Abstract
Chromatin insulator elements and associated proteins have been proposed to partition eukaryotic genomes into sets of independently regulated domains. Here we test this hypothesis by quantitative genome-wide analysis of insulator protein binding to Drosophila chromatin. We find distinct combinatorial binding of insulator proteins to different classes of sites and uncover a novel type of insulator element that binds CP190 but not any other known insulator proteins. Functional characterization of different classes of binding sites indicates that only a small fraction act as robust insulators in standard enhancer-blocking assays. We show that insulators restrict the spreading of the H3K27me3 mark but only at a small number of Polycomb target regions and only to prevent repressive histone methylation within adjacent genes that are already transcriptionally inactive. RNAi knockdown of insulator proteins in cultured cells does not lead to major alterations in genome expression. Taken together, these observations argue against the concept of a genome partitioned by specialized boundary elements and suggest that insulators are reserved for specific regulation of selected genes.
Insulator elements were first discovered in Drosophila melanogaster by biochemical (Udvardy et al. 1985) and genetic approaches (Holdridge and Dorsett 1991; Geyer and Corces 1992) as specialized chromatin structures that appeared to define boundaries between different chromatin states. It was soon found that such insulator elements have the ability to block enhancer action when interposed between enhancers and promoters and that this activity depended on specific DNA binding proteins that associate with the insulator element. In Drosophila, we now know of four well-defined insulator DNA binding proteins, SU(HW), ZW5 (also known as DWG), BEAF-32, and CTCF (Geyer and Corces 1992; Zhao et al. 1995; Gaszner et al. 1999; Moon et al. 2005), of which only CTCF has an ortholog in mammals (Baniahmad et al. 1990; Lobanenkov et al. 1990). Two other proteins, MOD(MDG4)67.2 and CP190, were found to associate with the SU(HW)-binding insulator element found in the gypsy transposon and are also required for its insulator function (Georgiev and Gerasimova 1989; Gerasimova et al. 1995; Pai et al. 2004). Both MOD(MDG4)67.2 and CP190 contain a POZ/BTB structural motif, known to mediate homotypic and heterotypic protein–protein interactions, which may drive the association between multiple insulator elements. Subsequent work has shown that CP190 also shares some of its chromatin binding sites with BEAF-32 and CTCF and can interact directly with the latter (Gerasimova et al. 2007; Mohan et al. 2007; Bushey et al. 2009; Nègre et al. 2010).
The mechanistic interdependencies between CP190 and the sequence-specific DNA binding proteins remain somewhat controversial. CP190 protein is recruited to gypsy insulator by SU(HW) but also binds directly to the endogenous SU(HW)-dependent insulator DNA from the y-achaete locus (Pai et al. 2004). CTCF binding to chromosomes was variously claimed to be either strictly (Gerasimova et al. 2007) or partially (Mohan et al. 2007) dependent on CP190 or, more recently, completely independent of CP190 (Wood et al. 2011). Despite the uncertainties, an influential model proposes that CP190 acts as a universal “glue” protein that mediates interactions between insulator elements of different classes, thereby generating chromatin loops, whose properties are postulated to be such that regulatory elements located on one loop are hindered from interacting with promoters or other elements on the adjacent loop (Gerasimova et al. 2007; Bushey et al. 2009).
Previous immunolocalization and ChIP-chip/ChIP-seq analyses have shown that insulator proteins have numerous binding sites in the Drosophila genome (Zhao et al. 1995; Gerasimova and Corces 1998; Mohan et al. 2007; Bartkuhn et al. 2009; Bushey et al. 2009; Nègre et al. 2010). Together with similar data for the mammalian CTCF protein (Kim et al. 2007; Cuddapah et al. 2009), these findings suggest that the genome is partitioned into domains delimited by boundaries that prevent spreading or influencing the chromatin state of flanking domains. According to this view, insulator elements would be expected to be very abundant in the genome and serve an essential function to protect genes from inappropriate action of enhancers, silencers, and other chromatin-modifying activities affecting gene function.
Another view of insulator function, not incompatible with the first, derives from the discovery that insulator protein binding sites play a critical role in several complex regulatory regions from mammalian and Drosophila genomes, where they bring together different components or juxtapose regulatory elements with the appropriate promoters (Kurukuti et al. 2006; Ling et al. 2006; Splinter et al. 2006; Li et al. 2011). Although these elements were originally discovered for their ability to separate genomic units, the view that insulators may bring parts of the genome together is consistent with the realization that chromatin looping is an essential feature of genome architecture and gene regulation (Lanctôt et al. 2007; Schoenfelder et al. 2010). In this view, however, such “linking” and folding is the basic function of “insulator” elements, and in principle, not every insulator protein binding site necessarily has an enhancer blocking insulator function.
Here we evaluate the two concepts by genome-wide analysis of insulator protein binding to Drosophila chromatin. We focus on the quantitative aspects of binding, which reveal classes of binding sites occupied by specific combinations of insulator proteins. We demonstrate that distinct rules govern the binding of an insulator protein to different classes of sites, which sometimes involve cooperation between several insulator proteins. We also describe a novel class of robust Drosophila insulator elements that in cultured cells bind CP190 but not any other known insulator proteins. We find that only a small fraction of insulator protein binding sites act as robust enhancer blockers in vivo and that significant depletion of insulator proteins in cultured cells has small effects on genome-wide expression or the spreading of the H3K27me3 mark. Our observations argue against the concept of a genome partitioned by specialized boundary elements and suggest, instead, that insulators are reserved for specific regulation of selected genes.
Results
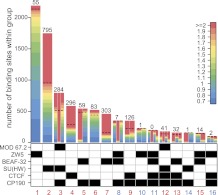
By use of chromatin immunoprecipitation analyzed by hybridization to Drosophila genomic tiling arrays (ChIP-chip), we have mapped the distributions of SU(HW), CTCF, BEAF-32, ZW5, CP190, and MOD(MDG4)67.2 proteins in cultured S2-DRSC and ML-DmBG3-c2 cells (hereafter referred to as S2 and BG3). As has been reported previously (Bushey et al. 2009; Nègre et al. 2010), the genomic distributions of insulator proteins overlap. To characterize the persistent co-binding groups, we first used a relaxed threshold of log2(IP/INPUT) > 0.7 to define genomic regions bound by each protein and record all possible types of overlapping combinations. Each region was further examined for the strength of binding of associated proteins, and only those regions in which all associated proteins bound with comparative strength were considered for further analysis (Fig. 1; Supplemental Tables S1–S18). The last step is critical to take into account differences in antibody strengths and discriminate between sites with robust co-binding of several proteins and sites at which one protein binds strongly but others are barely detectable. This approach shows clearly that some of the co-binding combinations reported earlier (Nègre et al. 2010) are in fact at the edge of computational detection (Fig. 1). For example, class 14 sites that would appear to co-bind CP190 and SU(HW) in the absence of MOD(MDG4)67.2 display exceedingly weak CP190 signals, which is in stark contrast to the robust CP190 binding to class 3 sites in the presence of SU(HW) and MOD(MDG4)67.2.
