Abstract
We take a comprehensive approach to the study of regulatory control of gene expression in melanocytes that proceeds from large-scale enhancer discovery facilitated by ChIP-seq; to rigorous validation in silico, in vitro, and in vivo; and finally to the use of machine learning to elucidate a regulatory vocabulary with genome-wide predictive power. We identify 2489 putative melanocyte enhancer loci in the mouse genome by ChIP-seq for EP300 and H3K4me1. We demonstrate that these putative enhancers are evolutionarily constrained, enriched for sequence motifs predicted to bind key melanocyte transcription factors, located near genes relevant to melanocyte biology, and capable of driving reporter gene expression in melanocytes in culture (86%; 43/50) and in transgenic zebrafish (70%; 7/10). Next, using the sequences of these putative enhancers as a training set for a supervised machine learning algorithm, we develop a vocabulary of 6-mers predictive of melanocyte enhancer function. Lastly, we demonstrate that this vocabulary has genome-wide predictive power in both the mouse and human genomes. This study provides deep insight into the regulation of gene expression in melanocytes and demonstrates a powerful approach to the investigation of regulatory sequences that can be applied to other cell types.
Regulation of gene expression at the level of transcription is an essential process in all organisms. In eukaryotes, DNA sequence elements known as enhancers play a major role in regulating transcription. Enhancers direct an increase in the expression of one or more target genes in cis, and their activity is typically restricted to specific cell types. Though the molecular mechanisms by which enhancers function are not fully understood, it is generally accepted that they are composed of short sequence motifs that bind transcription factors (TFs) (Noonan and McCallion 2010). These TFs recruit transcriptional coactivators such as EP300, CREBBP, and members of the Mediator complex (Panne et al. 2007; He et al. 2011b), and this enhancer-bound complex of TFs and coactivators then mediates an increase in the expression of a target gene at least in part through direct interaction with the basal transcriptional machinery present at the gene's promoter (Ong and Corces 2011; Chepelev et al. 2012). Epigenetic modifications are also thought to play a significant role in enhancer function, as it is well established that histones in the vicinity of enhancers bear characteristic modifications, including H3K4me1 (Heintzman et al. 2007, 2009) and H3K27Ac (Creyghton et al. 2010).
The identification of enhancers in eukaryotic genomes is challenging, due in large part to the expansive search space of noncoding sequence in which enhancers can reside. The recent integration of chromatin immunoprecipitation (ChIP) with massively parallel/next-generation sequencing (ChIP-seq) or microarray hybridization (ChIP-chip) facilitates the identification of loci bound by enhancer-related proteins and/or modified histones in a cell or tissue type of interest. Among the many ChIP targets used in this way to identify putative enhancers, the transcriptional coactivator EP300 and the histone modification H3K4me1 have proven particularly effective (Heintzman et al. 2007, 2009; Visel et al. 2009a; Blow et al. 2010; Bulger and Groudine 2011). Though ChIP-seq and ChIP-chip have dramatically increased the throughput with which putative enhancers can be identified, it remains important to thoroughly analyze the identified loci using bioinformatics-based tools and reporter assays before drawing conclusions about their function.
Enhancer identification is one step in a larger effort to understand the DNA sequence features that underlie enhancer function. These sequence features provide insight into the molecular mechanisms of transcriptional regulation and can be used to predict the impact of variation in regulatory sequence, much in the same way that the canonical genetic code can be used to predict the impact of variation in protein-coding sequence. We apply a recently developed strategy to identify sequence features predictive of enhancer function (Lee et al. 2011), which uses a supervised machine learning algorithm based on the concept of a support vector machine (SVM). Importantly, this strategy does not require any prior knowledge of TF binding specificities and thus has the potential to identify completely novel predictive sequence features. Furthermore, this strategy performs well on large sequence sets, allowing for accurate prediction of additional enhancer sequences genome-wide.
In this study we investigate regulatory control of gene expression in epidermal melanocytes, the pigment-producing cells that generate skin and hair color. These cells also play a central role in several pathological phenotypes, including melanoma, albinism, and vitiligo (for review, see Lin and Fisher 2007). These qualities, along with extensive knowledge about the key TFs and developmental origins of melanocytes (Silver et al. 2006; Hou and Pavan 2008; Thomas and Erickson 2008), make this lineage an attractive model system for the study of enhancers. We employ ChIP-seq for EP300 and H3K4me1 to identify melanocyte enhancers genome-wide. We use a novel set of criteria that takes into account both EP300 and H3K4me1 to define a single set of putative enhancers, and validate these enhancers through a series of in silico, in vitro, and in vivo analyses. Having validated the identified enhancers, we use them as a training set for a machine learning algorithm, developing a comprehensive vocabulary of 6-mers predictive of melanocyte enhancer function with power to predict additional melanocyte enhancers in the mouse and human genomes. Our data establish an extensive body of knowledge about regulatory control in melanocytes, which is relevant to phenotypic variation and disease. Moreover, we demonstrate a comprehensive approach that integrates ChIP-seq and machine learning to discover lineage-dependent enhancers and reveal the sequence vocabulary underlying their function.
Results
Previously characterized melanocyte enhancers are bound by EP300 and flanked by H3K4me1
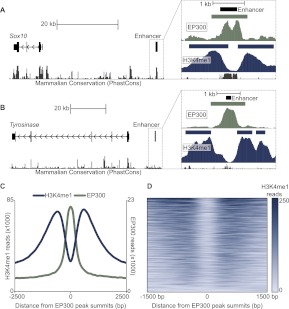
We sought to identify a large set of putative melanocyte enhancers from which we could derive a predicative sequence vocabulary. We began our enhancer identification by performing ChIP-seq for both EP300 and H3K4me1 in a line of immortalized melanocytes (melan-a) derived from Ink4a-Arf–null mice on a C57BL/6J background (Bennett et al. 1987; Sviderskaya et al. 2002). We identified 3622 and 59,965 ChIP-seq peaks for EP300 and H3K4me1, respectively. We expected a priori that both EP300 and H3K4me1 would be enriched at melanocyte enhancers loci, based on similar findings in other cell types (Barski et al. 2007; Heintzman et al. 2009; Visel et al. 2009a; Wang et al. 2009). Consistent with these observations, we confirmed the presence of enrichment for these factors at previously characterized melanocyte enhancers (Fig. 1A,B; Supplemental Table S1). More specifically, we observed that a central EP300 peak overlaps these enhancers and that this peak is flanked on both sides by strong H3K4me1 enrichment. To further assess the relationship between EP300 and H3K4me1 in melanocytes, we examined the distribution of H3K4me1 ChIP-seq reads relative EP300 peaks genome-wide. We found that H3K4me1 enrichment flanking EP300 peaks is a striking genome-wide trend (Fig. 1C,D), similar to observations made in other cell types (Heintzman et al. 2007, 2009; Ghisletti et al. 2010).
Figure 1.
EP300 and H3K4me1 ChIP-seq signature at melanocyte enhancers. (A, left) Schematic of chr15:78,984,500–79,034,500 (UCSC Genome Browser; mm9) showing Sox10 and previously characterized melanocyte enhancer Sox10 MSC#5. (Right) Detailed view of the region immediately surrounding Sox10 MSC#5 (chr15:79030709–79033709), showing ChIP-seq data for EP300 (green) and H3K4me1 (blue) in melan-a. Rectangles are ChIP-seq peaks, and colored vertical bars below peaks show density of ChIP-seq reads in 10-bp bins. Gray bars at the bottom of inset show the phastCons score (Euarchontoglires). (B) Same scheme as in A, but showing the interval chr7:94,575,283–94,662,322 containing the Tyr gene and previously characterized melanocyte enhancer Tyr DRE-15kb. Interval shown to the right is chr7:94655287–94658287. (C) Number of ChIP-seq reads for H3K4me1 (blue, left axis) and EP300 (green, right axis) in a 5-kb window around the summits of 3622 EP300 peaks (averaged in 100-bp bins). (D) Heatmap showing the number of H3K4me1 ChIP-seq reads in a 3-kb window around 3,622 EP300 peaks.
