Telomerase catalyzed synthesis of telomere DNA provides the foundation for DNA-protein structures called telomeres. Telomeres are required to evade DNA processing which may result in nucleolytic degradation and fusion of linear chromosomes. Human telomerase contains a protein reverse transcriptase (hTERT), telomerase RNA (hTR), and several additional proteins.[1] hTR provides the template sequence for hTERT during a novel reverse transcription reaction that produces short telomere DNA repeat sequences.[2] Additionally, hTR provides a flexible scaffold for the assembly and function of telomerase associated proteins[3] and has recently been suggested to contribute to enzyme catalysis.[4] Many pathogenic mutations have been mapped to hTR;[5] however the precise mechanisms of these mutations remain unclear.
In vitro reconstitution of hTR with hTERT into a catalytically active enzyme requires two separable RNA domains harboring several conserved structural motifs, including an RNA pseudoknot (PK) fold.[5–10] NMR studies of wild-type and mutant minimal PK constructs have revealed tertiary interactions in the folded pseudok-not that form a conserved triple helix, as well as a partially folded hairpin.[10–15] Single molecule experiments have used mechanical force to probe the folding/unfolding of a minimal PK construct[11], and a modified two color coincidence detection assay (TCCD-lex) to reconstruct the distribution of RNA conformations for a larger hTR PK domain construct in solution.[16] Due to the large size of the native hTR PK domain, analyses of its folding properties have thus far been largely limited to truncated variants of the natural RNA sequence. To circumvent this challenge we employ a single molecule Förster resonance energy transfer (smFRET) assay,[17] which measures the efficiency of excitation energy transfer between a donor (Cy3) and acceptor (Cy5) dye, to analyze the folding and dynamics of the minimal and a larger functional hTR PK domain. Using a combination of smFRET together with high resolution NMR measurements, we have dissected the contribution of Mg2+, RNA triplex formation, and the native hTR sequence to PK domain folding and stability.
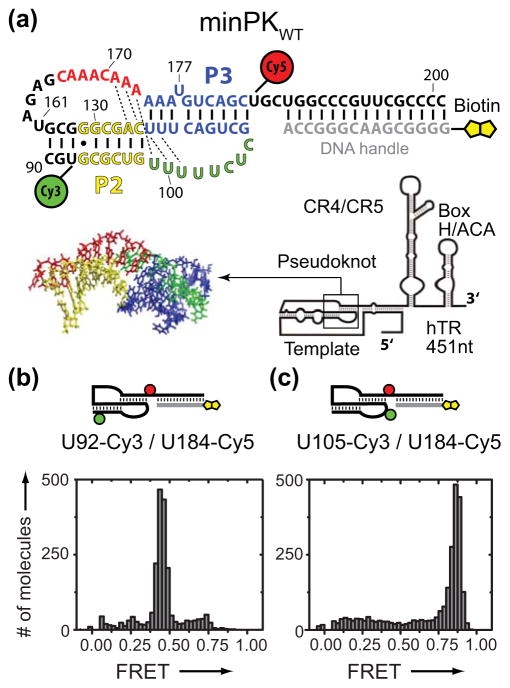
We initially designed a minimal hTR PK construct (minPKWT) similar to the hTR fragments used in previously reported NMR studies (Figure 1a).[13] FRET-labeled minPKWT was annealed to a biotinylated DNA handle, surface immobilized on a streptavidin coated microscope slide, and imaged using prism-type TIRF microscopy. The minPKWT with U92-Cy3 and U184-Cy5 modifications yielded a dominant RNA population centered at FRET = 0.45 (Figure 1b), a value consistent with the analogous NMR structure (Supplementary Figure 1).[13] We then altered the position of the donor Cy3 dye to U105, which shifts the FRET distribution to a higher value as predicted for the folded minPKWT (FRET = 0.87, Figure 1c). Taken together with additional control experiments (Supplementary Figure 2), these results demonstrate the smFRET system accurately reports on the folding of minPKWT.
Figure 1.
Design and validation of a minimal hTR pseudoknot smFRET construct. a) Schematic diagram of smFRET minPK construct. Nucleotide numbering corresponds to the native full-length hTR sequence. b) FRET distribution for minPKWT (U92-Cy3/U184-Cy5). c) FRET distribution for minPKWT (U105-Cy3/U184-Cy5). Data in (b) and (c) were acquired in the presence of telomerase imaging buffer containing 1 mM Mg2+.
