Abstract
Biologists possess the detailed knowledge critical for extracting biological insight from genome-wide data resources, and yet they are increasingly faced with nontrivial computational analysis challenges posed by genome-scale methodologies. To lower this computational barrier, particularly in the early data exploration phases, we have developed an interactive pattern discovery and visualization approach, Spark, designed with epigenomic data in mind. Here we demonstrate Spark's ability to reveal both known and novel epigenetic signatures, including a previously unappreciated binding association between the YY1 transcription factor and the corepressor CTBP2 in human embryonic stem cells.
A pressing challenge arising from the productivity of large-scale data-generating consortia, such as the Encyclopedia of DNA Elements (ENCODE) Project (The ENCODE Project Consortium 2012) or the Roadmap Epigenomics Project (Bernstein et al. 2010), is ensuring that these data are accessible to the biological community for analysis. While public repositories provide easy access to primary data, subsequent data processing and analysis can pose a significant computational hurdle to many biologists. In addition, the depth and breadth of these resources are unprecedented, and much of the initial analysis may be exploratory in nature. The biologically interesting signals may be too poorly understood at the outset to be identified and analyzed in an automated fashion. Visualization is a powerful approach in such cases. Not only does it lower the computational barrier for use, but also it is particularly effective in facilitating human reasoning about complex data, which is essential during this early exploration phase.
Genome browsers are one such class of visualization tool that have enjoyed widespread popularity among biologists and that frequently serve as the primary means of examining genome-wide data during the initial inspection and discovery phases. Part of their power comes from the ability to integrate diverse data sets by plotting them as vertically stacked ‘tracks’ across a common genomic x-axis. Genome browsers have played an important role in increasing the accessibility of large public data sets, for example, the ENCODE data resource is currently hosted by the UCSC Genome Browser (Kent et al. 2002).
However, the power of genome-wide data sets is in their ability to reveal global regulatory patterns that would be difficult, if not impossible, to extrapolate from studies of individual loci. Genome browsers inherently limit the data view to individual loci, and while invaluable for visualizing data patterns at specific regions of interest, they have limited power to facilitate global analysis. For many types of queries, there is a mismatch between the level of data abstraction at which the investigator wishes to interrogate the data set (e.g., gene set) and the level at which the data are displayed in a genome browser (e.g., individual gene). As a result, computational experts typically conduct such global analyses with custom tools. Recently, the Human Epigenome Browser (Zhou et al. 2011) enabled users to filter the genomic x-axis to only annotated genes involved in a pathway of interest, as queried by a KEGG identifier. This is an important step toward replacing the genome coordinate axis with a functional axis and enabling comparisons of data tracks across multiple loci within the genome browser framework, but depending on the size of the gene set, it can still be challenging to obtain an overview of the data patterns from such a view.
There are several good examples of computational methods that generate biologically meaningful genome-wide data summaries. One common approach used to interpret epigenomic data, such as histone modifications and DNA methylation, is to identify and functionally characterize combinatorial data patterns. For example, methylation of both lysine 4 and lysine 27 on histone H3 is an epigenetic signature characteristic of embryonic stem cells, termed a ‘bivalent domain,’ thought to silence developmental genes while keeping them poised for activation (Azuara et al. 2006; Bernstein et al. 2006). Early work in signature detection clustered well-annotated promoters on the basis of specific histone modification patterns derived from chromatin immunoprecipitation (ChIP) coupled microarray data (ChIP-chip) (Heintzman et al. 2007). Both seqMINER (Ye et al. 2011) and Cistrome (Liu et al. 2011) are analysis tools that include such a clustering approach and provide cluster visualization through static heatmaps. A probabilistic method, ChromaSig, subsequently eliminated the dependence on existing annotations and offered a way to discover chromatin signatures de novo by searching genome-wide using data from ChIP followed by sequencing (ChIP-seq) (Hon et al. 2008). More recently, hidden Markov model (HMM), and Bayesian network approaches have been applied to uncover recurrent chromatin states (Ernst and Kellis 2010; Hoffman et al. 2012). However, none of these approaches support interactive data exploration.
