Abstract
Epi-fluorescence microscopy is available in most life sciences research laboratories, and when optimized can be a central laboratory tool. In this chapter, the epi-fluorescence light path is introduced and the various components are discussed in detail. Recommendations are made for incident lamp light sources, excitation and emission filters, dichroic mirrors, objective lenses, and charge-coupled device (CCD) cameras in order to obtain the most sensitive epi-fluorescence microscope. The even illumination of metal-halide lamps combined with new “hard” coated filters and mirrors, a high resolution monochrome CCD camera, and a high NA objective lens are all recommended for high resolution and high sensitivity fluorescence imaging. Recommendations are also made for multicolor imaging with the use of monochrome cameras, motorized filter turrets, individual filter cubes, and corresponding dyes that are the best choice for sensitive, high resolution multicolor imaging. Images should be collected using Nyquist sampling and should be corrected for background intensity contributions and nonuniform illumination across the field of view. Photostable fluorescent probes and proteins that absorb a lot of light (i.e., high extinction co-efficients) and generate a lot of fluorescence signal (i.e., high quantum yields) are optimal. A neuronal immune-fluorescence labeling protocol is also presented. Finally, in order to maximize the utility of sensitive wide-field microscopes and generate the highest resolution images with high signal-to-noise, advice for combining wide-field epi-fluorescence imaging with restorative image deconvolution is presented.
Keywords: Epi-fluorescence, CCD camera, Light sources, Filters, Dichroic mirrors, Image collection, Objective lenses, Fluorophore, Fluorescent protein, Deconvolution, 3D rendering
1. Introduction
Epi-fluorescence microscopy is a tool used in virtually all fields of the life sciences. The high specificity of immuno-fluorescent antibodies enables targeting of specific proteins of interest and even specific phosphorylation sites within proteins (1, 2). Advances in filter, mirror, and camera technologies continue to make optical microscopes increasingly sensitive with the ability to detect single fluorescent molecules (3–5). In addition, many possibilities for researchers have been created by the advances in fluorescent protein development and optimization. This has led to the generation of an array of fluorescent proteins with colors spanning the visible spectrum. These probes are available for multicolor live imaging of protein dynamics within single cells, tissues, and organisms (6–8). Lastly, innovations in fluorescent probes allow researchers not only to specifically label cellular organelles and substructures but also to observe protein function in living cells. For example, probes are available for measuring dynamic changes in the concentrations of small molecules such as calcium (9). Taken together, these tools enable studies of the architecture of the cell, of both intra- and intercellular molecular interactions, and of protein function to be carried out with amazing specificity. Combining the images collected on an epi-fluorescence microscope with state-of-the-art image processing and analysis software allows researchers to study the cell in ways that were once inaccessible.
This chapter will present the epi-fluorescence microscope optical train (i.e., light path), details about the components within the light path, and recommendations for these components. The recommendations will focus on ways to develop the most sensitive epi-fluorescence microscope possible in order to minimize light damage to the sample and high background intensities due to auto fluorescence and light scattering. Furthermore, immunofluorescence labeling will be discussed along with an optimized protocol for labeling neurons. Fluorescent probes and proteins will be briefly introduced and readers will be referred to more comprehensive reviews for additional details. Proper techniques for image acquisition, analysis, and processing will be presented including 3D imaging, image deconvolution, and 3D rendering of data.
2. The Fluorescence Light Path
The key to high resolution and high signal-to-noise (S/N) epi-fluorescence imaging is to be able to excite the sample with a specific color of relatively bright incident light, while having the capacity to separate and detect the much dimmer fluorescence being emitted from the sample. If the wavelength (i.e., color) of the incident excitation light is not well separated from the wavelength of the fluorescence, then there can be reflections of incident light off of optical surfaces (e.g., filters, mirrors, lenses) within the microscope. These reflections will increase the background intensity of the image decreasing S/N which will lead to reduced image quality.
The fluorescence microscope light path is made up of many components (Fig. 1a). Incident light from a lamp is focused through a collector lens system, which may include heat and neutral density (ND) filters. Infrared wavelengths from the light source can generate a great deal of heat when they are focused by the objective lens onto the specimen. ND filters, which uniformly attenuate most visible wavelengths of light, reduce incident lamp power and any potential photo-bleaching (Fig. 1b). The light source is aligned (if required) and the light passes through a diffuser to provide an even illumination across the microscope's field of view. The field and aperture diaphragms are aligned and opened so that they are just larger than the field of view. This reduces stray incident light in the system that can lead to sample degradation and decreased S/N. A manual or automatic shutter should be present to block incident light when the sample is not being viewed. This will reduce photo-bleaching, and is essential for live cell time-lapse imaging to reduce photo-toxicity. Some shutters are built into the microscope while other systems require external shutters between the lamp and the microscope stand (10) (Fig. 1b).
Fig. 1.
(a) Cut through of an Olympus IX81 fully motorized epi-fluorescence microscope showing details of the fluorescence light path. (b) Detailed view of the epi-illumination arm for the IX81 microscope showing components of the fluorescence light path. Images (a, b) are courtesy of Michael Davidson, Florida State University, National High Magnetic Field Laboratory, Tallahassee, FL. (c) Schematic of a fluorescence cube for EGFP imaging. Blue light is selected from the white incident light from the lamp using a blue band pass excitation filter. The blue light reflects off the dichroic mirror and is directed upwards to the objective lens then focused on the sample. Green emission light from the sample passes through the dichroic mirror and through a green band pass emission filter to the detector. The emission filter also reduces image background by rejecting any reflected blue incident light.
The subsequent component in the optical train is the fluorescence filter cube (Fig. 1a, b). The first filter in the cube is the excitation filter (Fig. 1c). It is typically a band pass filter that selects a specific range of wavelengths of light from the white light source in order to excite the fluorophore of interest. For example, for EGFP excitation a 470/40 blue band pass filter could be used. The filter nomenclature gives the central transmitted wavelength (470 nm) followed by the band width (40 nm). Thus, the 470/40 filter will transmit light between 450 and 490 nm (Fig. 2a, solid line and solid line with open circles). Incident light then comes to a dichroic mirror (mounted at 45°), which is designed to reflect light below a certain wavelength and pass light above that same wavelength (Fig. 1c; Fig. 2a, dashed line and line with triangles). The selected excitation light is reflected off of the dichroic mirror and is directed at 90° towards the sample. For an inverted microscope, the incident excitation light travels up through the objective lens and is focused onto the sample. The fluorophores within the sample are excited and give off fluorescence. Some light energy is always lost during the fluorescence process, so the emission light is always of lower energy (longer wavelength) than the incident light (e.g., blue light is used to excite green probes). This shift to a longer wavelength is termed the Stokes shift of the fluorophore (11). Fluorescence will be emitted from the dye molecules within the sample in all directions. The emission light reaching the objective lens is focused down through the microscope and passes through the dichroic mirror (Fig. 1c).
Fig. 2.
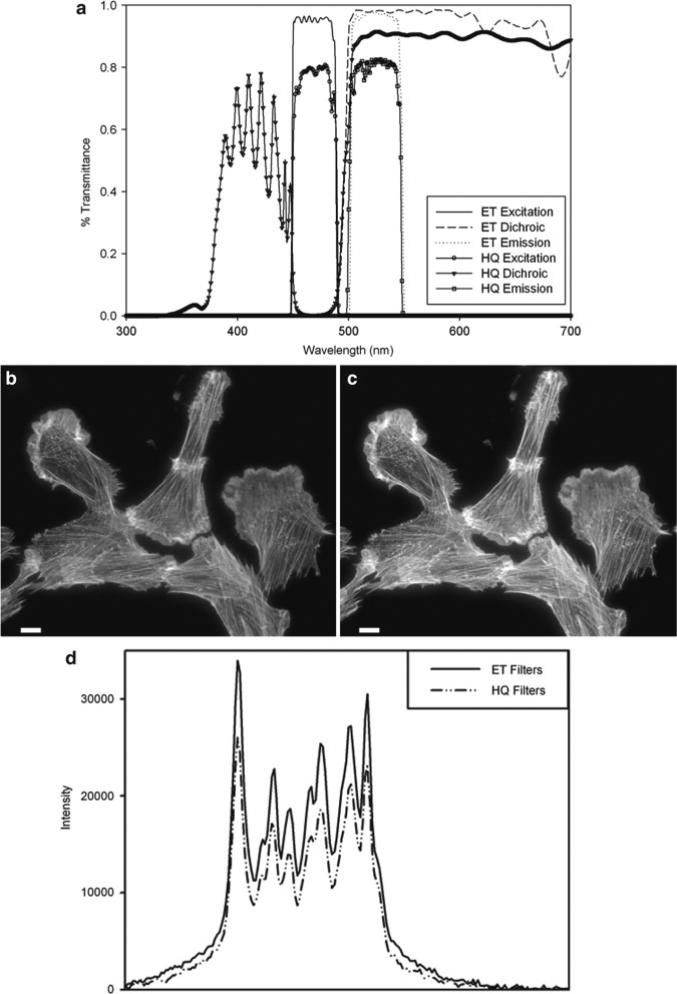
(a) Filter transmission curves for EGFP cubes using either standard soft coated filters (HQ series) or hard coated filters (ET series) from Chroma Technology. (b, c) Cells stained with phalloidin AlexaFluor® fluorophore 488 (Invitrogen) were imaged on a Zeiss Axiovert 200 M microscope with a 63×/1.4 NA oil immersion lens and Axiocam HR camera at full resolution with 500 ms exposure times. Images were taken with either (b) the Zeiss soft coated 13 FITC; or the (c) Chroma Technology ET-GFP 49002 filter cube sets. The mercury lamp was attenuated to 5% power with neutral density filters. The image contrast, brightness, and gamma factor were all adjusted to the same levels for images displayed in (b, c). (d) Intensity pro file from the white line shown in (b, c) for the hard coated ET (solid line) and soft coated HQ (dashed and dotted line) filter sets. The gamma factor was used to bring up dim features within the images so that they are visible by eye. Scale bar is 10 μm.