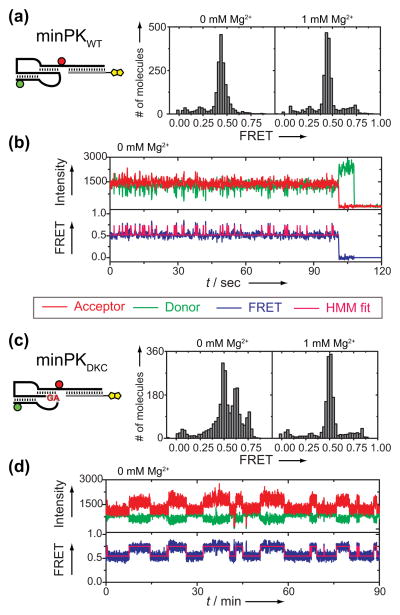
Next, we analyzed the impact of Mg2+ ions on minPKWT folding and dynamics. The dominant FRET distribution in the absence of Mg2+ centered on FRET = 0.44. However, close inspection of the time traces revealed rapid structural dynamics in the absence of Mg2+ between the PK fold and an alternative high FRET conformation in the majority (56%, n=312) of molecules (Figure 2b and Supplementary Figure 3). These structural dynamics are consistent with a transient sampling of the previously reported P2 hairpin.[13] In contrast, addition of 1 mM Mg2+ suppressed these structural dynamics completely at the time resolution of our measurements (Figure 2a, right panel and Supplementary Figure 4). We next analyzed the structural impact of the pathogenic GC(107-108)AG mutation that is genetically linked to the blood disorder Dyskeratosis Congenita (DKC).[19] In the absence of Mg2+, the minimal hTRDKC PK domain (minPKDKC) yielded a FRET distribution for the PK fold (FRET = 0.50) as well as multiple alternative conformations (FRET = 0.62 and 0.75, Figure 2c, left panel), in accord with the significant destabilization of the native PK fold expected for these mutations.[13–15] Introduction of a compensatory GA(182-183)CU mutation that rescues the Watson-Crick basepairs affected by the DKC mutant largely recovered the FRET distribution for the native PK fold in the presence or absence of Mg2+ (Supplementary Figure 5). Surprisingly, addition of 1 mM Mg2+ completely suppressed the DKC mutant folding defect (Figure 2c, right panel). Analysis of single molecule trajectories in the absence of Mg2+ revealed very slow structural dynamics for minPKDKC between the PK fold and alternative conformations in the majority (64%, n=214) of molecules (Figure 2d and Supplementary Figure 3). Importantly, the occurrence of alternative conformations in our experiments was not dependent on the presence or absence of Mg2+ during the initial folding of the RNA (Supplementary Figure 6).
Figure 2.
Mg2+-dependent folding and dynamics of minimal hTR pseudoknot domain. a) FRET distribution for minPKWT in the absence (left panel) or presence (right panel) of 1 mM Mg2+. b) Representative single molecule FRET trace of minPKWT in the absence of Mg2+. The abrupt loss of signal intensity at the end of the trace is due to photobleaching of the dye. Hidden Markov Modeling (HMM)[18] of the FRET trajectories yielded idealized state traces (magenta) which were used to calculate transition rates between observed FRET states (Supplementary Figure 3). c) FRET distribution for minPK DKC in the absence (left panel) or presence (right panel) of 1mM Mg2+. d) Representative single molecule trace of minPKDKC in the absence of Mg2+.