Figure 1.
The classes of insulator protein binding sites. The composition of 16 co-binding groups detected by initial overlap comparison is indicated by the checkerboard pattern under the bar plot. The color code in log2(IP/INPUT) units (indicated to the right) is used to show the number of sites of different binding strength within each class. For the multiprotein classes, the bars are divided from left to right corresponding to the top to bottom positions of the proteins in the chart below. The numbers of sites of each class that bind all corresponding proteins within 60% of their ChIP-chip signal dynamic range are indicated above the bars. Only those sites were used for further analysis. The dashed line on each bar indicates the position of the 60% cutoff. The classes representing robust co-binding combinations are numbered in red.
Consistent with a broad role of CP190 in the insulator network, ∼80% of robust CP190 binding sites are shared with SU(HW), CTCF, or BEAF-32 (Fig. 1). In contrast, more than half of the SU(HW), CTCF, and BEAF-32 sites are standalone, i.e., none of the other proteins tested are bound to these sites. This implies that the interaction of SU(HW), CTCF, and BEAF-32 with CP190 and other co-binding partners depends on additional factors. In addition, 83 robust standalone CP190 sites indicate that this protein can be recruited to chromatin independently of SU(HW), CTCF, and BEAF-32.
As expected, from polytene chromosome staining (Gerasimova and Corces 1998), we detect ∼300 sites with simultaneous robust binding of SU(HW), MOD(MDG4)67.2, and CP190, the combination of proteins associated with the gypsy insulator (Fig. 1, class 3). We will refer to this class of binding sites as gypsy-like, although none of them corresponds to gypsy retrotransposon insertions as all repetitive sequences were excluded from our analysis. We see no MOD(MDG4)67.2 binding in the absence of SU(HW) and CP190.
cis cooperation and motif coincidence govern the co-binding of CP190 with CTCF and SU(HW)
Since SU(HW) and CTCF interact directly with CP190 (Pai et al. 2004; Gerasimova et al. 2007), the large number of standalone sites for these two proteins requires explanation. The comparison of the genomic distributions of SU(HW), CTCF, and CP190 in S2 and BG3 cells and in whole embryos (from Nègre at al. 2010) shows that the distinction between the standalone and CP190 co-bound sites is well-preserved in all three sources of chromatin. We conclude that the co-binding of CP190 is an inherent property of a site rather than a product of tissue specific regulation.
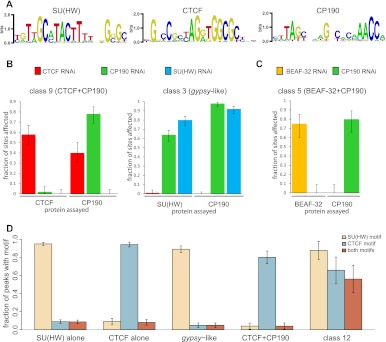
The analysis of DNA sequences in standalone SU(HW) and CTCF regions (class 2, 4) and those shared with CP190 (class 3, 9) indicates that SU(HW) and CTCF bind to DNA directly and with the same sequence specificity irrespective of CP190 presence. The most prominent motifs derived from the corresponding standalone and CP190 co-bound regions are essentially identical (Fig. 2A; Supplemental Fig. S1) and match the reported binding sequences of SU(HW) and CTCF in vivo (Adryan et al. 2007; Holohan et al. 2007; Nègre et al. 2010) and in in vitro (Spana and Corces 1990; Golovnin et al. 2003; Moon et al. 2005). The number of SU(HW) or CTCF motifs in the standalone sites does not differ significantly from that in the CP190 co-binding sites.
Figure 2.
The sequence determinants and interdependence of the insulator protein binding to chromatin. (A) The logo representations of sequence motifs characteristic of SU(HW), CTCF, and CP190 binding sites defined by the MEME algorithm and used in the analysis in D. (B) The effects of the RNAi knock-down on the target protein and its co-binding partners. The sites at which ChIP-chip signal was consistently reduced judged from the comparison of two replicate mock RNAi experiments and two specific RNAi experiments (z-scores < −3, unpaired t-test) were counted and their fractions plotted. Here and in C and D, the error bars indicate the 95% confidence interval. The bar plots show that the binding of CP190 to some of the class 9 but not at gypsy-like sites depends on CTCF. However, the binding of CTCF to class 9 sites does not depend on CP190. In contrast, the binding of SU(HW) and CP190 to gypsy-like sites is interdependent. (C) As illustrated by this bar plot, BEAF-32 and CP190 bind to common sites independently. (D) The presence of SU(HW) and CTCF recognition sequences within indicated classes of sites demonstrates that the coincidence of the two motifs is responsible for the co-binding of SU(HW) and CTCF to class 12 sites.
In addition to the canonical CTCF motif, the sequence analysis reveals a new motif enriched in class 9 (CTCF+CP190) but not class 4 (standalone CTCF) regions (Fig. 2A; Supplemental Fig. S1). Strikingly, this motif is also enriched at class 6 (standalone CP190) sites (Fig. 2A; Supplemental Fig. S1), suggesting that CP190 can bind to DNA directly or through an unknown DNA-binding protein(s) (for more details, see Supplemental Results) and that the binding of CTCF and CP190 to common sites results from the coincidence of the corresponding recognition sequences. It is clear, however, that at many sites CTCF is required for CP190 binding. RNAi depletion of CTCF that reduces its binding at class 9 (CTCF+CP190) sites (Fig. 2B) also reduces CP190 binding at most of those same sites but not at class 3 (gypsy-like) or other sites where CP190 is not accompanied by CTCF (Supplemental Fig. S2). The converse knock-down of CP190 has very little effect on CTCF binding (Fig. 2B; Supplemental Fig. S2), indicating that CTCF is recruited to class 9 (CTCF+CP190) sites independently of CP190.
