Abstract
MicroRNAs (miRNAs) are short noncoding RNAs that play crucial roles in tumorigenesis and tumor progression. Melanoma is the most aggressive skin cancer that is resistant or rapidly develops resistance to a variety of chemotherapeutic agents. The role of miRNAs in melanoma progression and drug resistance has not been well studied. Herein, we demonstrate that miR-200c is down-regulated in melanomas (primary and metastatic) compared with melanocytic nevi. Overexpression of miR-200c in melanoma cells resulted in significantly decreased cell proliferation and migratory capacity as well as drug resistance. miR-200c overexpression resulted in significant down-regulation of BMI-1, ABCG2, ABCG5, and MDR1 expression and in a concomitant increase in E-cadherin levels. Knockdown of BMI-1 showed similar effects as miR-200c overexpression in melanoma cells. In addition, miR-200c overexpression significantly inhibited melanoma xenograft growth and metastasis in vivo, and this correlated with diminished expression of BMI-1 and reduced levels of E-cadherin in these tumors. The effects of miR-200c on melanoma cell proliferation and migratory capacity and on self-renewal were rescued by overexpression of Bmi-1, and the reversal of these phenotypes correlated with a reduction in E-cadherin expression and increased levels of ABCG2, ABCG5, and MDR1. Taken together, these findings demonstrate a key role for miR-200c in melanoma progression and drug resistance. These results suggest that miR-200c may represent a critical target for increasing melanoma sensitivity to clinical therapies.
Melanoma is the most deadly skin cancer, and its incidence is steadily rising. Survival in patients with distant metastases (stage IV) is poor: 10-year survival ranges from ∼6% to 15% in patients with stage IV disease.1,2 There is, therefore, a critical need to identify clinically significant biochemical pathways central to the aggressive behavior of this disease and, in the process, unveil new opportunities for the design of rational therapeutic interventions in high-risk patients. A key contributor to the poor prognosis of advanced-stage melanoma is that most melanomas are refractory to systemic therapies.3 The newly developed targeted antisignaling therapies, such as vemurafenib and imatinib, showed great promise in the treatment of melanomas with BRAF or CKIT mutations, but these melanomas rapidly acquired resistance, and the median duration of response was only 6 to 10 months.4–6 A variety of mechanisms have been proposed to explain the observed resistance to systemic therapeutic agents, including reduced intracellular accumulation of drug and derangements in pathways controlling apoptosis, cell cycle checkpoints, and the repair of damaged cellular targets.3,7 Indeed, in melanoma, members of the ATP-binding cassette (ABC) transporters—a superfamily of transmembrane proteins that transport many varied substrates across biological membranes in an ATP-dependent manner—exhibit high levels of expression and mediate chemoresistance in melanoma cells.3,8 In particular, ABCG2 exhibits increased expression in melanoma cells with enhanced tumorigenic capabilities, including the capability for self-renewal and differentiation.9
The Polycomb group (PcG) of proteins comprises an important class of transcriptional repressors that orchestrate changes in chromatin structure to regulate gene activity, and many of the PcG proteins demonstrate altered expression in human cancers.10,11 BMI-1 is a PcG protein that has been shown to be an important transcriptional repressor of the Ink4a/Arf gene locus,12,13 which encodes two separate gene products—p16ink4a and p19Arf—from two distinct reading frames. p16ink4a inhibits CDK activity and, thereby, blocks entry into the cell cycle by preventing phosphorylation (and consequent inactivation) of the retinoblastoma protein (Rb) by cyclin D–CDK4/6 complexes. p19ARF arrests cell cycle progression and promotes apoptosis by promoting the stability of p53.14 BMI-1 also plays a critical role in the maintenance of stem cells.15 Consistent with these observations suggesting an important oncogenic role for BMI-1, BMI-1 overexpression has been demonstrated in numerous human cancers,10,11 including melanoma.16
MicroRNAs (miRNAs) are noncoding RNAs of approximately 20 to 22 nucleotides that function in posttranscriptional gene regulatory pathways. Alterations in miRNA expression have been described in many different human tumors, and numerous studies have demonstrated that miRNAs function as key pathogenic components, impacting cancer cell growth, survival, and the capacity to metastasize.17–22 In particular, the miR-200 family (including miR-200a, miR-200b, miR-200c, and miR-141) has been shown to repress Zinc finger E-box binding homeobox proteins 1 and 2 (ZEB1/ZEB2) in a variety of different cellular contexts, culminating in increased E-cadherin expression; in contrast, loss of miR-200, which occurs in many different human cancers, including breast cancer,23 ovarian cancer,24 prostate cancer,25 and endometrial carcinoma,26 results in increased ZEB1/ZEB2 and repression of E-cadherin and represents the hallmark of the so-called epithelial to mesenchymal transition pathway.27–29 This latter change is coincident with more aggressive biological behavior in cancer (increased migration and invasion).23,28,30,31
Herein, we demonstrate a delicate interaction among miR-200c, BMI-1, and drug resistance genes representing a pivotal cellular axis impacting not only the capacity of melanoma cells to proliferate and metastasize but also their sensitivity to systemic therapeutic agents.