The final filter within the cube is the emission filter which is green for an EGFP cube (525/50), and is designed to both pass fluorescence emission and block any potential reflections from the incident light that may have passed through the dichroic mirror (Figs. 1c and 2a, dotted line and line with open squares). Most modern fluorescence microscopes are equipped with a multi-position filter cube turret so that multiple fluorescence cubes can be moved in and out of place rapidly. With motorized microscopes these turrets can be automated to facilitate multicolor image acquisition over time (10). Fluorescence emission is then either directed to the eyepieces generating an image on the retina of the observer or sent through another microscope port to a camera detector. The key components of the optical train and how to choose them appropriately will be discussed in detail in the following sections.
3. Components
3.1. Light Sources
(a) Tungsten-Halogen
An incandescent tungsten-halogen bulb (50–100 W) is placed inside of a reflective housing. Lamp intensity is controlled by varying the applied voltage on the bulb. These lamps are typically used for bright field applications but can be used for fluorescence imaging as well (12). Tungsten-halogen lamps use inexpensive bulbs that are long lasting and do not need to be aligned. In addition, for fluorescence live cell imaging there is reduced photo-toxicity due to the low incident light power and the lack of a UV light component. However, since there is no UV component, these lamps cannot be used to image blue dyes, and sometimes the power levels are not high enough to image weak fluorophores such as cyan fluorescent protein (CFP).
(b) Mercury and Xenon Arc
The HBO mercury vapor arc lamp is commonly used for fluorescence microscopy. The lamp is a white light source, but the spectrum shows many bright intensity peaks. Thus, the power of the excitation light for different wavelengths is NOT uniform from the UV to the IR range of wavelengths (13). Bulbs are expensive, last only about 200 h, decay in brightness over time, and have to be disposed of in hazardous waste due to their mercury content. The bulbs also have to be precisely aligned in order to ensure an even illumination across the field of view. At lower magnifications there is typically nonuniform illumination, even if the lamp is well aligned, resulting in a field of view with a bright center. This nonuniformity should always be corrected for when performing quantitative imaging. Details on how to perform this correction will be presented later in this chapter. Arc lamps are intense and can be used to see even very dim fluorescent samples. Typically, it is best to attenuate these lamps using ND filters (to <10%). With reduced incident light, longer camera exposure times will be required in order to generate high S/N images; however, photo-bleaching will be reduced. The XBO xenon arc lamp has a much more even intensity profile across the visible spectrum and into the IR wavelengths (13). This makes it ideally suited for quantitative fluorescence imaging. In general, it is not as intense as the mercury lamp, especially at wavelengths where the mercury lamp spectrum shows intense peaks. Xenon bulbs suffer from the same issues as mercury bulbs. They are expensive, have a short lifetime, and their brightness decays over time.
(c) Metal Halide ( e.g., Chroma Technologies-Photofluor; Lumen Dynamics—X-cite® ; Zeiss-Illuminator HXP 120)
Metal halide lamps offer many advantages over mercury arc lamps. The bulbs last about ten times longer (>1,500 h) and come with the reflector attached, so no bulb alignment is required. The light is coupled to the microscope by a liquid light guide, providing uniform excitation intensity across the microscope field of view. The liquid light guides do have to be replaced every couple of years. However, if there is an internal shutter within the light source, the life of the light guide can be extended. This would also negate the need for an internal shutter within the microscope. The metal halide lamp emission spectrum is similar to the mercury arc lamp, but the intensity between peak illumination bands is up to 50% brighter. This allows for access to the excitation of a broader range of fluorophores (10).
(d) Light emitting diodes (LEDs)
The most recent light source technology is based on LEDs. Commercial systems that are currently available include the Colibri™ (Carl Zeiss, Jena, Germany) (14), PrecisExcite™ (CoolLED Ltd., Hampshire, UK), LED4C (Thorlabs, Newton, NY), X-Cite® X-LED1 (Lumen Dynamics, Mississauga, ON), DM IL LED (Leica Microsystems, Wetzlar, Germany), and AURA light engine (Lumencor Inc., Beaverton, OR). Highly affordable systems can also be custom built with LEDs that are commercially available (15, 16). Essentially, the lamp consists of a number of discrete LED light sources each of a different color with relatively narrow bands of wavelengths. However, many of the LED sources in the green to yellow region of the spectrum are relatively broad (>50 nm) and do require excitation filters in order to avoid the excitation of multiple dyes with one LED. The LEDs can be turned on and off electronically within milliseconds. This eliminates the need for a mechanical shutter for the lamp. In many cases, excitation band pass filters are no longer required. The lamp brightness does not decay over time and ND filters are not needed since LED intensities can be precisely controlled electronically. LED lifetimes are on the order of 10,000 h and they are only turned on during image acquisition. LEDs eliminate the bleed through of unwanted wavelengths (e.g., UV, IR) found with arc lamps and filter-based wavelength selection; they do not produce heat and do not require any alignment. The disadvantages are that systems directly coupled to the microscope often have only three to four different wavelength LEDs available at one time. Other systems combine seven to eight LEDs together making a white light source that can be coupled to the microscope with a liquid light guide. The LED power levels have been steadily increasing, making them suitable for photo-bleaching and photo-activation experiments.
(e) Monochromator
The DeltaRAM X™ (Optical Building Blocks Corp.) and the RatioMaster™ (Photon Technology International, NJ) are commercially available monochromators with ~2 ms switching times between excitation wavelengths. These slit-based systems allow the width of the band pass to be adjusted giving a lot of flexibility so that virtually any dye excited from the UV to the visible range (250–650 nm) can be imaged with a few dichroic mirror sets. These systems eliminate the need for excitation filter wheels, making them ideal for rapid ratio imaging. Nonetheless, monochromators suffer from the same drawbacks as the light sources they use, which are typically Xenon Arc lamps.
3.2. Objective Choice
Phase contrast objectives should not be used in combination with fluorescence microscopy because the phase ring in the objective blocks some of the fluorescence emission light. DIC objectives are preferred when combined with fluorescence imaging. However, the DIC prism and analyzer should be removed from the light path during fluorescence imaging to avoid large losses in light throughput and resolution (17). The brightness (ef ficiency of transmission of excitation light and collection of emission light) of any objective lens is proportional to the numerical aperture (NA) of the lens to the fourth power and to the inverse of the magnification (M) squared (18, 19).
This should be taken into consideration when choosing between objective lenses. For example, a 60×/1.4 NA oil immersion lens will have a higher light throughput than a 100×/1.4 NA lens with the same resolving power. In fact, a 40×/1.2 NA water immersion lens will have ~20% higher light throughput than a 60×/1.4 NA lens at the expense of some resolution. Since the NA dependence is to the fourth power, even small changes in NA can lead to significant changes in lens brightness. For example, when comparing the intensities of the images collected in Figure 3 using three different 20× lenses, the 0.75 NA lens has the highest brightness (Fig. 3c) while the 0.5 NA lens collects only 40% of the relative signal (Fig. 3b) and the 0.4 NA lens only 23% (Fig. 3a). In general, the lowest magnification needed to see the features of interest should be used because higher magnification decreases brightness with a squared dependence. Ordinarily, plan-apochromatic lenses are recommended because they are corrected for field curvature, spherical aberrations, and chromatic aberrations. In other words, images are in focus in the center and at the edges of the field of view, there is little distortion when focusing through the sample, and focus differences between different colors are minimized.
Fig. 3.