All of the above tools produce static summary images, typically in the form of heatmaps and there are few or no mechanisms by which to dynamically guide the analysis based on human knowledge of the biological system under study. Here we present Spark, a visualization approach that employs clustering to create a global data overview and high-level entry point for analysis, while also enabling interactive drill-down to the supporting data at the level of individual loci. It is intended to facilitate responsive exploratory navigation through a genome-wide data set and to be used as a complement to genome browsing. Its novelty over existing tools lies in its support of user-guided clustering, specifically enabling users to split existing clusters into subclusters and thus direct the clustering algorithm toward patterns of interest. Given that the clusters are generated across a set of user-specified input regions, Spark supports the analysis of both well-annotated regions and potential novel elements, such as those identified as having enrichments in a particular ChIP-seq experiment. The tool is connected to popular external resources, for example, the display links individual loci to the corresponding view in the UCSC Genome Browser, and gene ontology (GO) analysis is available at the cluster level by interfacing with the DAVID suite of tools (Huang et al. 2009) and thus minimizes the need for programmatic data manipulation. Spark employs a very general clustering technique with few parameters and can therefore flexibly handle diverse data sets. The ENCODE and Human Epigenome Atlas data sets are directly accessible through the Spark user interface, and initial results suggest that Spark will be a valuable exploratory tool for these communities.
Results
Availability and installation
Spark is a Java application for all platforms and is currently available from http://www.sparkinsight.org. A sample clustering analysis is packaged with Spark and can be loaded from the initial launch screen or from the Help menu. We provide a built-in user guide and tutorial video, also linked from the initial launch screen and Help menu. All of these supporting resources are additionally available from the above Spark website.
The preprocessing and clustering steps of Spark are available as command-line utilities to facilitate batch processing if desired. For convenience, we have run the Spark preprocessing step on all 1800 Epigenome Atlas files (Release 7; http://www.epigenomeatlas.org) using the set of reference regions available in Spark and default parameters. This enables Spark to load these resources in a much shorter time.
In addition to being deployed as a standalone package, Spark is also available as a service within the Epigenome toolset of the Genboree Workbench (http://www.genboree.org) (Challis et al. 2012). The Genboree deployment enables analysis of any private or public data hosted at Genboree. It also supports simultaneous processing of several Spark clustering analyses, which is not possible with the standalone tool. A tutorial video demonstrating these features is available from the Spark website.
Questions and comments about Spark can be directed to the Spark Google Group: http://groups.google.com/group/spark_users/.
Overview
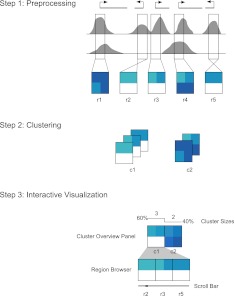
A Spark analysis begins with two user inputs: (1) one or more data files and (2) a set of regions. Wiggle/bigWig and GFF3 formats are accepted for these two inputs, respectively. Within Spark's graphical user interface (GUI), a user can either select files from the listed ENCODE and Epigenome Atlas data resources or can specify their own data files either as URLs or by browsing their local file system. The user-specified regions can be any set of genomic coordinates, for example, the regions flanking known transcriptional start site (TSS) annotations or defined by a set of ChIP-seq enrichment peaks. Several human reference region sets are also available through the GUI. Spark extracts data matrices from the specified regions, which are then binned and normalized (Fig. 1, step 1). These values form the basis of the clustering and are written to a binary file for faster future reloading.
Figure 1.