In contrast to class 9 (CTCF+CP190) sites, the sequence analysis of class 3 (gypsy-like) and class 2 (standalone SU(HW)) sites revealed no characteristic motifs other than the SU(HW) recognition sequence. The RNAi knock-down of SU(HW) results in its efficient depletion from chromosomes and also in depletion of CP190 from the majority of gypsy-like sites, but not from other kinds of CP190 sites (Fig. 2B; Supplemental Fig. S2). Unexpectedly, CP190 depletion also causes loss of both proteins from class 3 (gypsy-like) sites (Fig. 2B), indicating that the binding of SU(HW) and CP190 to these sites is mutually dependent. CP190 RNAi has little effect on the binding of SU(HW) to standalone sites (Supplemental Fig. S2), arguing that the dependence is direct. Although other explanations are possible, these results suggest that SU(HW) and CP190 proteins cooperate, allowing SU(HW) to bind to a recognition sequence of a quality or in an environment inadequate to recruit on its own. Supporting this notion, we find that the consensus scores of the SU(HW) motifs at class 3 (gypsy-like) sites are markedly lower than those at class 2 (standalone SU(HW)) sites (Supplemental Fig. S3).
Finally, although more than half of SU(HW) or CTCF sites are standalone, SU(HW) and CTCF never bind a common region unless together with CP190 and MOD(MDG4)67.2 (Fig. 1; class 12 sites). The apparent co-binding of CTCF and SU(HW) to class 12 sites might be attributed to the crosslinking of complexes recruited to distinct insulator elements and bridged in trans by interactions between CP190 and MOD(MDG4)67.2 proteins. This model predicts that the DNA sequences recognized by CTCF and SU(HW) would rarely group together at class 12 sites. Contrary to this prediction, class 12 sites show a high coincidence of SU(HW) and CTCF motifs, a feature absent from sites that bind only SU(HW) or only CTCF (Fig. 2D). This points to the DNA sequence as the primary determinant of the common binding to class 12 sites and argues against their being the product of crosslinking of distinct trans-interacting regions (although such trans-interactions are not excluded).
BEAF-32 is dispensable for the recruitment of CP190 to chromatin
BEAF-32 was suggested to act as a DNA binding recruiter of CP190 (Bushey et al. 2009). Indeed the comparison of BEAF-32 and CP190 regions defined at low threshold [log2(IP/INPUT) > 0.7] shows extensive overlap (Fig. 1). It is immediately obvious, however, that the binding of CP190 to these sites is often weak and disproportional to BEAF-32 (hence the relatively small number of regions listed as robustly bound by both proteins, classes 5 and 8). RNAi depletion of BEAF-32 causes a reduction of its binding to the majority of the sites shared with CP190 (class 5 sites) (Fig. 2C). However, it has no effect on the binding of CP190 to these sites. Conversely CP190 depletion reduces its binding to the majority of class 5 sites but does not affect BEAF-32 binding (Fig. 2C). We conclude that BEAF-32 and CP190 bind chromatin independently of each other and that their coincidence may result from a bias of both proteins toward active transcription start sites (TSSs).
RNAi-knockdown discriminates between low- and high-affinity binding sites
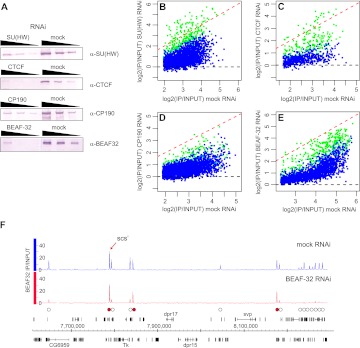
The loss of a chromatin protein from its genomic binding sites upon RNAi knock-down is sometimes used to validate the genome-wide mapping. The 10-fold reduction of nuclear protein levels in the above RNAi experiments (Fig. 3A; Supplemental Fig. S4) results in the complete loss of binding at many chromosomal sites and reduction of binding to the majority of them (Figs. 3B–E). Yet in all cases, we see a number of strong sites that remain unaffected by RNAi. Immunoprecipitations using two antibodies independently raised against different parts of the proteins strongly suggest that these are genuine high-affinity binding sites, and not false positives. For example, one of the sites with persistent BEAF-32 binding is scs′, the prototype BEAF-32–dependent insulator (Fig. 3F). We conclude that immunoprecipitation with two independently derived antibodies is a better validation criterion than the use of RNAi depletion, which in our case would reject sites with the highest affinity and therefore with the best functional potential.
Figure 3.
The effects of RNAi knock-down on the binding of insulator proteins to chromatin. BG3 cells were subjected to RNAi against key insulator proteins followed by ChIP-chip. (A) Western blots of threefold serial dilutions of nuclear protein from cells treated with specific and mock dsRNA (indicated above the panels) show 10-fold or greater knock-down of the corresponding proteins. The antibodies used for detection are indicated to the right, and the loading controls are shown in Supplemental Figure S4. The comparison of average binding for (B) SU(HW), (C) CTCF, (D) CP190, and (E) BEAF-32 after mock and specific RNAi shows that the binding is reduced at the majority of sites (data points below red dashed line). (Blue dots) The sites with consistent reduction in both replicate experiments (estimated conservatively with unpaired t-test; z-scores < −3); (green dots) others. (F) scs′ is one of the BEAF-32 high-affinity binding sites resistant to RNAi. The BEAF-32 ChIP-chip signals after BEAF-32 and mock RNAi are plotted along the segment of chromosome 3R. (White circles) Peaks affected by BEAF-32 knock-down; (red circles) peaks that remain unchanged. The genes shown above the coordinate scale are transcribed from left to right, those below the scale from right to left.
Analysis of insulator function
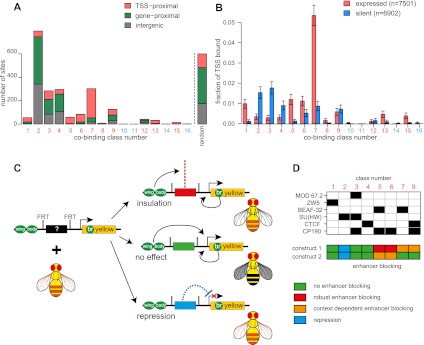
A comparison of the genomic distributions of different classes of binding sites to genes and gene activity shows very clear differences. The majority of class 3 (gypsy-like), standalone SU(HW) (class 2), and CTCF (class 4) sites and about a half of class 9 (CTCF+CP190) sites are situated within introns of transcriptionally inactive genes or in intergenic regions (Fig. 4A,B). In contrast, the other half of class 9 (CTCF+CP190), as well as ZW5, BEAF-32, and standalone CP190 sites, tend to reside within 2 kb of transcriptionally active TSSs (Fig. 4A,B). None of the classes of binding sites have significant preference for positions situated between an active and a silent gene.
Figure 4.