A specific EP300/H3K4me1 ChIP-seq signature identifies melanocyte enhancers genome-wide
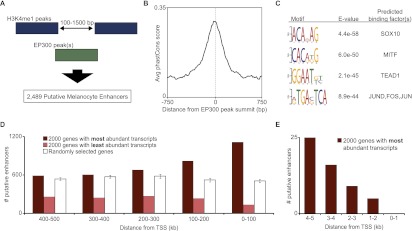
To identify a finite set of putative enhancers, we looked genome-wide for loci bearing the signature observed at previously characterized melanocyte enhancers, i.e., at which an EP300 peak is flanked by H3K4me1 enrichment. First, we identified a set of H3K4me1-flanked regions at which adjacent H3K4me1 peaks are separated by between 100 and 1500 bp (n = 21,189). This distance of 100–1500 bp was chosen based on the range of intervals between adjacent H3K4me1 peaks at known melanocyte enhancers (Supplemental Fig. S1). Next, we identified all EP300 peaks that overlap an H3K4me1-flanked region. This approach, represented schematically in Figure 2A, yields 2489 loci at which an EP300 peak falls in a region flanked by H3K4me1 peaks. Hereafter we refer to these 2489 loci as “putative melanocyte enhancers” (Supplemental Tables S4–S7). These putative melanocyte enhancers include previously reported enhancers at Tyr and Sox10 (Murisier et al. 2007; Antonellis et al. 2008), as well as novel enhancers at a number of other genes central to melanocyte biology, including Mitf, Tyrp1, Kit, and Mc1r (Supplemental Fig. S2). For downstream analysis, we use the summit of the EP300 peak as a surrogate for the center of a given enhancer, and where necessary, we use the boundaries of the EP300 peak as surrogates for the enhancer's boundaries.
Figure 2.
H3K4me1-flanked EP300 peaks have multiple characteristics of melanocyte enhancers. (A) Visual representation of our approach to identify putative melanocyte enhancers (described in text and Methods). (B) Average phastCons score (vertebrate, mm9) in a 1.5-kb window around the summit of 2489 putative melanocyte enhancers. (C) Top four motifs enriched in sequences of putative enhancers, with corresponding E-values (enrichment P-value times number of motifs tested; calculated by DREME) and factors predicted to bind to these motifs. (D) Number of putative enhancers in 1-MB window (100-kb bins) around the TSS of 2000 genes with the most abundant transcripts in melan-a (dark red), 2000 genes with least abundant transcripts in melan-a (light red), and 2000 randomly selected genes (white; average of five sets with SD represented by error bars). (E) Similar analysis to D, but using a 10-kb window and 1-kb bins.
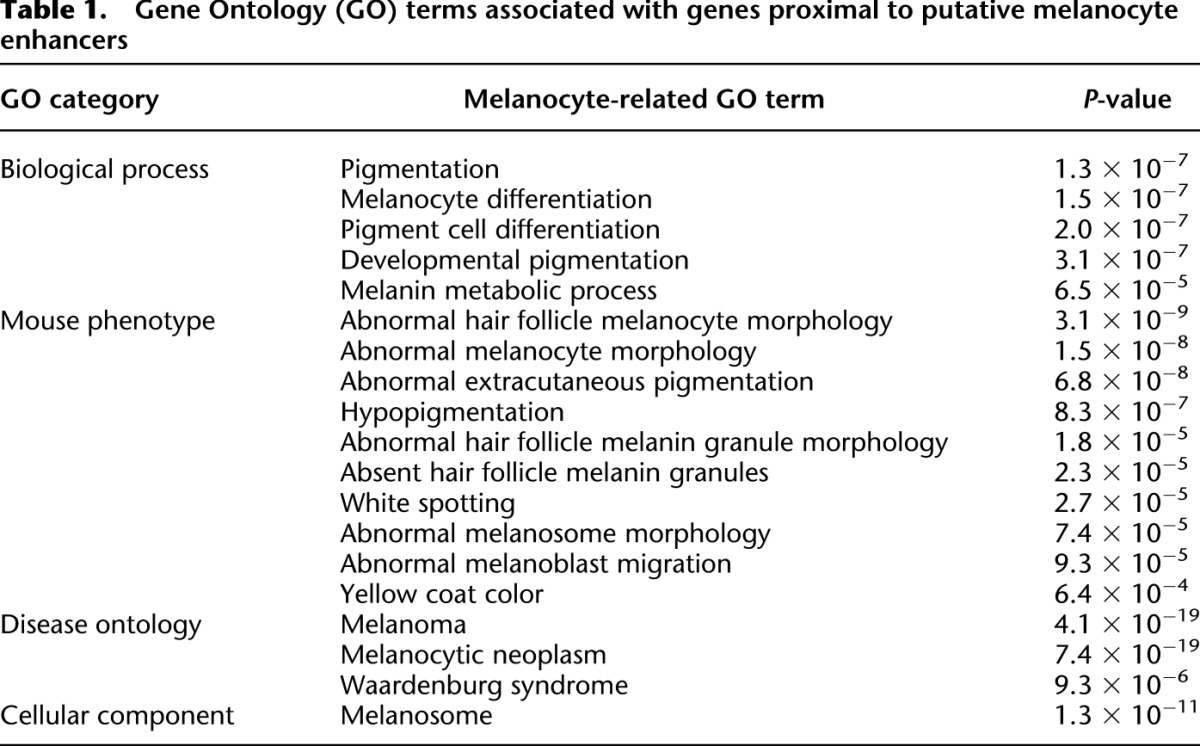
Several additional lines of evidence support the imputed function of these 2489 putative melanocyte enhancers. First, the putative melanocyte enhancers show evolutionary sequence constraint (Fig. 2B), providing independent evidence of their functional significance. Second, these putative melanocyte enhancers are enriched for sequence motifs predicted to bind key melanocyte TFs, including SOX10 and MITF, as detected by DREME (Fig. 2C; Bailey 2011). Mutations in SOX10 and MITF in humans cause Waardenburg syndrome (WS), a pleiotropic neural crest disorder with characteristic pigmentary defects (SOX10 mutations cause WS type 2E OMIM:611584 and 4C OMIM:613266; MITF mutations cause WS type 2A OMIM:193510) (McKusick 1998; http://omim.org/), and both TFs are involved in the pathogenesis of melanoma (Cronin et al. 2009; Harris et al. 2010). Third, analysis with GREAT (McLean et al. 2010) reveals that genes proximal to the putative melanocyte enhancers (within ∼50 kb; see GREAT methods) are significantly associated with Gene Ontology (GO) terms relevant to melanocyte biology, including melanoma, melanosome, pigmentation, and melanocyte differentiation (Table 1). Furthermore, using previously reported gene expression data for the melan-a line (Buac et al. 2009), we found that putative melanocyte enhancers are enriched near the most highly expressed genes and depleted near genes that are not expressed at appreciable levels (Fig. 2D), reflecting the expected distribution of active melanocyte enhancers.
Table 1.
