Abstract
Understanding the relationship between genome organization and expression is central to understanding genome function. Closely apposed genes in a head-to-head orientation share the same upstream region and are likely to be coregulated. Here we identify the Drosophila BEAF-32 insulator as a cis regulatory element separating close head-to-head genes with different transcription regulation modes. We then compare the binding landscapes of the BEAF-32 insulator protein in four different Drosophila genomes and highlight the evolutionarily conserved presence of this protein between close adjacent genes. We find that changes in binding of BEAF-32 to sites in the genome of different Drosophila species correlate with alterations in genome organization caused by DNA rearrangements or genome size expansion. The cross-talk between BEAF-32 genomic distribution and genome organization contributes to new gene-expression profiles, which in turn translate into specific and distinct phenotypes. The results suggest a mechanism for the establishment of differences in transcription patterns during evolution.
Eukaryotic genomes are not organized randomly. Rather, genes and their regulatory elements are arranged in a manner that allows for the correct function of the genome. Genes that show similar function or expression tend to be clustered on the chromosomes (Kamath et al. 2003; Pal and Hurst 2003; Hurst et al. 2004; Batada and Hurst 2007), but not all adjacent genes are coregulated. One interesting feature of eukaryotic genomes is the head-to-head juxtaposition of genes with two adjacent transcription start sites (TSS). Approximately 10% of genes in vertebrates are arranged in a head-to-head orientation and located closer than 1 kb from each other (Adachi and Lieber 2002; Yang et al. 2008). The proportion of close head-to-head gene pairs in the genome correlates with gene density (Li et al. 2006; Yang and Yu 2009), and Drosophila shows a higher than expected proportion of genes in this type of arrangement (Koyanagi et al. 2005; Yang and Yu 2009). The intergenic regions of close head-to-head gene pairs are referred to as bidirectional promoters, indicating that the two TSS's are close enough to share the same upstream regulatory region (Adachi and Lieber 2002; Koyanagi et al. 2005). Genes positioned in a head-to-head orientation show overall higher correlation of expression than those arranged in other orientations (Herr and Harris 2004; Yang and Yu 2009). However, there are also head-to-head gene pairs whose expression is not correlated or is negatively correlated in both humans and Drosophila (Herr and Harris 2004; Li et al. 2006). Interestingly, close head-to-head gene pairs in Drosophila species tend to have higher rearrangement rates during evolution (Weber and Hurst 2011), suggesting that they are not constrained in their genomic location and that they do not share common regulatory sequences. Drosophila must then possess mechanisms to functionally separate closely apposed genes in a head-to-head orientation in order for these genes to be independently regulated.
Insulators have been shown to contribute to the establishment of specific patterns of chromatin organization important for regulation of transcription by, at least in part, regulating interactions between enhancers and promoters (Phillips and Corces 2009; Handoko et al. 2011; Yang and Corces 2011). In Drosophila there are several types of insulators differentially distributed throughout the genome in a manner suggestive of distinct functions in gene expression (Bushey et al. 2009; Nègre et al. 2010). BEAF-32 is the DNA-binding protein for one of these insulators with a role in the recruitment of other components to specific sites in the genome. In D. melanogaster, BEAF-32 associates preferentially with actively transcribed genes, although the specific mechanism by which it affects gene expression is not known (Bushey et al. 2009; Jiang et al. 2009). Here we identify the BEAF-32 insulator as a cis element located between head-to-head genes to attain differential regulation of transcription. Changes in cis regulatory sequences represent an important source of variability necessary for divergence between species (Borneman et al. 2007; Odom et al. 2007; Schmidt et al. 2010). A large number of chromosome rearrangements have taken place during Drosophila speciation that have resulted in changes in the location of genes in the genome (Drosophila 12 Genomes Consortium 2007). Given the presence of the BEAF-32 insulator between close head-to-head gene pairs, we mapped the binding profiles of BEAF-32 in different Drosophila species and investigated changes in the pattern of BEAF-32 localization during the evolution of Drosophila species. Comparison between changes in BEAF-32 insulator distribution and gene location in different Drosophila species enabled us to establish correlations between changes in genome organization and function.
Results
BEAF-32 specifically associates with close head-to-head gene pairs
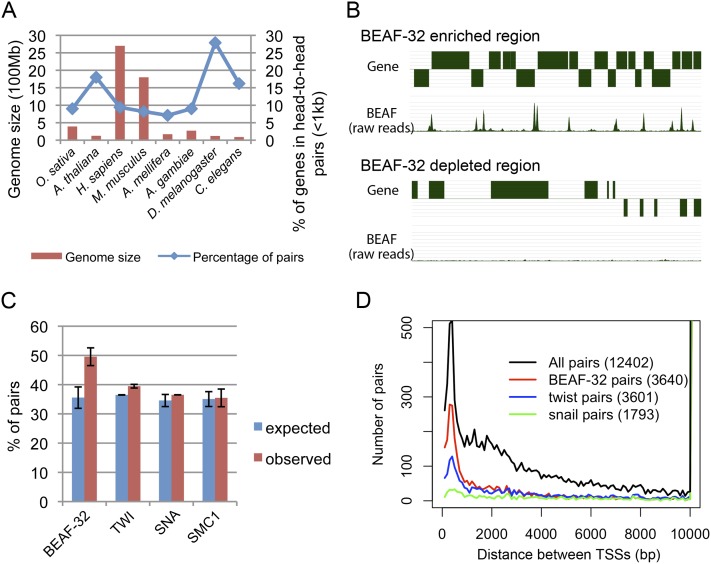
We first used the latest annotation of the D. melanogaster genome to examine the frequency of gene pairs. We found that 28% of genes are in a head-to-head orientation with intergenic regions shorter than 1 kb. This fraction is much higher than that found in other eukaryotes, including other insect species, in which the proportion of close head-to-head gene pairs ranges between 8% and 18% (Fig. 1A; Supplemental Table S1; Li et al. 2006; Dhadi et al. 2009). It is unlikely that such large numbers of genes are coregulated in Drosophila but not in other species, suggesting the existence of Drosophila-specific mechanisms to maintain independent regulation of close head-to-head gene pairs. Insulators are good candidates to perform such function given their ability to regulate enhancer–promoter interactions. More specifically, the BEAF-32 insulator protein is highly conserved in Drosophila and its presence appears to be restricted to this genus (Schoborg and Labrador 2010). We therefore examined the genome-wide distribution of BEAF-32 in D. melanogaster embryos using ChIP-seq, and found BEAF-32 frequently located between close adjacent genes oriented head-to-head (Fig. 1B). This is consistent with previous reports suggesting that ∼50% of BEAF-32-associated genes are arranged in a head-to-head orientation (Jiang et al. 2009). Based on the genomic distribution of BEAF-32 relative to genes, 50% is significantly greater than expected (P < 1 × 10−4) (Fig. 1C; Supplemental Fig. S1). This enrichment is unique to BEAF-32, but not to transcription factors, factors for general transcription, other promoter-associated factors, or other insulator proteins (Fig. 1C; Supplemental Fig. S2).