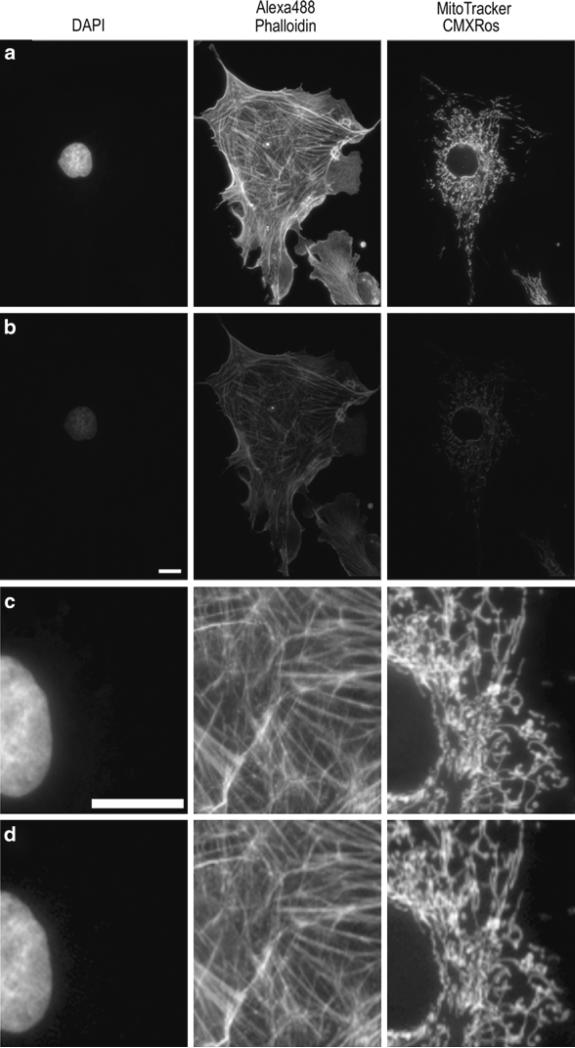
Images of BPAE cell slide from Molecular Expressions labeled with DAPI, AlexaFluor® 488 Phalloidin, and MitoTracker® CMXRos. Images were collected on an Olympus IX71 microscope coupled with a mercury HBO lamp and using DAPI, FITC, and Texas Red BrightLine® hard coated filter sets (Semrock). Images were collected on a Retiga 2000R camera at full resolution with the same settings using three different 20× lenses. The lamp intensity was attenuated to 6%, with exposure times of 250 ms for DAPI, 1,000 ms for AlexaFluor® 488, and 500 ms for MitoTracker® used. Lenses used were (a) Plan 20×/0.4 NA; (b) UPlanFL N 20×/0.5NA; (c) UPlanSApo 20×/0.75NA. Images of the same sample as in (a–c) were collected using a 40×/0.6NA LWD, U PLAN FLlens with a coverslip thickness correction collar. Images were collected with the collar set correctly to 0.17 mm (d) or incorrectly to 1 mm (e). The image contrast, brightness, and gamma factor were all adjusted to the same levels for the images displayed in (a–c) as well as and for (d, e). The gamma factor was used to bring up dim features so they are visible within the images by eye. Scale bar is 10 μm.
All high NA immersion objectives are corrected for imaging through a glass coverslip that is ~170 μm thick; therefore, it is important to mount samples using #1.5 coverslips and not the thinner #1 or thicker #2 coverslips. Some objectives actually have a correction collar that can be adjusted such that samples on glass or thick plastic tissue culture plates (~2 mm thick) can be imaged with the same lens. It is important to adjust the correction collar for the correct substrate thickness or images will be of low resolution and light throughput will be significantly reduced (Fig. 3d, e). If imaging more than a cell layer or two, it is also important to match the index of refraction of the media to the lens. For example, water immersion lenses when working with thick biological samples, because there is a reduction in spherical aberrations when imaging into tissue with a high water content (20).
3.3. Filter Choice
Traditionally, produce higher resolution images fluorescence filters and mirrors have been designed using “soft” coatings whereby layers of low-optical-index material are evaporated onto optical quality glass surfaces. Blocking and transmission of different wavelengths of light to different degrees depends on the thickness, number of layers, and the order that the different types of materials are coated onto the filter. These coatings are often made on colored glass to further block undesired wavelengths of light, but this causes a reduction in transmission of specific wavelengths as well. “Soft” coated filters are not very durable, are difficult to clean without damaging the coatings, and degrade over time due to exposure to high intensity light and humidity. Technologies for reliably producing “hard” coated filters by depositing metal-oxide films on optical quality glass have been developed (Chroma Technology—ET Series, Omega Optical—QMAX, Semrock—Brightline) (21, 22). These filters are very robust, do not degrade under normal conditions within the microscope, and are atomically flat thereby reducing nonspecific light scatter. Since the metal-oxide layer thickness can be precisely controlled, these filters have much better blocking and transmission properties as well as sharper cutoffs between blocking and transmission wavelengths (Fig. 2a, compare ET (Hard coated) with the HQ (Soft coated)). Images taken with traditional soft coated filters (Fig. 2b, d) are ~30% dimmer than images taken with exactly the same imaging conditions using a hard coated filter (Fig. 2c, d). Importantly, since the efficiency of fluorescence light transmission is nearing 100%, the incident lamp power can be reduced, minimizing photo-bleaching, photo-toxicity, and background. It is recommended to replace soft coated filters with “hard” coated filters. The new filter sets can be mounted in existing filter cubes so the cost of upgrading is minimal. This is especially important for live cell applications in order to reduce photo-toxicity.
For multicolor imaging, if time is not an issue, it is best to image each fluorophore separately with single fluorophore fluorescence cubes. If speed is an issue, or the filter turret is not motorized, then multiband mirrors can be used in place of dichroic mirrors. There are two main disadvantages when using multicolor cubes: (1) to avoid excitation and emission cross-talk between different dyes, the band pass filtering for both the excitation and emission filters has to be very narrow; (2) the coating on the multi-chroic mirror for excitation of one dye often reflects fluorescence emission from another dye. For example, red fluorescence is excited by green light, but some of the light coming from the green dye will also reflect off the mirror and will not be detected. Taken together, these factors result in multicolor cubes having a much lower light throughput.
3.4. Cameras
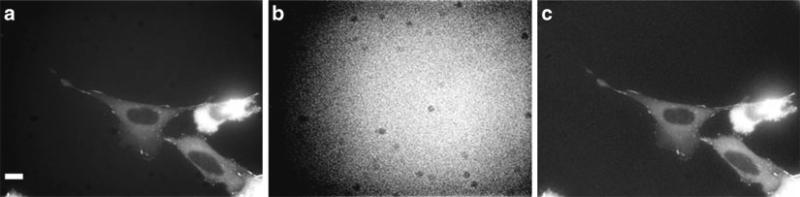
Charge-coupled device (CCD)-based cameras are most commonly used for high resolution fluorescence imaging. In general, color cameras should not be used for fluorescence imaging because they have lower resolution and lower sensitivity than monochrome cameras. Most color CCD cameras use a Bayer filter placed over the CCD pixel array making 25% of the pixels red, 50% green, and 25% blue. Therefore, much of the emitted light is not collected (Compare Fig. 4a, b) and the resolution is lower (Compare Fig. 4c, d). In this example, images were collected with exactly the same conditions, but with the color camera only ~12% of the blue light, ~20% of the green light, and ~11% of the red light is detected relative to the monochrome camera (Fig. 4). Therefore, it is preferable to use red, green, and blue filter sets, take three images on a monochrome camera, and use software to apply false color post-acquisition.
Fig. 4.
Images of the same sample collected on the same microscope as in Fig. 2. Images were captured with a Retiga 2000R monochrome (a, c) or color (b, d) camera at full resolution with a 60×/1.42 NA, PlanApoN objective. All images were collected with the same settings with the lamp intensity attenuated to 6% and exposures of 200 ms for DAPI, 500 ms for AlexaFluor® 488, and 50 ms for MitoTracker®. Images in (a, b) are presented with the same image display settings. The gamma factor is used to bring up dim features so they are visible within the images. Images in (c, d) are zoomed in areas of the images in (a, b). Images in (d) were further adjusted for brightness and contrast so they appear to have similar brightness when compared to (c) in order to provide a fair comparison of the image resolution. Scale bars are 10 μm.
There are many ways to automate multicolor imaging. For maximum sensitivity, it is best to put the different filter cube sets for each dye in a motorized microscope turret. For increased speed, a multiband dichroic mirror can be used in combination with motorized excitation and emission filter wheels. LED- or monochromator-based light sources can be used instead of excitation filter wheels. Liquid crystal filters (e.g., QImaging Corporation, Tucson, AZ) can be used to select for red, green, or blue emission; however, a lot of light is also lost. For maximal sensitivity and speed, there are color cameras equipped with three CCD chips and filters within the camera head to split the light to red, green, and blue components with minimal light loss and no resolution loss (e.g., ORCA-3CCD, Hamamatsu, Japan). Many of the high resolution scientific grade CCD cameras on the market are based on the Sony ICX285 sensor with 1,392 × 1,040, 6.45 × 6.45 μm pixels (e.g., Andor Technologies Clara; Hamamatsu OrcaER; Photometrics CoolSNAP ES2; QImaging Retiga EXi). This chip has a quantum efficiency (QE) of 60–70% in the visible range meaning 60–70% of the photons that reach the camera will be detected. What differs between the manufacturers is the electronics surrounding the chip and the acquisition software. All cameras should be tested for a given application before a purchase is made.
For high speed applications, electron multiplied (EM) CCD cameras are recommended. The EM-CCD sensor array includes an electron multiplication gain register that all pixel signals pass through before being read out, thus signal, but not read noise, is amplified. The top of the line EM-CCD cameras use back thinned sensors with >90% QE in the visible range (e.g., Andor Technologies iXON; Hamamatsu ImagEM; Photometrics Evolve; QImaging Rolera MGi Plus).
There are also high resolution EM-CCD models with a 1,024 × 1,024 pixel array with 13 × 13 μm pixels; however, these chips are not back-thinned so the QE is in the 60–70% range (Andor Technology LucaEM; Hamamatsu ImageEM 1K; Photometrics Cascade II:1024). Nevertheless, if speed is not an issue, standard CCD sensors still have at least twice the resolution when compared to EM-CCDs. A nice option is to have a high resolution CCD camera on one detector port of the microscope for high resolution imaging and an EM-CCD on another detector port for high speed imaging. Devices are also available to split multiple colors of light onto the two halves/quarters of the CCD array allowing for simultaneous two/four color imaging (Dual-View, Quad-View—Photometrics, Tuzson, AZ), which is great for high speed applications.
