
Insulator proteins, including MOD(MDG4), are necessary for the maintenance of H3K27me3 levels at several loci. (A–C) ChIP-seq profiles for insulator proteins and H3K27me3 levels before and after dCTCF knockdown at the even skipped, eyes absent, and Ods-site homeobox gene loci, respectively. (D–F) ChIP–PCR levels determined by qRT–PCR against H3K27me3 ChIP before and after knockdown of several insulator proteins. Control sample is represented as SEM from three independent biological replicates.
The even-skipped gene provides a model for dCTCF alignment at H3K27me3 domain borders
In mammals, broad domains of repressive H3K27me3 characterized by Polycomb have been shown to silence clusters of developmentally important genes (Bracken et al. 2006; Pauler et al. 2009). Recent findings have demonstrated similar repression of developmental genes in stable, cell-stage independent H3K27me3 domains in D. melanogaster (Negre et al. 2011). Genes within H3K27me3 domains are highly enriched for developmental genes in Kc cells, including the even-skipped (eve) gene (Supplemental Table S1), consistent with previous results. The eve gene thus provides an excellent model to analyze the role of tandemly aligned dCTCF sites and chromatin organization.
eve is an early pair-rule gene encoding a homeodomain-containing transcription factor involved in segmentation (Macdonald et al. 1986). Expression of eve peaks within the first 6 h of embryogenesis and is essentially nonexpressed in late embryonic/adult Drosophila tissues (Gelbart and Emmert 2010), including late-embryonic Drosophila Kc cells (Celniker et al. 2009). eve is one of several hundred genes targeted by Polycomb (Pc), and recent data suggest that Pleiohomeotic, a Pc DNA-binding protein, negatively regulates eve during embryogenesis (Kwong et al. 2008; Kim et al. 2011). Analysis of the eve locus in Drosophila Kc cells reveals H3K27me3 mediated repression in the form of a 15-kb H3K27me3 domain (Supplemental Fig. S9). The domain is flanked immediately downstream by dCTCF aligned with both BEAF-32 and SU(HW), and immediately upstream by a dCTCF site aligned with two SU(HW) elements. In both cases, dCTCF binding sites are marked by the secondary target sequence identified by MEME-ChIP and weeder1.3 (Pavesi et al. 2004; Machanick and Bailey 2011). These aligned dCTCF sites overlap with ChIP-seq profiles for CP190, MOD(MDG4), L(3)MBT, and Chromator, consistent with genome-wide enrichments for insulator-associated proteins. Knockdown of insulator proteins has no effect on the expression of domain-flanking genes CG12134 and TER94 (Supplemental Table S2), nor does it significantly affect the expression of eve. However, knockdown of dCTCF results in H3K27me3 depletion within the repressed eve domain (Supplemental Fig. S9), and knockdown of additional insulator proteins, including MOD(MDG4), produces similar results (Fig. 6A,D). Despite loss of insulators and H3K27me3 depletion, eve appears to remain repressed, suggesting that insulator proteins contribute to appropriate chromatin domain structure but are not essential for maintenance of gene silencing in these domains. Importantly, this model for insulator alignment at H3K27me3 domain borders is consistent throughout the genome, including early-stage developmental gene eyes absent (eya) and hybrid sterility gene Odysseus-site homeobox (OdsH) (Fig. 6B,C,E,F).
Discussion
Improvements in genomic strategies for mapping genome-wide interactions have allowed recent studies to probe basic genome folding principles as well as insulator-mediated chromatin interactions (Lieberman-Aiden et al. 2009; Handoko et al. 2011; Yaffe and Tanay 2011; Dixon et al. 2012; Nora et al. 2012; Sexton et al. 2012). Results consistently support current models proposing roles for insulator proteins in chromosome organization (Phillips and Corces 2009) and challenge the basic barrier and enhancer-blocking activities that classically defined these proteins. Instead, the ability of insulators to block the spread of heterochromatin and impede enhancer-promoter interactions may simply be consequences of a more paramount role in chromosome organization. New findings in Drosophila also suggest that insulators are required to mediate long-range interactions important for Polycomb (Pc) repression (Comet et al. 2011; Li et al. 2011), and the recent identification of CTCF in transcription factories (Melnik et al. 2011) suggests that insulators may direct the localization of specific genomic loci to discrete nuclear subcompartments for gene regulation (Pirrotta and Li 2011).