Materials and Methods
Reagents and Cell Culture
We compared the miRNA expression profiles of benign nevi (n = 10), primary melanomas (n = 10), and metastatic melanomas (n = 10) by microarray as previously described.32 Total RNA extraction was performed as previously described.33 Tissue samples were obtained from archives in the Department of Pathology and Laboratory Medicine, Hospital of the University of Pennsylvania (Philadelphia). The protocol was approved by the University of Pennsylvania Institutional Review Board. Human melanoma cell lines (WM35, WM793, WM115A, WM3523A, and 1205Lu) were gifts from Meenhard Herlyn at The Wistar Institute, Philadelphia. Human melanoma cell lines were maintained in 2% MCDB medium as described.34 Human 293T cell line was a gift from Frank Lee at the University of Pennsylvania, and they were maintained in high-glucose Dulbecco's modified Eagle's medium with 10% fetal bovine serum and penicillin/streptomycin (100 U/mL and 100 mg/mL). Nude mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All the animal protocols were approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania.
The following antibodies were used: anti-goat BMI-1 (c1510; Santa Cruz Biotechnology, Santa Cruz, CA; 1:200 dilution), monoclonal mouse anti-human E-cadherin antibody (Dako, Carpinteria, CA; 1:500 dilution), rat monoclonal anti-human ABCG2 antibody (sc- sc-69989; Santa Cruz Biotechnology; 1:200 dilution), rabbit polyclonal to ABCG5 antibody (ab69713; Abcam Inc, Cambridge, MA; 1:500 dilution), rabbit polyclonal MDR1 antibody (LS-B1448; LifeSpan Biosciences Inc., Seattle, WA; 1:200 dilution), and mouse anti-β-actin monoclonal antibody (010M4816; Sigma-Aldrich, St. Louis, MO; 1:2500 dilution).
pEIZ–HIV–ZsGreen–miR-200c, pEIZ-HIV-ZsGreen-vector, c-cherry vector, and c-cherry–Bmi-1 were gifts from M. Clarke at Stanford University (Stanford, CA)23; the WST-1 cell proliferation kit was purchased from Roche Diagnostics GmbH (Mannheim, Germany) (11974100) and was performed according to the manufacturer's instructions; and FuGENE6 transfection reagent was purchased from Roche Diagnostics GmbH (12132000) and was used according to the manufacturer's instructions.
RNA Isolation and Quantitative RT-PCR
Total RNA was isolated using the RNeasy kit (Qiagen Inc., Valencia, CA), followed by cDNA synthesis using the SuperScript first-strand synthesis system (Invitrogen, Carlsbad, CA). Quantitative PCR was performed using iQ SYBR green supermix (Bio-Rad Laboratories, Hercules, CA) with the specific primers listed below. cDNA corresponding to 1 μg of RNA was added to the iQ SYBER green supermix and was analyzed using an iCycler thermal cycler (Bio-Rad Laboratories) according to the manufacturer's instructions. The thermal profiles were 95°C for 30 seconds and 56°C for 30 seconds. Melting curve analysis was performed for each PCR to confirm the specificity of amplification. At the end of each phase, fluorescence was measured and quantified. Hsa–miR-200c TaqMan miRNA assay and GAPDH were purchased from Applied Biosystems (Foster City, CA). BMI-1 RT-PCR was performed as described.35 Real-time PCR primers were as follows: E-cadherin forward primer, 5′-TTCCCTGCGTATACCCTGGT-3′; E-cadherin reverse primer, 5′-GCCATCTCTTGCTCGAAGTCC-3′; ABCG2 forward primer, 5′-CAATGGGATCATGAAACCTG-3′; ABCG2 reverse primer, 5′-GAGGCTGATGAATGGAGAA-3′; ABCG5 forward primer, 5′-TGGGACATCACATCTTGCCG-3′; ABCG5 reverse primer, 5′-CCGTTCACATACACCTCCCC-3′; MDR1 forward primer, 5′-AGGAAGCCAATGCCTATGACTTTA-3′; MDR1 reverse primer, 5′-CAACTGGGCCCCTCTCTCTC-3′; β-actin forward primer, 5′-TGACTGACTACCTCATGAAGATCC-3′; and β-actin reverse primer, 5′-GCCATCTCTTGCTCGAAGTCC-3′. β-Actin was used as an internal control, and iCycler iQ software version 3.1.0.7050 (Bio-Rad Laboratories) was used for analyzing the relative values for E-cadherin, ABCG2, ABCG5, and MDR1.