Functional evaluation of classes of insulator protein binding sites. (A,B) Sites bound by different combinations of insulator proteins show distinct biases in their distribution relative to genes and gene activity. Class 2–4 sites are rarely close to TSSs, while class 5–7 sites are primarily TSS-proximal. In rare cases when class 2–4 sites are TSS proximal, these promoters tend to be inactive. In contrast, BEAF-32 (classes 5 and 7) binds predominantly next to active TSSs. While many standalone CP190 sites are next to active TSSs, some are not. The proximity to TSSs and genes in A is defined based on a 2-kb margin, and the binding to TSSs in B is defined on a 1-kb margin. The background distribution expected by chance is shown as the rightmost bar in A and is derived from 10 times the number of positions sampled randomly but with the same chromosome representation. (C) The schematic of the transgenic enhancer blocking assay. A DNA fragment of interest (black rectangle) is cloned in the FRT cassette positioned between the upstream wing and body enhancers (green ovals) and the promoter of the reporter yellow gene (yellow rectangle). The resulting construct is injected into yellow minus flies. DNA fragments capable of enhancer blocking (red rectangle) prevent the activation of the reporter yellow gene by upstream enhancers but allow the activation of the gene by the downstream bristle enhancer (“br” green oval). This yields transgenic flies with pigmented bristles but a yellow body and wings. Ineffectual DNA fragments (green rectangle) allow activation of the reporter gene in all tissues and yield wild-type transgenic flies. The fragments harboring repressive activity (blue rectangle) block the expression of transgenic yellow in all tissues, which results in flies devoid of any pigmentation. The results of transgenic tests are summarized in D.
The distinct genomic location of different classes of binding sites raises the question of whether they have the same functions or insulating properties. To examine insulator function, we selected two representative 1-kb DNA fragments from each major class and measured their ability to block the activation of the yellow reporter gene by the upstream wing- and body-specific enhancers when placed between these enhancers and the promoter. Unlike general repressors, insulators are expected to block the upstream enhancers without affecting the activation of the yellow promoter by the downstream bristle specific enhancer (Fig. 4C, S5; Geyer and Corces 1992). Five randomly chosen 1-kb genomic fragments that showed no association with any of the insulator proteins in our ChIP-chip experiments and the 680-bp gypsy insulator element were tested in the same reporter assay as negative and positive controls. Initially all reporter constructs were integrated in the same 51D landing site by targeted φC31 att recombination (Bischof et al. 2007), which allowed direct comparison of the effects produced by different test fragments in the same chromosomal environment. Subsequently, the transgenes were mobilized from the 51D site using P-element–mediated transposition to assess the robustness of the enhancer blocking effect in different chromosomal contexts.
As summarized in Figure 4D, Table 1, and Supplemental Tables S19 and S20, the transformation of flies lacking yellow function with negative control constructs restores their phenotype to nearly wild type. The pigmentation of the body and wings in these flies varies somewhat depending on the site of insertion but is always much stronger than in flies transformed by the positive control construct that carries the gypsy insulator, which have black bristles but completely yellow wings and a very light body cuticle. Of the 16 insulator protein binding sites tested, only two (BC1, class 5; CP1901, class 6) block the upstream enhancers to the same extent as the gypsy insulator construct. The enhancer block is robust, evident at all tested chromosomal locations, and completely reversed upon FLP-mediated excision of tested fragments. In addition, we find four fragments (CTCFC1, class 9; B1, class 7; CP1902, class 6; and BC2, class 5) whose enhancer blocking ability is less strong than that of BC1 (class 5) and CP1901 (class 6) and varies depending on the surrounding chromatin context. Fragment BC2 (class 5) represents the most striking example of variability, with extremely good enhancer blocking at some insertion sites and complete lack of it at others.
Table 1.
The results of enhancer blocking assay
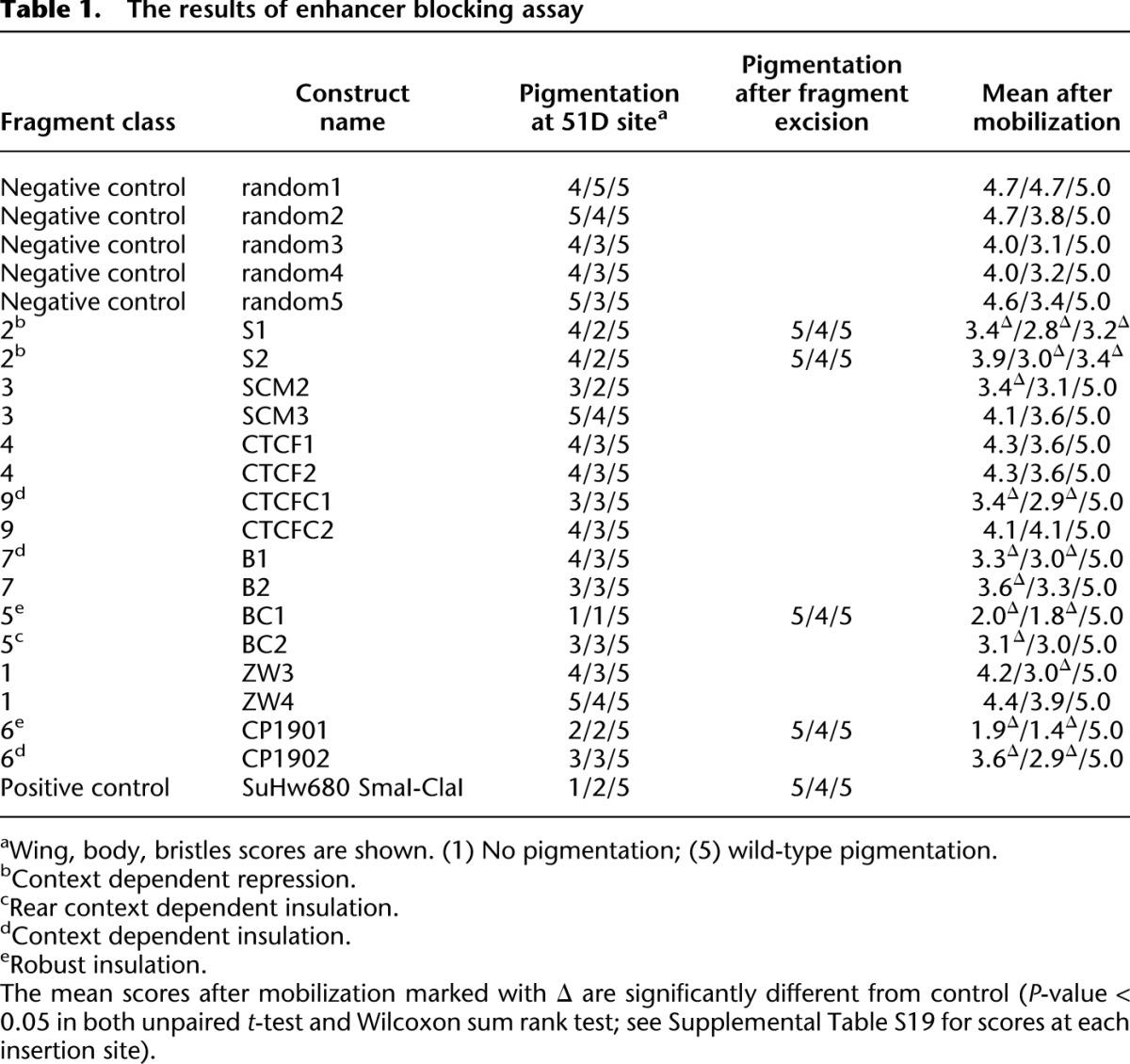
None of the class 3 (gypsy-like) sites tested displayed enhancer blocking activity, in agreement with the results of Nègre et al. (2011), who tested several other fragments of this class using a different enhancer-blocking assay based on the eve stripe 2 and 3 enhancers. On the other hand, the class 3 (gypsy-like) binding sites from the yellow-achaete and 62D regions have been shown to robustly block yellow enhancers in transgenic tests (Golovnin et al. 2003; Parnell et al. 2003; Kuhn-Parnell et al. 2008). This suggests that the simple recruitment of CP190, MOD(MDG4)67.2, and SU(HW) to a chromosomal site is not sufficient for robust enhancer blocking and that additional unknown factors or specific chromatin configurations are required for gypsy-like binding sites to have this function.