Nevertheless, our finding that heterochromatin does not spread into flanking chromatin domains in response to disruption of insulator proteins is surprising based on numerous examples of insulator-mediated barrier function. Though individual insulator elements may indeed serve to prevent the spread of silencing chromatin, our disruption of total insulator protein levels instead significantly affected the levels of H3K27me3 within rather than outside of repressive chromatin domains. Knockdown of insulator proteins had no effect on the expression of E(z) or total H3K27me3 levels (Supplemental Fig. S7). Therefore, the loss of H3K27me3 within Pc domains genome wide suggests that insulators play a critical role necessary for the maintenance of appropriate chromatin architecture at these specific loci. Given the requirement for insulators in long-range Pc interactions (Comet et al. 2011; Li et al. 2011), we speculate that long-range interactions mediated by dCTCF and other Drosophila insulator proteins are ultimately disrupted by RNAi depletion of insulator proteins, and that H3K27me3 depletion likely reflects a defect in Pc-mediated compaction and maintenance of H3K27me3 at developmental loci. Interestingly, however, expression of genes within repressive H3K27me3 domains was not significantly affected (Supplemental Fig. S8), suggesting that Pc-mediated gene silencing was not abrogated or that additional steps are required to activate these developmental genes. Future studies investigating the role of insulators in Pc-mediated repression, and the effects of insulator disruption in nuclear organization, will provide valuable insight into the relationship between insulator proteins and chromatin architecture.
The diverse activities of CTCF in gene expression and chromatin organization require exploration of the proteins with which it functions and the target sequences associated with specific functions. By combining the resolution conferred by high-throughput sequencing (ChIP-seq), with mapping of core target sequences, we provide a stringent but exhaustive map of direct binding sites for Drosophila insulators and extend our previous analyses of dCTCF, SU(HW), BEAF-32, and CP190 to include the insulator protein MOD(MDG4). We show that dCTCF aligns with both the SU(HW) and BEAF-32 insulators, where dCTCF becomes enriched for additional insulator and insulator-associated proteins. The presence of aligned dCTCF sites at the borders of H3K27me3 domains provides an excellent system to query the importance of insulator proteins at the boundaries of discrete chromatin domains. Recently identified correlations for insulator proteins at the boundaries of physical domains mapped in Drosophila melanogaster (Sexton et al. 2012) provide evidence for why only a subset of aligned dCTCF localize to H3K27me3 domain borders (Fig. 4), and clearly demonstrate that insulators are also involved in the organization of other, distinct chromatin domains. Whereas Pc-repressed domains are relatively easily identifiable in the form of H3K27me3 signatures, future characterization of discrete physical domains and domain boundaries will require genome-wide interrogation of chromosome interactions in individual cell types of interest. Nearly 40% of aligned dCTCF sites (∼355) localize to physical domain boundaries mapped in late embryos by Sexton et al. (2012), suggesting that physical domains and insulator localization may be conserved at many loci across cell types.
Interestingly, dCTCF appears to target three different sequences in D. melanogaster, including the highly conserved core motif for which dCTCF has been described as binding in both Drosophila and mammals (Holohan et al. 2007). The secondary motif appears highly similar to the conserved core consensus (AGGNGGC) with an insertion between the first pair of guanines (AGTGTGGC), and average dCTCF levels suggest that this represents a low occupancy and potentially lower-affinity binding site. These novel dCTCF sites are highly enriched for insulator protein CP190 when compared with its primary target sequence. This finding, combined with previous data indicating that CP190 is essential for dCTCF binding to a subset of its target sites, suggests that CP190 might facilitate dCTCF binding to these secondary sites. The absence of CP190 in vertebrates may explain why these sequences have not been identified as mammalian target sequences, raising the possibility that these binding sites are a Drosophila-specific phenomenon.