Gene Ontology (GO) terms associated with genes proximal to putative melanocyte enhancers
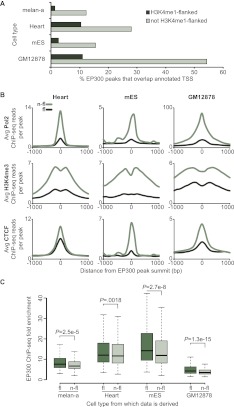
Although the 2489 putative melanocyte enhancers are enriched within 100 kb of highly expressed genes, they are not enriched in a 1-kb window immediately adjacent to the transcription start site (TSS) of these genes (Fig. 2E). This suggests that the enhancers identified are truly distal-acting and include very few, if any, proximal promoter elements. In contrast, EP300 peaks that are not flanked by H3K4me1 are far more likely to overlap annotated TSSs (Fig. 3A). This trend is also true for additional cell types in which data are available from the ENCODE and modENCODE Project consortia (The ENCODE Project Consortium 2007; The modENCODE Project Consortium 2009). Furthermore, in these cell types the non-H3K4me1-flanked EP300 peaks show markedly higher levels of ChIP-seq enrichment for RNA polymerase II and the promoter-associated modification H3K4me3 (Fig. 3B). Consistent with these observations, we noted several melan-a EP300 peaks at the promoters of melanocyte-related genes that have H3K4me1 enrichment on one side (upstream of the TSS) but are not flanked (Supplemental Fig. S3). Somewhat surprisingly, EP300 peaks that are not flanked by H3K4me1 also show higher levels of binding for CTCF in the ENCODE cell types examined (Fig. 3B). CTCF plays a central role in the function of insulator elements (Bell et al. 1999) and in physical organization of chromatin (Phillips and Corces 2009). In further comparing H3K4me1-flanked and non-H3K4me1-flanked EP300 peaks, we found that H3K4me1-flanked peaks have higher levels of EP300 enrichment than non-H3K4me1-flanked peaks (P = 2.5 × 10−5) (Fig. 3C).
Figure 3.
EP300 peaks that overlap H3K4me1-flanked regions have distinct properties. (A) Percent of EP300 peaks that directly overlap an annotated TSS (UCSC Genes; [dark green] peaks that overlap H3K4me1-flanked regions; [light green] peaks that do not overlap H3K4me1-flanked regions). Data for Heart (C57bl/6 mouse tissue taken at 8 wk), mES (Mouse ES-Bruce 4), and GM12878 generated by ENCODE and modENCODE consortia. (B) Average number ChIP-seq reads per peak for Pol2 (top row), H3K4me3 (middle row), and CTCF (bottom row) in a 2-kb window around the summits of indicated EP300 peaks (H3K4me1-flanked indicated by fl and dark green; non-H3K4me1-flanked, n-fl and light green). Three columns show data from the heart, mES, and GM12878, respectively. (C) EP300 ChIP-seq fold enrichment (determined by MACS) of EP300 peaks that overlap H3K4me1-flanked regions (fl; darker green), and EP300 peaks that do not overlap H3K4me1-flanked regions (n-fl; lighter green). Corresponding P-values calculated by two-tailed t-test. Numbers of peaks are as follows: melan-a fl, 2489; n-fl, 1133; heart fl, 3324; heart n-fl, 23,236; mES fl, 1258; mES n-fl, 20,062; GM12878 fl, 3404; and GM12878 n-fl, 6703.
Collectively, these data show that our set of 2489 candidate loci is highly enriched for bona fide melanocyte enhancers. By selecting only those EP300 peaks that overlap H3K4me1-flanked regions, we obtain a set of putative melanocyte enhancers with stronger EP300 binding that includes fewer regions containing sequence features of nonenhancer regulatory elements such as promoters and insulators. Importantly, these characteristics of our approach are particularly well suited to the creation of a training set from which key sequence features of enhancers can be extracted. Furthermore, our results add to a growing body of evidence linking EP300 and H3K4me1 to enhancer function and suggest the existence of functionally distinct subsets of EP300 peaks that can be distinguished to some extent by proximal histone modifications.
Identified melanocyte enhancers direct reporter expression in melanocytes in vitro and in vivo
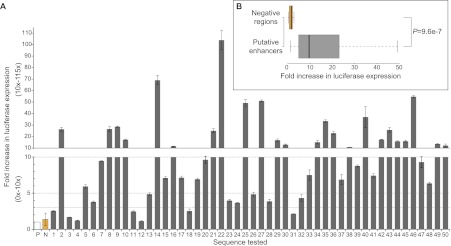
Given the evidence already supporting the role of the identified putative melanocyte enhancers in melanocyte regulatory control, we next sought to validate their biological activity in reporter assays. To this end, we first selected 50 putative enhancers at random from the full set of 2489 and analyzed each one for its ability to direct expression of a luciferase reporter gene in the melan-a line. We found that 86% (43/50) of enhancers tested increase reporter expression greater than threefold relative to the minimal promoter alone (Fig. 4A; Supplemental Table S2). Moreover, 72% (36/50) of enhancers tested increase reporter expression more than fivefold, and 48% (24/50) increase expression more than 10-fold relative to the minimal promoter alone. As there is considerable variation in the activity of melanocyte enhancers in this assay, we tested an additional 10 regions as negative controls. These regions were matched to our putative enhancers in average size and GC content but did not have significant EP300 or H3K4me1 ChIP-seq enrichment. None of these negative control regions increase reporter expression more than threefold relative to promoter alone (Supplemental Fig. S4A). As expected, the difference in reporter expression between putative enhancers and negative control regions is highly significant (P = 9.6 × 10−7 by two-tailed t-test) (Fig. 4B). We also assayed three previously characterized melanocyte enhancers for reference, which directed expression at levels 11-fold, 42-fold, and 51-fold higher than the minimal promoter alone, respectively (Supplemental Fig. S4B). However, it should be noted that these three enhancers are not directly comparable to our test sequences because the critical regions of these enhancers have been refined in previous studies. In this assay, a given enhancer will show highest activity when the amplified region contains the motifs critical for enhancer function with as little additional sequence as possible.
Figure 4.
Putative melanocyte enhancers direct reporter expression in melan-a. (A) Fold increase in luciferase reporter expression directed by indicated sequence relative to promoter-only control (P; white bar). Gray bars show fold increase of randomly selected putative enhancers (numbered 1–50). N (orange bar) represents the average of 10 negative regions. (Error bars) SD of three biological replicates, except in the case of N, where error bars show the standard deviation of 10 different negative regions. Note the difference in scale between bottom panel (onefold to 10-fold by one) and top panel (10-fold to 115-fold by 10). (Dotted lines) 10-fold, fivefold, and threefold thresholds (top to bottom). (B) Box plot summarizing results of reporter assays for 10 negative regions (top, orange) and 50 putative enhancers (bottom, gray). P = 9.564 × 10−7 by two-tailed t-test. Four outliers in putative enhancer group not shown in box plot (nos. 14, 22, 27, 46).
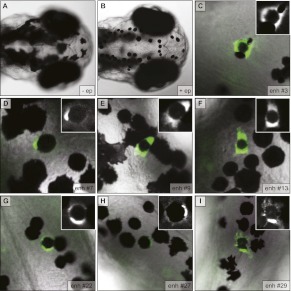
To further validate the biological activity of the putative enhancers, we tested the ability of a subset (n = 10) to appropriately direct melanocyte expression of a GFP reporter in vivo in transgenic zebrafish. We used an established pipeline for analyzing putative enhancers in zebrafish (Fisher et al. 2006a,b; McGaughey et al. 2008; Prasad et al. 2011), which we have previously used to analyze melanocyte regulatory elements at Sox10 (Antonellis et al. 2008) and GPNMB (Loftus et al. 2009). The 10 putative enhancers tested were chosen at random from the 50 analyzed in vitro as described above. We found that 70% (7/10) of enhancers tested direct GFP expression in the melanocytes of mosaic transgenic zebrafish (Fig. 5; Supplemental Table S3). The observed reporter expression is consistent with what we have seen previously when assaying melanocyte enhancers (Loftus et al. 2009) and is highly specific to melanocytes (Supplemental Fig. S5). We do not see consistent expression in other tissues with any of the seven positive constructs, with two exceptions that result from inherent artifacts of the assay: (1) We always see background GFP expression in the yolk (into which the construct is injected at day 0); and (2) we often see expression in skeletal muscle, which we suspect is caused by a cryptic regulatory sequence in the backbone of the reporter construct that we have been unable to locate. One melanocyte-negative construct (putative enhancer 25) did drive consistent expression in ganglia of the peripheral nervous system (PNS). Interestingly, the PNS and melanocytes both arise from the neural crest during embryonic development.