Overall, we conclude (1) that, unlike the prototype insulators, the majority of insulator protein binding sites are not robust enhancer blockers; (2) that the complement of binding proteins at each class of sites is a poor predictor of whether a site can act as an enhancer blocker; and (3) that a given site can act as an enhancer blocker in one genomic context but not in another. The latter implies the possibility that a site that does not appear to act as an enhancer blocker might become such if the chromatin environment changes. Some functional regulatory elements can be pinpointed based on their high DNA sequence conservation. This appears not to be the case for insulator protein binding sites. Thus the sequences of BC1 (class 5) and CP1901 (class 6) fragments, which show the best enhancer blocking in the transgenic test, and the sequences of the sites from these classes in general show surprisingly low conservation (Supplemental Fig. S6–S8; Supplemental Results).
Unaddressed by our functional test is the question whether the sites occupied by a combination of DNA binding insulator proteins have properties markedly different from their simpler counterparts. Future experiments should reveal, e.g., whether class 12 sites have poor enhancer-blocking ability like class 3 (gypsy-like) sites or can block enhancer–promoter communications as well or better than class 9 (CTCF+CP190) sites.
Finally, we note that both fragments tested to represent class 6 (standalone CP190 sites) and class 5 (BEAF-32+CP190) sites display a degree of enhancer blocking and include the only robust enhancer blockers found in our tests (Table 1). Considering the fact that the CP190 binds to class 5 (BEAF-32+CP190) sites independently of BEAF-32, this underscores the importance of the novel pathway of CP190 recruitment to chromatin and suggests that it may be utilized at the majority of robust enhancer blocking elements in Drosophila.
Standalone SU(HW) binding sites act as general repressors
In these assays we found no evidence of enhancer blocking by class 2 and 4 (standalone CTCF or SU[HW]) binding sites, although we cannot exclude the possibility that some may be active in specific tissues where they acquire CP190, as recently proposed by Wood et al. (2011). Instead, at many chromosomal locations, the two representative class 2 (standalone SU[HW]) binding fragments S1 and S2 cause loss of yellow expression not only in wings and body but also in bristles, indicative of general promoter repression rather than enhancer blocking activity (Supplemental Tables S19, S20). Such behavior is reminiscent of the repressive properties of the gypsy insulator upon loss of mod(mdg4) function (Gerasimova et al. 1995) and suggests that transcriptional repression is a general feature of SU(HW) protein when not associated with MOD(MDG4)67.2.
The impact of insulator proteins on gene expression
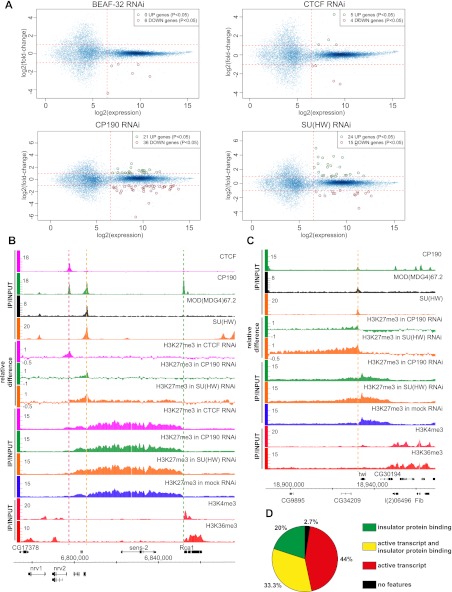
It is surprising that most of the insulator protein binding sites tested appear to lack robust enhancer blocking activity, raising the possibility that the transgenic assay may underestimate the fraction of functional binding sites because they require their native genomic context. Therefore, as an independent measure of an insulator protein impact on the genome, we evaluated genomic changes in gene expression after depletion of SU(HW), CTCF, BEAF-32, or CP190 in BG3 cells. Consistent with the notion that only a small fraction of insulator protein binding sites corresponds to functional insulators, significant depletion of any single insulator protein does not lead to major alterations in gene expression (Fig. 5A). We see no widespread switching on of the inactive genes by the adjacent “active” chromatin environment or repression of active genes by encroaching “repressive” chromatin states
Figure 5.
Functional effects of insulator protein withdrawal. (A) Affymetrix GeneChip expression analysis of cells from the RNAi experiments described in Figure 3. The average fold change between the two specific and two mock RNAi experiments (y-axes) was plotted against the highest average expression value detected in the mock or specific RNAi samples (x-axes). Each graph point represents one transcript interrogated by the microarray. Transcripts robustly expressed before or after specific RNAi treatment are to the right of the vertical dashed lines. Of these, those showing consistent twofold or greater change after specific RNAi treatment in both replicate experiments are circled. (B) As evident from ChIP-chip of H3K27me3 from mock RNAi-treated BG3 cells, the sens-2 gene is repressed by PcG. The right border of the corresponding H3K27me3 domain is sharp and coincides with a standalone CP190 site (marked by a vertical green dashed line) and with the Rca1 transcript. The ChIP-chip with H3K4me3 and H3K36me3 indicates that Rca1 is transcriptionally active. The left side of the H3K27me3 domain declines gradually with no obvious border. It harbors gypsy-like and CTCF+CP190 binding sites marked by orange and purple dashed lines, respectively. The knock-downs of insulator proteins have no effect on the position of the right border of the H3K27me3 domain but change the shape of its left tail. The changes in histone methylation profile are best seen on the relative difference browser tracks. (C) twi is also repressed by PcG mechanisms in BG3 cells. The right border of the corresponding H3K27me3 domain is set by the presence of an active transcript. The left border is maintained by a gypsy-like (class 3) insulator (vertical orange dashed line), as evident from the extension of K27 trimethylation after SU(HW) or CP190 knock-down. (D) The pie chart shows the frequencies of various genomic features associated with definable H3K27me3 domain borders.