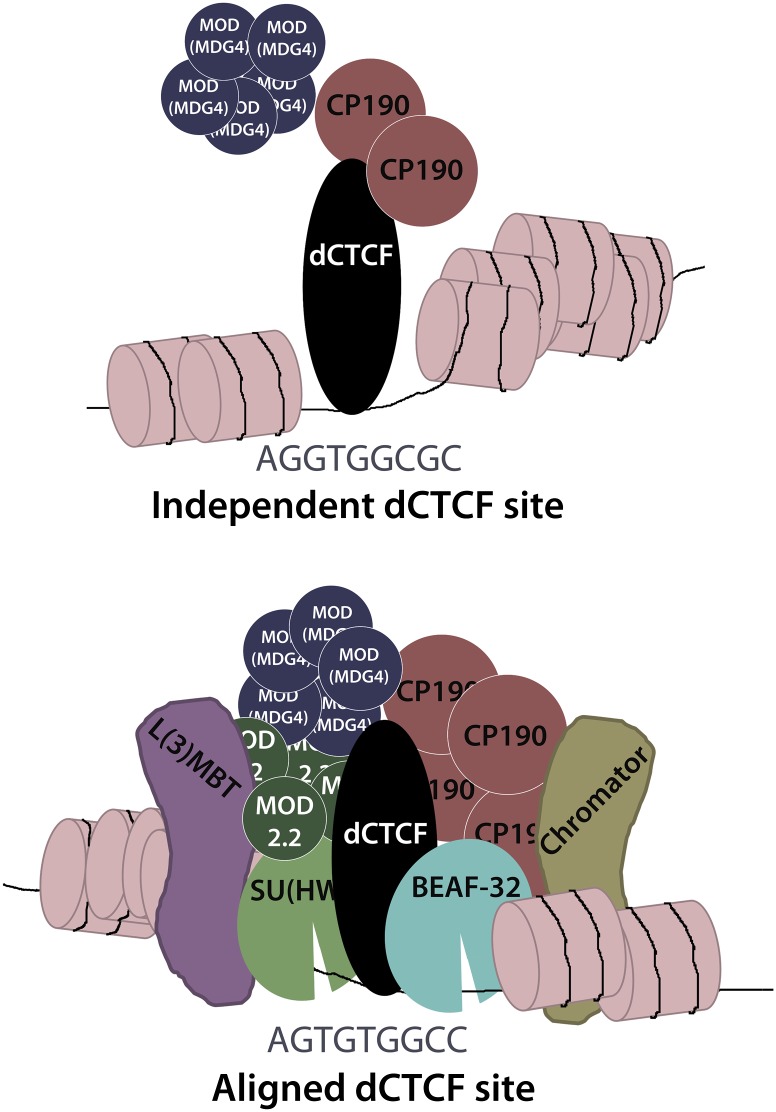
Analysis of dCTCF insulator alignment at the eve locus and genome wide uncovers a tight association with BEAF-32 and SU(HW), which may provide dCTCF with numerous advantages for effectively establishing a functional insulator. First, alignment of multiple insulator DNA elements may increase the likelihood of sequence accessibility at important loci, as insulator-binding sites have been characterized by reduced nucleosome density (Negre et al. 2010). For example, an insulator-binding protein may access its cognate sequence, thereby creating an accessible landscape for other, potentially different insulator proteins to bind their respective targets. Second, by aligning in close proximity, recruitment of essential insulator proteins [i.e., CP190 and MOD(MDG4)] by one insulator-binding protein may facilitate recruitment by others, given that CP190 and MOD(MDG4) may be recruited as multimers. Third, given that dCTCF binds secondary sites that potentially require CP190, recruitment of CP190 by a neighboring insulator [i.e., SU(HW) or BEAF-32] may preclude dCTCF binding, thereby providing a regulatory step in dCTCF recruitment to DNA. Finally, by aligning with SU(HW) and BEAF-32, dCTCF establishes a unique identity compared with independent dCTCF sites, where it becomes enriched for additional cofactors, including L(3)MBT and Chromator (Fig. 7).