Infection of pEIZ–HIV–ZsGreen–miR-200C and c-Cherry–Bmi-1
pEIZ-HIV-ZsGreen-vector control, pEIZ-HIV-ZsGreen containing miR-200c, c-cherry–vector control, and c-cherry–vector containing Bmi-1 (gifts from M. Clarke), sh control, and sh-Bmi-136 were transfected into 293T cells with packing vector (pCMV-dR8.2-dupr and pCMV-VSV), and viral supernatants were collected 72 hours after transfection to infect human melanoma cells (WM115A, WM35, WM793, WM115A, and 1205Lu). After a 48-hour infection period, miR-200c–infected green fluorescent protein–positive (green) cells were sorted by fluorescence-assisted cell sorting (FACS). For the rescue assay, we co-infected the BMI-1 virus into the miR-200c–overexpressing cells. miR-200c and BMI-1 expression were confirmed in the sorted cells by PCR or Western blot analysis.
Cell Proliferation Assay
A total of 5 × 104 cells per well were seeded into a 24-well plate. After 48 hours, the absorbance at 450 nm (A450) was detected using an iQuant universal microplate spectrophotometer (BioTek Instruments Inc., Winooski, VT) as previously described.37
Wound-Healing Assay
WM115A control cells, WM115A cells overexpressing miR-200c, WM35 control cells, WM35 cells overexpressing Bmi-1, WM115A control cells, WM115A cells with Bmi-1 knock down, and WM115A cells overexpressing BMI-1 and miR-200c were grown to confluence and were wounded by dragging a 1-mL pipette tip across the monolayer. The cells were washed to remove cellular debris and then were allowed to migrate for 24 or 36 hours. Images were taken 0, 24, or 36 hours after wounding using a DMI6000 inverted microscope (Leica Microsystems GmbH, Wetzlar, Germany). Three replicates each of two independent experiments were performed.37
Western Blot Analysis
Western blot analysis was performed as previously described.37
Cell Cycle Analysis
Cell viability was detected by trypan blue exclusion assay. A total of 1 × 106 miR-200c overexpression and control-WM115A cells were harvested, rinsed twice with Ca2+, Mg2+-free cold PBS, and fixed with 70% ethanol overnight at 4°C. Fixed cells were then washed twice with Ca2+, Mg2+-free cold PBS and stained with 20 μg/mL propidium iodide. The analysis was performed on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) using CellQuest Pro software (BD Biosciences). Cell cycle analysis was performed using ModFit software version 3.2.1 (Verity Software House, Topsham, ME).
Soft Agar Colony Formation Assay
A total of 1 × 104 WM115A control cells, WM115A cells overexpressing miR-200c, or WM115A cells overexpressing BMI-1 and miR-200C were resuspended in 3 mL of 1.8% (w/v) Bacto Agar solution containing MCDB with 20% fetal bovine serum. The mixtures were overlaid onto a 3.3% (w/v) Bacto Agar solution in 6-well dishes. After 24 hours, 0.5 mL of MCDB supplemented with 2.0% fetal bovine serum was added. Colonies were counted under a microscope after 15 days. Colony-forming efficiency was calculated by the number of colonies × 100/the number of cells plated.37
Limiting Dilution Assay
WM115A control cells, WM115A cells overexpressing miR-200c, or WM115A cells overexpressing BMI-1 and miR-200c were trypsinized, washed, and resuspended at a concentration of 100 cells/mL. A total of 10 μL of this cell suspension was dispensed into individual wells of 96-well plates containing 90 μL of MCDB media supplemented with 2% fetal bovine serum. All the plates were examined to confirm that each well contained only a single cell. The cells were incubated for another 7 days, with changes in media every other day. Colonies were counted in single-cell seeded wells.
Xenograft Tumor Model
Four- to five-week-old male athymic nu/nu mice were used in this study. Control or pEZX-miR-200c–infected WM115A cells (2 × 106 cells per animal) were injected subcutaneously into nude mice (eight mice per group). After 35 days, the animals were sacrificed, and necropsy was performed on each animal for further analysis of the primary tumor along with possible metastases.
Statistical Analysis
A t-test or one-way analysis of variance was used to analyze gene expression and cell viability. Statistical significance was set at two-sided P < 0.05.