Figure 1.
BEAF-32 specifically associates with close head-to-head gene pairs. (A) Genome size and percentage of genes in head-to-head gene pairs in different eukaryotic genomes. There is a high proportion of head-to-head gene pairs in the compact D. melanogaster genome compared with other species. (B) Snapshot of two regions of the D. melanogaster genome showing BEAF-32 binding sites associate with close head-to-head gene pairs. The top track represents genes. Genes above the line are transcribed from the plus strand and genes below the line are transcribed from the minus strand. The bottom track represents sites of BEAF-32 localization in the region; signal corresponds to the number of raw reads from ChIP-seq analysis. (C) Percentage of head-to-head gene pairs flanking different proteins. BEAF-32 associated pairs are significantly enriched for head-to-head gene pairs compared with the genome-wide expectation as well as compared with other proteins. The error bars are from the results of different data sets. The expected and observed fraction of gene pairs was calculated independently for each data set, and the mean and standard deviation were then determined. For BEAF-32, we used data sets obtained using embryos from this study and modEncode, and S2 cells. For TWI or SNA, we used data sets from different biological repeats. For SMC1, we used data sets for cell lines Kc, S2, and Bg3. (D) Distribution of distances between TSS's for genes flanking BEAF-32 and transcription factors. The number in parentheses is the total number of gene pairs in each category. BEAF-32 frequently associates with adjacent gene pairs close to each other.
One consequence of the arrangement of genes in pairs in a head-to-head orientation is shorter distances between the TSS's compared with other possible orientations (head-to-tail, tail-to-tail, or tail-to-head) (Supplemental Fig. S3A–B). We thus examined the distance between TSS's flanking BEAF-32 binding sites in D. melanogaster and confirmed that BEAF-32-associated TSS's have a close neighboring TSS. The distance between the two TSS's peaked at 300–400 bp (Fig. 1D; Supplemental Fig. S3C). A total of 66% (1042/1563) of close head-to-head gene pairs (distance <500 bp) contain BEAF-32 binding sites, while only 36% (506/1400) distant head-to-head gene pairs (distance >1 kb) contain BEAF-32 binding sites between the two genes (P < 1 × 10−4). Thus, BEAF-32 preferentially associates with close head-to-head gene pairs.
BEAF-32-associated close head-to-head gene pairs are not coexpressed
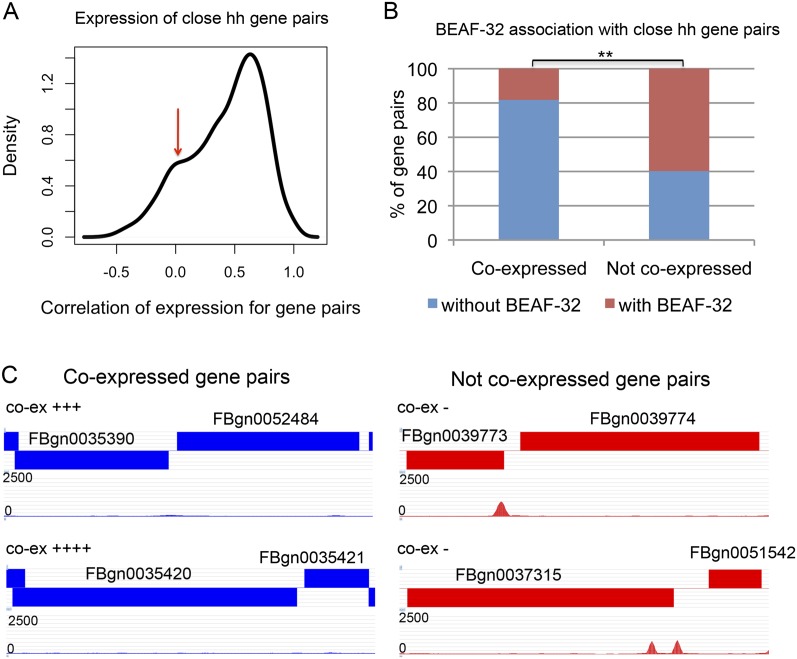
Close head-to-head genes tend to be coregulated in D. melanogaster compared with distant head-to-head genes or genes not in a head-to-head orientation, as there is a higher proportion of coexpression for close head-to-head gene pairs (Fig. 2A; Supplemental Fig. S4). However, the correlation in expression for the two genes in close head-to-head gene pairs is spread over a broad range. In addition to a peak of high correlation, the distribution also shows a second peak at a value indicative of no correlation (Fig. 2A). Therefore, there are close head-to-head gene pairs whose expression is not correlated or is negatively correlated (correlation <0.1). For these gene pairs, ∼60% (150/251) have BEAF-32 binding sites between the genes. In contrast, <20% (6/33) of highly coexpressed gene pairs (correlation >0.9) have BEAF-32 (P < 4 × 10−5). Over 80% (27/33) of highly correlated head-to-head genes do not harbor BEAF-32 binding sites between them. Herr and colleagues have examined the coexpression of eight head-to-head gene pairs spatially and temporally during embryonic stages of Drosophila development (Brogiolo et al. 2001; Renault et al. 2002; Herr et al. 2003, 2004; Herr and Harris 2004). For the two gene pairs found to be highly coexpressed, we examined the presence of BEAF-32 and found that there is no BEAF-32 binding signal between the genes. A similar analysis shows that BEAF-32 is present in the two gene pairs containing genes that are expressed differently (Fig. 2C; Supplemental Table S2). These results suggest a correlation between the presence of BEAF-32 between two close adjacent genes and their ability to be independently regulated.
Figure 2.
BEAF-32-associated close head-to-head genes are not coexpressed. (A) Distribution of the correlation of expression for the two genes in close head-to-head gene pairs (distance <500 bp). (Red arrow) Secondary peak for enrichment of genes that are not coregulated. (B) Percentage of gene pairs associated or not associated with BEAF-32 binding sites present between coexpressed and non-coexpressed genes in close head-to-head gene pairs. (C) Examples of BEAF-32 location in coexpressed and non-coexpressed gene pairs. The blocks indicate genes with FlyBase IDs. Blocks on top of the track are transcribed from the plus strand, and blocks at the bottom of the track are transcribed from the minus strand. The tracks under the gene tracks show the location of BEAF-32 signal with raw reads from ChIP-seq. The symbol “co-ex” represents the level of coexpression between the two genes. Detailed information about the expression of these genes is presented in Supplemental Table S2.