Figure 7.
Diagram comparing independent dCTCF sites and dCTCF sites aligned with BEAF-32 and SU(HW). dCTCF, BEAF-32, and SU(HW) function similarly through the recruitment of CP190 and MOD(MDG4). Aligned dCTCF sites are enriched for the secondary DNA sequence and CP190. Ultimately, alignment may allow for cooperative recruitment of CP190 and MOD(MDG4), ensuring that dCTCF establishes a functional insulator complex at domain borders. Recruitment of additional proteins, such as L(3)MBT and Chromator, may also contribute to insulator activities at these loci.
Though our data shed new and valuable insight into what appears to be cooperative insulator function in Drosophila melanogaster, many questions remain. Given current models that insulators function via intra- and interchromosomal interactions, it is plausible that aligned dCTCF sites and their enrichment for CP190 and MOD(MDG4) allow for stable chromosomal interactions. Current locus- and genome-wide interaction assays may effectively answer this question in the near future. While BEAF-32 has been defined as lineage specific (Schoborg and Labrador 2010), and SU(HW) appears to lack a counterpart in mammals, our results suggest that mammalian CTCF may align with other, unique DNA-binding proteins important for appropriate insulator function at the boundaries of Pc domains.
Methods
ChIP-seq
Chromatin immunoprecipitation was performed as previously described (Bushey et al. 2009). For Re-ChIP assays, chromatin was eluted in 1% SDS, 0.1 M NaHCO3, diluted 10-fold in IP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, 167 mM NaCl), and ChIP repeated using antibodies against BEAF-32 or SU(HW). ChIP of MOD(MDG4) was carried out with antibodies against the mod2.2 isoform (α-Rabbit; gift from Elissa Lei [NIDDK, NIH]) and against the region shared by all isoforms as previously described (Bushey et al. 2009). ChIP for L(3)MBT in Drosophila Kc cells was carried out using a previously described antibody (α-Guinea-pig; gift from Jurgen Knoblich) (Richter et al. 2011), and ChIP against H3K27me3 was performed using a commercially available polyclonal antibody (Millipore Cat# 07-449). To generate sequencing libraries, ChIP DNA was prepared for adaptor ligation by end repair (End-It DNA End Repair Kit, Epicenter Cat# ER0720) and addition of “A” base to 3′ ends (Klenow 3′–5′ exo–, NEB Cat# M0212S). Illumina adaptors (Illumina Cat# PE-102-1001) were titrated according to prepared DNA ChIP sample concentration and ligated with T4 ligase (NEB Cat# M0202S). Ligated ChIP samples were PCR-amplified using Illumina primers and Phusion DNA polymerase (NEB Cat# F-530L) and size selected for 200–300 bp by gel extraction. ChIP libraries were sequenced at the HudsonAlpha Institute for Biotechnology, using an Illumina HiSeq 2000. Sequences were mapped to the dm3 genome with Bowtie 0.12.3 (Langmead 2010) using default settings. Peaks were then called with MACS 1.4.0alpha2 (Zhang et al. 2008) using equal numbers of unique reads for input and ChIP samples and a P-value cutoff of 1 × 10−10.