In previous work, the K. lactis PK was reported to require Mg2+ for compaction into the conserved triplex structure,[20] similar to DNA triplexes.[21] However, triplex formation in the hTR PK cannot strictly require Mg2+ since the structure that identified the tertiary interactions was solved in the absence of Mg2+.[13] This structure revealed an H-type pseudoknot with P2 and P3 stems stabilized by tertiary interactions with the two loops to form a central triplex containing five base triples flanking a junction A173-U99 Hoogsteen basepair.[13] Our smFRET approach does not permit differentiation between a PK lacking tertiary interactions and a PK plus triplex structure. Therefore, we investigated the folding of a PK with UU(99-100)CC substitutions (minPKC99C100), which disrupt the tertiary interactions by abolishing the junction base pair and one base triple but would still in principle allow the P2 and P3 stems to form.[14, 22]
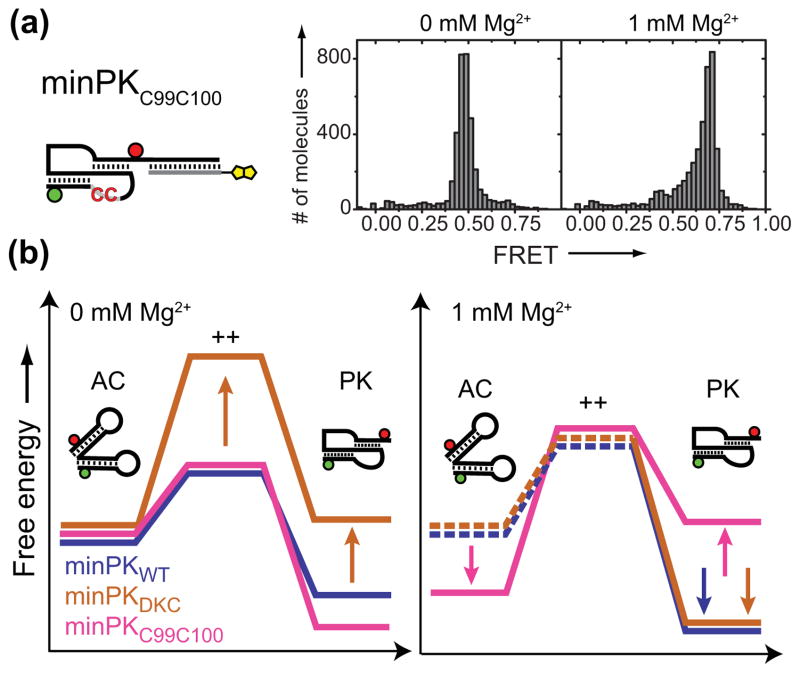
In the absence of Mg2+, FRET distributions generated with minPKC99C100 centered on a value consistent with a folded PK (FRET = 0.48) (Figure 3a, left panel). Indeed, NMR experiments with a similar construct bearing the UU(99-100)CC mutant in the absence of Mg2+ indicated that the P2 and P3 stems are partially formed, but the tertiary triplex interactions are disrupted (Supplementary Figure 7). In striking contrast, FRET distributions of minPKC99C100 in the presence of 1 mM Mg2+ shifted to a substantially higher FRET value (FRET = 0.69) (Figure 3a, right panel, and Supplementary Figure 8), indicative of an alternative RNA conformation.
Figure 3.
a) FRET distribution for minPKC99C100 in the absence (left panel) or presence (right panel) of 1 mM Mg2+. b) Schematic energy landscapes. AC: alternative conformation, PK: pseudoknot fold, ++: transition state. Mg2+-dependent folding and dynamics of minimal hTR pseudoknot domain. Dashed lines represent states not directly detected in our experiments.
Taken together, these experiments reveal that both minPKWT and minPKDKC exhibit structural dynamics in the absence of Mg2+, albeit with markedly different kinetics (Figure 3b, left panel): minPKWT is predominantly in the PK state, but transiently samples an alternative conformation (AC). Note, the structure drawn for AC is not intended to represent the actual conformation and is based upon secondary structure calculations (Supplementary Figure 9). In contrast, minPKDKC shows a significantly destabilized PK fold and exhibits structural inter-conversion with AC on much slower time scales, as indicated by a significantly higher energy transition state. The stability of the PK fold for minPKC99C100 is very similar to minPKWT (Supplementary Figure 8). In the presence of 1 mM Mg2+ (Figure 3b, right panel), both minPKWT and minPKDKC are stabilized in the PK fold. In contrast, minPKC99C100 is stabilized in the AC. Moreover, near physiological Mg2+ levels stabilize both minPKWT and minPKDKC folding into the PK structure, raising the question of how the pathogenic DKC mutation disrupts telomerase activity in vivo. Lastly, experiments with minPKC99C100 demonstrate that destabilization of the RNA triplex increases the probability of the minimal PK domain to become misfolded under physiological conditions.