BEAF-32 separates close head-to-head genes with different patterns of transcription regulation
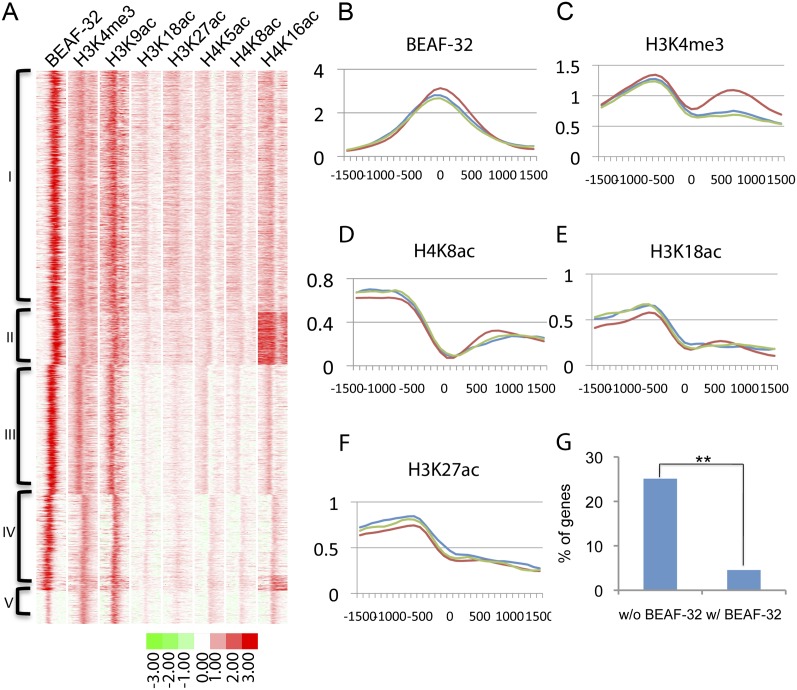
To understand the mechanisms by which the presence of BEAF-32 allows genes to be differentially regulated, we compared the distribution of BEAF-32 binding sites with the mapped landscape of histone modifications in the D. melanogaster genome (Kharchenko et al. 2011; Negre et al. 2011). We aligned the map of BEAF-32 binding sites with histone modification data, both obtained in S2 cells (Fig. 3A; Supplemental Fig. S5). Consistent with the association between BEAF-32 and active genes, we found that BEAF-32 clusters with active histone marks and not with repressive marks. Histone marks for active TSS's, such as H3K4me3, are present on both sides of BEAF-32 binding sites between pairs of genes oriented head to head (Fig. 3B,C). Interestingly, histone modifications such as H4K8ac, H3K18ac, and H3K27ac are present at only one of the TSS's of the two genes in each pair, and the signal is reduced to background levels at the other TSS (Fig. 3D–F). In Drosophila, H3K18ac and H3K27ac are thought to be produced by the acetyltransferase CREB binding protein (CBP), which is present at enhancers and promoters (Tie et al. 2009). The presence of these histone modifications adjacent to TSS's is suggestive of interactions between the enhancer and promoter that lead to activation of transcription. Thus, the asymmetric distribution of histone marks at the two TSS's suggests that BEAF-32 may separate two genes that are differentially transcribed, even though they share the same upstream region.
Figure 3.
BEAF-32 separates close head-to-head gene pairs to achieve differential regulation of transcription. (A) Alignment and clustering of BEAF-32 and histone modifications in D. melanogaster S2 cells. All sites were identified and aligned using ChromaSig. Each lane represents a 3-kb region. Clusters I–V grouped here are clusters with BEAF-32 binding from Supplemental Figure S5. (B–F) Mean value of enrichment for sites in cluster I. Site 0 is the site where BEAF-32 is enriched. Each figure represents a 3-kb region flanking site 0. Each colored line represents a different type of gene pair arrangement case in cluster I: head-to-head (red), tail-to-head (blue), and head-to-tail (green). (G) Fraction of gene pairs whose expression changes significantly in the same up or down direction in Sox14 mutant animals. Changes are considered significant when the difference is at least threefold.
If this conclusion is true, changing the effect of putative regulatory sequences located in the intergenic region would only affect one of the genes but not the other. However, if the two genes in a pair are not separately regulated, they are likely to respond in the same way to changes in regulation. To test this hypothesis, we examined changes in the transcription profile resulting from mutations in SOX14, which is a D. melanogaster transcription factor (Ritter and Beckstead 2010). Among the 271 genes not associated with BEAF-32, 68 (25%) change their transcription in the same direction as their neighbor when SOX14 is mutant. However, only four out of 88 (4.5%) of BEAF-32-associated genes change simultaneously with their neighbor (P < 2 × 10−5), a fivefold difference with respect to genes not associated with BEAF-32 (Fig. 3G).
Other histone modifications characteristic of transcription activation, such as H4K5ac, H4K8ac, and H4K16ac, are also distributed differently at the two sides of BEAF-32 binding sites (Fig. 3A,D). H4K5ac has been reported as a histone modification present in genes bookmarked during mitosis (Zhao et al. 2011) and H4K16ac is the product of the acetyltransferase MOF, which functions at enhancers and promoters of X-linked and autosomal genes (Zippo et al. 2009). Both modifications are indicative of transcription activation, enforcing the conclusion that BEAF-32 is present between close head-to-head genes in small genomes, such as those of Drosophila species, to separate the TSS's of two different genes that need to be differentially regulated.
Conservation and diversity of BEAF-32 insulators across the Drosophila species
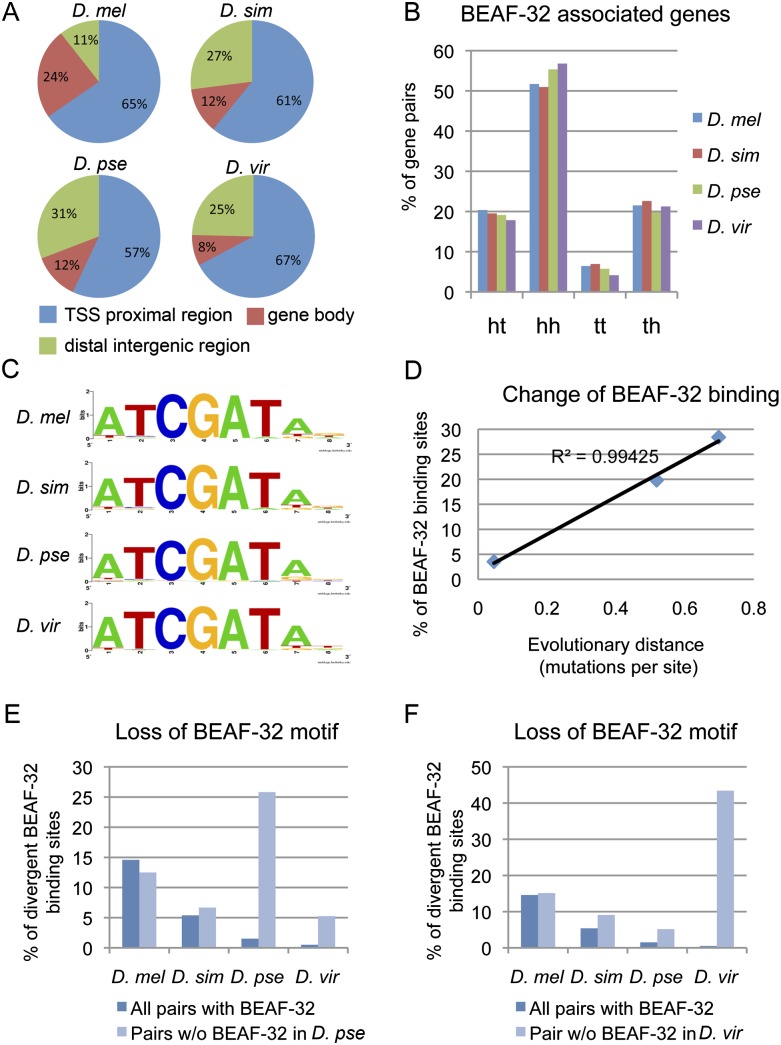
Since BEAF-32 appears to functionally separate close head-to-head genes, gain or loss of BEAF-32 binding during evolution may prevent or allow adjacent genes to be affected by neighboring regulatory sequences, leading to changes in gene expression. In order to investigate the role of BEAF-32 during the evolution of the Drosophila species, we systematically compared its binding site distribution in four Drosophila genomes, D. melanogaster, D. simulans, D. pseudoobscura, and D. virilis. For the larger genome of species such as D. virilis, we sequenced twice the number of tags as in D. melanogaster to reach equal coverage for all of the genomes studied (Supplemental Table S3). Genome wide, BEAF-32 shows a similar binding distribution with respect to TSS's, gene bodies, and intergenic regions across the four species, with a preference for sequences close to TSS's (Fig. 4A). The association of BEAF-32 with head-to-head gene pairs is conserved in all four species (Fig. 4B), suggesting a conserved function in Drosophila. The consensus motifs identified for BEAF-32 binding sites are virtually identical among the four species (Fig. 4C), consistent with the protein conservation, particularly in the DNA-binding domain.