ChIP-seq and bioinformatics analyses
Previously published ChIP-seq data are available from GEO accession GSE30740 (Wood et al. 2011). DNA sequence motifs present in binding sites for dCTCF, BEAF-32, and SU(HW) were identified using commonly called peaks from three independent biological samples (and thus represent insulator binding sites of highest confidence), Drosophila Kc cells treated with ecdysone at 0, 3, and 48 h (Wood et al. 2011). Primary motifs were identified by MEME-ChIP using default settings (Machanick and Bailey 2011). dCTCF motif 2 was identified in both MEME-ChIP and Weeder 1.3 (Pavesi et al. 2004), and motif 3 in MEME-ChIP by excluding peaks containing the primary conserved motif. Insulator peaks were then trimmed to include only those containing core consensus sequences for each protein using ambiguity codes specified by MEME-ChIP. For dCTCF this included both the motifs described (AG[GA][TG]GGCGC (allowing for one mutation), [AG]GTGT[GT][GA]CC (allowing for one mutation), and GGT[TG][TGC][GA][TA][GA][TA]C[TC][TC][CGT]GCTA (allowing for one mutation). For BEAF-32, this included the previously identified motif [ATG][TGC]CGATA with no mutations allowed and for SU(HW) the motif GC[AC]TA[CT]TTT allowing for one mutation. Direct insulator binding sites were thus finally called as summits identified by MACS in three independent biological samples ±150 bp that contain the described consensus sequence specific for each insulator protein. Overlap between insulators and associated proteins were identified using publicly available tools on Galaxy (Giardine et al. 2005; Blankenberg et al. 2010; Goecks et al. 2010).
H3K27me3 domains were called using H3K27me3 ChIP-seq data obtained here in Drosophila Kc cells, with comparison to publicly available H3K27me3 ChIP-seq data in late embryos and the added requirement for Polycomb occupancy/signal in Drosophila Kc cells (Celniker et al. 2009). Domain borders were called as 0th nucleotide of peaks called, and organized by K-means clustering by Cluster 3.0 (de Hoon et al. 2004). Genes within H3K27me3 domains were called as intersecting (>300 bp) H3K27me3 domains using publicly available tools on Galaxy (Giardine et al. 2005; Blankenberg et al. 2010; Goecks et al. 2010). Comparisons between histone H3K27me3 before and after dCTCF knockdown were performed after rank order normalization, as recently described (Whyte et al. 2012). Briefly, these ChIP-seq data sets are rank-ordered in 10-bp bins across the Drosophila genome, from highest to lowest read intensity. Averages between the two data sets are then assigned to each bin—from highest to lowest read values.
Enrichments for insulator-associated proteins at aligned dCTCF clusters were calculated as percentage of co-occurrence between dCTCF and BEAF-32, SU(HW), CP190, MOD(MDG4), MOD(MDG4)2.2, L(3)MBT, and Chromator at independent dCTCF sites, aligned dCTCF sites, and sites hosting dCTCF + BEAF-32 + SU(HW). Results were hierarchically clustered using cluster 3.0 (de Hoon et al. 2004) and visualized by Java Treeview (Saldanha 2004).
Immunofluorescence microscopy
Immunofluorescence microscopy of polytene chromosomes was done as previously described (Ivaldi et al. 2007). Cells were stained with primary antibodies in antibody dilution buffer (1× PBS, 0.1% Tween20, 1% BSA) overnight at 4°C (1:100 rabbit α-MOD(MDG4)2.2) (gift from Elissa Lei), 1:100 rat α-MOD(MDG4) (Pai et al. 2004).
Real-time PCR analysis
Real-time PCR analyses for H3K27me3 levels in insulator knockdown experiments and Re-ChIP were performed with independent ChIP samples. Fermentas Life Sciences Maxima qPCR SYBR Green ROX Mix (#K0222) was used and percent input was calculated with a three-point standard curve from the input sample. ChIP DNA and input DNA concentrations were calculated using a Qubit 2.0 fluorometer HS assay (Invitrogen Q32851). ChIP DNA concentrations were consistently lower in insulator knockdown conditions, and thus normalized by equal ChIP/input DNA ratios before qRT–PCR. Primers used for both analyses are provided in Supplemental Table S3.
Gene expression analyses
RNAi knockdown in Drosophila Kc cells culture was conducted as per the Drosophila RNAi Screening Center (DRSC) protocol (Armknecht et al. 2005), with the exception that dsRNA was added every day for 3 d and the cells were then collected on the fourth day. Also, multiple amplicons targeting each gene for knockdown were used, with the exception of BEAF-32, which only used one. RNA was then isolated from the Kc cells using the Qiagen RNeasy kit (catalog #74104) with on-column DNA digestion (catalog #79254) following the manufacturer's protocols. cDNA synthesis was then performed using the Applied Biosystems High Capacity cDNA Reverse Transcription Kit (catalog #4368814). cDNA was hybridized to a NimbleGen D. melanogaster Gene Expression 12X135K Array based on D. melanogaster annotation DM5.45 at the Florida State University–NimbleGen Microarray Facility. Expression analysis of variance was performed using Partek software, version 6.5. A list of primers used for amplicon formation are provided in Supplemental Table S4.