Results
miR-200 Expression Is Decreased in Melanoma Tissues and Cell Lines
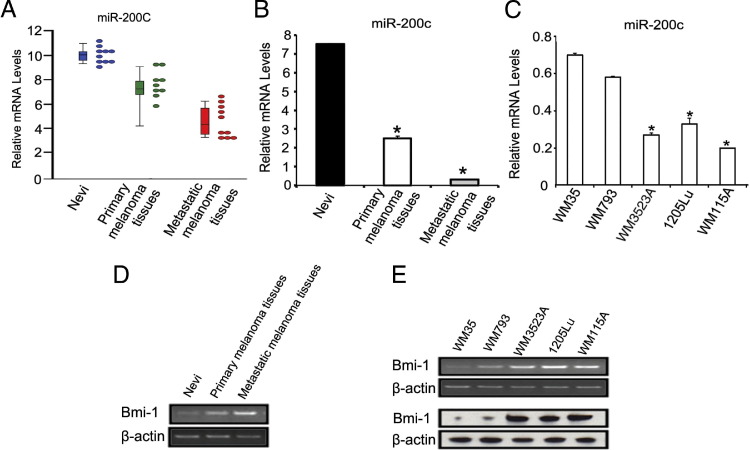
We previously demonstrated that formalin-fixed, paraffin-embedded tissue is a suitable resource for miRNA expression profiling.33,38 We, therefore, used formalin-fixed, paraffin-embedded tissues consisting of 10 nevi, 10 primary melanomas, and 10 metastatic melanomas using a microarray platform to examine miRNA expression. We demonstrate a striking reduction in miR-200c expression in melanomas compared with nevi and a trend toward reduced expression in metastatic compared with primary melanomas (Figure 1, A and B). We then examined miR-200c expression in five melanoma cell lines isolated from various stages of melanoma progression. 1205Lu, WM3523A, and WM115A were derived from metastatic melanomas. These cells consistently expressed lower levels of miR-200c by quantitative RT-PCR compared with WM35 (derived from radial growth phase primary melanoma) and WM793 (derived from vertical growth phase primary melanoma) (Figure 1C). Bmi-1 has been found to be a target for miR-200c.23 Indeed, the expression of Bmi-1 inversely correlated with miR-200c expression in tissue samples (Figure 1D) and cell lines (Figure 1E) at mRNA and protein expression levels. Together, these results demonstrate a progressive diminution of miR-200c expression in melanoma compared with nevi and suggest a further reduction in expression during melanoma progression, and Bmi-1 expression correlates inversely with miR-200c expression.
Figure 1.
miR-200c mRNA expression levels decrease in metastatic melanoma tissues and cell lines. A: Relative expression of miR-200c by microarray analysis (P = 1.09 × 10−4). Each dot represents relative expression level from one clinical specimen and each bar represents mean of relative expression level with standard deviation. B: Quantitative RT-PCR analysis of miR-200c expression in tissue from 10 nevi, 10 primary melanomas, and 10 metastatic melanomas (n = 3 replicate experiments). Data are given as mean ± SD. *P < 0.05 compared with control. C: Quantitative RT-PCR analysis of miR-200c expression in WM35, WM793, WM115A, WM3523A, and 1205Lu cells (n = 3 replicate experiments). Data are given as mean ± SD. *P < 0.05 compared with control. D: RT-PCR analysis of BMI-1 expression in tissue from 10 nevi, 10 primary melanomas, and 10 metastatic melanomas (n = 3 replicate experiments). E: RT-PCR analysis (top panel) and Western blot analysis (bottom panel) of BMI-1 expression in WM35, WM793, WM115A, WM3523A, and 1205Lu cells (n = 3 replicate experiments).
miR-200c Inhibits Melanoma Cell Proliferation
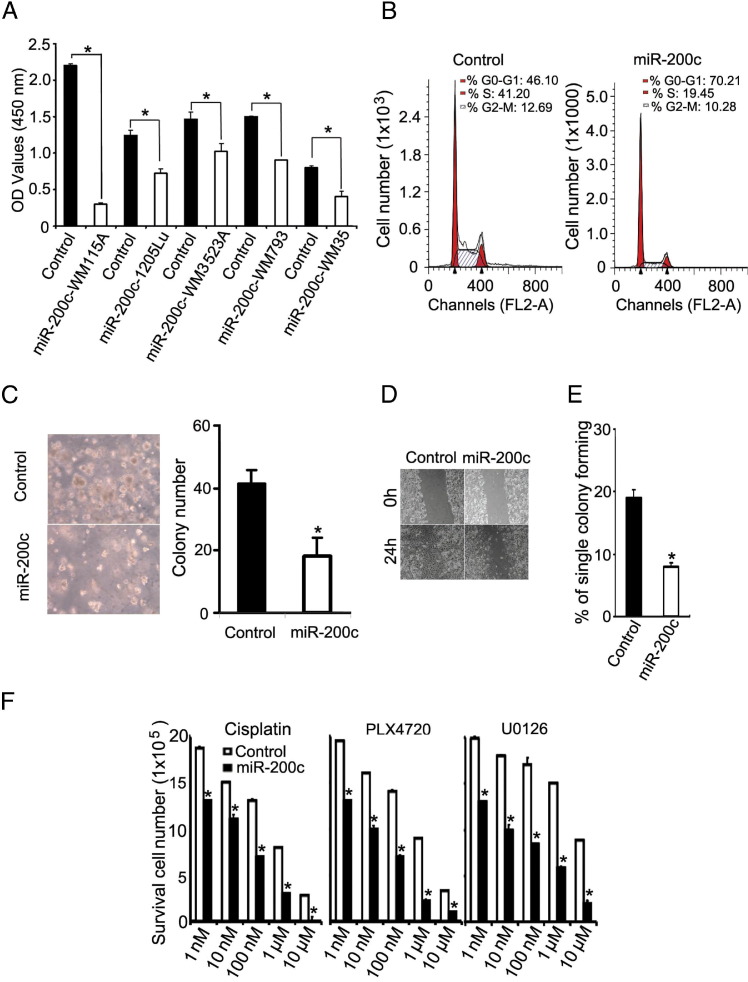
To characterize the function of miR-200c in melanoma cells, we examined the effects of miR-200c overexpression in human melanoma cell lines. We infected WM115A, 1205Lu, WM793, WM3523A, and WM35 cells with lentivirus carrying miR-200c with a green fluorescent protein tag. Green fluorescent protein–expressing cells were sorted out by FACS 48 hours after infection (data not shown). Cell proliferation was examined by the WST-1 proliferation assay. Enforced expression of miR-200c caused a significant reduction in cell proliferation compared with the control group (Figure 2A). To further characterize the nature of this defect, we performed a cell cycle progression study using FACS analysis in WM115A cells infected with miR-200c. Overexpression of miR-200c results in fewer cells in the S and G2-M phases (mean ± SD: 53.89% ± 8.19% in control cells compared with 29.73% ± 2.37% in WM115A cells overexpressing miR-200c) with a concomitant increase in G0-G1 (mean ± SD: 46.10% ± 4.98% in control cells compared with 70.21% ± 11.07% in WM115A cells overexpressing miR-200c; Figure 2B). Together with the results of the WST-1 cell proliferation assay, this finding is consistent with a model in which enforced expression of miR-200c compromises progression through G0-G1.