The few changes in gene expression that we can detect are consistent with the results of the transgene tests (Fig. 5A). Of 39 genes affected by SU(HW) RNAi, the expression of 24 genes is up-regulated. The magnitude of the expression increase at these genes is much higher than the reduction at genes where the expression goes down, which fits well with the repressive properties of standalone SU(HW) sites seen in the transgenic assay. The transgenic assay also suggests that CP190 bound sites, especially standalone, tend to block enhancer–promoter communications. If they function as pure insulator elements, one would expect that in some cases their loss might cause inappropriate activation of a gene, while in other cases, it might lead to inappropriate repression. We find instead that the genes affected by CP190 knock-down tend to reduce their expression. It is possible that this apparent stimulatory role of CP190 stems from a consistent bias for using CP190-dependent insulators to block long-range transcriptional repression. However, we favor an alternative explanation that the most frequent role of CP190 complexes is to aid chromatin folding to bring distant activators to their appropriate targets.
Insulators and Polycomb silencing
Broad domains of histone H3 trimethylated at K27 (H3K27me3) mark loci repressed by Polycomb group (PcG) proteins (Schwartz et al. 2006). In Drosophila, these proteins are recruited to the target genes by Polycomb response elements (PREs), from which the H3K27me3 mark spreads by a chromatin looping mechanism (Kahn et al. 2006; Comet et al. 2011). The gypsy insulator can interfere with the looping of PRE-bound complexes and block the spreading of H3K27me3 (Kahn et al. 2006; Comet et al. 2011). Visual inspection shows that half of the H3K27me3 domain edges (110 of 221) display a gradual decline to background level (exemplified by the left edge of the sens-2 domain in Fig. 5B). The remaining 111 edges are sharp enough to define a distinct domain boundary (Fig. 5C), which we will refer to as H3K27me3 domain borders. Two genomic features correlate with the presence of domain borders (Fig. 5D); 54% of the domain borders coincide with robust insulator protein binding sites, and 78% of the borders coincide with the 5′ or 3′ ends of active transcripts. At least one of the two features is present at 97% (108 of 111) of definable borders, suggesting that both may contribute to limiting the spread of H3K27 trimethylation.
Forty-four percent of the borders are marked only by the presence of adjacent transcriptional activity, and 33% of the borders coincide with both the ends of active transcription and insulator protein binding sites (Fig. 5D), consistent with the tendency of BEAF-32 and CP190 proteins to bind in the 5′ region of active genes. Transcriptional activity itself may be sufficient to prevent the spread of H3K27me3 by the associated histone H3 replacement, H3K27 acetylation, or inhibition of the histone methyltransferase activity of PcG complexes by H3K4me3 (Schmitges et al. 2011). This appears to be the case as only three H3K27me3 domains bordered by active transcripts are extended after RNAi knockdown of insulator proteins (3% of all domain borders associated with transcriptional activity). We conclude that if putative insulator elements contribute to the establishment of the borders adjacent to active loci, they are dispensable for their maintenance in most cases.
In contrast, at 75% (18 out of 24) of domain borders that contain insulator protein binding sites but no active genes, RNAi knock-down of the corresponding insulator proteins results in expansion of the H3K27me3 domains. The affected boundaries coincide with class 12, class 3 (gypsy-like), class 9 (CTCF+CP190), and class 6 (standalone CP190) binding sites, consistent with the idea that such sites can act as functional insulators.
In ∼5% (five out of 110) of cases, the knockdown of insulator proteins leads to the extension or changes in the shape of the gradually declining H3K27me3 domains. In these cases, exemplified by the left tail of the sens-2 domain (Fig. 5B), the affected domains contain class 3 (gypsy-like) and/or class 9 (CTCF+CP190) sites within their tails and change their length and/or shape upon RNAi knockdown of CP190, SU(HW), or CTCF. We interpret this to indicate that the block imposed by class 3 (gypsy-like) or class 9 (CTCF+CP190) insulator elements is not always robust: Some H3K27me3 may bleed through the insulator, but in its absence, spreading of the histone methylation is more efficient and longer-range. Finally, we note that standalone CTCF sites are never found at H3K27me3 domain borders, supporting the idea that these are not insulators and cannot block the spreading of histone methylation.
Overall, we conclude that insulators participate in shaping the genomic distribution of H3K27me3. However, in cultured cells, their contribution is small and used primarily to prevent the extensive H3K27 trimethylation of transcriptionally inactive genes adjacent to PcG target regions. This role may be most important when Polycomb repression is first established in the embryo.
Discussion
The binding sites of insulator proteins are often taken to represent elements that partition the genome into independent regulatory domains and demarcate chromosomes into regions of “active” and “repressed” chromatin. The results presented here give little support to this view as a general principle of genome organization, although it may be true in certain regions. Instead we would like to argue that: (1) Insulator proteins bind to genomic sites in specific combinatorial patterns; (2) the properties of sites bound by key insulator proteins SU(HW) and CTCF are markedly different depending on whether the two co-bind with CP190; (3) many of the known insulator proteins sites do not function as robust enhancer blockers; and (4) at least in cultured cells the depletion of insulator proteins has a limited impact on genome-wide gene expression.
Combinatorial binding patterns
Classifications of combinatorial binding of insulator proteins have been described previously (Bushey et al. 2009; Nègre et al. 2010). These classifications relied on the overlapping of bound regions defined according to arbitrary statistical thresholds and the position of these regions relative to TSSs. Because they did not take into account the relative strengths of binding, such classifications grouped together binding sites with very different biochemical and functional properties.
In contrast, we define the persistent co-binding patterns based on the strength of binding of the associated proteins, treating regions strongly bound by a combination of proteins differently from regions at which the same proteins are detected according to a statistical threshold but where the extent of their binding is disproportional. We argue that our approach retains the information on biochemical interrelations between the co-bound proteins and separates the sites with different functional properties. The strongest support for our argument comes from RNAi knock-down experiments, which demonstrate that the effect of the loss of one insulator protein on the binding of another insulator protein is constrained to a specific class of co-bound regions. For example, the knock-down of SU(HW) results in the loss of CP190 from class 3 (gypsy-like) sites but not from class 9 (CTCF+CP190) or class 5 (BEAF-32+CP190) sites.