Figure 5.
Putative enhancers direct reporter expression in melanocytes of transgenic zebrafish embryos. (A) Dorsal view of melanocytes on the head of wild-type zebrafish embryo at 3 d post-fertilization (dpf). (B) Same view as A after treatment with epinephrine, which causes contraction of pigment granules to the center of the cell and enables the visualization of GFP at the periphery of melanocytes in transgenic embryos. (C–I) Representative images for all seven enhancers positive in this assay showing GFP-positive melanocytes in transgenic (mosaic) embryos at 3 dpf after treatment with epinephrine. Numbering is consistent with Figure 4.
The results of these functional assays demonstrate that the majority of putative melanocyte enhancers can direct gene expression in melanocytes both in vitro and in vivo, providing strong additional evidence that the identified loci function as melanocyte enhancers.
Machine learning reveals sequence features that underlie melanocyte enhancer function
To more thoroughly investigate the sequence composition of melanocyte enhancers, we used the putative enhancers identified by ChIP-seq as a training set for a supervised machine learning algorithm based on the statistical framework of a SVM (Lee et al. 2011). This approach as applied to embryonic mouse enhancers from other tissues is presented in detail by Lee et al. (2011). Briefly, the SVM finds an optimal decision boundary to distinguish the set of enhancers from random genomic regions using sequences of length k (k-mers) as features. Here, we used the putative melanocyte enhancers as positive sequences, a 50× larger set of random genomic regions as negative sequences, and the full set of 2080 distinct 6-mers as features. We have previously found that 6-mers and 7-mers are more informative in these analyses than are k-mers of other lengths, and we prefer 6-mers for robustness and ease of interpretation (Lee et al. 2011). SVM training assigns a weight, w, to each feature (6-mer), which determines its relative contribution to the decision boundary. The SVM discriminatory function, fSVM(x) = wx + b, represents the distance of a sequence x from the decision boundary and determines the predicted class, enhancer or nonenhancer, of the sequence x. This approach, which we call the kmer-SVM classifier, has three major advantages: (1) It identifies the specific sequences recognized by TFs active in melanocytes and provides independent support for our putative melanocyte enhancers based on previously known biology; (2) it allows the identification of additional melanocyte enhancers outside the original set of 2489 putative enhancers; and (3) it allows an indirect assessment of the quality of our putative enhancer set based on its sequence properties.
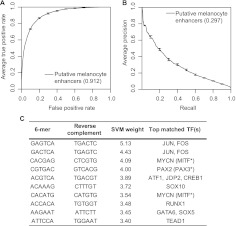
After training, we assessed the kmer-SVM classifier by its ability to accurately predict the class of reserved test sets via fivefold cross validation, as shown by the area under (au) the receiver operating characteristic curve (ROC) and precision-recall curves (PRCs) in Figure 6, A and B. The kmer-SVM trained on putative melanocyte enhancers achieved auROC of 0.912 and auPRC of 0.297, providing independent verification of the quality of our experimental enhancer identification.
Figure 6.
Analysis of kmer-SVM classifier trained with putative melanocyte enhancers. (A) Receiver operating characteristic curve for kmer-SVM classifier trained on putative melanocyte enhancers, with overall area under curve (auROC = 0.912) (B) Precision-recall curve with area under curve (auPRC = 0.297). (C) 6-mers with highest positive predictive value to kmer-SVM classifier, and factors predicted to bind each 6-mer. (*) No PWM in queried databases. Match based on similarity to published binding specificities (see Methods).
A key feature of the kmer-SVM is that it produces a list of features—in this case all unique 6-mers (n = 2080)—and the corresponding weight assigned to each feature by the SVM (Supplemental Table S10). The SVM weight represents the relative contribution of a given 6-mer to the overall predictive power of the classifier. Collectively, the list of weighted 6-mers provides a sequence vocabulary that is useful in interpreting the primary sequence of melanocyte enhancers. Importantly, the most predictive 6-mers (i.e., those assigned the largest SVM weights) correspond to binding sites for TFs known to be directly involved in melanocyte biology, including MITF, SOX10, and FOS/JUN (Fig. 6C; Supplemental Fig. S6). These 6-mers, and the 6-mer predicted to bind TEAD1, are in agreement with motifs found by DREME to be enriched in the training set (see Fig. 2C). It is also notable that one of the top 6-mers (ranked fourth) is predicted to bind PAX3, a key regulator of melanocyte differentiation (Lang et al. 2005) which can cause Waardenburg syndrome type 1 and type 3 when mutated (OMIM:193500 and OMIM:148820, respectively). In addition, CREB1, SOX5, and RUNX-family TFs (predicted to bind 6-mers ranked fifth, eighth, and ninth, respectively) have been shown to play roles in regulating gene expression in melanocytes (Tada et al. 2002; Raveh et al. 2005; Saha et al. 2006; Kingo et al. 2008; Stolt et al. 2008; Kanaykina et al. 2010; Mizutani et al. 2010).
Sequenced-based predictions identify additional enhancers in the mouse and human genomes
Having trained the kmer-SVM classifier, we next sought to determine whether we could use it to predict additional melanocyte enhancers genome-wide from primary sequence alone. Though these computational predictions are not likely to be as accurate as ChIP-seq, demonstrating that the kmer-SVM can predict bona fide enhancers is a powerful validation of the sequence vocabulary of weighted 6-mers on which the predictions are based. Furthermore, the ability to make enhancer predictions from sequence is particularly useful in genomes for which ChIP-seq data are not readily available. To make enhancer predictions genome-wide, we first segmented the mouse genome into 400-bp regions with 300 bp overlap and scored all regions with the kmer-SVM. The top 10,000 regions were chosen for further analysis, corresponding to an SVM cut-off score of 1.0 and yielding a precision of 0.74 and recall of 0.05 estimated from the PR curve in Figure 6B. We then eliminated any predicted regions overlapping the original training set (508 regions overlapping 348 enhancers from the original training set) and merged any overlapping regions. None of the six previously characterized melanocyte enhancers in Supplemental Table S1 overlap a kmer-SVM prediction, though it should be noted that four are included in the training set as they were bound by EP300 and flanked by H3K4me1 (Tyr DRE-15kb, Sox10 MCS4, Sox10 MCS5, Sox10 MCS9).
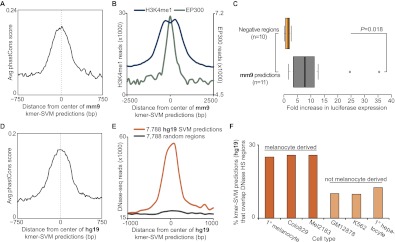
Ultimately, we obtained a set of 7361 predicted melanocyte enhancers (Supplemental Table S8). These predicted enhancers show strong sequence constraint (Fig. 7A), albeit to a lesser extent than the original set of putative enhancers. In addition, the predicted enhancers also show an EP300 and H3K4me1 ChIP-seq signature reminiscent of the original enhancer set (Fig. 7B). This suggests that the kmer-SVM predictions share underlying biology with the original set of 2489 putative enhancers, though the ChIP-seq signal at these loci is much lower than at regions detected by peak calling (Supplemental Fig. S7). We further analyzed the ability of a subset of the kmer-SVM–predicted enhancers to direct expression of a luciferase reporter in vitro in melanocytes (n = 11). We found that majority of enhancers tested direct luciferase expression in vitro more than threefold higher than the minimal promoter alone (8/11; 73%), and several drove expression more than fivefold (6/11; 55%) and 10-fold higher (3/11; 27%) (Fig. 7C; Supplemental Fig. S8A). We also tested the enhancer activity of three predicted enhancers in vivo using the same assay described above for ChIP-identified enhancers, and found that two of the three sequences assayed directed expression of GFP in the melanocytes of transgenic zebrafish (Supplemental Fig. S8B; Supplemental Table S3). GFP expression was mostly specific to melanocytes, though one predicted enhancer (no. 1) also directed expression in the CNS and otic vesicle. It should be noted that the predicted enhancers assayed here were chosen from among the predictions with the highest SVM scores rather than at random (see Methods).