Figure 4.
Conservation and divergence of BEAF-32 sites in Drosophila species. (A) Distribution of BEAF-32 binding sites with respect to various gene landmarks. (B) Percentage of various gene pair combinations flanking BEAF-32 binding sites; (ht) head-to-tail; (hh) head-to-head; (tt) tail-to-tail; (th) tail-to-head. BEAF-32 association with head-to-head gene pairs is conserved. (C) Consensus motif for BEAF-32 occupied sequences in the four species. (D) Correlation of BEAF-32 binding divergence with evolutionary distance between D. melanogaster and other species. (Y-axis) Percentage of BEAF-32 binding sites that are not conserved in the other three species with respect to all BEAF-32 binding sites in D. melanogaster. (E,F) Species-specific loss of BEAF-32 binding associates with species-specific loss of the BEAF-32 motif in the DNA sequence. (Dark blue bars) Background absence of the BEAF-32 motif for all BEAF-32-associated gene pairs in each species. (Light blue bars) Absence of the BEAF-32 motif for a group of gene pairs with BEAF-32 binding lost only in D. pseudoobscura (E) or only in D. virilis (F).
Since BEAF-32 is significantly associated with gene pairs, in order to investigate changes in the profile of BEAF-32 binding in the genome of different Drosophila species we developed a gene-pair centric analysis pipeline. To do so, we examined whether a BEAF-32 binding site present in the intergenic region of a gene pair in one species was also present in the corresponding intergenic region of the gene pair in the second species (see Methods). With this pipeline, we pooled all of the BEAF-32 binding sites from the four species and scored the presence of BEAF-32 at each site in each of the species (Supplemental Table S4). Using Cluster 3.0, we then clustered the pattern of BEAF-32 binding among the four species. The results of this clustering agree well with the evolutionary tree of these species (Supplemental Fig. S6A).
To quantitatively investigate differences in the distribution of BEAF-32 sites between species using this pipeline, we then analyzed the conservation of BEAF-32 binding between D. melanogaster, which has the best-annotated genome, and other species. In this D. melanogaster centric comparison we examined the occupancy of BEAF-32 on orthologous chromosomal regions between D. melanogaster and each of the three other species (Supplemental Table S5). The fraction of nonconserved binding sites ranges from 3% in D. simulans to 29% in D. virilis (Supplemental Fig. S6B). The difference increases appropriately with the molecular distance between these genomes, suggesting that the divergence follows the molecular clock. This relationship fits a simple linear regression, with an estimated divergence rate of BEAF-32 binding of 0.6% per Myr (R2 > 0.99) (Fig. 4D). This estimate may be affected by the quality of the data, although ChIP-seq gives relatively low false-positive or negative results. This divergence rate is higher than the nonsynonymous nucleotide substitution rate of 0.4% per Myr, but lower than the synonymous nucleotide substitution rate of 6.34% per Myr (Drosophila 12 Genomes Consortium 2007), suggesting that the binding of BEAF-32 in the genome is under selection. We thus examined possible changes in the DNA sequence at BEAF-32 binding sites. Since the motif for BEAF-32 binding is conserved in the four Drosophila species (Fig. 4C), we searched for the presence of this motif at the orthologous regions in their genomes. The results confirm changes in the DNA sequence consistent with the loss of the BEAF-32 binding motif, specifically in the species where BEAF-32 binding is lost (Fig. 4E,F). Thus, the function of BEAF-32 is conserved in the Drosophila species, but gain or loss of specific binding sites is under selection during the evolution of these species.