Data access
Gene expression and ChIP-seq data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE36944 [L(3)MBT and MOD(MDG4)2.2/MOD(MDG4) in Drosophila Kc cells], GSE37444 (H3K27me3 in Drosophila Kc cells—control and dCTCF knockdown), and GSE36393 (Gene expression in Drosophila Kc cells before and after insulator knockdown).
Acknowledgments
We thank Dr. Elissa Lei and Dr. Jurgen Knoblich for sharing antibodies. We also thank The Genomic Services Lab at the HudsonAlpha Institute for Biotechnology for their help in performing Illumina sequencing of ChIP-seq samples. This work was supported by U.S. Public Health Service Awards to V.G.C. (GM35463) and E.R. (CA133106) from the National Institutes of Health.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.136788.111.
References
- Armknecht S, Boutros M, Kiger A, Nybakken K, Mathey-Prevot B, Perrimon N 2005. High-throughput RNA interference screens in Drosophila tissue culture cells. Methods Enzymol 392: 55–73 [DOI] [PubMed] [Google Scholar]
- Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R 2009. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J 28: 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J 2010. Galaxy: A web-based genome analysis tool for experimentalists. Current Protoc Mol Biol 19: 19.10.1–19.10.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonchuk A, Denisov S, Georgiev P, Maksimenko O 2011. Drosophila BTB/POZ domains of “ttk group” can form multimers and selectively interact with each other. J Mol Biol 412: 423–436 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Dorman ER, Corces VG 2008. Chromatin insulators: Regulatory mechanisms and epigenetic inheritance. Mol Cell 32: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Ramos E, Corces VG 2009. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev 23: 1338–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. 2009. Unlocking the secrets of the genome. Nature 459: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT 2002. CTCF, a candidate trans-acting factor for X-inactivation choice. Science 295: 345–347 [DOI] [PubMed] [Google Scholar]
- Comet I, Schuettengruber B, Sexton T, Cavalli G 2011. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc Natl Acad Sci 108: 2294–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S 2004. Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn R, Krauss V 2003. The modifier of mdg4 locus in Drosophila: Functional complexity is resolved by trans splicing. Genetica 117: 165–177 [DOI] [PubMed] [Google Scholar]
- Eggert H, Gortchakov A, Saumweber H 2004. Identification of the Drosophila interband-specific protein Z4 as a DNA-binding zinc-finger protein determining chromosomal structure. J Cell Sci 117: 4253–4264 [DOI] [PubMed] [Google Scholar]
- Essien K, Vigneau S, Apreleva S, Singh LN, Bartolomei MS, Hannenhalli S 2009. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol 10: R131 doi: 10.1186/gb-2009-10.11-r131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV 1996. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol 16: 2802–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan M, Moebus S, Eggert H, Saumweber H 2011. The Chriz-Z4 complex recruits JIL-1 to polytene chromosomes, a requirement for interband-specific phosphorylation of H3S10. J Biosci 36: 425–438 [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G 2006. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7: 703–713 [DOI] [PubMed] [Google Scholar]
- Gelbart WM, Emmert DB 2010. FlyBase high throughput expression pattern data beta version A Database of Drosophila Genes &Genomes. [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG 1995. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82: 587–597 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Lei EP, Bushey AM, Corces VG 2007. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell 28: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Gerasimova TI, Corces VG 2001. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J 20: 2518–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. 2005. Galaxy: A platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. 2003. A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736 [DOI] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J 2010. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86 doi: 10.1186/gb-2020.11-8-r86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero PA, Maggert KA 2011. The CCCTC-binding factor (CTCF) of Drosophila contributes to the regulation of the ribosomal DNA and nucleolar stability. PLoS ONE 6: e16401 doi: 10.1371/journal.pone.0016401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, et al. 2011. CTCF-binding elements mediate control of V(D)J recombination. Nature 477: 424–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurudatta BV, Corces VG 2009. Chromatin insulators: Lessons from the fly. Brief Funct Genomics Proteomics 8: 276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, et al. 2011. A protein complex network of Drosophila melanogaster. Cell 147: 690–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. 