Figure 7.
kmer-SVM classifier predicts additional enhancers in the mouse and human genomes. (A–C) Mouse predictions; (D–F) human. (A) Average phastCons score (vertebrate, mm9) in a 1.5-kb window around the centers of the 7361 kmer-SVM-predicted mouse melanocyte enhancers. (B) Number of ChIP-seq reads for H3K4me1 (blue line, left axis) and EP300 (green, right axis) in a 5-kb window around the centers of 7361 predicted melanocyte enhancers (averaged in 100-bp bins). (C) Box plot summarizing results of reporter assays for 10 negative regions (top, orange) and 11 predicted enhancers (bottom, gray). P = 0.01803 by two-tailed t-test. (D) Average phastCons score (vertebrate, hg19) in a 1.5-kb window around the centers of the 7788 kmer-SVM–predicted human melanocyte enhancers. (E) Number of DNase-seq reads from human primary melanocytes in a 2-kb window (averaged in 100-bp bins) around the centers of predicted enhancers (orange) and randomly selected regions matched in size and GC content (gray). (F) Percentage of predicted human melanocyte enhancers (n = 7788) that overlap DNase HS peaks in six cell types, which are either derived from melanocytes (orange) or not (beige).
To further demonstrate the power of this approach, we also made genome-wide enhancer predictions in the human genome in the same way as described above for mouse. We identified 7788 predicted melanocyte enhancers in the human genome (Supplemental Table S9). Like the mouse predictions, the human predictions show strong sequence constraint (Fig. 7D), even though conservation was not taken into account when making predictions. The predicted human enhancers display elevated levels of DNase I hypersensitivity (HS) in human primary melanocytes (data generated by The ENCODE Project Consortium) (Fig. 7E), which is a feature of active enhancers (Song and Crawford 2010; Song et al. 2011). Moreover, the degree of overlap between the kmer-SVM predictions and DNase I HS peaks is markedly higher in primary melanocytes and melanoma cell lines than in unrelated cell types (Fig. 7F), suggesting that the activity of the predicted enhancers is largely specific to the melanocyte lineage.
The ability of the kmer-SVM classifier to make valid genome-wide predictions in the mouse and human genomes clearly demonstrates the high information content of the 6-mer vocabulary derived from our original training set. The kmer-SVM predictions also augment the catalog of putative melanocyte enhancers identified in this study by adding an additional 7361 predicted enhancers in the mouse and 7788 in humans. Furthermore, the fact that a classifier trained on mouse sequences can make accurate predictions in the human genome clearly demonstrates the utility of this approach in identifying enhancers in genomes for which ChIP-seq data are not available, and provides direct proof of regulatory sequence vocabulary conserved between mouse and human.
Discussion
In this study, we demonstrate an approach to the investigation of regulatory sequences that integrates ChIP-based enhancer discovery with computational interrogation of sequence composition. The comprehensive nature of this approach represents a significant step forward in our ability to decipher the sequence basis of regulatory control of gene expression. Importantly, our strategy can be applied to any cell type of interest for which ChIP-seq and functional validation are feasible. We began this study by employing ChIP-seq for EP300 and H3K4me1 to discover a large set of previously unidentified putative melanocyte enhancers. In our melan-a ChIP-seq data, we observed a striking relationship between EP300 and H3K4me1, similar to that observed in other cell types (The ENCODE Project Consortium 2007; Heintzman et al. 2007, 2009; Ghisletti et al. 2010). The bimodal pattern of H3K4me1 ChIP-seq signal around EP300 peaks likely reflects the tendency of enhancers to be nucleosome depleted (Boyle et al. 2008; Song et al. 2011), and thus the flanking H3K4me1 signal arises from positioned nucleosomes marked by H3K4me1 on either side of the enhancer. A similar phenomenon was elegantly demonstrated in the case of nucleosomes at androgen-responsive enhancers in pancreatic cancer cells by He et al. (2010).
Though other studies have employed ChIP-seq for EP300 alone to identify putative enhancers with notable success (Visel et al. 2009a; Blow et al. 2010), we chose to focus specifically on EP300 peaks flanked by H3K4me1 peaks as this approach minimized the inclusion of nonenhancer sequence features with the potential to obscure the sequence vocabulary underlying enhancer function. Though not the primary focus of this study, we show that there are significant differences between the subset of EP300 peaks that are flanked by H3K4me1 and those that are not and that these differences are consistent across unrelated cell types. These differences suggest that there is considerable value in using both EP300 and H3K4me1 data sets together for enhancer discovery, and that future studies to further unravel the relationship between EP300 and H3K4me1 are likely to yield important insights into enhancer biology.
The rates of functional validation we observed (86% in vitro and 70% in vivo) are consistent with validation rates of ChIP-seq identified enhancers reported previously, though there is considerable variation between studies (Heintzman et al. 2009; Visel et al. 2009a; Blow et al. 2010; Ghisletti et al. 2010). There was general agreement in activity between the in vitro and in vivo assays used here. Six of seven elements showing activity in vivo also showed activity in vitro (threefold threshold). In addition, the enhancer with the strongest activity in vitro (no. 22) clearly had the strongest activity in vivo as well, as judged by the level of fluorescence in GFP-expressing melanocytes, the number of positive embryos observed, and the number of positive melanocytes per positive embryo. However, putative enhancer 3 drove melanocyte expression in vivo even though its enhancer activity was not significant in vitro, and conversely, three enhancers that drove expression in vitro did not drive expression in vivo in mosaic transgenic zebrafish (nos. 20, 25, and 30). These discrepancies between the results of the in vitro and in vivo functional assays used here could be the result of differences among the model organisms (mouse and zebrafish, respectively), the minimal promoters in the reporter constructs (E1B and FOS, respectively), or other limitations of the respective reporter assays. Our results demonstrate the importance of using multiple complimentary assays to assess the function of putative enhancers.
We observed that the orientation of the amplicon tested relative to the minimal promoter had a dramatic impact on the enhancer activity of sequences assayed in vitro. This is not likely to reflect an orientation dependence of the enhancer in its native genomic context. Rather, it is likely an artifact of the placement of the sequence in the synthetic context of a reporter construct. The orientation effect likely arises from the fact that the distance between an enhancer and minimal promoter in a reporter construct can strongly influence its functional output. This distance effect can be observed with as little 50 bp separating the two components (Nolis et al. 2009) and can manifest as orientation-dependent activity when testing an amplicon in which the critical sequence components (TF binding sites) are skewed to one side. In such a case, a given amplicon will show higher activity in the orientation that places its critical components closest to the minimal promoter, and lower (in some cases even undetectable) activity in the orientation that places its critical components furthest from the minimal promoter. Indeed, our strongest putative enhancer (no. 22), which mediates an increase of >100-fold reporter expression in the “forward” orientation and drives strong melanocyte expression in vivo, does not drive detectable expression in vitro in the “reverse” orientation (Supplemental Table S2).
The similarity between the motifs identified by DREME (Fig. 2C) and the 6-mers identified by the kmer-SVM classifier (Fig. 6C) is strong evidence that these sequences are binding motifs for TFs that play significant roles in melanocyte biology. The identification of motifs predicted to bind SOX10 and MITF is consistent with the well-characterized roles for these TFs in the melanocyte lineage. JUN and FOS are major effectors of the MAP kinase signaling cascade, which is critical to the proliferation of melanocyte cells in culture (Swope et al. 1995). In addition, constitutive activation of the MAP kinase pathway is a hallmark of melanoma (Dutton-Regester and Hayward 2012). The enrichment for a motif predicted to bind members of the TEAD family may reflect an as yet unappreciated role for TEAD TFs in melanocytes. To our knowledge, no TEAD family member has been previously shown to play a specific biological role in melanocytes. However, TEAD2 has been shown to bind an enhancer active in neural crest, the developmental precursor to melanocytes (Degenhardt et al. 2010). This binding causes an increase in the expression of Pax3, itself a TF that is predicted to bind one of the most highly weighted 6-mers (Fig. 6C).