Changes of BEAF-32 insulator localization correlate with alterations in genome organization during Drosophila evolution
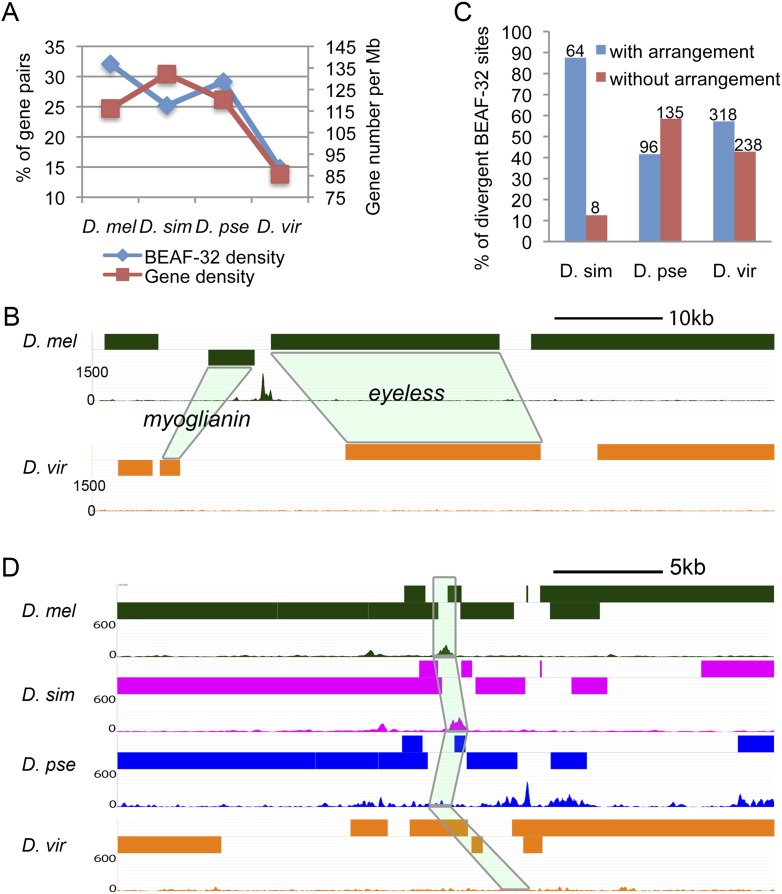
Two obvious changes affecting Drosophila genomes during evolution are alterations in genome size and chromosome rearrangements. How does BEAF-32 contribute to the function of the genome after such changes? Since BEAF-32 is preferentially located between close divergently transcribed genes, BEAF-32 binding sites may change along with variations of distance between the genes. The four Drosophila species examined show differences in gene density across their genomes. Compared with D. melanogaster, the genome size of D. virilis is 46% larger and gene density decreases from 116 to 85 genes per megabase (Drosophila 12 Genomes Consortium 2007). At the same time, 32% of all gene pairs have BEAF-32 binding sites in D. melanogaster, whereas the fraction is reduced to 15% in D. virilis (Fig. 5A). For example, the intergenic region between the genes myoglianin and eyeless contains a functional BEAF-32 binding site in D. melanogaster (Sultana et al. 2011), but not in D. virilis. The distance between the two TSS's increased 10 times in D. virilis, and this change correlates with the loss of the BEAF-32 binding site in this species or the gain in D. melanogaster (Fig. 5B). For D. simulans and D. pseudoobscura, the fraction of gene pairs remains around 28%, and their gene density is similar to that of D. melanogaster (Fig. 5A). Therefore, BEAF-32 may be recruited to intergenic regions between close TSS's when the distance between the two genes decreases, or may be lost when the distance between genes increases.
Figure 5.
Changes in BEAF-32 binding correlate with changes in genome organization and function. (A) BEAF-32 density decreases as gene density declines. (Left y-axis) Percentage of gene pairs containing BEAF-32 with respect to all well-mapped gene pairs and is a measure of BEAF-32 density. (Right y-axis) Number of genes per megabase as a measure of gene density. (B) Arrangement of the myoglianin and eyeless genes and location of BEAF-32 binding sites. Light-green shadowing indicates the orthologous genes in D. virilis. (C) Percentage of divergent BEAF-32 binding sites that either associate or do not associate with chromosome rearrangement between D. melanogaster and the species listed. The numbers above the bar indicate the number of cases in each category. (D) An example of gene arrangement and location of BEAF-32 binding sites in a region whose mutation affects body size in D. melanogaster is shown for four different species. The mutation affecting body size results in alteration of sequences in the intergenic region encompassing the BEAF-32 binding site in D. melanogaster. Light-green shadowing represents the four orthologous regions in each of the Drosophila species.
When we examined the association between nonconserved BEAF-32 sites and chromosome rearrangements we found two types of nonconserved BEAF-32 binding sites. For the first type, the changes of BEAF-32 binding co-occur with chromosomal rearrangements, since the genes flanking these BEAF-32 binding sites have different neighbors in the two species. In this case, BEAF-32 binding is gained or lost when the arrangement between gene pairs is altered. There are 87%, 41%, and 55% nonconserved BEAF-32 binding sites at regions where genes have been rearranged in D. simulans, D. pseudoobscura, and D. virilis, respectively (Fig. 5C). Most nonconserved binding sites in D. simulans are of the first type. For the second type of nonconserved BEAF-32 binding sites, gain/loss of binding sites does not associate with changes in chromosomal organization, as they are located at intergenic regions between the same gene pairs in the two species being compared. There are only nine (13%) nonconserved binding sites of the second type in D. simulans. However, D. pseudoobscura and D. virilis show a higher frequency of changes in BEAF-32 binding not associated with rearrangements compared with D. melanogaster; the number of these events are 145 (59%) and 308 (45%), respectively (Fig. 5C). Phenotypically, D. simulans looks more like D. melanogaster, while the other two species are more different. The results may suggest that the first type of nonconserved binding sites may help maintain proper expression patterns in newly rearranged genes, whereas the second type may result in alterations in the regulation of transcription of flanking genes that may contribute to phenotypic differences between the species.
Alterations in BEAF-32 insulator localization correlate with changes of genome function during Drosophila evolution
Genes flanking BEAF-32 sites are preferentially involved in metabolic processes that are also known to affect body size (Carreira et al. 2008; Bushey et al. 2009). Thus, it is possible that changes in gene expression arising as a consequence of the gain or loss of BEAF-32 binding may affect body size, which is also one of the most obvious phenotypic differences among the Drosophila species studied. In a screen for mutations that alter body size in D. melanogaster, 26 mutations were identified containing P-element insertions in intergenic regions (Carreira et al. 2008). We examined these mutations and found that eight map to regions containing BEAF-32 binding sites. Out of these eight regions, six (75%) show loss of BEAF-32 binding in D. virilis (Fig. 5D; Supplemental Table S6,). These changes in BEAF-32 binding cannot be explained solely by the increase in genome size and distance between genes that took place in D. virilis, as the 75% difference is significantly higher than the overall genome-wide difference for BEAF-32 binding (29%) between the two species (P < 4 × 10−3). For these intergenic regions, the distance between genes has not changed appreciably, but BEAF-32 binding is lost in D. virilis compared with D. melanogaster. Gain or loss of BEAF-32 binding may alter the expression of one or more genes flanking BEAF-32 sites in this region, which may lead to changes in body size.
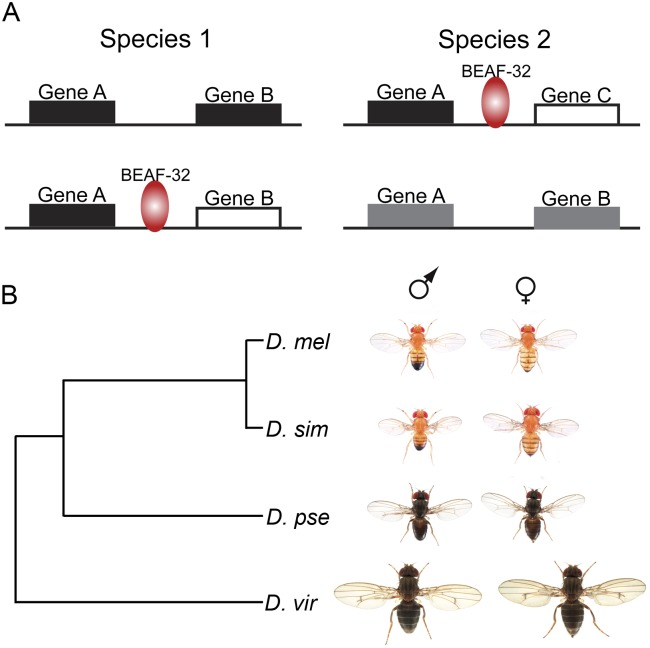
Thus, during Drosophila evolution, BEAF-32 binding sites are gained or lost with or without change in gene location, to either maintain transcription or allow for diversity. After the genomic location of genes is altered, genes may be brought close to a new neighboring gene, and the proximity to new regulatory sequences in the adjacent gene may alter their expression pattern. The presence of BEAF-32 binding sites may permit the two new neighboring genes to maintain their original expression patterns (Fig. 6). In addition, in the absence of chromosome rearrangements, alterations in BEAF-32 binding may result in changes in the expression profile of one or more genes, resulting in the appearance of new complex traits, such as those affecting body size (Fig. 6). Therefore, other than evolutionary changes of protein structure and function, which are certainly contributing to phenotypic divergence among species, gene expression in general also underlies phenotypic diversity.