2011. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 43: 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R 2007. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet 3: e112 doi: 10.1371/journal.pgen.0030112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivaldi MS, Karam CS, Corces VG 2007. Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila. Genes Dev 21: 2818–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SN, Shim HP, Jeon BN, Choi WI, Hur MW, Girton JR, Kim SH, Jeon SH 2011. The pleiohomeotic functions as a negative regulator of Drosophila even-skipped gene during embryogenesis. Mol Cells 32: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH, Neiman PE, Lobanenkov VV 1993. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol 13: 7612–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong C, Adryan B, Bell I, Meadows L, Russell S, Manak JR, White R 2008. Stability and dynamics of polycomb target sites in Drosophila development. PLoS Genet 4: e1000178 doi: 10.1371/journal.pgen.1000178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador M, Corces VG 2003. Extensive exon reshuffling over evolutionary time coupled to trans-splicing in Drosophila. Genome Res 13: 2220–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B 2010. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics 32: 11.7.1–11.7.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HB, Muller M, Bahechar IA, Kyrchanova O, Ohno K, Georgiev P, Pirrotta V 2011. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol Cell Biol 31: 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald PM, Ingham P, Struhl G 1986. Isolation, structure, and expression of even-skipped: A second pair-rule gene of Drosophila containing a homeo box. Cell 47: 721–734 [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL 2011. MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnik S, Deng B, Papantonis A, Baboo S, Carr IM, Cook PR 2011. The proteomes of transcription factories containing RNA polymerases I, II or III. Nat Methods 8: 963–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RA, Renkawitz-Pohl R, Saumweber H, et al. 2007. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J 26: 4203–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet 6: e1000814 doi: 10.1371/journal.pone.1000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Oliver D, Sheehan B, South H, Akbari O, Pai CY 2010. The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions. BMC Cell Biol 11: 101 doi: 10.1186/1471-2121-11-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Lei EP, Ghosh D, Corces VG 2004. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell 16: 737–748 [DOI] [PubMed] [Google Scholar]
- Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP 2009. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res 19: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, Mereghetti P, Mauri G, Pesole G 2004. Weeder Web: Discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res 32: W199–W203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG 2009. CTCF: Master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V, Li HB 2011. A view of nuclear Polycomb bodies. Curr Opin Genet Dev 22: 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos E, Ghosh D, Baxter E, Corces VG 2006. Genomic organization of gypsy chromatin insulators in Drosophila melanogaster. Genetics 172: 2337–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Oktaba K, Steinmann J, Muller J, Knoblich JA 2011. The tumour suppressor L(3)mbt inhibits neuroepithelial proliferation and acts on insulator elements. Nat Cell Biol 13: 1029–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ 2004. Java Treeview–extensible visualization of microarray data. Bioinformatics 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
- Schoborg TA, Labrador M 2010. The phylogenetic distribution of non-CTCF insulator proteins is limited to insects and reveals that BEAF-32 is Drosophila lineage specific. J Mol Evol 70: 74–84 [DOI] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 1488: 458–472 [DOI] [PubMed] [Google Scholar]
- Weth O, Renkawitz R 2011. CTCF function is modulated by neighboring DNA binding factors. Biochem Cell Biol 89: 459–468 [DOI] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA 2012. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482: 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, Jones KC, Corces VG 2011. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol Cell 44: 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES 2007. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci 104: 7145–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Tanay A 2011. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet 43: 1059–1065 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, Caiafa P 2009. CTCF and its protein partners: Divide and rule? J Cell Sci 122: 1275–1284 [DOI] [PubMed] [Google Scholar]