Motifs predicted to bind other TFs involved in melanocyte biology could have escaped detection due to high variation in consensus sequence, low enrichment relative to negative control sequences, or inherent biases in the algorithms used here for motif detection. Additionally, our EP300/H3K4me1-based approach likely identifies only a subset of enhancers active in melanocytes. This particular subset of enhancers may be more highly enriched for some TF binding sites than for others. Mechanistically distinct subsets of enhancers have been reported in other cell types (He et al. 2011a). Though beyond the scope of this study, ChIP-seq for additional factors and in additional melanocyte-related cellular substrates would likely help to distinguish potential differences between subsets of enhancers.
Taken collectively, the melanocyte enhancers and corresponding sequence vocabulary described here greatly enhance our understanding of the regulation of gene expression in melanocytes. Furthermore, they are relevant to human phenotypes and disease risk caused by variation in regulatory sequences. To date, at least 18 distinct genome-wide association studies (GWAS) have identified 52 SNPs associated with melanocyte-related phenotypes, including skin color, hair color, freckling, tanning response, number of cutaneous nevi, melanoma risk, and vitiligo (Hindorff et al. 2011). Many of these associations are likely to reflect causative variants that impact regulatory sequences (Hindorff et al. 2009; Visel et al. 2009b). This study, and others like it, promises to aid the identification of causative variants underlying genome-wide associations, as well as the molecular mechanisms by which they act.
Methods
ChIP-seq
Melan-a cells were propagated according to guidelines from Sviderskaya et al. (2002). ChIP was performed according to the method previously described (Lee et al. 2006). Alternative lysis buffers to those in the referenced protocol were used as follows: lysis buffer 1 (5 mM PIPES, 85 mM KCl, 0.5% NP-40, and 1× Roche Complete, EDTA-free protease inhibitor), lysis buffer 2 (50 mM Tris-HCl, 10 mM EDTA, 1% SDS, and 1× Roche Complete, EDTA-free protease inhibitor), and lysis buffer 3 (16.7 mM Tris-HCl, 1.2 mM EDTA, 167 mM NaCl, 0.01% SDS, 1.1% Triton X-100, and 1× Roche Complete, EDTA-free protease inhibitor). Sonication was performed using a Bioruptor (Diagenode) with the following settings: high output; 30-sec disruption; 30-sec cooling; total sonication time of 35 min with addition of fresh ice and cold water to water bath every 10 min. Four micrograms of ab8895 (Abcam) and 10 μg of antibody sc-585 (Santa Cruz Biotechnology) were used for H3K4me1 and EP300 ChIP, respectively. IP wash conditions were adjusted from the protocol referenced above as follows: Each immunoprecipitation (IP) was washed twice with low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 150 mM NaCl), twice with high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, 500 mM NaCl), and twice with LiCl wash buffer (0.25 M LiCl, 1% IGEPAL CA630, 1% deoxycholic acid [sodium salt], 1 mM EDTA, 10 mM Tris-HCl) and rinsed once with PBS (pH 7.4). At least two biological replicates were performed for each antibody, with each replicate consisting of a ChIP sample and an input (pre-IP) sample. Each replicate was performed with ∼1 × 108 melan-a cells. ChIP libraries were submitted to NIH Intramural Sequencing Center, and each was sequenced on one lane of an Illumina GA2 yielding >20 million reads per sample, with the exception that each EP300 ChIP library was sequenced on two lanes for increased coverage depth.
Analysis of ChIP-seq data: peak calling
EP300 peaks were called using the Model-based Analysis for ChIP-seq (MACS) algorithm (Zhang et al. 2008). Peaks were called for each replicate independently, and only those that were called in both replicates (n = 3622) were selected for further analysis. Coordinates reported are from Replicate 1. H3K4me1 peaks were called using cisGenome (Ji et al. 2008) because it tends to call separate peaks corresponding to each apex of the bimodal distribution of H3K4me1 signal flanking enhancers, whereas MACS tends to call the entire bimodal distribution as a single peak. The Two Sample Peak Calling option in cisGenome was used, which allows both replicates to be entered simultaneously to produce a single set of output files. Default settings were used for both peak callers, except that ‘half window size W’ was set to 4 for cisGenome.
Distribution of ChIP-seq reads relative to features of interest
The total number of sequencing reads covering each base in a window of indicated size (x-axis) around the summit/center of the set of genome regions of interest (ChIP-seq peaks/kmer-SVM predictions) was calculated with a custom script. The total number of reads covering each base in the window was then smoothed in 100 bp bins, and is represented as ‘reads’ (y-axis) in Figures 1C and 7, B and E. For Figure 3B and Supplemental Figure S7, a subsequent calculation was performed in which the total reads in each bin was divided by the number of genome regions in the set of interest, to facilitate comparison between sets of different sizes. This normalized measure is represented as “Avg reads per peak” (y-axis) in Figure 3B and Supplemental Figure S7. The heatmap in Figure 2D was generated with the heatmap tool in the Cistrome Analysis Pipeline (Liu et al. 2011) using a bed file of 3622 EP300 peaks (300-bp regions centered the peak summits), and a wig file of H3K4me1 ChIP enrichment generated by MACS as standard output from peak calling.
ENCODE data
ENCODE data in Figures 3 and 7 were processed as described above for melan-a data. Much of the data handling for these analyses was performed with Galaxy (Giardine et al. 2005; Blankenberg et al. 2010; Goecks et al. 2010).
In silico analysis of putative enhancers:
Average phastCons score
Average phastCons score plots (Figs. 2B, 7A,D) were generated with the Conservation Plot tool as part of the Cistrome Analysis Pipeline using an interval file of H3K4me1-flanked EP300 peaks (300-bp intervals around peak summits) (Fig. 2B) or kmer-SVM predicted enhancers (Fig. 7A,D).
Motif analysis
DREME (Bailey 2011) was used to identify enriched motifs (Fig. 2C). Sequences of 2489 putative melanocyte enhancers (centered on the EP300 ChIP-seq peak summit and extending ±150 bp) were used as input. Default settings for motif size (mink = 3, maxk = 7) were used. Motifs were submitted to TOMTOM (Gupta et al. 2007) as part of the MEME Suite (Bailey et al. 2009) to predict binding factors corresponding to each enriched motif, and the top vertebrate TF match was reported unless otherwise indicated in text. In the case of MITF and PAX3 (Figs. 2C, 6C; Supplemental Fig. S6), match was made based on high similarity to published binding specificities (Bentley et al. 1994; Chalepakis and Gruss 1995; Yasumoto et al. 1995), as there is no position weight matrix (PWM) for either of these TFs in the databases queried by TOMTOM (JASPER and UniProbe).
GO analysis
GREAT (McLean et al. 2010) was used to identify GO terms enriched among genes proximal to putative enhancers. The association rule was set as follows: proximal, 50 kb upstream and 50 kb downstream (any gene in this interval relative to input regions is included); plus distal, up to 500 kb (if no gene is present in the proximal interval, the closest gene in this distal interval is included). For details, see McLean et al. (2010).
Distribution of enhancers relative to genes expressed at different levels in melan-a
Previously published melan-a microarray data were used (Buac et al. 2009). For analyses in Figure 2, only genes represented on the array with a corresponding TSS in RefSeq (n = 17,957) were used. These genes were ranked by raw expression level in melan-a (probes averaged, mean of three replicates). Custom scripts were used to calculate the number of putative enhancers within 500 kb (in bins of 100 kb) (Fig. 2D) and 5 kb (in bins of 1 kb) (Fig. 2E) of TSSs of the top 2000 and bottom 2000 genes on the ranked list, as well as for five sets of 2000 genes selected randomly from this list.