Figure 6.
Simplified models for the role of BEAF-32 during evolution of Drosophila species. (A) Two alternative possibilities explaining how alterations in BEAF-32 binding may affect transcription. Blocks indicate genes. (Black and white) Different transcription regulatory modes of the genes. (Gray) Converged regulation for the two genes. Gain or loss of BEAF-32 binding when gene pairs are reorganized to maintain proper transcription (top). Gain or loss of BEAF-32 binding when gene organization does not change to create transcription diversity (bottom). (B) Phylogeny and phenotype of Drosophila species analyzed in this study.
Discussion
Here we show that the presence of BEAF-32 between close adjacent genes arranged in a head-to-head orientation correlates with different transcription regulatory patterns in the two genes of the pair in Drosophila. Close head-to-head gene pairs exist in almost all eukaryotes, but it is not known whether other species also use this strategy in order to maintain independent regulation of adjacent genes. In humans, genes present in head-to-head gene pairs also show a bimodal distribution in the correlation of expression (Li et al. 2006). In addition to the peak indicative of high correlation, there is also a peak of enrichment of gene pairs whose expression is not correlated. For these pairs, it is reasonable to predict the existence of regulatory mechanisms that functionally separate the two genes in order to attain the observed differential transcription. BEAF-32 is restricted to the Drosophila species (Schoborg and Labrador 2010), and mammalian cells may use other insulator proteins to accomplish this goal. In Drosophila there are several types of insulator elements that show different genomic distributions with respect to genes (Bushey et al. 2009; Nègre et al. 2010). The distribution of the dCTCF insulator partially overlaps that of BEAF-32. Since CTCF is conserved between Drosophila and humans (Moon et al. 2005; Schoborg and Labrador 2010), it is possible that this protein functionally replaces BEAF-32 in maintaining differential transcription programs in genes located in close head-to-head gene pairs. When the human genome was specifically examined for the organization of close head-to-head gene pairs, those containing CTCF showed lower correlation of expression, suggesting that this mechanism may be also conserved in humans (Xie et al. 2007).
The organization of the genome that provides the highest fitness should be selected during evolution. If coexpression of close head-to-head gene pairs provides lower fitness, selection should favor rearrangements that result in physical or functional separation of the two genes. A comparative analysis of head-to-head gene pairs in different species revealed that these pairs are more conserved in vertebrate lineage than in Drosophila species (Yang and Yu 2009; Weber and Hurst 2011). Drosophila has more close head-to-head gene pairs than mammals, but the conservation of these pairs is threefold lower (Yang and Yu 2009). This suggests that some of the head-to-head gene pairs in Drosophila arise from genome compaction rather than selection for this specific organization. For these gene pairs, maximum fitness will select for separation of the genes in order to attain differential expression of the two genes in the pair. One strategy to accomplish this is functional separation by recruiting insulator proteins. Alternatively, chromosomal rearrangements may physically separate the two genes. However, in an already compact genome like that of Drosophila, it may be difficult to organize all non-coexpressed genes apart from each other. Thus, a strategy relying on functionally separating the members of head-to-head gene pairs may be more effective. Our analysis has concentrated on close adjacent genes that are divergently transcribed, because this arrangement facilitates analysis of the correlation between the location of BEAF-32 and transcription patterns of the two genes. Nevertheless, 38% of BEAF-32 binding sites associate with non-head-to-head gene pairs. It is possible that BEAF-32 plays a similar role in this situation in order to control interactions between regulatory sequences located in the 3′ region or introns of genes and adjacent promoters from other genes. Although information on the location of regulatory sequences in the Drosophila genome is becoming available, it is not yet known which sequences regulate which genes. In the absence of this information, it is not possible at this time to evaluate the possible role of BEAF-32 in maintaining independent regulation of genes that are far apart and not in a head-to-head orientation.
The organization of head-to-head gene pairs in both humans and Drosophila is conserved during evolution, but the two members of each pair are not precisely coregulated. The distribution of expression correlation suggests that most gene pairs do not show either high correlation or no correlation, but rather a relative level of correlation (Fig. 2A; Li et al. 2006), suggesting that they may be coregulated in certain developmental stages or specific tissues. Coexpression is still important for the genes, but they are not coregulated all of the time. Thus, the head-to-head orientation needs to be maintained for coexpression, but it is also necessary to separate genes when they are not coregulated. The profiles of genome distribution of different insulator proteins in different cell types suggest a certain degree of cell-type specificity in both humans and Drosophila (Kim et al. 2007; Bushey et al. 2009; Cuddapah et al. 2009). These observations point to a role for insulators in coordinating genome organization and function during evolution.
Methods
Fly stocks and other reagents
Oregon-R was used as the wild-type strain for D. melanogaster. Strains for other species were obtained from the UC San Diego Drosophila Species Stock Center. Stock numbers are ID 14021-0251.195 for D. simulans, ID 14011-0121.94 for D. pseudoobscura, and ID 15010-1051.87 for D. virilis. Flies were grown at 25°C. BEAF-32B is the main BEAF-32 isoform and its sequence is highly conserved (Supplemental Fig. S7). BEAF-32B antibodies were generated against amino acids 1–83 of BEAF-32B in D. melanogaster (Bushey et al. 2009). The polyclonal antibody cross-reacts with BEAF-32 orthologs in other Drosophila species and recognizes specific bands on polytene chromosome squashes from salivary glands of the species examined (Supplemental Fig. S8).
ChIP-seq
Chromatin IP was carried out using embryos. To match specific developmental stages for embryos from each species, we determined the collecting time based on the length of the life cycle of the various species (Markow and O'Grady 2005). The age of the embryos used for chromatin immunoprecipitation was 0–8 h for D. melanogaster, 0–8 h for D. simulans, 0–10 h for D. pseudoobscura, and 0–12 h for D. virilis. ChIP was performed following published procedures (Sandmann et al. 2007) with the following adjustments. Two grams of embryos were used for two chromatin preparations and extracts were sonicated 20 cycles (10 sec on/30 sec off) on a Branson Sonifier 250 with output control set at 1.5. Libraries were prepared with the Illumina TruSeq DNA Sample Preparation Kit and sequenced at the HudsonAlpha Institute for Biotechnology.