Cooling of cameras to 0°C or slightly below is usually significant for the exposure times used with biological samples. Cooling to below –30°C is typically not necessary as dark currents (caused by thermal noise) are not significant with exposure times of a few seconds or less. More information on cameras for fluorescence microscopy is also available (23–25).
Finally, the latest development in camera technology is the advent of scientific CMOS (sCMOS) camera technology (Andor Technology—Fairchild Imaging—PCO.imaging sCMOS; Hamamatsu-Orca-Flash2.8). Early results suggest that these cameras have the potential to replace CCD devices with similar sensitivity, higher imaging speeds, lower noise, and higher resolution.
4. Fluorescence Probes and Immuno-fluorescence
Introduction to Fluorescence
This section will serve as a brief introduction to the process of fluorescence. Readers are referred to other publications for more in-depth discussions (11, 26, 27). Incident lamp excitation light is focused by the objective onto a fluorescently labeled specimen. Fluorophores within the specimen absorb light energy and consequently enter the singlet excited state. The singlet excited state is unstable and therefore the molecules want to emit excess energy and relax back to the ground state. Some of the excited molecules can relax back to the ground state by emitting light energy in the form of fluorescence. Some energy is inevitably lost through vibrations of the molecule during this process, so emitted fluorescent light is always of lower energy, or longer wavelength, than the incident light. This process is termed the Stokes shift (11, 27) and allows for the separation of the excitation light and the emission light using filters. During epi-fluorescence microscope imaging, incident light is continuously exciting fluorophores within the specimen. Each fluorophore will cycle between the ground state and the excited state, repeatedly giving off many photons of light with each cycle.
The quality of a fluorophore can be characterized by three fundamental properties: (1) the extinction co-efficient or the ability of the fluorophore to absorb light energy at a given wavelength; (2) the quantum yield or the percentage of absorbed light that produces fluorescence emission vs. other non-radiative processes (e.g., intersystem crossing, vibrational relaxation); and (3) the photo-stability of the fluorophore which is dependent on how stable the chemical structure of the dye is in the excited state. Optimization of all three of these properties is important; for example, a molecule may absorb a lot of light energy, but very little of it may result in fluorescence, or it may photo-bleach rapidly. In turn, if a molecule does not absorb a lot of light, but most of it results in fluorescence, and the molecule does not photo-bleach, it could be a very robust fluorophore.
Most photo-bleaching occurs when excited molecules undergo intersystem crossing and enter a long lived highly reactive excited triplet state (11, 26, 27). A molecule in the triplet state can absorb a second photon of light, and this excess energy can induce chemical reactions between the dye and other molecules. Triple state reactions can cause the nonreversible destruction of the fluorophore so that it can no longer cycle between the ground and excited states and give off fluorescent light. Triplet state reactions can also cause the generation of oxygen radicals which in turn can cause the destruction of the fluorophore and/or cause photo-toxicity to living cells and tissues. The best way to combat photo-bleaching is to use a stable fluorophore that is not likely to enter the triplet state in combination with minimal light exposure. For fixed samples, photobleaching can be minimized by using mounting medium containing anti-fade reagents.
An example of the importance of considering the extinction co-efficient, the quantum yield and the photo-stability of fluorophores is provided from the AlexaFluor® fluorophore series (Life Technologies–Molecular Probes). AlexaFluor® 633 is often a fluorophore of choice because the excitation is well matched with the 633 nm LASER excitation found on many confocal systems. However, AlexaFluor® 633 has a relatively low quantum yield and photo-bleaches quite readily when compared to AlexaFluor® 647. In addition, the extinction co-efficient for AlexaFluor® 647 is 237,000, while for AlexaFluor® 633 it is 100,000; thus AlexaFluor® 647 absorbs ~2.4 times more light when compared to AlexaFluor® 633. Therefore, the AlexaFluor® 647 is a much better fluorophore even when using a 633 nm excitation source.
4.1. Fluorescent Proteins (FP)
The importance of green fluorescent protein (GFP) was recognized with a Nobel prize in 2008 for Osamu Shimomura who first isolated the protein (28), Martin Chalfie who first demonstrated that GFP could be used as a genetic tag (29), and Roger Y. Tsien who characterized the photochemistry and photophysics of GFP as well as extended the color palette, generating new FPs with an array of colors (30). These FPs can be characterized by the same three parameters as any fluorophore, namely absorption, quantum yield, and photo-stability. There are several reviews that discuss and compare the rapidly changing array of available fluorescent proteins (6, 8, 31–33). It is also important that the FPs are not causing protein–protein interactions via dimerization sites in the FP itself. Thus, monomeric versions of the proteins (e.g., mEGFP) should be used (34), and expression levels should always be kept low to avoid over-expression artifacts. Aside from EGFP, some of the brightest and most photostable FPs include mCitrine (35) or venus-YFP (36); killer orange (KO) (37); mCherry (8); and teal FP (TFP) (38), although this list is constantly evolving (39–41).
4.2. Fluorescent Probes and Biosensors
Thousands of fluorescent probes are commercially available, and it is not a trivial task to make the best choice for labeling cellular tissue. Publications are available, that provide advice about choosing the appropriate fluorescent probes (42, 43). Fluorescent probes are also available for many organelle and subcellular compartments (e.g., lysosomes—LysoTracker; Life Technologies), for measuring calcium concentrations (e.g., Fura-2AM, Premo-Cameleon; Life Technologies), for measuring intracellular pH (e.g., pH sensitive FPs (44), BCECF; Life Technologies), or for chloride ion concentrations (45). More sophisticated FP-based biosensor probes are also available for activation of the Rho family of proteins (46–48). Quantum dots are nanoparticles that are very photostable, absorb a lot of light, and have large fluorescence quantum yields. Recent developments have begun to solve some of the problems associated with the use of these heavy metal particles in living cells, and they are emerging as a useful fluorescence tool, particularly in the field of single molecule imaging (49, 50).
4.3. Immuno-fluorescence
Immuno-fluorescence antibody availability has literally exploded over the last decade with commercial constructs available for thousands of proteins, including direct targets such as phospho-specific and kinase-specific antibodies (Biosource, Cell Signaling Technology, Affinity BioReagents). Immuno-fluorescence is particularly valuable for detecting endogenous proteins, which can serve as molecular markers in many cell types, including fibroblasts and neurons. In hippocampal neurons, immuno-fluorescence staining is especially useful for detecting synaptic proteins on both pre- and postsynaptic terminals. A commonly used presynaptic marker is the synaptic vesicle protein SV2, which is found in the presynaptic terminals of both excitatory and inhibitory synapses. The postsynaptic density (PSD) protein PSD-95 localizes to the postsynaptic side of the shaft and the spine excitatory endings. As such, it is an excellent marker for the PSD of excitatory synapses.
5. Sample Immuno-Fluorescence Staining Protocol
- Fix cells grown in vitro on coverslips by incubating as follows:
- Presynaptic Proteins: 4% paraformaldehyde (PFA)/4% sucrose in phosphate buffered saline (PBS) for 15 min at room temperature.
- Postsynaptic Density Proteins: methanol at –20°C for 20 min.
- PSD and EGFP Fluorescence: 1% PFA/4% sucrose in PBS for 3 min at room temperature, followed by methanol at –20°C for 10 min.
Rinse three times with PBS.
Permeabilize cells with 0.2% Triton X-100 for 5 min at room temperature.
Rinse three times with PBS.
Place coverslips in a humid environment to prevent them from drying out. To create a humid environment, wet Kimwipes can be placed in the incubation chamber.
Incubate with 100 μL of 20% goat serum at room temperature for 1 h to block nonspecific binding.
Dilute primary antibody in 5% goat serum at a dilution of 1:100. Note: This is a good starting concentration but lower or higher concentrations may be fine depending on the antibody. Always test a number of concentrations and use the lowest concentration that gives good specific binding and little nonspecific binding.
Incubate the coverslips with 100 μL of primary antibody for 1 h at room temperature or overnight at 4°C.
Rinse three times with PBS.
Dilute a fluorophore-conjugated secondary antibody 1:500 in 5% goat serum, and store in the dark. Again, the concentration of secondary antibody should be tested and the lowest concentration that gives good specific and minimal nonspecific labeling should be used.
Incubate the coverslip with 100 μL of secondary antibody for 1 h at room temperature. Do not incubate with secondary antibodies overnight, as it typically increases nonspecific binding without any significant improvement in specific binding. This may not be the case for thick tissue samples where it may take time for the secondary antibody to penetrate the tissue.
Rinse three times with PBS.
Place ~10 μL of mounting media containing an anti-fade agent (e.g., Aquamount; ThermoFisher) on a microscope slide and invert the coverslip onto the slide. Use a cotton swab to press down gently on the coverslip and direct any air bubbles to the edges.
Coverslips can be sealed with clear nail polish to prevent evaporation. However, there is evidence that the solvents in nail polish can alter the specimen integrity or the fluorescence of certain dyes. A more benign sealant is VALAP, which is a 1:1:1 mixture of Vaseline, lanolin, and paraf fin. This mixture can be made by melting the three components in a beaker on a hot plate, letting it cool and then cutting it into small blocks. These blocks can be melted and dripped onto the edges of coverslips using a glue gun or soldering iron. A third option is to use a product called Twinseal available from Picodent, Wipperfurth, Germany. The yellow and blue components are mixed 1:1 and spread around the coverslip and left for 5 minutes to harden.