Figure 2.
miR-200c overexpression inhibits melanoma cell growth and cell mobility. A: Effect of miR-200c on WM115A melanoma cell proliferation by WST-1 cell proliferation assay. The relative density of WM115A, 1205Lu, WM3523A, WM793, and WM35 melanoma cells infected with control vector and pHIV–miR-200c were detected at 450 nm (A450) of absorbance (n = 3 replicate experiments). Data are given as mean ± SD. *P < 0.05 compared with control. B: Effect of miR-200c on WM115A melanoma cell cycle progression. Control and miR-200c–WM115A cells in the G0-G1, G2-M, and S phases underwent FACS analysis (representative flowchart from 3 replicate experiments). C: Effect of miR-200c on WM115A melanoma cell soft agar colony formation assay. Control and miR-200c–WM115A were cultured in soft agar. The clones were counted using a microscope at ×100 magnification. The values (colony number) are expressed as mean ± SD from three separate measurements. *P < 0.05 compared with control. D: Effect of miR-200c on WM115A melanoma cell migration. Wound healing of control and miR-200c WM115A cells 0 and 24 hours after scratch formation. Shown are representative images from 3 experiments. E: Effect of miR-200c on WM115A melanoma cell self-renewal. A limiting dilution assay was performed, and single cells were followed for 14 days using control and miR-200c. The number of colonies formed was counted and averaged in three replicate experiments. Data are given as mean ± SD. *P < 0.05 compared with control. F: Effect of miR-200c on WM115A melanoma cell survival in different drug environments: cisplatin, PLX4720, and U0126 (n = 3 replicate experiments). *P < 0.05 compared with control.
To assess the tumorigenic effects of miR-200c in WM115A cells, we performed soft agar colony formation assays in cells overexpressing miR-200c. Compared with control cells, enforced expression of miR200c resulted in a significant reduction in the number of colonies formed in soft agar (Figure 2C). Since enforced expression of miR-200c impedes cell proliferation, we asked whether enforced expression of miR-200c would affect cell survival in the presence of therapeutic agents. WM115A melanoma cells were incubated with varying concentrations of cisplatin (a DNA damaging agent), PLX4720 (a specific inhibitor of B-RAFV600E), and U0126 (a selective inhibitor of MEK1/2 kinase) for 24 hours. miR-200c overexpression resulted in a significant decrease in cell survival in all therapeutic agents tested (Figure 2F).
Since miR-200 family members are known to be key regulators of E-cadherin expression in the epithelial to mesenchymal transition, we also examined the impact of enforced expression of miR-200c on WM115A cell migration. In wound-healing assays, enforced expression of miR200c in WM115A cells resulted in a significant reduction in their capacity to close an artificial wound compared with control cells (Figure 2D). Finally, we assessed the effects of enforced miR-200c expression on the self-renewal capacity of WM115A cells by limiting dilution assay and demonstrated a significant reduction in colony-forming units (Figure 2E).