Sequence analysis
For analysis of sequence data, we used genome sequence and annotations released on FlyBase, dmel_r5.39, dsim_r1.3, dpse_r2.22, and dvir_r1.2. Sequences were aligned to genomes using Bowtie with indexes built for each genome. The output map files were converted to bed format for each chromosome arm using the VancouverShort package. Only aligned reads on the main chromosomes were used to call peaks, as the small chromosome segments are not well annotated. The main chromosomes include D. melanogaster: chr2L, chr2LHet, chr2R, chr2RHet, chr3L, chr3LHet, chr3R, chr3RHet, chr4, and chrX; D. simulans: chr2L, chr2R, chr3L, chr3R, chr4, and chrX; D. pseudoobscura: chr2, chr3, chr4_group1-5, chrXL_group1a/1e/3a/3b, chrXR_group3a/5/6/8; D. virilis: scaffolds with more than 1000 reads. Peaks were called using CCAT3.0. BEAF-32 associated genes were defined as genes closest to each peak, and BEAF-32 associated pairs were defined as nonoverlapping gene pairs flanking each peak. Both genes in a gene pair are defined as BEAF-32 associated genes if they flank BEAF-32 binding sites and are arranged in a head-to-head orientation. For other orientations, only the closest gene is defined as a BEAF-32 associated gene. Overlapping gene pairs were discarded. Associated genes or pairs were called using a custom script (available upon request). Only pairs with well-mapped intergenic regions and a gap of <10% of the length of the region or 300 bp were defined as well-mapped pairs.
Fraction of genes in head-to-head gene pairs in different species
Genome annotations for each species were downloaded from the UCSC Genome Browser. For genes with alternative transcripts, only the longest transcript was considered for analysis. We then created a list of all possible nonoverlapping gene pairs and counted the number of unique genes in all gene pairs as A. We then selected the gene pairs in head-to-head combination and closer than 1 kb. We counted the number of unique genes in these head-to-head gene pairs as B. The fraction of genes in head-to-head gene pairs is B/A. For D. melanogaster, we used four different annotation versions. The flybase-dmel5.39 annotation includes both coding and noncoding genes, and the others include only coding genes. The results are the same for all different versions. We carried out a similar analysis with the latest genome annotation for H. sapiens and M. musculus. The results are comparable to the values previously reported (Supplemental Table S1). Values for genome size and percentage of genes in head-to-head orientation shown in Figure 1A for H. sapiens, M. musculus, O. sativa, and A. thaliana were obtained from the literature.
Calculation of the fraction of gene pair combinations associated with various proteins
To calculate the expected fraction, we call P1 the fraction of TSS's containing binding sites for a specific protein located in the 500-bp upstream region of a gene in the genome, and P2 the fraction of protein binding sites 500-bp downstream from the TTS. For all of the gene pairs in the genome, there are N1, N2, N3, and N4 pairs for the head-to-tail, head-to-head, tail-to-tail, and tail-to-head combinations, respectively. The expected number of gene pairs bound by a specific protein is N1 × [1−(1 − P1) × (1 − P2)], N2 × [1−(1 − P1) × (1 − P1)], N3 × [1−(1 − P2) × (1 − P2)], N4 × [1−(1 − P2) × (1 − P1)] for each combination. The expected fraction for each combination is then determined as the expected numbers divided by the total number of gene pairs. To calculate the observed fraction, we count the number of gene pairs (X) in each category (head-to-tail, head-to-head, tail-to-tail, and tail-to-head) present in the pool of total gene pairs associated with the different proteins (T). Then, the observed fraction is obtained by dividing X by T for each category.
Gene coexpression analysis
The expression value for each gene in each gene pair was extracted from the table in modENCODE_3305 (http://submit.modencode.org/submit/public/download/modENCODE_3305?root=data), which includes expression scores for different cell lines and developmental stages from embryo to adult. Pearson correlations were calculated for the expression scores for the two genes in each pair across the cell lines and developmental stages.
Alignment of BEAF-32 and histone modifications
The clustering of BEAF-32 sites and alignment of BEAF-32 with histone modifications were carried out using ChromaSig (Hon et al. 2008). Since Drosophila genomes are smaller than the mammalian genomes for which this program was originally written, we changed several parameters as follows: STAT_HALF_WINDOW_SIZE = 1000 and OVERLAP_HALF_WINDOW_SIZE = 1000. The output of ChromaSig was viewed using custom scripts (available upon request) and TreeView. To distinguish the differences between the two sides flanking BEAF-32 binding sites, the direction information from the ChromaSig output was also incorporated for graphical viewing.
Gene-pair-centric conservation analysis
BEAF-32 associated pairs are two nonoverlapping genes flanking a BEAF-32 binding site. For a BEAF-32-associated pair composed of gene1 and gene2 in species A, orthologous genes are found in table gene_orthologs_fb_2011_07.tsv from FlyBase. Then the BEAF-32 binding signal is examined for the corresponding intergenic region for gene1 or gene2 in the second species-species B. The term “corresponding intergenic region” signifies that this region should be downstream from gene1 or upstream of gene2 in species B if it is downstream from gene1 and upstream of gene2 in species A. If BEAF-32 is found at the corresponding intergenic region in species B, it is determined to be conserved. For clustering analysis, all BEAF-32 binding sites from the four species were pooled together. Each site in each species is assigned a value of 1 if BEAF-32 is present, −1 if BEAF-32 is not present, 0 if no ortholog is found, and NA if the site is not mapped by ChIP-seq. The created matrix is then clustered using hierarchical clustering in Cluster 3.0, and the results were viewed using TreeView. For comparisons among species, the conservation score was calculated based on the peaks called by CCAT 3.0 using default parameters (enrichment value of 5). For the quantitative comparison between D. melanogaster and other species, peaks used were called with an enrichment of 10 for D. melanogaster and an enrichment of three for other species. Thus, the regions called as nonconserved are the ones with at least 10-fold enrichment in D. melanogaster and at most threefold enrichment in other species. At least a threefold difference was required to call a gain or loss of protein binding. To count the co-occurrence of nonconserved BEAF-32 sites and chromosome rearrangements, gene pairs flanking nonconserved BEAF-32 sites in D. melanogaster are searched for their orthologous presence in other species. If the two genes in the gene pair are still next to each other and in the other species, it is counted as nonrearranged. Otherwise, it is counted as having undergone a rearrangement.
Motif analysis
Consensus sequences were discovered using Weeder to analyze BEAF-32 binding sequences obtained from peak files called with CCAT 3.0 (Pavesi et al. 2004). Changes of sequences in the BEAF-32 binding sites were determined based on the 5-bp motif sequence CGATA or its reverse complementary sequence TATCG in intergenic regions.