In general, a mounting media that cures and hardens is the best for specimen preservation (e.g., Shandon Immu-Mount-ThermoFisher; CytoSeal-Edmund Scientific; Prolong Gold-Invitrogen). Slides can be kept for weeks or months when stored in the dark at 4°C.
Neurons were stained for synapses with an SV2 (Fig. 5a) primary antibody and AlexaFluor® 488 (Life Technologies) secondary antibody. Filamentous actin was stained with phalloidin-TRITC (Fig. 5b). Phalloidin-TRITC was incubated in the same step as the primary anti-SV2 antibody after goat serum blocking. Phalloidin is commonly used to visualize dendritic spines since filamentous actin is abundant in these structures (Fig. 5b) (51, 52). Note: phalloidin staining does not work well with methanol fixation. For live cell work, transfection with plasmids for the membrane localized GAP-GFP protein conjugate (the palmitoylation and membrane targeting domain of neuromodulin (GAP-43)) provides a nice outline of dendrites and axons (53).
Fig. 5.
Neurons were fixed with 4% paraformaldehyde/4% sucrose in phosphate buffered saline (PBS) for 15 min at room temperature and permeabilized with 0.2% Triton X-100 for 5 min at room temperature. (a) Cells were stained with a SV2 mouse monoclonal antibody (1:250 dilution) from the Developmental Studies Hybridoma Bank (The University of Iowa, Iowa City, IA) and anti-mouse Alexa Fluor® 488 (1:500 dilution). (b) Cells were co-stained with phalloidin-TRITC (1:250 dilution) from Sigma (St. Louis, MO) by incubating for 1 h at room temperature in the same step as the primary SV2 antibody. Neurons were imaged with a Retiga EXi CCD camera (QImaging, Surrey, BC) attached to an Olympus IX71 inverted microscope (Melville, NY) with a 60×, PlanApo/1.45 NA objective (Olympus). Image acquisition was controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA) interfaced with a Lambda 10-2 automated controller (Sutter Instruments, Novato, CA). For SV2, an Endow GFP Band pass filter cube (excitation HQ470/40, emission HQ525/50, Q495LP dichroic mirror) (Chroma, Brattleboro, VT) was used. Phalloidin was imaged with a Chroma TRITC/Cy3 cube (excitation HQ545/30, emission HQ610/75, Q570LP dichroic mirror). Scale Bar is 10 μm.
6. Image Collection, Corrections, and Display
6.1. Collecting Images
When collecting fluorescence images, it is important that the camera settings are optimized. Camera pixels can be thought of as “buckets” that are filled up with photo-electrons. For fixed specimens, it is possible to use most of the dynamic range of the camera, namely for each image acquisition fill up each pixel nearly to capacity with photo-electrons. Under these conditions, high S/N images with many intensity values (i.e., gray levels) are generated. However, it is critical to make sure that no pixels, of the camera are saturated (over filled with photo-electrons). Saturated pixels no longer contain quantitative data. In addition, photons can spill over to neighboring pixels, affecting the quantitative nature of the intensity data within them as well (54). It is critical to make sure the camera and the objective resolutions match so that the pixel size is ~2–3 times higher than the resolution of the objective lens for a given wavelength. Set the exposure time to fill the camera dynamic range to ~75% with your brightest sample in order to leave room for sample variability. In general, it is better to use lower incident light powers and longer camera exposure times. This will increase the signal while reducing camera noise and fluorophore photo-bleaching. When imaging living samples light exposure needs to be reduced, so it is rare that the full dynamic range of the detector can be used.
It is also important to let your incident light source warm up. You should use a fluorescent plastic test slide in order to measure the light source intensity during warm up and the intensity stability over time to ensure it does not vary during your experimental timeframe (55). Some light sources may need more time to warm up (mercury arc lamps), while others may not need to be warmed up at all (LED light sources).
6.2. Saving Images
To perform accurate quantitative image analysis, it is crucial that images are saved in a format that maintains the raw data intact. Often software programs will default to formats such as .jpg which use compression algorithms that result in data loss. If a 12-bit camera has 4,096 gray levels, but the image is saved as an 8 bit image with 256 gray levels, a great deal of image detail will be lost, potentially losing the ability to differentiate between subtle changes in fluorescence intensity. In general, images should always be saved in the format of the software platform to retain the “meta” data about the image acquisition (e.g., exposure time, pixel size, objective used). Then, if need be, images can also be saved in .tif or .raw format for use in other software programs. Other image formats such as jpg, bmp, or png are useful for visualization but not for quantification. Always retain a copy of the original images when performing any data processing or analysis.
6.3. Correcting for Nonuniformity and Background
With any quantitative fluorescence imaging, it is important to do proper corrections for background intensities within the images (54). Simply taking the intensity of a region of interest (ROI) within the image where there are no cells and subtracting it from the image is often not sufficient. Mercury- or xenon-based light sources deliver a nonuniform illumination of the field of view. This results in a bright region in the center of the field of view with darker regions at the edges (Fig. 6a; background intensity values vary by 200–300 intensity units). To correct for this nonuniform incident light, an image should be acquired in a region of the sample where there are no cells (Fig. 4b), or by using a fluorescence plastic slide (Chroma Technology Corp; Rockingham, VT) that is the same color as the dye you are imaging (e.g., yellow-green if imaging EGFP). The fluorescent slide will give a better result because images will have higher S/N and will not add noise to the specimen images when performing corrections. Shading correction features within image processing software can be used to correct images. However, if these programs are not available the basic process is the following: (1) normalize the image of the fluorescent plastic slide and the image of the specimen by dividing by the maximum intensity and then multiplying a scale factor of 100 (8 bit images) or 1,000 (12 bit images) to maintain integer intensity values; (2) divide each normalized specimen image by the normalized background image to remove field nonuniformity. Ensure no pixel values read below 1; if they do, the scaling factor will need to be adjusted; (3) take the corrected specimen image and subtract the average intensity of an ROI where there are no cells from each pixel in the image (Fig. 4c). Background images should be taken for each color being acquired, and more importantly, for each objective lens being used. If these corrections are not done, cells in the center of the field will appear to have more fluorescent protein or dye than those at the edges, producing erroneous intensity information.
Fig. 6.
Images of living CHO-K1 cells stably expressing paxillin-EGFP were collected with a CoolSNAP EZ camera with 2 × 2 binning on an Olympus IX71 microscope equipped with a 60×/1.45 NA oil immersion objective. Excitation was from a mercury lamp using a custom EGFP cube from Chroma Technologies (hq480/20×, z488rdc, hq525/50 m). (a) Image corrected for the background intensity of an ROI. (b) Image of the background intensity in a region of the sample with no cells collected with the same parameters as in (a). (c) Image corrected using a normalized background intensity image of (b), followed by subtraction of the average background intensity of an ROI with no cells. The brightness, contrast, and gamma factor are set to the same levels for (a, c), but were adjusted in order to visualize the background intensity in (b). Scale bar is 10 μm.
6.4. Image Display
Many of the problems with obtaining fluorescence images result from default image display settings within the acquisition or image processing and analysis software. Often images are displayed with auto-contrast on and samples that have very different intensities appear to have the same brightness. Other times, an image appears black, but the data are simply not filling up a large part of the dynamic range of the camera. This is often the case when taking images from a 12-bit camera in .tif format into Photoshop. The images are saved on the computer as 16-bit and therefore they appear black in Photoshop because most of the 16-bits are empty. This can be corrected by adjusting the image brightness and contrast so the data can be observed. Care must be taken, however, as Photoshop, unlike most scientific image analysis programs, will change the raw intensity data within the images when brightness and contrast are changed. The gamma factor is a nonlinear adjustment of the image intensity that can be useful when observing both bright and dim image features, such as actin filaments and soluble actin at the edge of the cell (54). When displaying images for figures it is best to use monochrome or grayscale look up tables (LUTs) as the human eye is not very sensitive to blue and red. Specialized rainbow, fire, or spectrum LUTs are very useful when trying to display subtle differences in intensity within or between images. When displaying images for publication, it is important to disclose any image manipulations including brightness, contrast, and gamma factor adjustments in the figure captions. It is also important to manipulate all images within a panel or sequence in the same way if the images are to be compared. Furthermore, all quantitative analysis must be done on the raw image data, not on images that have been manipulated.