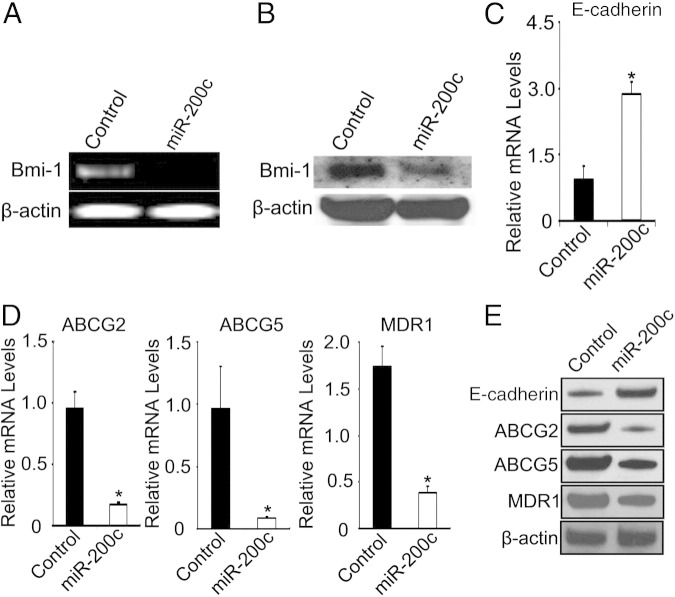
miR-200c Inhibits BMI-1 and Increases E-Cadherin Expression Levels in WM115A Cells
In breast cancer cells, miR-200c directly binds to BMI-1 and regulates its expression.23 Therefore, we asked whether enforced expression of miR-200c would also affect BMI-1 expression in melanoma cells. Enforced expression of miR-200c in WM115A cells resulted in a marked reduction of BMI-1 mRNA (Figure 3A) and protein (Figure 3B) expression compared with controls. Furthermore, enforced expression of miR-200c in WM115A cells resulted in significantly increased expression of E-cadherin (Figure 3, C and E). Finally, since enforced expression of miR-200c results in decreased survival of cells grown in the presence of cisplatin, PLX4720, and U0126, we studied whether enforced expression of miR-200c impacted ABC transporter expression, which is well-known to be involved in drug resistance.8,9,39 We demonstrated that enforced expression of miR-200c significantly reduces the expression of ABC transporters ABCG2, ABCG5, and MDR1 (Figure 3, D and E) compared with control cells.
Figure 3.
miR-200c overexpression correlates with decreased BMI-1, ABCG2, ABCG5, and MDR1 and increased E-cadherin expression levels in WM115A cells. A: RT-PCR analysis of BMI-1 expression in control and miR-200c–WM115A (n = 3 replicate experiments). B: Expression of BMI-1 by Western blot analysis. Cell lysates from control and miR-200c–WM115A were used for Western blot analyses with anti-Bmi1 antibody, β-actin was used as loading controls (n = 3 replicate experiments). C: Quantitative RT-PCR analysis of E-cadherin expression in control and miR-200c–WM115A (n = 3 replicate experiments). Data are given as mean ± SD. *P < 0.05 compared with control. D: Quantitative RT-PCR analysis of ABCG2, ABCG5, and MDR1 expression in control and miR-200c–WM115A (n = 3 replicate experiments). Data are given as mean ± SD. *P < 0.05 compared with control. E Expression of E-cadherin, ABCG2, ABCG5, and MDR1 by Western blot analysis in control and miR-200c–WM115A (n = 3 replicate experiments).
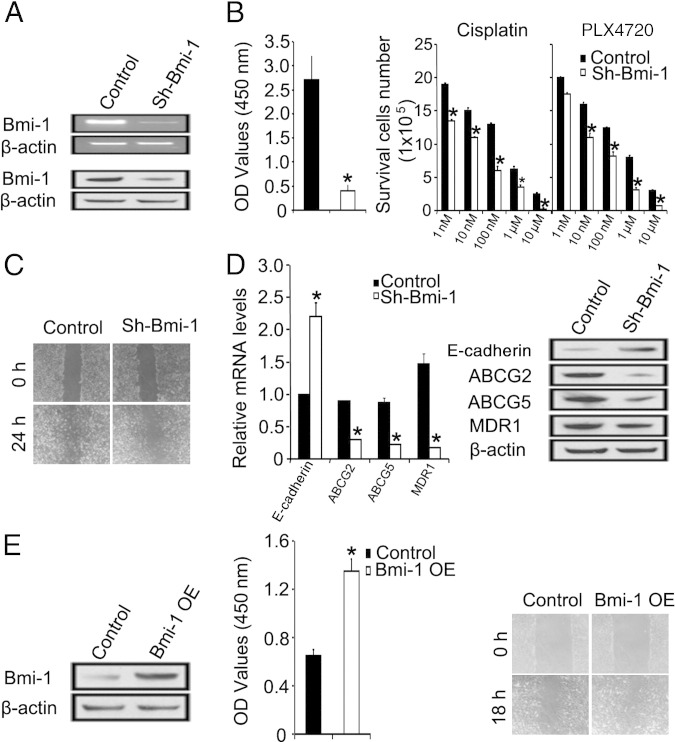
Bmi-1 Knockdown Phenocopies miR-200c Overexpression
To further assess the relationship between Bmi-1 and miR-200c in melanoma cells, we asked whether depletion of Bmi-1 would have similar effects as miR-200c overexpression. We knocked down Bmi-1 expression in WM115A cells as previously described.36 Decreased expression of Bmi-1 mRNA and protein was confirmed by RT-PCR and Western blot analysis, respectively (Figure 4A). Bmi-1 knockdown in WM115A cells resulted in decreased cell proliferation (Figure 4B), increased sensitivity to varying concentrations of cisplatin and PLX4720 (Figure 4B), and diminished cell migration (Figure 4C). These changes correlated with down-regulation of ABCG2, ABCG5, and MDR1 (Figure 4D), findings similar to those seen after miR-200c overexpression. Furthermore, Bmi-1 overexpression in WM35, a radial growth phase melanoma cell line, promoted cell proliferation and migration (Figure 4E).