The Spark workflow. In step 1, the user's input data and regions of interest are preprocessed to enable rapid data retrieval in later steps. (Gray) Data enrichment peaks for two data samples; (vertical black boxes) user's regions of interest (r1–r5) centered on transcriptional start sites (TSSs). A data matrix is extracted for each input region and oriented according to strand. Rows in these matrices correspond to data samples, while the columns represent data bins along the genomic x-axis; two bins per region are used in this diagram. The values are then normalized to be between 0 and 1, represented here by white and dark blue, respectively. In step 2, the matrices are clustered. k = 2 in this diagram, resulting in two clusters (c1 and c2). In step 3, the clusters and their region members are viewed in the Spark interactive visualization interface.
The preprocessed data are then clustered using k-means clustering (Fig. 1, step 2) using a user specified number of clusters (k). This technique was chosen for its effectiveness, its relative simplicity, and runtime speed. Clusters are also written to text files for reuse.
Finally, the analysis output is displayed in the Spark GUI (Fig. 1, step 3). The interactive visualization encompasses two core components: a cluster overview panel, which provides a summary of each cluster, and the region browser. For a video demonstration of the interface, see the Spark website. In the cluster overview panel, clusters are initially sorted from left to right by decreasing number of regions. Each cluster is represented by a heatmap computed by averaging the data matrices from the cluster's member regions. A histogram immediately above the cluster panel indicates the number of regions per cluster. When a user selects a given cluster, data matrices from the cluster's regions are displayed as heatmaps in the region browser, where they are sorted by chromosome position. The genome coordinates are displayed below each individual region in the browser, and a context menu provides a hyperlink to that region in the UCSC Genome Browser. The interface is also equipped with search functionality, enabling a user to easily locate a region of interest within the clustering.
Interactive cluster refinement
The general problem of finding a globally optimal partitioning of d-dimensional data into k sets is known to be NP-hard. Heuristic algorithms, such as k-means clustering, are therefore used to efficiently find a local optimum and come with the risk of reporting poor solutions. Even if a globally optimal solution was attainable, clustering involves minimizing some mathematical criterion, and it is very possible that such a criterion will not sufficiently capture the features a biologist would use to categorize their data.
The philosophy behind Spark is to employ a simple and computationally efficient clustering algorithm (k-means) and to augment it by allowing the user to interactively guide the output according to their expert biological knowledge. This is done by enabling interactive cluster splitting whereby a user can run a k-means clustering using k = 2 on only the subset of regions contained within the selected cluster. An additional discussion of the initial choice of k is provided in the Supplemental Material. This approach synergizes automated clustering with user feedback to produce a more powerful exploration tool.
Interactive GO analysis
The functional classification of regions bearing interesting data signatures is a natural and common next analysis step. Spark supports the interactive analysis of gene ontology (GO) term enrichments for each cluster within the GUI. This is achieved through interfacing with the DAVID suite of web-based tools (Huang et al. 2009).
Applications
Epigenetic patterns flanking TSSs
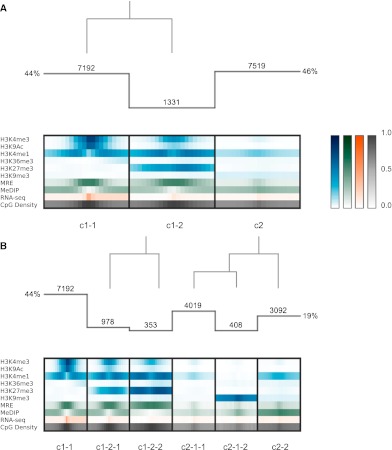