Luciferase assays
All tested sequences (putative enhancers, negative regions, kmer-SVM predictions, and previously characterized enhancers) were PCR amplified from mouse genomic DNA (Promega, no. G309A) and TA-cloned with the pCR8/GW/TOPO TA Cloning kit (Life Technologies). The luciferase reporter construct contains the firefly luciferase gene downstream from a minimal E1B promoter (Antonellis et al. 2006). Test sequences were inserted into a gateway cloning site upstream of the promoter with a directional LR reaction (Gateway cloning from Life Technologies). All sequences were tested in both orientations, and data from the orientation with the highest expression were used for downstream analysis to give the most accurate representation of the potential of each sequence to drive expression in melanocytes. For negative control regions, a set of 2000 regions was generated in which the regions were matched to the putative enhancers in size, GC%, and repeat fraction, but with a read count below for EP300 and H3K4me1. Ten regions were selected at random from this set for functional testing. For all luciferase assays, melan-a cells were plated in 24-well format (40,000 cells/well) and transfected next day with 400 ng of luciferase reporter and 8 ng of pCMV-RL Renilla expression vector (Promega) using 2 μL Lipofectamine 2000 per well (Life Technologies). Cell lysate was collected at 48 h post-transfection and assayed with the Dual-Luciferase Reporter Assay System (Promega) using a Tecan GENiosPro Microplate Reader (Tecan Group). Three biological replicates were performed for each construct.
Zebrafish transgenesis
All tested sequences were PCR amplified and TA-cloned as described above (see Luciferase Assays). The GFP reporter construct, described previously (Fisher et al. 2006b), contains a gateway recombination cassette (Life Technologies) upstream of a minimal (FOS) promoter and EGFP. The reporter used here was modified slightly by insertion of an eye-specific regulatory element from the zebrafish crybb1 locus (chr10:45,529,501–45,530,122; Zv9) downstream from EGFP to facilitate screening for successful transgenesis independent of the test sequence. Zebrafish transgenesis was performed as previously described (Fisher et al. 2006b). Briefly, each construct was injected into >150 wild-type (AB) embryos at the one- to two-cell stage with Tol2 transposase mRNA to facilitate efficient and random integration of the reporter construct (flanked by tol2 recombination arms) into the zebrafish genome. Embryos were screened for GFP expression at 3 d post-fertilization (dpf), a timepoint at which melanocytes are well developed and the embryos are most amenable to comprehensive screening. Embryos were also screened at 2, 4, and 5 dpf, albeit less thoroughly, and no significant differences in expression from 3 dpf were observed. At least 10 positive embryos were imaged at 3 dpf for each positive construct. For high-magnification fluorescent images of melanocytes (Fig. 5; Supplemental Fig. S8), zebrafish were treated with epinephrine 5–10 min prior to imaging (4 mg/mL) in order to contract pigment granules toward the center of the cell and thus facilitate visualization of GFP at the periphery. For full-body lateral images (Supplemental Fig. S5) embryos were raised in 1-phenyl 2-thiourea (PTU) from 24 hpf until imaging to inhibit melanin synthesis. All Images were taken on a Nikon AZ100 Multizoom microscope with NIS-elements software. All zebrafish work was performed under an approved protocol (FI10M369), reviewed by the Johns Hopkins Institutional Animal Care and Use Committee.
Kmer-SVM classifier
To generate a high-confidence training set, we defined a new set of 400-bp regions that maximizes the overall EP300 ChIP-seq signal within each of the 2489 putative melanocyte enhancers after removing any enhancers which were >70% repeats. We used repeat masked sequence data (mm9) from the UCSC Genome Browser to calculate repeat fractions. For negative sequences, we found a 50× larger set of random genomic 400-bp sequences by matching GC and repeat fraction of the positive set. Additionally, we excluded any potential EP300-bound regions with Poisson test P-value <0.1 (10 ChIP-seq reads). At each sampling step, we randomly selected a region from the positive set, calculated the GC content and the repeat fraction, sampled a genomic sequence that matched these properties, and repeated sampling until we obtained 50× sequences. We performed standard fivefold cross validation to assess the performance of our kmer-SVM classifier. We measured the quality of the classifier by calculating the auROC, which plots the true positive rate vs. the false-positive rate of the predictions. The PRC is a more reliable measure of performance than the ROC when positive and negative sets are unbalanced, as in our case. Precision is the ratio of true positives to predicted positives, and recall is identical to the true positive rate in the ROC. The PRCs can be quantified by the auPRC, or average precision. TFs predicted to bind top 6-mers were determined as described above for DREME motifs (see Motif Analysis). Predictions for functional validation (n = 11) were chosen from the top of a list of regions ranked by SVM score. These are not the top 11 ranked predictions overall however, because the list they were chosen from was generated by an earlier version of the classifier trained on a slightly different input set. In the final set of predictions, the 11 regions tested are ranked by SVM score as 13, 15, 1, 9, 2, 44, 21, 108, 24, 273, and 203, respectively.
Data access
The data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE38498.
Acknowledgments
We thank Barbara Migeon, Kirby Smith, Hongkai Ji, George Wu, Megana Prasad, Zachary Stine, Samantha Maragh, and Amanda Price for helpful comments and discussion. This work was funded in part by the National Institute of General Medical Sciences (GM071648) and the National Institute of Neurological Disease and Stroke (NS062972) to A.S.M., by the National Human Genome Research Institute's (NHGRI) Intramural Research Program (W.J.P., S.K.L.), by an NSF Graduate Research Fellowship to D.U.G., and by a predoctoral training grant (GM07814) to X.R. M.A.B. was supported by the Searle Scholars Program and in part by NS062972.
Author contributions: The study was conceived by D.U.G., A.S.M., S.K.L., and W.J.P. ChIP-seq was performed by D.U.G. and X.R. Peak calling and downstream analysis of ChIP-seq data was performed by D.U.G., in vitro validation by D.U.G. and C.F-B, and in vivo validation by D.U.G. and S.L.B. D.L. and M.A.B. developed and applied the kmer-SVM model. Kmer-SVM predictions were generated by D.L. and validated by D.U.G. and C.F-B. Experimental data were interpreted by D.U.G. and A.S.M. The manuscript was written by D.U.G., A.S.M., D.L., M.A.B., S.K.L., and W.J.P.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.139360.112.