Figure 4.
Knockdown of Bmi-1 phenocopies miR-200c overexpression in WM115A melanoma cells. A: RT-PCR analysis of BMI-1 expression in control and sh-Bmi-1–WM115A (n = 3 replicate experiments) (top panel) and expression of BMI-1 by Western blot analysis (bottom panel). Cell lysates from control and sh-Bmi-1–WM115A were used for Western blot analyses with anti-Bmi1 antibody. β-Actin was used as loading controls (n = 3 replicate experiments). B: Left panel: Effect of sh-Bmi-1 on WM115A melanoma cell proliferation by WST-1 cell proliferation assay. The relative density of WM115A cells infected with control vector and sh-Bmi-1 were detected at 450 nm (A450) of absorbance (n = 3 replicate experiments). *P < 0.05 compared with control. Right panel: Effect of Bmi-1 knock down on WM115A melanoma cell survival in different drug environments (cisplatin and PLX4720) (n = 3 replicate experiments). Data are given as mean ± SD. *P < 0.05 compared with control. C: Effect of sh-Bmi-1 on WM115A melanoma cell migration. Wound healing of control and sh-Bmi-1 WM115A cells 0 and 24 hours after scratch formation. Shown are representative images from three experiments. D: Quantitative RT-PCR (left panel) and Western blot (right panel) analyses of E-cadherin, ABCG2, ABCG5, and MDR1 expression in control and sh-Bmi-1–WM115A (n = 3 replicate experiments). Data are given as mean ± SD. *P < 0.05 compared with control. E: Left panel: Expression of BMI-1 by Western blot analysis. Cell lysates from control and Bmi-1–WM35 were used for Western blot analyses (n = 3 replicate experiments). Middle panel: Wst-1 assay analysis of control and Bmi-1–WM35 cell proliferation at 450 nm. Data are given as mean ± SD. *P < 0.05 compared with control. Right panel: Wound-healing assay showed the increased cell motility ability in Bmi-1–overexpressed (OE) WM35 cells compare with control (n = 3 replicate experiments).
Bmi-1 Rescues the Effects of miR-200c on WM115A Cells
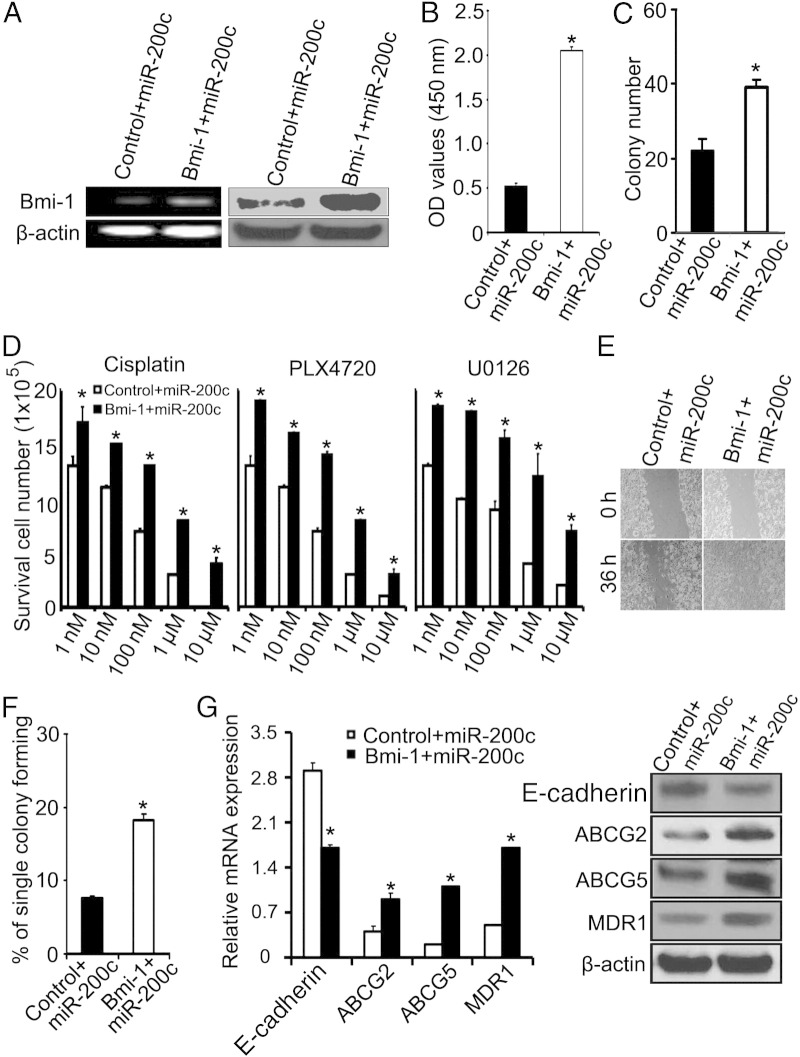
To confirm that the effects of miR-200c are mediated through Bmi-1 in melanoma, we introduced Bmi-1 (with a cherry-red tag) into miR-200c–overexpressing WM115A cells. Increased expression of BMI-1 mRNA and protein was confirmed by RT-PCR and Western blot analysis, respectively (Figure 5A). Enforced expression of BMI-1 reversed the inhibitory effects of miR-200c on cell proliferation, restoring WM115A cell proliferation to endogenous levels (WST-1 assay; Figure 5B). BMI-1 also reversed the negative effects of miR-200c overexpression on the capacity of WM115A cells to undergo self-renewal, restoring their ability to form colonies in a limiting dilution assay to wild-type levels (Figure 5, C and F). In