6.5. Correcting for Excitation and Emission Cross-talk
As discussed above, there are many ways to collect multicolor image data. The method of choice depends on the hardware available and the need for sensitivity vs. speed. In all cases; for quantitative imaging it is critical to perform proper controls and correct for any excitation or emission cross-talk within the system. Excitation cross-talk occurs when a dye is excited by the same color of incident light as another dye. One common example is RFP or DsRed and EGFP, because RFP and DsRed absorb a substantial amount of light at 488 nm. Emission cross-talk is far more common and results from the fact that most organic dyes have a fluorescence emission curve with a long tail out into the red end of the visible spectrum. Therefore, cross-talk usually occurs from shorter wavelength dyes into the longer wavelength detector. In this case, the EGFP signal would be detected in the RFP imaging channel. Both types of cross-talk can be minimized by imaging dyes sequentially rather than simultaneously. However, if speed is required, this is not an option. In any case, images should be collected for control samples independently labeled for each dye as well as for unlabelled cells to assess any contributions from cellular autofluorescence. Control images should be collected with exactly the same settings that are used during experimentation on multiple labeled samples. If no significant cross-talk is seen (e.g., no image in the RFP channel with a sample only containing EGFP), corrections may not need to be done. Otherwise, corrections are relatively straightforward. Take the EGFP and RFP example. A sample with EGFP alone is imaged for EGFP (green channel) and RFP (red channel). If there is significant emission cross-talk, there will be some signal in the RFP channel even though the sample does not contain RFP. The EGFP and RFP images should be corrected for field nonuniformity and background. Then, bright features within the EGFP image can be selected using the thresholding feature of any image analysis program. A mask of the EGFP staining can be created and the average intensity of these regions can be measured. This mask can then be transferred onto the RFP image and the average intensity of the emission cross-talk can be measured. Cells can also be manually outlined or several regions of interest can be drawn and the intensities can be averaged. This analysis should be done for 10–20 cells, and the average percentage of cross-talk can then be calculated. For instance, let's say the cross-talk is high at 25%. For the experimental data, the corrected EGFP images should be multiplied by the cross-talk percentage and then this image should be subtracted from the corrected RFP image. In other words, 25% of the signal from the EGFP would be removed from the RFP image. These types of corrections need to be done for all of the fluorophores within a given experiment. Although it is preferable to use dyes that have well-separated excitation and emission spectra, in general, as long as control images are available, it is relatively straight forward to correct or “un-mix” the fluorescence signals (56).
7. 3D Imaging and Deconvolution
When 3D imaging and deconvolution are applied to widefield images, they make the epi-fluorescence microscope a much more powerful research tool. In fact, widefield microscopes can be much more sensitive than confocal microscopes because out-of-focus light that is rejected by the confocal pinhole is detected (57). This combined with the use of highly sensitive CCD cameras (QE >60%) make the widefield microscope the preferred choice for low light applications, such as single molecule or livecell microscopy (58). Traditionally, two things have made image deconvolution difficult: (1) the requirement of powerful computers and hours of computation time; and (2) the need to wade through the many algorithms—published and proprietary—for performing image deconvolution. Recent improvements in computer power such as quad processors have helped to overcome the computing power and time issues, while many references have been written to try and guide researchers through deconvolution algorithms (59–65). An introduction to the concepts and utility of deconvolution will be presented here, thus showing that a more affordable epi-fluorescence microscope can perform just as well as a laser scanning confocal microscope for many applications. Of course, for thick samples (>20–30 μm) where there is a high amount of light scattering and out-of-focus light, confocal microscopes will still be required.
7.1. Collecting 3D image series
When collecting a 3D image series, it is important to use the least amount of incident light required to achieve enough S/N in order to see the objects of interest. This is especially true if many image planes need to be imaged so that photo-bleaching is minimized. In general, to make the most of the imaging system, sampling in x, y, and z should be consistent with Nyquist sampling, or the sampling rate (pixel size) should be ~2.3× higher than the resolving power of the system (66). For example, when imaging with a 60×/1.4 NA oil immersion objective and 550 nm emission light, the resolution of the system is ~0.2 μm. Therefore, the pixel size in x and y should be ~90 nm. The resolution along the z-axis is typically ~3 times lower than in the image plane. Therefore, an image spacing of ~250 nm along the z-axis would be optimal for this objective lens. However, for image deconvolution the specimen is often oversampled with slightly higher x, y, and z-axis sampling. The specimen should be imaged in z to a distance equal to the depth-of-field for the objective lens above and below the focal point (59). In practice, this distance can be estimated using a fast of live camera mode and moving the z focus to a position where the image minimum and maximum intensity values do not change between successive z-planes (59). For live cell imaging, undersampling along the z-axis will reduce photo-toxicity and help to maintain cell viability.
7.2. Image deconvolution
When imaging point sources through a microscope, the image that is measured is actually the result of light diffraction. As light from the point sources travels through the microscope, it spreads out due to diffraction so that sub-resolution objects within images appear to be much bigger than they are. Therefore, all sub-resolution objects will appear to be the same size within the image regardless of their actual size (62). Because the light is spread out by the optics of the system, the 3D image of a point source is termed the point spread function (PSF). The shape of the PSF is determined by the wavelength of light, the objective lens, the refractive index of the medium, and many other factors. Sub-resolution fluorescent micro-spheres can be used to measure the PSF of the microscope. This PSF can then be used to test the quality of the microscopes objective lens and to determine the microscope resolution (67). When a more complex fluorescently labeled specimen is imaged with the epi-flourescence microscope, the image that is generated can be considered to be the sum of thousands or millions of PSFs generated by each fluorophore molecule. Due to the spreading out of the fluorescence light along the z-axis, there are contributions within each image plane from objects that are out-of-focus. This “blur” reduces the image S/N and contrast. Image deconvolution is essentially a mathematical technique whereby images are processed post acquisition in order to remove the “blur.”
7.3. Deblurring or Nearest-Neighbor Non-quantitative Deconvolution
Some deconvolution techniques are designed to work very quickly, generating “deblurred” images within seconds. These algorithms typically rely on the assumption that the out-of-focus light in the image plane is mainly derived from light coming from the two adjacent image planes, one above and one below. Out-of-focus light from other planes is assumed to be negligible. These algorithms remove the out-of-focus light by creating blurred images from the planes directly above and below a selected image plane and subtracting them from the in-focus image. These techniques use subtraction, so sensitivity is lost, and because they use estimates of the out-of-focus light and ignore light coming from other image, planes, they are only useful for visualization and are not quantitative (60, 61).
7.4. Quantitative Restorative Deconvolution
If the PSF of the optical system has been measured, then 3D image stacks can be de-convolved in order to make an estimate of what the actual specimen looks like before being convolved with the microscope optics (59–65). The basic process is an iterative one. The theoretical PSF is calculated, or the PSF of the microscope is measured using sub-resolution fluorescent microspheres. An initial estimate of what the specimen looks like is generated based on the resulting fluorescence image. This estimated image of the “object” is then mathematically convolved with the PSF to generate a “theoretical” image. This add quotes around theoretical theoretical image is then compared by statistical analysis to the actual image. This comparison is used to refine the “theoretical” image and the process is repeated difference between the “theoretical” image and the actual image reaches a user defined minimum (59–65). A choice has to be made with restorative deconvolution as to whether the algorithm operates using a theoretical PSF calculated from the optical properties of the microscope (i.e., objective lens, numerical aperture, x, y, z sampling frequency, color of light), or a PSF measured on the actual optical platform using sub-resolution (~100 nm) fluorescent microspheres. In general, if the measured PSF is of poor quality the theoretical PSF will give better results. However, there are subtle changes in the PSF for each objective lens, sample preparation, mounting media, and so on, and thus, a well-measured PSF will provide the most accurate results. It is important to collect the PSF using the same imaging conditions as for the sample. Therefore, measuring the PSF with micro-spheres in mounting media is not appropriate if images will be collected from cells mounted in aqueous buffer. The optimal preparation is to make an unlabelled sample and place microspheres on the surface of the sample, for example, microspheres on top of a cellular monolayer. It is even better if microspheres are within the sample (i.e., inside the cell), but this can be difficult to do without introducing other artifacts, such as microsphere aggregation or accumulation in internal vesicles. Commercial software packages, such as Huygens software (Scientific Volume Imaging) and AutoQuant (Media Cybernetics), are available for performing restorative deconvolution.
7.5. Blind Deconvolution
Blind deconvolution is a modification of restorative deconvolution that minimizes the need to accurately measure the PSF of the microscope. In simple terms, the algorithm starts with a calculated theoretical PSF; however, during successive iterations not only is the original guess at the object modified, but the PSF itself is also modified. In a sense the PSF is determined from the specimen images themselves, or the PSF is “found” in the data (64). The images can even be broken up into small subsections for deconvolution so that different regions of the cell (e.g., cytosol vs. nucleus), which likely have different optical properties, can be treated separately. This type of localized PSF image restoration is not available using traditional restorative deconvolution software platforms. To ensure accurate image restoration, it is always advisable to have a specimen containing sub-resolution fluorescent microspheres in order to ascertain that the algorithms accurately restore the microsphere shape.
One software platform that performs blind deconvolution is the AutoQuant AutoDeblur software (Media Cybernetics). Many standard image processing software packages use AutoQuant as their deconvolution solution including ImagePro Plus (Media Cybernetics), Imaris (Bitplane Inc.), and MetaMorph (Molecular Devices). Images of the nucleus, actin fibers, and mitochondria in BPAE cells were collected at high resolution with an oil immersion objective (Fig. 7). Maximum projections of nine images of the raw data, including the focal plane and four image planes above and below, are shown in the widefield panel (Fig. 7). These images were then deconvolved using AutoQuant X AutoDeblur software for widefield microscopes resulting in the maximum projections shown in the deconvolution panel (Fig. 7). Details within the nucleus are restored following deconvolution (Fig. 7a). All of the diffuse actin staining through the cell has been reassigned to the filamentous structures from where it originated, revealing beautiful high contrast actin filaments (Fig. 7b). Similarly, the mitochondrial staining is highly localized and the diffuse staining throughout the cell has been reassigned (Fig. 7c). An important bene fit of restorative deconvolution is that out-of-focus light is not simply removed as it is with confocal microscopy, or with simple nearest neighbor deconvolution, but rather reassigned to the location from whence it originated. This results in deconvolved images that have a much higher S/N when compared to raw widefield data (compare intensity of second and third columns in Fig. 7a–c).