To validate our approach, we applied Spark to sequencing-based histone modification, DNA methylation, and expression data in H1 human embryonic stem cells (hESCs) (Harris et al. 2010) across transcriptional start sites (TSSs) where epigenetic signatures have been previously characterized (Lister et al. 2009; Hawkins et al. 2010). Trimethylation of Histone H3 Lys4 (H3K4me3) or Lys27 (H3K27me3) have positive and negative regulatory effects on transcription, respectively (for review, see Schuettengruber et al. 2007). These two modifications collocate to form ‘bivalent’ domains at the promoters of developmentally important genes in embryonic stem cells, serving to silence these genes while keeping them poised for lineage-specific activation (Azuara et al. 2006; Bernstein et al. 2006). These modifications therefore discriminate three main classes of promoters in embryonic stem cells: active, repressed, and poised (Mikkelsen et al. 2007). Spark successfully recapitulates these classes of TSSs in hESCs (Fig. 2A): From left to right, the first cluster is clearly marked with H3K4me3 and possesses an RNA-seq signal indicative of transcriptional activity, the second cluster bears the bivalent signature of both H3K4me3 and H3K27me3, and the third cluster appears transcriptionally inactive. Only the transcriptionally active and poised clusters (Fig. 2A) have notable CpG densities, consistent with previous observations that H3K4me3 predominantly localizes to CpG-rich promoters, suggesting important regulatory differences between promoters at the two extremes of CpG density (Mikkelsen et al. 2007). Using Spark's option to launch DAVID's Functional Annotation Tool (Huang et al. 2009), we find that the poised cluster shows significant enrichment in the terms ‘homeobox’ (P < 1 × 10−59), ‘regulation of transcription’ (P < 1 × 10−17), and ‘embryonic morphogenesis’ (P < 1 × 10−31), consistent with earlier characterizations of bivalent domains overlaying developmentally important transcription factors (Bernstein et al. 2006).
Figure 2.
Clustering analysis at annotated TSSs. (A) Histogram indicates the number of regions in each cluster, and the overlaid dendrogram traces the interactive cluster splitting events (initial clustering with k = 2, followed by one manual split of cluster c1 into c1-1 and c1-2). Chromatin modification (blue), DNA methylation (green; MeDIP and MRE indicate methylated and unmethylated CpGs, respectively), and RNA-seq (orange) data from H1 hESCs together with genomic CpG density values (gray) were clustered using a bin size of 300 bp across 6-kb windows centered on RefSeq transcriptional start sites (TSSs). (B) Further exploration and interactive refinement of the clusters from A.
These data can be further explored using Spark's interactive cluster splitting mechanism. For example, we can interactively split the poised cluster to produce two groups, one bearing a much broader H3K27me3 signal than the other (Fig. 2B, c1-2-1 and c1-2-2). This refined clustering is consistent with a report suggesting that the minority of bivalent sites contain ‘wide’ H3K27me3 signals extending over regions of at least 5 kb, while the majority shows punctate H3K27me3 signatures (Mikkelsen et al. 2007). Bivalent regions have been reported to be hypomethylated (Brunner et al. 2009; Meissner et al. 2008) and in this study, we employed a methylation-sensitive restriction enzyme assay (MRE) to detect unmethylated CpGs, and a methylation-dependent IP procedure (MeDIP) to enrich for methylated CpGs. Intriguingly, Spark highlights how closely the absence of DNA methylation, indicated by the strong MRE sequencing (MRE-seq) and weak MeDIP sequencing (MeDIP-seq) signals, tracks with H3K27me3 localization at bivalent sites.
In a similar fashion, cluster splitting can be used to explore the transcriptionally inactive class of TSSs (Fig. 2A, c2). This group appears to be heterogeneous, with a subcluster displaying a strong H3K9me3 signal (Fig. 2B, c2-1-2). This H3K9me3 containing group includes several gene clusters, for example, the olfactory receptors (ORs) and the late cornified envelope (LCE) gene family, as reported recently (Hawkins et al. 2010). The users' ability to direct the subclustering in this way allows them to take advantage of their biological knowledge to isolate interesting subsets that may not have been immediately produced by an automated clustering using default parameters and the same end k value.