Other data sets
ChIP-chip results for BEAF-32, other insulator proteins, JIL1, and histone modifications in S2 cells were obtained from modENCODE (www.modencode.org/publications/integrative_fly_2010/) (The modENCODE Consortium et al. 2010). ChIP-seq results for Twist and Snail in embryo were obtained from the EMBL-EBI website under accession code E-MTAB-376 (He et al. 2011). ChIP-chip data for SMC1 was obtained from GEO under accession number GSE9248 (Misulovin et al. 2008). Expression data for Sox14 mutant animals is from GSE23355 (Ritter and Beckstead 2010).
Data access
ChIP-seq data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE35648.
Acknowledgments
We thank Dr. Nicolas Gompel for the images of the Drosophila species shown in Figure 6B. We thank Chunhui Hou for enlightening discussions, Naomi Takenaka for help with the ChIP-seq libraries, and the Corces lab for moral support. We also thank the Genomic Services Lab at the HudsonAlpha Institute for Biotechnology for their help in performing Illumina sequencing of ChIP-seq samples. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM035463 to V.G.C. and National Cancer Institute award number K01CA133106 to E.R. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.142125.112.
References
- Adachi N, Lieber MR 2002. Bidirectional Gene Organization: A common architectural feature of the human genome. Cell 109: 807–809 [DOI] [PubMed] [Google Scholar]
- Batada NN, Hurst LD 2007. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat Genet 39: 945–949 [DOI] [PubMed] [Google Scholar]
- Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M, Snyder M 2007. Divergence of transcription factor binding sites across related yeast species. Science 317: 815–819 [DOI] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol 11: 213–221 [DOI] [PubMed] [Google Scholar]
- Bushey AM, Ramos E, Corces VG 2009. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev 23: 1338–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira VP, Mensch J, Fanara JJ 2008. Body size in Drosophila: Genetic architecture, allometries and sexual dimorphism. Heredity 102: 246–256 [DOI] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh T-Y, Cui K, Zhao K 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhadi SR, Krom N, Ramakrishna W 2009. Genome-wide comparative analysis of putative bidirectional promoters from rice, Arabidopsis and Populus. Gene 429: 65–73 [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218 [DOI] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CWH, Ye C, Ping JLH, Mulawadi F, et al. 2011. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 43: 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Bardet AF, Patton B, Purvis J, Johnston J, Paulson A, Gogol M, Stark A, Zeitlinger J 2011. High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat Genet 43: 414–420 [DOI] [PubMed] [Google Scholar]
- Herr DR, Harris GL 2004. Close head-to-head juxtaposition of genes favors their coordinate regulation in Drosophila melanogaster. FEBS Lett 572: 147–153 [DOI] [PubMed] [Google Scholar]
- Herr DR, Fyrst H, Phan V, Heinecke K, Georges R, Harris GL, Saba JD 2003. Sply regulation of sphingolipid signaling molecules is essential for Drosophila development. Development 130: 2443–2453 [DOI] [PubMed] [Google Scholar]
- Herr DR, Fyrst H, Creason MB, Phan VH, Saba JD, Harris GL 2004. Characterization of the Drosophila sphingosine kinases and requirement for Sk2 in normal reproductive function. J Biol Chem 279: 12685–12694 [DOI] [PubMed] [Google Scholar]
- Hon G, Ren B, Wang W 2008. ChromaSig: A probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput Biol 4: e1000201 doi: 10.1371/journal.pcbi.1000201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pal C, Lercher MJ 2004. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet 5: 299–310 [DOI] [PubMed] [Google Scholar]
- Jiang N, Emberly E, Cuvier O, Hart CM 2009. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol Cell Biol 29: 3556–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green Roland D, Zhang MQ, Lobanenkov VV, Ren B 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi KO, Hagiwara M, Itoh T, Gojobori T, Imanishi T 2005. Comparative genomics of bidirectional gene pairs and its implications for the evolution of a transcriptional regulation system. Gene 353: 169–176 [DOI] [PubMed] [Google Scholar]
- Li Y-Y, Yu H, Guo Z-M, Guo T-Q, Tu K, Li Y-X 2006. Systematic analysis of head-to-head gene organization: Evolutionary conservation and potential biological relevance. PLoS Comput Biol 2: e74 doi: 10.1371/journal.pcbi.0020074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow TA, O'Grady P, ed. 2005. Drosophila: A guide to species identification and use. Academic Press, NY [Google Scholar]
- Misulovin Z, Schwartz Y, Li X-Y, Kahn T, Gause M, MacArthur S, Fay J, Eisen M, Pirrotta V, Biggin M, et al. 2008. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma 117: 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The modENCODE Consortium, Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, et al. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, et al. 2005. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep 6: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RAH, et al. 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet 6: e1000814 doi: 10.1371/journal.pgen.1000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E 2007. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 39: 730–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Hurst LD 2003. Evidence for co-evolution of gene order and recombination rate. Nat Genet 33: 392–395 [DOI] [PubMed] [Google Scholar]
- Pavesi G, Mereghetti P, Mauri G, Pesole G 2004. Weeder Web: Discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res 32: W199–W203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG 2009. CTCF: Master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault AD, Starz-Gaiano M, Lehmann R 2002. RETRACTED: Metabolism of sphingosine 1-phosphate and lysophosphatidic acid: A genome wide analysis of gene expression in Drosophila. Gene Expr Patterns 2: 337–345 [DOI] [PubMed] [Google Scholar]
- Ritter AR, Beckstead RB 2010. Sox14 is required for transcriptional and developmental responses to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Dev Dyn 239: 2685–2694 [DOI] [PubMed] [Google Scholar]
- Sandmann T, Jakobsen JS, Furlong EEM 2007. ChIP-on-chip protocol for genome-wide analysis of transcription factor binding in Drosophila melanogaster embryos. Nat Protoc 1: 2839–2855 [DOI] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoborg T, Labrador M 2010. The phylogenetic distribution of non-CTCF insulator proteins is limited to insects and reveals that BEAF-32 is Drosophila lineage specific. J Mol Evol 70: 74–84 [DOI] [PubMed] [Google Scholar]
- Sultana H, Verma S, Mishra RK 2011. A BEAF dependent chromatin domain boundary separates myoglianin and eyeless genes of Drosophila melanogaster. Nucleic Acids Res 39: 3543–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ 2009. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136: 3131–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C, Hurst L 2011. Support for multiple classes of local expression clusters in Drosophila melanogaster, but no evidence for gene order conservation. Genome Biol 12: R23 doi: 10.1186/gb-2011-12-3-r23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES 2007. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci 104: 7145–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Corces VG 2011. Chromatin insulators: A role in nuclear organization and gene expression. Adv Cancer Res 110: 43–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Yu J 2009. A comparative analysis of divergently-paired genes (DPGs) among Drosophila and vertebrate genomes. BMC Evol Biol 9: 55 doi: 10.1186/1471-2148-9-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Taylor J, Elnitski L 2008. Comparative analyses of bidirectional promoters in vertebrates. BMC Bioinformatics 9: S9 doi: 10.1186/1471-2105-9-S6-S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL 2011. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat Cell Biol 13: 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S 2009. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138: 1122–1136 [DOI] [PubMed] [Google Scholar]