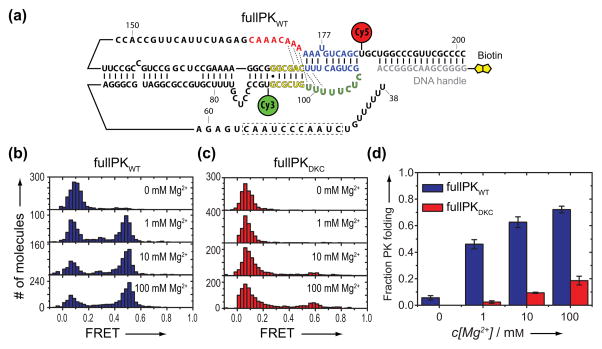
A powerful attribute of smFRET is the ability to analyze RNA folding within the larger native hTR PK domain. Using DNA-splinted RNA ligation, we generated a smFRET construct comprising hTR nucleotides 39-201 with U92-Cy3 and U184-Cy5 modifications (Figure 4a and Supplementary Figure 10). This nearly full-length hTR PK WT domain (fullPKWT) can be reconstituted with the CR4/CR5 RNA domain and hTERT protein in rabbit reticulocyte lysates to produce catalytically active telomerase enzyme.[23] When compared to an in vitro transcribed fullPKWT, the smFRET fullPKWT supported ~60% of wild type levels of catalytic activity and showed a slight reduction in repeat addition processivity (Supplementary Figure 11).
Figure 4.
Mg2+-dependent folding of the nearly full-length hTR PK domain. a) Schematic illustration of the fullPKWT smFRET construct sequence with FRET dye attachments and DNA handle attached as described for the minPK WT. b) FRET distributions for fullPKWT as a function of [Mg2+].c) FRET distributions for the fullPKDKC as a function of [Mg2+]. d) Quantification of pseudoknot formation at varying Mg2+ concentrations. The fraction of fullPKWT (blue) or fullPKDKC (red) exhibiting PK folding. Error bars represent the standard deviation of experiments done in triplicate.
In the absence of Mg2+, we observed a dominant misfolded population of RNA molecules centered on FRET = 0.1 and no detectable folding of fullPKWT, as evidenced by the absence of a population centered at FRET ≈ 0.5 (Figure 4b). This result contrasts with our observations of minPKWT folding in the absence of Mg2+ (Figure 2a) and reveals the folding requirements for fullPKWT are more stringent, as expected for a larger RNA. Interestingly, in a near physiological buffer (1 mM Mg2+) ~50% of the fullPKWT molecules fold into a conformation consistent with a folded PK domain (FRET = 0.48) (Figure 4b and Supplementary Figure 12). To further validate the designation of the 0.48 FRET state as the folded fullPKWT, we measured the FRET distribution for a fullPKWT construct containing the GC(107-108)AG DKC mutation (fullPKDKC). Interestingly, whereas small amounts of Mg2+ were sufficient to rescue the folding defects of minPKDKC, we observed no PK domain folding for fullPKDKC in the near physiological buffer (1 mM Mg2+) (Figure 4c). Moreover, whereas the fraction of folded fullPKWT molecules continued to increase at elevated Mg2+ concentrations (Figure 4d), only 20% of fullPKDKC folded into a PK conformation at the highest Mg2+ tested (100 mM Mg2+). Taken together, these results support the assignment of the 0.48 FRET state to the native PK fold and reveal that within the context of the fullPKDKC, physiological levels of Mg2+ are not sufficient to suppress the folding defects incurred by the GC(107-108)AG DKC mutation, suggesting that the pathogenic phenotype observed for this mutation is due to an RNA folding defect (Supplementary Figure 13).
In summary, our results directly demonstrate the hTR PK domain has the intrinsic ability to form alternative conformations distinct from the native pseudoknot fold. Experiments with the minPK suggest a model for PK domain folding, wherein the P2 and P3 stems of the hTR PK form in a Mg2+-independent manner together with a Mg2+-induced stabilization of tertiary triplex interactions. The unique capability of smFRET to probe fullPK folding exposed for the first time the extreme ruggedness of the hTR folding landscape, and highlights the potentially critical importance of Mg2+ in stabilizing the native PK fold. Moreover, the inability of the fullPKDKC construct to fold under near physiological conditions provides unique insights into the structural basis of this pathogenic RNA mutation. Recent experiments analyzing the folding of the RNA pseudoknot in Tetrahymena thermophila telomerase RNA demonstrated a requirement for telomerase proteins to achieve the native pseudoknot fold.[24] The propensity of the hTR PK domain to fold under near physiological conditions reported here highlights an important difference in the intrinsic stabilities of the human and Tetrahymena RNA pseudoknot domains.[16] Finally, the system established in this study will provide an essential experimental framework for future investigation of hTR PK structure and dynamics within the functional telomerase ribonucleoprotein complex.