Epigenetic patterns around YY1 binding sites
After validating Spark using previously published histone modification and DNA methylation data from hESCs, we sought to apply it to explore the genome-wide profiles of three transcription regulatory factors and their relationships with particular epigenetic signatures. This analysis was motivated by the hierarchical recruitment model in Drosophila, which suggests that the sequence-specific transcription factor, pleiohomeotic (PHO), recruits the polycomb repressive complex 2 (PRC2), which in turn trimethylates H3K27 and leads to the binding of the polycomb repressive complex 1 (PRC1) (Wang et al. 2004). Polycomb group (PcG) proteins, which include PHO and members of PRC1 and PRC2, typically function in maintaining transcriptional repression and play essential roles in normal development in most multicellular organisms (Morey and Helin 2010). While the human ortholog of PHO, YY1 transcription factor (also known as Yin Yang 1) (YY1), has identical DNA binding specificities to PHO in vitro (Brown et al. 1998) and can functionally compensate for loss of PHO in pho mutant flies (Atchison et al. 2003), it remains unclear whether YY1 plays a role in triggering a regulatory cascade that results in H3K27 trimethylation and subsequent transcriptional silencing in mammalian cells.
To investigate this model, we profiled three factors in hESCs using ChIP-seq: (1) YY1; (2) a component of PRC2, suppressor of zeste 12 (SUZ12); and (3) the corepressor C-terminal binding protein 2 (CTBP2), which is thought to play a role in YY1 binding and PcG recruitment in fly (Srinivasan and Atchison 2004). Using Spark, these ChIP-seq profiles were explored and integrated with the previously described DNA methylation and histone modification data from hESCs. To avoid limiting our analysis to annotated promoter or enhancer regions, we adopted a data-driven approach and took advantage of Spark's flexibility to use the data peaks themselves to define the input region set for clustering. For this, region boundaries were defined as ±3 kb from each YY1 peak center using the top 5% of peaks sorted by peak height.
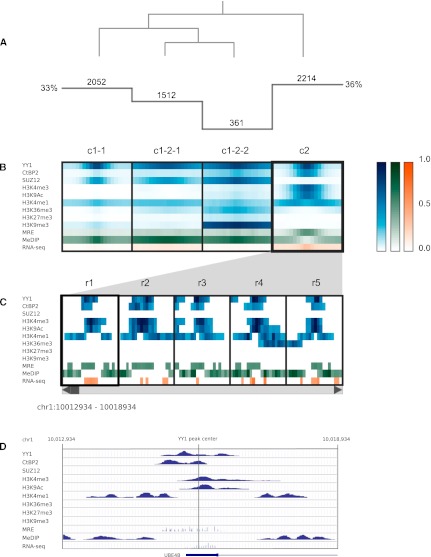
The hierarchical recruitment model introduced above predicts colocalization of YY1 with H3K27me3. Strikingly, no YY1-centered cluster shows a strong H3K27me3 signal (Fig. 3). In fact, only 1% of the YY1 peaks share an overlap with H3K27me3. This trend is robust even when the peak threshold is relaxed (<5% of YY1 peaks overlap a H3K27me3 peak in the full set). Our results indicate that, at most, only a minority of YY1 binding events are involved in H3K27me3 deposition.
Figure 3.
Clustering analysis of YY1 binding sites. (A) Histogram indicates the number of regions in each cluster, and the overlaid dendrogram traces the interactive cluster splitting events. (B) ChIP-seq data for YY1, CTBP2, SUZ12, and histone modifications (blue) together with MRE-seq and MeDIP-seq (green) and RNA-seq (orange) data from H1 hESCs were clustered using a bin size of 300 bp across 6-kb windows centered on sites of YY1 ChIP-seq enrichment. (C) Scrollable region browser: Data from individual regions within the currently selected cluster (c2) can be interactively viewed (five regions displayed at one time, r1–r5). (D) A context menu provides a hyperlink to the corresponding region display within the UCSC Genome Browser (view of r1 shown).