Our approach to select the sites representative of each co-binding class is conservative and inevitably excluded a fraction of binding sites from downstream analyses. For example, strong SU(HW) binding sites assigned to class 14 by initial overlap comparison (Fig. 1) were not analyzed further due the uncertainty of their co-binding by CP190. We therefore caution readers that our selection of representative binding sites (Supplemental Tables S1–S18) is not a complete genomic catalog, and advise to use the ChIP-chip binding profiles, deposited to GEO and modMINE, to gauge whether their locus of interest has a strong insulator protein binding site.
The role of CP190 in insulation
The prevailing model in the field suggests that CP190 is recruited to different insulator elements by DNA binding proteins where it serves as a universal adapter that mediates interactions between different insulator elements (Bushey et al. 2009). Our results present a more complex picture. First, RNAi knock-down experiments demonstrate that the binding of SU(HW) protein to class 3 (gypsy-like) sites is dependent on CP190, indicating that CP190 is not passively tethered to common sites by SU(HW) and instead plays an active role in recruitment and/or stabilization of the bound complex. Second, the sequence analysis of class 9 (CTCF+CP190) sites suggests that the binding of both proteins to these sites is likely due to the coincidence of cognate recognition sequences. Third, RNAi knock-down experiments indicate that BEAF-32 is dispensable for CP190 binding at shared sites. Clearly CP190 plays an active role in the selection of sites shared with SU(HW), CTCF, or BEAF-32. It is still possible that once it co-binds, or binds sufficiently close to another insulator protein, it may mediate the trans-interactions of the bound sites. However, such interactions would have to be rather transient, at least in cultured cells, as they are not easily detected in our ChIP-chip data.
The class of sites in which CP190 is not accompanied by any of the insulator proteins tested indicates the existence of a novel pathway of CP190 recruitment to chromatin.
Notably, our functional tests suggest that the sites employing this pathway (i.e., class 6 [standalone CP190] and class 5 [CP190+BEAF-32] sites) may constitute the major pool of robust insulator elements in flies. This conclusion is supported by functional analyses of Nègre et al. (2011), who found enhancer blocking activity by three DNA fragments that we would classify as class 6 (standalone CP190) sites and one fragment that we classify as a class 5 (CP190+BEAF-32) site. Interestingly, Nègre et al. (2011) found some degree of CTCF binding at these sites in embryos. Our results show that these sites bind no CTCF in S2 or BG3 cultured cells. Furthermore, unlike most CTCF binding sites (Fig. 2D), these regions contain no CTCF recognition motif. Whether and how such sites can actually recruit CTCF in embryonic cells but not in cultured cells will require further investigation.
Is transcriptional repression the primary function of SU(HW) protein?
SU(HW) is not required for Drosophila viability, but mutant flies display defective oogenesis and female sterility (Parkhurst et al. 1988). As follows from the experiments presented here and previously (Golovnin et al. 2003; Parnell et al. 2003; Kuhn-Parnell et al. 2008), the class 3 (gypsy-like) sites do not have a direct impact on gene promoters, and some can act as enhancer blockers. In contrast, our results show that standalone SU(HW) protein binding sites tend to repress transcription rather than insulate. Remarkably, a recent study indicates that neither CP190 nor MOD(MDG4)67.2 is required for oogenesis (Baxley et al. 2011), which suggests that the role of SU(HW) in the control of this process is distinct from its enhancer-blocking function. We hypothesize that the SU(HW) function critical for oogenesis is transcriptional repression exerted at standalone binding sites, consistent with the up-regulation of gene expression we observe after depletion of SU(HW) in cultured cells.
Insulator proteins and chromatin states
Previously, we have shown that the fly genome can be partitioned based on nine combinatorial patterns of 18 histone modifications (Kharchenko et al. 2011). Contrary to initial expectations, we find little correlation between the positions of insulator protein binding sites and the boundaries of these combinatorial chromatin states (data not shown). In agreement with this result, the transgenic tests suggest that only a small fraction of insulator protein binding sites can robustly block enhancer–promoter communication, and we see no major changes in gene expression after RNAi knock-down of insulator proteins. Taken together, these observations suggest that insulator proteins are unlikely to play a general role in partitioning of the fly genome into distinct domains of different chromatin states.
We realize that the incomplete loss of insulator proteins after RNAi knock-downs may cause an underestimate of the potential changes in gene expression and the extent of H3K27me3 domains. We believe the underestimated changes are likely to be few, and their accounting would not influence our overall conclusions. First, we note that even partial loss of insulator proteins from a site is sufficient to impair its ability to block the spreading of H3K27 methylation (Supplemental Fig. S9). Conversely, if all H3K27me3 domain borders at which the lack of expansion can be explained by the lack of significant reduction of insulator protein binding after corresponding RNAi are disregarded, the fraction of affected borders remains essentially the same. Second, it is clear that our statistical definition of the consistent reduction of binding to a site is conserved. As illustrated by Figure 3, B through E, most of the sites, even those not deemed to reduce the binding significantly (green dots on the scatter-plots), bind less insulator proteins after the corresponding RNAi. The sites that remain truly unaffected (data points on or above scatter-plot diagonals) are very few: 81, SU(HW) binding sites; 27, CTCF binding sites; one, CP190 binding site; and seven, BEAF-32 binding sites. These numbers are at least two orders of magnitude lower than the number of chromatin state partitions (Kharchenko et al. 2011) or active genes in BG3 cells (Cherbas et al. 2011). The disparity between the actual binding reduction and its statistical significance is greatest in the case of BEAF-32 and CTCF RNAi (Fig. 3C,3E) and is explained by the higher variability between the corresponding replicate experiments. Since our test to detect significant expression changes relies on replicate comparison, it may have underestimated the number of changes in these two cases. In an attempt to account for this, we have relaxed the detection criteria and looked for all measurable twofold changes irrespective of their statistical significance. As illustrated in Supplemental Figure S10, the numbers of expression changes increase but remain small (BEAF-32 RNAi, 16 up/36 down; CTCF RNAi, 28 up/34 down).
Perhaps not so important in the global scale, insulators may still be critical to restrict chromatin states at a limited set of sites. In fact, the ability of the gypsy insulator to shield reporter genes from Polycomb repression is well documented (Sigrist and Pirrotta 1997; Kahn et al. 2006; Comet et al. 2011, and the extension of endogenous H3K27me3 domains in CTCF and CP190 mutants has been reported (Bartkuhn et al. 2009). The results of genome-wide assays presented here confirm that insulators restrict the spreading of H3K27me3, but only at a small number of Polycomb target regions and only to prevent the repressive histone methylation of adjacent genes that are already transcriptionally inactive. While this has no obvious consequences in cultured cells, it may be important in the context of the developing embryo to ensure that genes in the vicinity of Polycomb targets do not become permanently repressed.