Fig. 7.
Images of (a) DAPI (Zeiss 49 DAPI cube; 250 ms), (b) phalloidin AlexaFluor® 488 (Zeiss 13 FITC cube; 400 ms), and (c) MitoTracker® Red CMXRos (Chroma Technology ET-Texas Red cube, 600 ms) taken on the same microscope with the same camera settings as in Fig. 2. The mercury lamp was attenuated to 5% power with neutral density filters. Z-axis images were taken every 0.1 μm for a total of 82 image frames. (a–c) Images are maximum projections of nine images centered about the focal plane for the raw widefield images (first column), and images deconvolved with AutoQuant X (third column). The second and third columns are displayed with the same brightness, contrast, and gamma factor settings to show the increase in S/N with deconvolution. The gamma factor was used to bring up dim features so they are visible in the images. The first column is displayed with enhanced brightness and contrast so that image resolution can be visualized. Scale bar is 10 μm.
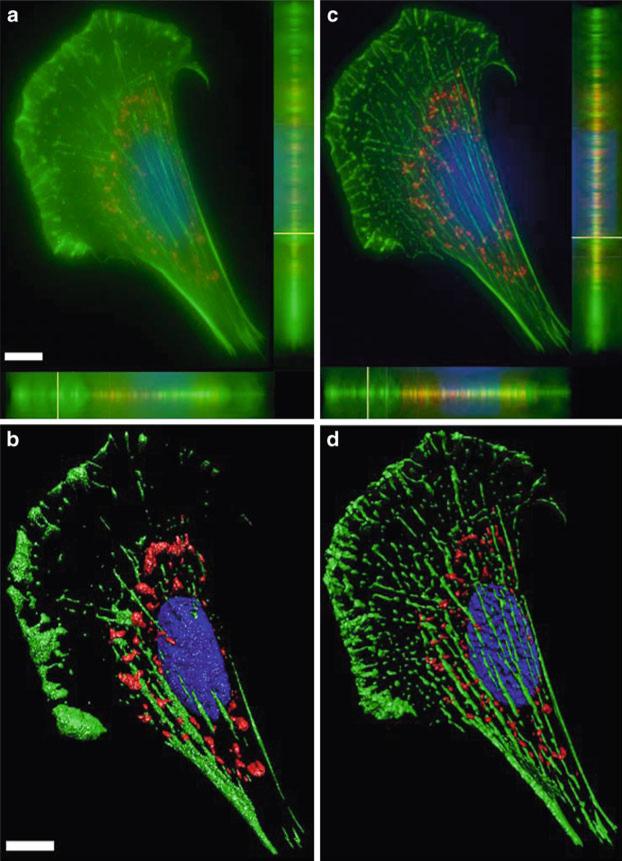
The significant improvement of image quality and S/N with image deconvolution can also be seen in color overlays of the image data, once again comparing widefield (Fig. 8a) with deconvolved (Fig. 8c) images. In fact, 3D visualization software (Imaris, Bitplane Inc.) has a difficult time even generating iso-surfaces of widefield data (Fig. 8b). In contrast, deconvolved data not only generates beautiful iso-surfaces (Fig. 8d), but these images can also be used to perform intensity analysis, as well as measurements of filament volumes and lengths. These types of measurements would be difficult to perform on the raw widefield data. As a cautionary note, deconvolved algorithms should always be thoroughly tested to ensure that no feature within the original data set is lost, no new features are generated and that quantitative relationships are maintained (e.g., if one cell is twice as bright as another before deconvolution, it should also be twice as bright after deconvolution) (59).
Fig. 8.
A color overlay of the maximum projection of the same data from Fig. 7 with blue (DAPI), green (actin phalloidin AlexaFluor® 488), and red (MitoTracker® Red) using AutoQuant X (Media Cybernetics) software for the (a) raw data and (c) deconvolved data. (a, c) also show x-z (bottom) and y-z (right side) axial profiles through the image stack emphasizing reduction in out-of-focus light following deconvolution. The same images were processed with Imaris 6.1.5 software (Bitplane Inc.) and 3D iso-surfaces were automatically produced from the (b) raw and (c) deconvolved images. Scale bar is 10 μm.
8. Conclusions
Epi-fluorescence microscopy is an important tool for life sciences research enabling the visualization of cellular compartments and very specific cellular markers. Advances in microscope optics, filters, light sources, and cameras allow researchers to peer into the microscopic workings of cells, tissues, and organisms. The availability of an array of highly specific immuno-fluorescence agents as well as cellular markers, sensors, and fluorescent proteins will continue to light up the architecture and complex biochemical pathways that regulate cellular function. Improved sensitivity has enabled the use of epi-fluorescence microscopy on the most precious of tissues, for example, primary neurons, by immuno-fluorescence imaging and also live-cell imaging. When combined with restorative image deconvolution to generate beautiful high resolution quantitative images, the epi-fluorescence microscope can be a fundamental tool in any life sciences research lab.
Acknowledgements
This work was supported by the McGill Life Sciences Complex Imaging Facility (CMB) and by grants GM092914 and MH071674 from NIH to DJW. Thank you goes out to the following people: Loralei Dewe formerly of Media Cybernetics and David Hitrys formerly of QImaging Corporation for their work performing the deconvolution of raw widefield images using AutoQuant X software for Fig. 7 and for generating the images used in Fig. 8a, b; Cory Glowinski formerly of Bitplane Inc. for generating the 3D iso-surfaces for Fig. 8c, d using Imaris Software; Adam Wegner for collecting the neuronal images in Fig. 5; Michael Davidson for providing the microscope and fluorescence illuminator figures in Fig. 1a, b; and Olympus Canada for providing the equipment in order to generate the images and Ji-Sook Lee for Figs. 2 and 4; Brady Eason and Ji-Sook Lee for critically reading the chapter.
Footnotes
Company names and brands are mentioned throughout the chapter. This information is meant to facilitate scientists who are new to the field and wish to find out more information about the various components of the epi-fluorescence microscope. The authors in no way endorse specific products and apologize for any company who may have inadvertently been left out.
References
- 1.Blaydes JP, Vojtesek B, Bloomberg GB, Hupp TR. The development and use of phosphospecific antibodies to study protein phosphorylation. Methods Mol Biol. 2000;99:177–189. doi: 10.1385/1-59259-054-3:177. [DOI] [PubMed] [Google Scholar]
- 2.DiGiovanna MP, Roussel RR, Stern DF. Production of antibodies that recognize specific tyrosine-phosphorylated peptides. Curr Protoc Cell Biol. 2002 doi: 10.1002/0471143030.cb1606s13. Chapter 16:Unit 16 16. [DOI] [PubMed] [Google Scholar]
- 3.Xu N, Xu M, Zhang YY. Optical detection of single molecules in living cells. Sheng Li Xue Bao. 2005;57:271–277. [PubMed] [Google Scholar]
- 4.Lang E, Baier J, Kohler J. Epifluorescence, confocal and total internal reflection microscopy for single-molecule experiments: a quantitative comparison. J Microsc. 2006;222:118–123. doi: 10.1111/j.1365-2818.2006.01579.x. [DOI] [PubMed] [Google Scholar]
- 5.Triller A, Choquet D. New concepts in synaptic biology derived from single-molecule imaging. Neuron. 2008;59:359–374. doi: 10.1016/j.neuron.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 6.Day RN, Schaufele F. Fluorescent protein tools for studying protein dynamics in living cells: a review. J Biomed Opt. 2008;13:031202. doi: 10.1117/1.2939093. [DOI] [PubMed] [Google Scholar]
- 7.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 8.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 9.Allen GJ, Kwak JM, Chu SP, Llopis J, Tsien RY, Harper JF, Schroeder JI. Cameleon calcium indicator reports cytoplasmic calcium dynamics in Arabidopsis guard cells. Plant J. 1999;19:735–747. doi: 10.1046/j.1365-313x.1999.00574.x. [DOI] [PubMed] [Google Scholar]
- 10.Dailey ME, Khodjakov A, Rieder CL, Swedlow JR, Andrews PD, Waters JC, Claxton NS, Olenych SG, Grif fi n JD, Davidson MW. 2007 The automated microscope http://www. microscopyu.com/articles/livecellimaging/automaticmicroscope.html.
- 11.Wolf DE. Fundamentals of fluorescence and fluorescence microscopy. Methods Cell Biol. 2007;81:63–91. doi: 10.1016/S0091-679X(06)81005-8. [DOI] [PubMed] [Google Scholar]
- 12.Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 13.Abramowitz M, Davidson MW. 2003 Light sources for optical microscopy http://micro.magnet.fsu.edu/primer/anatomy/sources.html.