Further exploration of the YY1-centered clusters in Spark suggests that YY1 forms mutually exclusive complexes in hESCs with two important coregulators, SUZ12 and CTBP2 (Fig. 3B). SUZ12 and YY1 were found to colocalize within intergenic regions (cluster c1-1) and gene bodies (clusters c1-1 and c1-2-1) and at centromeres and telomeres (cluster c1-2-2). The absence of H3K27me3 in these clusters was initially surprising given that SUZ12 is a component of PRC2, which has known histone methyltransferase activity. However, this activity is mediated by EZH2, which may be absent at these sites. Alternatively, PRC2 can also methylate H1K26 (Xu et al. 2010), and it is possible that these sites display this mark. Alternatively, YY1 and SUZ12 may colocate with a histone demethylase, which has converted the H3K27me3 to H3K27me2 or H3K27me1. In subsequent motif analysis performed outside of Spark (see Methods), none of these clusters show enrichment for the canonical YY1 motif, suggesting that further investigation is needed to determine whether these patterns arise from direct YY1 binding or as a result of an alternate YY1 recruitment mechanism.
In contrast, YY1 motif enrichment (P < 0.0001) is observed at sites of colocalization with CTBP2 (cluster c2). These regions display strong H3K4me3, H3K9Ac, and RNA expression signals in Spark characteristic of transcriptionally active promoters, and subsequent comparison to known annotations outside of Spark reveals that the majority (88%) of these YY1 peak centers are within 2 kb of an annotated TSS. Individual regions can be viewed in the region browser (Fig. 3C) or via links to the UCSC Genome Browser (Fig. 3D). CTBP2, absence of which is embryonic lethal in mice (Hildebrand and Soriano 2002), is typically considered to function as a corepressor in mammalian cells (Chinnadurai 2003). There exists some evidence that the Drosophila CtBP homolog possesses a context-dependent transcriptional activation function (for review, see Chinnadurai 2003); however, the observed colocalization with YY1 at transcriptionally active TSSs has not been previously reported. GO analysis points to these genes being enriched in roles of RNA binding and processing, suggesting potential novel regulatory roles for CTBP2 and YY1 in hESCs.
Discussion
Spark is motivated by the need for data exploration tools that facilitate initial investigation of genome-wide data sets by the biology community. We recognize that the current paradigm of delegating analysis to a comparatively small community of computational experts will not effectively scale to the analysis demands of the current and ever-growing data resources. It is essential that the broader biology community is able to actively conduct initial inquiries and thus formulate the more detailed and biologically motivated hypotheses that warrant in-depth investigation. Visualization techniques are ideal for such applications in that they effectively lower the computational barrier for use while providing a powerful mechanism to facilitate human reasoning about complex data. We propose a visualization method that blends automated clustering with user interaction to provide a navigational tool that offers both meaningful data overviews and access to the relevant data details on demand.
The approach embodied in Spark has several strengths: (1) it employs a very general clustering technique with few parameters, which can flexibly handle diverse data sets; (2) it is not dependent on existing annotations, but rather clusters data across a user-specified set of input regions that can be known or novel elements; (3) it provides an interactive visual interface that enables simultaneous viewing of both genome-scale data signatures and patterns at individual loci, providing information about content and variation; and (4) it offers users interactive cluster refinement capabilities, enabling them to dynamically guide the clustering.
To facilitate using Spark with existing public resources, we have integrated the data inventory of the ENCODE Project and the Roadmap Epigenomics Project directly into the Spark GUI. We also support import of a user's own data in standard formats (wig/bigwig). Following the design philosophy to leverage existing and widely used tools, we link each locus in the Spark display to the corresponding view in the UCSC genome browser and also interface with the DAVID GO analysis tools to enable downstream functional analysis without the need for programmatic manipulation. In addition to being available as a standalone software package, Spark is also deployed as a service within the Epigenome toolset of the Genboree Workbench.