Supplementary Material
Acknowledgments
We thank Prof. Kathleen Collins for the gift of FLAG-tagged hTERT plasmid and Prof. Julian Chen for the gift of the hTR expression plasmid. This work was financially supported by the following grants: NIH GM095850 to M.D.S. and NIH GM04813 to J.F.. M.H. is supported by a German Research Foundation fellowship (DFG HE6181/1-1).
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Martin Hengesbach, Department of Chemistry and Biochemistry, Center for Molecular Biology of RNA, University of California Santa Cruz, Santa Cruz, CA 95064 (USA), Fax: (+1) 831-459-2935.
Nak-Kyoon Kim, Department of Chemistry and Biochemistry, University of California Los Angeles, Los Angeles, CA 90095 (USA).
Prof. Juli Feigon, Department of Chemistry and Biochemistry, University of California Los Angeles, Los Angeles, CA 90095 (USA)
Prof. Michael D. Stone, Email: mstone@chemistry.ucsc.edu, Department of Chemistry and Biochemistry, Center for Molecular Biology of RNA, University of California Santa Cruz, Santa Cruz, CA 95064 (USA), Fax: (+1) 831-459-2935.
References
- 1.Collins K. Nat Rev Mol Cell Biol. 2006;7:484. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greider CW, Blackburn EH. Nature. 1989;337:331. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 3.Egan ED, Collins K. Mol Cell Biol. 2010;30:2775. doi: 10.1128/MCB.00151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiao F, Cech TR. Nat Struct Mol Biol. 2008;15:634. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Theimer CA, Feigon J. Curr Opin Struct Biol. 2006;16:307. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Chen JL, Blasco MA, Greider CW. Cell. 2000;100:503. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell JR, Collins K. Mol Cell. 2000;6:361. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 8.Tesmer VM, Ford LP, Holt SE, Frank BC, Yi X, Aisner DL, Ouellette M, Shay JW, Wright WE. Mol Cell Biol. 1999;19:6207. doi: 10.1128/mcb.19.9.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q, Kim NK, Feigon J. Proc Natl Acad Sci U S A. 2011;108:20325. doi: 10.1073/pnas.1100279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Q, Kim NK, Peterson RD, Wang Z, Feigon J. Proc Natl Acad Sci U S A. 2010;107:18761. doi: 10.1073/pnas.1013269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G, Wen JD, Tinoco I., Jr RNA. 2007;13:2175. doi: 10.1261/rna.676707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comolli LR, Smirnov I, Xu L, Blackburn EH, James TL. Proc Natl Acad Sci U S A. 2002;99:16998. doi: 10.1073/pnas.262663599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim NK, Zhang Q, Zhou J, Theimer CA, Peterson RD, Feigon J. J Mol Biol. 2008;384:1249. doi: 10.1016/j.jmb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theimer CA, Blois CA, Feigon J. Mol Cell. 2005;17:671. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 15.Theimer CA, Finger LD, Trantirek L, Feigon J. Proc Natl Acad Sci U S A. 2003;100:449. doi: 10.1073/pnas.242720799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeoman JA, Orte A, Ashbridge B, Klenerman D, Balasubramanian S. J Am Chem Soc. 2010;132:2852. doi: 10.1021/ja909383n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stone MD, Mihalusova M, O’Connor M, Prathapam CR, Collins K, Zhuang X. Nature. 2007;446:458. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKinney SA, Joo C, Ha T. Biophys J. 2006;91:1941. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I. Nature. 2001;413:432. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 20.Shefer K, Brown Y, Gorkovoy V, Nussbaum T, Ulyanov NB, Tzfati Y. Mol Cell Biol. 2007;27:2130. doi: 10.1128/MCB.01826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank-Kamenetskii MD, Mirkin SM. Annu Rev Biochem. 1995;64:65. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- 22.Chen JL, Greider CW. Proc Natl Acad Sci U S A. 2005;102:8080. doi: 10.1073/pnas.0502259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beattie TL, Zhou W, Robinson MO, Harrington L. Curr Biol. 1998;8:177. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 24.Mihalusova M, Wu JY, Zhuang X. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1017686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.