- 14.Hohman B. LED light source: major advance in fluorescent microscopy. Biomed Instrum Technol. 2007;41:461–464. doi: 10.2345/0899-8205(2007)41[461:LLSMAI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 15.Albeanu DF, Soucy E, Sato TF, Meister M, Murthy VN. LED arrays as cost effective and efficient light sources for wide field microscopy. PLoS One. 2008;3:e2146. doi: 10.1371/journal.pone.0002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole RW, Turner JN. Light-emitting diodes are better illumination sources for biological microscopy than conventional sources. Microsc Microanal. 2008;14:243–250. doi: 10.1017/S1431927608080288. [DOI] [PubMed] [Google Scholar]
- 17.Frigault MM, Lacoste J, Swift JL, Brown CM. Live-cell microscopy — tips and tools. J Cell Sci. 2009;122:753–767. doi: 10.1242/jcs.033837. [DOI] [PubMed] [Google Scholar]
- 18.Abramowitz M, Spring KR, Keller HE, Davidson MW. Basic principles of microscope objectives. Biotechniques. 2002;33:772–774. 776–778, 780–771. doi: 10.2144/02334bi01. [DOI] [PubMed] [Google Scholar]
- 19.Keller HE. Objective lenses for confocl microscopy. In: Pawley J, editor. Handbook of biological confocal microscopy. 3rd edn. Springer; New York: 2006. pp. 145–161. [Google Scholar]
- 20.North AJ. Seeing is believing? A beginners’ guide to practical pitfalls in image acquisition. J Cell Biol. 2006;172:9–18. doi: 10.1083/jcb.200507103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Standish B. High performance with fluorescence optical filters. BioOptics World. 2008;1 5:35–37. [Google Scholar]
- 22.Erdogan T, Pradhan A, Mizrahi V. Optical fi lters impact fluorescence fidelity. Biophotonics Int. 2003;10:34–38. [Google Scholar]
- 23.Spring KR, Fellers TJ, Davidson MW. Introduction to charged-coupled devices (CCDs) http://www.microscopyu.com/articles/digitalimaging/ccdintro.html.
- 24.Spring KR. Cameras for digital microscopy. Methods Cell Biol. 2007;81:171–186. doi: 10.1016/S0091-679X(06)81010-1. [DOI] [PubMed] [Google Scholar]
- 25.Tran P. CCD cameras for fluorescence imaging of living cells. In: Goldman RD, Spector DL, editors. Live cell imaging: a laboratory manual. Cold Spring Harbor Laboratory Press; New York: 2005. pp. 87–100. [Google Scholar]
- 26.Tsien RY, Ernst L, Waggoner A. Fluorophores for confocal microscopy: photophysics and photochemistry. In: Pawley J, editor. Handbook of biological confocal microscopy. 3rd edn. Springer; New York: 2006. pp. 338–352. [Google Scholar]
- 27.Lichtman JW, Conchello JA. Fluorescence microscopy. Nat Methods. 2005;2:910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 28.Shimomura O, Johnson FH, Saiga Y. Extraction, puri fication and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 29.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 30.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein http://www.invitrogen.com/site/us/en/home/References/Molecular-Probes-The-Handbook.html.
- 31.Day RN, Davidson MW. The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev. 2009;38:2887–2921. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsien RY. Building and breeding molecules to spy on cells and tumors. FEBS Lett. 2005;579:927–932. doi: 10.1016/j.febslet.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 33.Goldman RD, Spector DL. Live cell imaging: a laboratory manual. Cold Spring Harbor Laboratory Press; New York: 2005. [Google Scholar]
- 34.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 35.Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 36.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 37.Karasawa S, Araki T, Nagai T, Mizuno H, Miyawaki A. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem J. 2004;381:307–312. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ai HW, Henderson JN, Remington SJ, Campbell RE. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kremers GJ, Gilbert SG, Cranfill PJ, Davidson MW, Piston DW. Fluorescent proteins at a glance. J Cell Sci. 2011;124:157–160. doi: 10.1242/jcs.072744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rizzo MA, Davidson MW, Piston DW. Fluorescent protein tracking and detection: applications using fluorescent proteins in living cells. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.top64. pdb top64. [DOI] [PubMed] [Google Scholar]
- 41.Rizzo MA, Davidson MW, Piston DW. Fluorescent protein tracking and detection: fluorescent protein structure and color variants. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.top63. pdb top63. [DOI] [PubMed] [Google Scholar]
- 42.Johnson ID. Practical considerations in the selection and application of fluorescent probes. In: Pawley J, editor. Handbook of biological confocal microscopy. 3rd edn. Springer; New York: 2006. pp. 353–367. [Google Scholar]
- 43.The Molecular Probes Handbook—A Guide to Fluorescent Probes and Labeling Technologies
- 44.Patterson G, Day RN, Piston D. Fluorescent protein spectra. J Cell Sci. 2001;114:837–838. doi: 10.1242/jcs.114.5.837. [DOI] [PubMed] [Google Scholar]
- 45.Jayaraman S, Haggie P, Wachter RM, Remington SJ, Verkman AS. Mechanism and cellular applications of a green fluorescent protein-based halide sensor. J Biol Chem. 2000;275:6047–6050. doi: 10.1074/jbc.275.9.6047. [DOI] [PubMed] [Google Scholar]
- 46.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 47.Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 48.Pertz O, Hahn KM. Designing biosensors for Rho family proteins—deciphering the dynamics of Rho family GTPase activation in living cells. J Cell Sci. 2004;117:1313–1318. doi: 10.1242/jcs.01117. [DOI] [PubMed] [Google Scholar]
- 49.Jaiswal JK, Goldman ER, Mattoussi H, Simon SM. Use of quantum dots for live cell imaging. Nat Methods. 2004;1:73–78. doi: 10.1038/nmeth1004-73. [DOI] [PubMed] [Google Scholar]
- 50.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Allison DW, Gelfand VI, Spector I, Craig AM. Role of actin in anchoring postsynaptic receptors in cultured hippocampal neurons: differential attachment of NMDA versus AMPA receptors. J Neurosci. 1998;18:2423–2436. doi: 10.1523/JNEUROSCI.18-07-02423.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fischer M, Kaech S, Knutti D, Matus A. Rapid actin-based plasticity in dendritic spines. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, Fisher DA, Storm DR. Analysis of the palmitoylation and membrane targeting domain of neuromodulin (GAP-43) by site-specific mutagenesis. Biochemistry. 1993;32:10714–10719. doi: 10.1021/bi00091a023. [DOI] [PubMed] [Google Scholar]
- 54.Brown CM. Fluorescence microscopy—avoiding the pitfalls. J Cell Sci. 2007;120:1703–1705. doi: 10.1242/jcs.03433. [DOI] [PubMed] [Google Scholar]
- 55.Stack RF, Bayles CJ, Girard AM, Martin K, Opansky C, Schulz K, Cole RW. Quality assurance testing for modern optical imaging systems. Microsc Microanal. 2011;17:598–606. doi: 10.1017/S1431927611000237. [DOI] [PubMed] [Google Scholar]
- 56.Kraus B, Zeigler M, Wolff H. Linear fluorescence unmixing in cell biological research. In: Méndez-Vílas A, Diaz, editors. Modern research and educational topics in microscopy. Formatex Microscopy Book Series; Badajoz, Spain: 2007. pp. 863–873. [Google Scholar]
- 57.Swedlow JR, Hu K, Andrews PD, Roos DS, Murray JM. Measuring tubulin content in Toxoplasma gondii: a comparison of laser-scanning confocal and wide- field fluorescence microscopy. Proc Natl Acad Sci U S A. 2002;99:2014–2019. doi: 10.1073/pnas.022554999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swedlow JR, Platani M. Live cell imaging using wide-field microscopy and deconvolution. Cell Struct Funct. 2002;27:335–341. doi: 10.1247/csf.27.335. [DOI] [PubMed] [Google Scholar]
- 59.McNally JG, Karpova T, Cooper J, Conchello JA. Three-dimensional imaging by deconvolution microscopy. Methods. 1999;19:373–385. doi: 10.1006/meth.1999.0873. [DOI] [PubMed] [Google Scholar]
- 60.Swedlow JR. Quantitative fluorescence microscopy and image deconvolution. Methods Cell Biol. 2007;81:447–465. doi: 10.1016/S0091-679X(06)81021-6. [DOI] [PubMed] [Google Scholar]
- 61.Wallace W, Schaefer LH, Swedlow JR. A workingperson's guide to deconvolution in light microscopy. Biotechniques. 2001;31:1076–1078. 1080, 1082. doi: 10.2144/01315bi01. passim. [DOI] [PubMed] [Google Scholar]
- 62.Murray JM. Confocal microscopy, deconvolution, and structured illumination methods. In: Goldman RD, Spector DL, editors. Live cell imaging: a laboratory manual. Cold Spring Harbor Laboratory Press; New York: 2005. pp. 239–279. [Google Scholar]
- 63.Shaw PJ. Comparison of widefield/deconvolution and confocal microscopy for three-dimensional imaging. In: Pawley J, editor. Handbook of biological confocal microscopy. 3rd edn. Springer; New York: 2006. pp. 453–467. [Google Scholar]
- 64.Holmes TJ, Biggs D, Abu-Tarif A. Blind deconvolution. In: Pawley J, editor. Handbook of biological confocal microscopy. 3rd edn. Springer; New York: 2006. pp. 468–487. [Google Scholar]
- 65.Wallace W, Schaefer LH, Swedlow JR, Fellers TJ, Davidson MW. Deconvolution in optical microscopy.
- 66.Nyquist H. Certain topics in telegraph transmission theory. Trans AIEE. 1928;47:617–644. [Google Scholar]
- 67.Cole RW, Jinadasa T, Brown CM. Measuring and interpreting point spread functions to determine confocal microscope resolution and ensure quality control. Nat Protoc. 2011;6(12):1929–1941. doi: 10.1038/nprot.2011.407. [DOI] [PubMed] [Google Scholar]