One natural direction for future work would be to incorporate additional clustering techniques into Spark. In particular, methods that first identify the subset of data tracks that are most informative for clustering may be valuable as the number of input data tracks grows. However, one insight that emerged while using Spark for analysis is that the criteria for defining similarity between data patterns can vary greatly depending on the application. A researcher may be most interested in regions that show distinct positional distributions of data across the query regions, or they may be primarily interested in regions with different signal amplitudes. There is unlikely to be an optimal distance metric or clustering algorithm for all features of biological interest. Rather, what seems most promising is to provide easy-to-understand clustering methods and then exploit the biologist's knowledge and judgment to guide the clustering to construct subsets of interest for further inquiry. The interactive cluster manipulation functionality currently in Spark is only a first step in this direction and warrants further investigation.
Through our application examples using data from the ENCODE and Human Epigenome Atlas projects, we have demonstrated Spark's ability to discover novel data patterns from a diverse collection of genome-wide data types. These signatures were not readily apparent through a genome browser view and would otherwise have required custom computational manipulation to obtain. We anticipate that Spark will be of widespread use in exploring these large public data sets and will increase the accessibility of these resources to the broader biology community. It is also our hope that the navigational paradigm captured in Spark will inspire other visualization methods that complement traditional genome browsers by offering interactive, high-level, functional summaries of genomic data as an entry point for exploratory analysis.
Methods
ChIP-seq
Human embryonic stem cells (hESCs) were obtained from Cellular Dynamics as part of a large batch of cells prepared for the ENCODE Consortium and the RoadMap Epigenome Consortium. Cell growth and crosslinking conditions can be found at http://www.genome.ucsc.edu/ENCODE/cellTypes.html. ChIP-seq experiments for the histone modifications have been described previously (Harris et al. 2010). The YY1 and SUZ12 ChIP assays were performed using 5 × 107 cells per assay, and 28 μg chromatin was used for the CTBP2 ChIP assay. ChIP assays were performed following the protocol provided at http://farnham.genomecenter.ucdavis.edu/pdf/FarnhamLabChIP%20Protocol.pdf, except that StaphA cells were blocked only with BSA before use and the preclearing step was omitted. The antibodies used were as follows: SUZ12 (Kirmizis et al. 2004), YY1 (Santa Cruz Biotechnology, sc-1703X), and CTBP2 (BD Biosciences 612044). All ChIP and input samples (10% of the amount of chromatin used per ChIP) were purified using the QIAquick PCR purification kit (QIAGEN) according to manufacturer's instructions, and purified eluates were dissolved in 50 μL of water. ChIP libraries were created and sequenced according to the method described previously (Harris et al. 2010) with the YY1, SUZ12, and CTBP2 libraries sequenced by the DNA Technologies Core Facility at the University of California-Davis (http://genomecenter.ucdavis.edu/dna_technologies/).
DNA methylation assays and RNA-seq
Methylation dependent immunoprecipitation and sequencing (MeDIP-seq), methylation sensitive restriction enzyme sequencing (MRE-seq), and RNA-seq were performed as previously described (Harris et al. 2010).
Data processing
Illumina read sequences (75 bp) were aligned to the reference human genome (hg18) using BWA (Li and Durbin 2009). FindPeaks 4.0.15 (Fejes et al. 2008) was subsequently used to detect enrichment peaks at an FDR of 0.01.
Spark
Input data files were provided in wig format and input region coordinates specified in GFF3 format. k-means clustering was performed on 6-kb windows centered on Refseq TSSs. Any TSS having a neighboring TSS within 3 kb was removed from the set prior to clustering. For the YY1 analysis, clustering was performed on 6-kb windows centered on high-confidence YY1 peaks (the top 5% sorted by maximal peak height). Data values were normalized to be between 0.0 and 1.0, according to the method described by Hon et al. (2008), and k-means clustering was computed using Euclidean distance. Spark version 1.1.0 was used for all analyses.
Motif analysis
Motif finding was performed using the W-ChIPMotifs web application (http://motif.bmi.ohio-state.edu/ChIPMotifs/) (Jin et al. 2009), and Bonferroni-corrected P-values are reported.