Methods
Cell culture conditions and RNAi
Cell lines were obtained from the Drosophila Genome Resource Center (DGRC) S2-DRSC cells (stock 181) and ML-DmBG3-c2 cells (DGRC, stock 68) and grown according to recommendations. The RNAi was performed as described by Schwartz et al. (2010). The sequences of PCR primers used to produce DNA template for dsRNA synthesis are indicated in the Supplemental Table S21.
Genome-wide mapping
The mapping of each protein was initially done in the chromatin of S2 cells using two different independently raised antibodies when available (for technical details, see Supplemental Table S22; Supplemental Fig. S11; Supplemental Text). Because of the high congruence between the two independent antibodies (Supplemental Fig. S12), just one was used to map the corresponding proteins in BG3 cells. Chromatin preparation, immunoprecipitation, microarray hybridization, and sequencing were done according to the method described by Kharchenko et al. (2011).
Additional details of experimental procedures and data analyses are indicated in Supplemental Methods.
Data access
All data sets reported in this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE32775, GSE32773, GSE32774, GSE20811, GSE20812, GSE20760, GSE32816, GSE32777, GSE32780, GSE32776, GSE32778, GSE20815, GSE20766, GSE20814, GSE32781, GSE32783, GSE32782, GSE20767, GSE32749, GSE20768, GSE32750, GSE20802, GSE23489, GSE32808, GSE32812, GSE32813, GSE32810, GSE20808, GSE20833, GSE20809, GSE32853, GSE25373, GSE32791, GSE32788, GSE32789, GSE32790, GSE32792) and modMINE (http://intermine.modencode.org/; Supplemental Table S23).
Acknowledgments
We thank D. Acevedo, S. Gadel, C. Kennedy, and Rutgers BRTC for excellent technical assistance. We thank R. Renkawitz, V. Corces, F. Karch, R. Dorn, and S. Small for generous gifts of antibodies, fly stocks, and plasmids. This work was supported by NIH modENCODE grant U01HG004258 (G.H.K., M.I.K., P.J.P., S.C.R.E., V.P.), Wenner-Gren Foundations (Y.B.S.), and the Russian Academy of Sciences program for Molecular and Cellular Biology (M.S.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.138156.112.
References
- Adryan B, Woerfel G, Birch-Machin I, Gao S, Quick M, Meadows L, Russell S, White R 2007. Genomic mapping of Suppressor of Hairy-wing binding sites in Drosophila. Genome Biol 88: R167 doi: 10.1186/gb-2007-8-8-r167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniahmad A, Steiner C, Köhne AC, Renkawitz R 1990. Modular structure of a chicken lysozyme silencer: Involvement of an unusual thyroid hormone receptor binding site. Cell 61: 505–514 [DOI] [PubMed] [Google Scholar]
- Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R 2009. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J 28: 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxley RM, Soshnev AA, Koryakov DE, Zhimulev IF, Geyer PK 2011. The role of the Suppressor of Hairy-wing insulator protein in Drosophila oogenesis. Dev Biol 356: 398–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K 2007. An optimized transgenesis system for Drosophila using germ-line-specific ϕC31 integrases. Proc Natl Acad Sci 104: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Ramos E, Corces VG 2009. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev 23: 1338–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas L, Willingham A, Zhang D, Yang L, Zou Y, Eads BD, Carlson JW, Landolin JM, Kapranov P, Dumais J, et al. 2011. The transcriptional diversity of 25 Drosophila cell lines. Genome Res 21: 301–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comet I, Schuettengruber B, Sexton T, Cavalli G 2011. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc Natl Acad Sci 108: 2294–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Vazquez J, Schedl P 1999. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev 13: 2098–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev PG, Gerasimova TI 1989. Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol Gen Genet 220: 121–126 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG 1998. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92: 511–521 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG 1995. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82: 587–597 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Lei EP, Bushey AM, Corces VG 2007. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell 28: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Corces VG 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 6: 1865–1873 [DOI] [PubMed] [Google Scholar]
- Golovnin A, Biryukova I, Romanova O, Silicheva M, Parshikov A, Savitskaya E, Pirrotta V, Georgiev P 2003. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development 130: 3249–3258 [DOI] [PubMed] [Google Scholar]
- Holdridge C, Dorsett D 1991. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol Cell Biol 11: 1894–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R 2007. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet 3: e112 doi: 10.1371/journal.pgen.0030112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn TG, Schwartz YB, Dellino GI, Pirrotta V 2006. Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J Biol Chem 281: 29064–29075 [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn-Parnell EJ, Helou C, Marion DJ, Gilmore BL, Parnell TJ, Wold MS, Geyer PK 2008. Investigation of the properties of non-gypsy suppressor of hairy-wing-binding sites. Genetics 179: 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci 103: 10684–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T 2007. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat Rev Genet 8: 104–115 [DOI] [PubMed] [Google Scholar]
- Li HB, Müller M, Bahechar IA, Kyrchanova O, Ohno K, Georgiev P, Pirrotta V 2011. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol Cell Biol 31: 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312: 269–272 [DOI] [PubMed] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH 1990. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5: 1743–1753 [PubMed] [Google Scholar]
- Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RA, Renkawitz-Pohl R, Saumweber H, et al. 2007. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J 26: 4203–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, et al. 2005. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep 6: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet 6: e1000814 doi: 10.1371/journal.pgen.1000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Lei EP, Ghosh D, Corces VG 2004. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell 16: 737–748 [DOI] [PubMed] [Google Scholar]
- Parkhurst SM, Harrison DA, Remington MP, Spana C, Kelley RL, Coyne RS, Corces VG 1988. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev 2: 1205–1215 [DOI] [PubMed] [Google Scholar]
- Parnell TJ, Viering MM, Skjesol A, Helou C, Kuhn EJ, Geyer PK 2003. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc Natl Acad Sci 100: 13436–13441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Clay I, Fraser P 2010. The transcriptional interactome: Gene expression in 3D. Curr Opin Genet Dev 20: 127–133 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V 2010. Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet 6: e1000805 doi: 10.1371/journal.pgen.1000805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJ, Pirrotta V 1997. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana C, Corces VG 1990. DNA bending is a determinant of binding specificity for a Drosophila zinc finger protein. Genes Dev 4: 1505–1515 [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W 2006. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev 20: 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardy A, Maine E, Schedl P 1985. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol 185: 341–358 [DOI] [PubMed] [Google Scholar]
- Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, Jones KC, Corces VG 2011. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol Cell 44: 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Hart CM, Laemmli UK 1995. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81: 879–889 [DOI] [PubMed] [Google Scholar]