References
- Antonellis A, Bennett WR, Menheniott TR, Prasad AB, Lee-Lin SQ, Green ED, Paisley D, Kelsh RN, Pavan WJ, Ward A 2006. Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg-Shah (WS4) syndrome. Hum Mol Genet 15: 259–271 [DOI] [PubMed] [Google Scholar]
- Antonellis A, Huynh JL, Lee-Lin SQ, Vinton RM, Renaud G, Loftus SK, Elliot G, Wolfsberg TG, Green ED, McCallion AS, et al. 2008. Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet 4: e1000174 doi: 10.1371/journal.pgen.1000174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL 2011. DREME: Motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS 2009. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396 [DOI] [PubMed] [Google Scholar]
- Bennett DC, Cooper PJ, Hart IR 1987. A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. Int J Cancer 39: 414–418 [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Eisen T, Goding CR 1994. Melanocyte-specific expression of the human tyrosinase promoter: Activation by the microphthalmia gene product and role of the initiator. Mol Cell Biol 14: 7996–8006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol 89: 19.10.1–19.10.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2010. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet 42: 806–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE 2008. High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buac K, Xu M, Cronin J, Weeraratna AT, Hewitt SM, Pavan WJ 2009. NRG1/ERBB3 signaling in melanocyte development and melanoma: Inhibition of differentiation and promotion of proliferation. Pigment Cell Melanoma Res 22: 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalepakis G, Gruss P 1995. Identification of DNA recognition sequences for the Pax3 paired domain. Gene 162: 267–270 [DOI] [PubMed] [Google Scholar]
- Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K 2012. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res 22: 490–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin JC, Wunderlich J, Loftus SK, Prickett TD, Wei X, Ridd K, Vemula S, Burrell AS, Agrawal NS, Lin JC, et al. 2009. Frequent mutations in the MITF pathway in melanoma. Pigment Cell Melanoma Res 22: 435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt KR, Milewski RC, Padmanabhan A, Miller M, Singh MK, Lang D, Engleka KA, Wu M, Li J, Zhou D, et al. 2010. Distinct enhancers at the Pax3 locus can function redundantly to regulate neural tube and neural crest expressions. Dev Biol 339: 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton-Regester K, Hayward NK 2012. Reviewing the somatic genetics of melanoma: From current to future analytical approaches. Pigment Cell Melanoma Res 25: 144–154 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS 2006a. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 312: 276–279 [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS 2006b. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc 1: 1297–1305 [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. 2005. Galaxy: A platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J 2010. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS 2007. Quantifying similarity between motifs. Genome Biol 8: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ML, Baxter LL, Loftus SK, Pavan WJ 2010. Sox proteins in melanocyte development and melanoma. Pigment Cell Melanoma Res 23: 496–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. 2010. Nucleosome dynamics define transcriptional enhancers. Nat Genet 42: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, Pu WT 2011a. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci 108: 5632–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Ye J, Cai Y, Riquelme C, Liu JO, Liu X, Han A, Chen L 2011b. Structure of p300 bound to MEF2 on DNA reveals a mechanism of enhanceosome assembly. Nucleic Acids Res 39: 4464–4474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci 106: 9362–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff L, MacArthur J, Wise A, Junkins H, Hall P, Klemm A, Manolio T 2011. A catalog of published genome-wide association studies. www.genome.gov/gwastudies
- Hou L, Pavan WJ 2008. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: Do all roads lead to Mitf? Cell Res 18: 1163–1176 [DOI] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH 2008. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol 26: 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaykina N, Abelson K, King D, Liakhovitskaia A, Schreiner S, Wegner M, Kozlova EN 2010. In vitro and in vivo effects on neural crest stem cell differentiation by conditional activation of Runx1 short isoform and its effect on neuropathic pain behavior. Ups J Med Sci 115: 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingo K, Aunin E, Karelson M, Ratsep R, Silm H, Vasar E, Koks S 2008. Expressional changes in the intracellular melanogenesis pathways and their possible role in the pathogenesis of vitiligo. J Dermatol Sci 52: 39–46 [DOI] [PubMed] [Google Scholar]
- Lang D, Lu MM, Huang L, Engleka KA, Zhang M, Chu EY, Lipner S, Skoultchi A, Millar SE, Epstein JA 2005. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature 433: 884–887 [DOI] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 1: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Karchin R, Beer MA 2011. Discriminative prediction of mammalian enhancers from DNA sequence. Genome Res 21: 2167–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Fisher DE 2007. Melanocyte biology and skin pigmentation. Nature 445: 843–850 [DOI] [PubMed] [Google Scholar]
- Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, et al. 2011. Cistrome: An integrative platform for transcriptional regulation studies. Genome Biol 12: R83 doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus SK, Antonellis A, Matera I, Renaud G, Baxter LL, Reid D, Wolfsberg TG, Chen Y, Wang C, Prasad MK, et al. 2009. Gpnmb is a melanoblast-expressed, MITF-dependent gene. Pigment Cell Melanoma Res 22: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaughey DM, Vinton RM, Huynh J, Al-Saif A, Beer MA, McCallion AS 2008. Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res 18: 252–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKusick VA 1998. Mendelian inheritance in man. A catalogue of human genes and genetic disorders. Johns Hopkins University Press, Baltimore, MD. [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani Y, Hayashi N, Kawashima M, Imokawa G 2010. A single UVB exposure increases the expression of functional KIT in human melanocytes by up-regulating MITF expression through the phosphorylation of p38/CREB. Arch Dermatol Res 302: 283–294 [DOI] [PubMed] [Google Scholar]
- The modENCODE Project Consortium 2009. Unlocking the secrets of the genome. Nature 459: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murisier F, Guichard S, Beermann F 2007. The tyrosinase enhancer is activated by Sox10 and Mitf in mouse melanocytes. Pigment Cell Res 20: 173–184 [DOI] [PubMed] [Google Scholar]
- Nolis IK, McKay DJ, Mantouvalou E, Lomvardas S, Merika M, Thanos D 2009. Transcription factors mediate long-range enhancer-promoter interactions. Proc Natl Acad Sci 106: 20222–20227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan JP, McCallion AS 2010. Genomics of long-range regulatory elements. Annu Rev Genomics Hum Genet 11: 1–23 [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG 2011. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat Rev Genet 12: 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D, Maniatis T, Harrison SC 2007. An atomic model of the interferon-beta enhanceosome. Cell 129: 1111–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG 2009. CTCF: Master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad MK, Reed X, Gorkin DU, Cronin JC, McAdow AR, Chain K, Hodonsky CJ, Jones EA, Svaren J, Antonellis A, et al. 2011. SOX10 directly modulates ERBB3 transcription via an intronic neural crest enhancer. BMC Dev Biol 11: 40 doi: 10.1186/1471-213X-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh E, Cohen S, Levanon D, Groner Y, Gat U 2005. Runx3 is involved in hair shape determination. Dev Dyn 233: 1478–1487 [DOI] [PubMed] [Google Scholar]
- Saha B, Singh SK, Sarkar C, Bera R, Ratha J, Tobin DJ, Bhadra R 2006. Activation of the Mitf promoter by lipid-stimulated activation of p38-stress signalling to CREB. Pigment Cell Res 19: 595–605 [DOI] [PubMed] [Google Scholar]
- Silver DL, Hou L, Pavan WJ 2006. The genetic regulation of pigment cell development. Adv Exp Med Biol 589: 155–169 [DOI] [PubMed] [Google Scholar]
- Song L, Crawford GE 2010. DNase-seq: A high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc doi: 10.1101/pdb.prot5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. 2011. Open chromatin defined by DNase I and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res 21: 1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Hillgartner S, Wegner M 2008. The transcription factor Sox5 modulates Sox10 function during melanocyte development. Nucleic Acids Res 36: 5427–5440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sviderskaya EV, Hill SP, Evans-Whipp TJ, Chin L, Orlow SJ, Easty DJ, Cheong SC, Beach D, DePinho RA, Bennett DC 2002. p16Ink4a in melanocyte senescence and differentiation. J Natl Cancer Inst 94: 446–454 [DOI] [PubMed] [Google Scholar]
- Swope VB, Medrano EE, Smalara D, Abdel-Malek ZA 1995. Long-term proliferation of human melanocytes is supported by the physiologic mitogens α-melanotropin, endothelin-1, and basic fibroblast growth factor. Exp Cell Res 217: 453–459 [DOI] [PubMed] [Google Scholar]
- Tada A, Pereira E, Beitner-Johnson D, Kavanagh R, Abdel-Malek ZA 2002. Mitogen- and ultraviolet-B-induced signaling pathways in normal human melanocytes. J Invest Dermatol 118: 316–322 [DOI] [PubMed] [Google Scholar]
- Thomas AJ, Erickson CA 2008. The making of a melanocyte: The specification of melanoblasts from the neural crest. Pigment Cell Melanoma Res 21: 598–610 [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009a. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA 2009b. Genomic views of distant-acting enhancers. Nature 461: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumoto K, Mahalingam H, Suzuki H, Yoshizawa M, Yokoyama K 1995. Transcriptional activation of the melanocyte-specific genes by the human homolog of the mouse Microphthalmia protein. J Biochem 118: 874–881 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]