Data access
Data used in this article have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers CTBP2, GSM935463; H3K27me3, GSM428295; H3K36me3, GSM428296; H3K4me1, GSM434762; H3K4me3, GSM410808; H3K9Ac, GSM410807; H3K9me3, GSM428291; MRE-seq, GSM428286; MeDIP-seq, GSM456941; RNA-seq, GSM484408; SUZ12, GSM935352; and YY1, GSE39096. These data are also available from the Human Epigenome Atlas (http://www.epigenomeatlas.org) and the ENCODE data listings at the UCSC Genome Browser site (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeSydhTfbs/).
Acknowledgments
We thank the US National Institutes of Health (NIH) Roadmap Epigenomics Program; sponsored by the National Institute on Drug Abuse (NIDA) and the National Institute of Environmental Health Sciences (NIEHS). C.B.N. is a Canadian Institutes for Health Research (CIHR) and Michael Smith Foundation for Health Research (MSFHR) Post-doctoral Fellow. Funding for H.Y. was provided in part by a PGS-D scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC). A.M. and A.R.J. are supported by NIH grant U01 DA025956. J.F.C. and M.H. are supported by NIH grant 5U01ES017154-02. Funding for P.J.F., H.O., and X.X. was provided in part by NHGRI (HG004558). S.J.M.J. is a senior scholar of the Michael Smith Foundation for Health Research. We thank Richard Varhol and Dorothy Cheung for data processing assistance, Brett Johnson, Hunter Richards, Brad Hoffman, and Cory McLean for tool testing and commentary.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.140665.112.
References
- Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML 2003. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J 22: 1347–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. 2006. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. 2010. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28: 1045–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell 1: 1057–1064 [DOI] [PubMed] [Google Scholar]
- Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, Anton E, Medina C, Nguyen L, Chiao E, et al. 2009. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res 19: 1044–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, Milosavljevic A, Gibbs RA, Yu F 2012. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics 13: 8 doi: 10.1186/1471-2105-13-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G 2003. CtBP family proteins: More than transcriptional corepressors. Bioessays 25: 9–12 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M 2010. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 28: 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes AP, Robertson G, Bilenky M, Varhol R, Bainbridge M, Jones SJ 2008. FindPeaks 3.1: A tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics 24: 1729–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. 2010. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol 28: 1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. 2010. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6: 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Hildebrand JD, Soriano P 2002. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol 22: 5296–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MM, Buske OJ, Wang J, Weng Z, Bilmes JA, Noble WS 2012. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods 9: 473–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon G, Ren B, Wang W 2008. ChromaSig: A probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput Biol 4: e1000201 doi: 10.1371/journal.pcbi.1000201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DA, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Jin VX, Apostolos J, Nagisetty NS, Farnham PJ 2009. W-ChIPMotifs: A web application tool for de novo motif discovery from ChIP-based high-throughput data. Bioinformatics 25: 3191–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ 2004. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev 18: 1592–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, et al. 2011. Cistrome: An integrative platform for transcriptional regulation studies. Genome Biol 12: R83 doi: 10.1186/gb-2011-12-8-r83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L, Helin K 2010. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci 35: 323–332 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G 2007. Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Srinivasan L, Atchison ML 2004. YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev 18: 2596–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS 2004. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell 14: 637–646 [DOI] [PubMed] [Google Scholar]
- Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, Qiu W, Liu H, Jones AE, MacKenzie F, et al. 2010. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2). Proc Natl Acad Sci 107: 19266–19271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, Davidson I, Tora L 2011. seqMINER: An integrated ChIP-seq data interpretation platform. Nucleic Acids Res 39: e35 doi: 10.1093/nar/gkq1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Maricque B, Xie M, Li D, Sundaram V, Martin EA, Koebbe BC, Nielsen C, Hirst M, Farnham P, et al. 2011. The Human Epigenome Browser at Washington University. Nat Methods 8: 989–990 [DOI] [PMC free article] [PubMed] [Google Scholar]