addition, BMI-1 restored the sensitivity of miR-200c–overexpressing WM115A cells to varying concentrations of cisplatin, PLX4720, and U0126 (Figure 5D), and this correlated with a reversal of the miR-200c–induced down-regulation of ABCG2, ABCG5, and MDR1 (Figure 5G). Finally, enforced expression of BMI-1 in miR-200c–overexpressing WM115A cells restored their capacity to undergo cell migration in a wound-healing assay (Figure 5E), and this correlated with a reversal of the miR-200c–induced up-regulation of E-cadherin mRNA and protein (Figure 5G).
Figure 5.
BMI-1 rescues the effects of miR-200c on WM115A melanoma cells. WM115A cells were co-transfected with c-cherry control and miR-200c or with c-cherry–BMI-1 and miR-200c as indicated (n = 3 replicate experiments). A: Left panel: RT-PCR analysis confirms increased BMI-1 mRNA expression in c-cherry BMI-1–miR-200c–WM115A cells compared with c-cherry control–miR-200c–WM115A (β-actin loading control, n = 3 replicate experiments). Right panel: Western blot analysis confirms increased BMI-1 protein expression in c-cherry BMI-1–miR-200c–WM115A cells compared with c-cherry control–miR-200c–WM115A (β-actin loading control, n = 3 replicate experiments). B: BMI-1 overexpression reverses the negative impact of miR-200c on cell proliferation. WST cell proliferation assay: c-cherry control–miR-200c–WM115A and c-cherry BMI-1–miR-200c–WM115A cells were detected at 450 nm (A450) of absorbance (n = 3 replicate experiments). *P < 0.05 compared with control. C: BMI-1 overexpression reverses the negative impact of miR-200c on the ability of WM115A cells to form colonies in soft agar. C-cherry control– miR-200c––WM115A and c-cherry BMI-1–miR-200c–WM115A were cultured in soft agar. Colonies were counted using a microscope at ×100 magnification. The values (colony number) are expressed as mean ± SD from three separate measurements. *P < 0.05 compared with control cells. D: BMI-1 expression reverses chemosensitivity phenotype of enforced miR-200c expression in WM115A cells. Cell survival in cisplatin, PLX4720, and U0126 in c-cherry control–miR-200c–WM115A and c-cherry BMI-1–miR-200c–WM115A cells (n = 3 replicate experiments). *P < 0.05 compared with control. E:BMI-1 overexpression reverses the negative impact of miR-200c on cell migration. Wound healing of c-cherry control–miR-200c–WM115A and c-cherry BMI-1–miR-200c–WM115A cells 0 and 36 hours after scratch formation. Shown are representative images from three experiments. F:BMI-1 overexpression reverses the negative impact of miR-200c on the capacity of WM115A cells to undergo self-renewal. A limiting dilution assay was performed, and single cells were followed for 14 days using c-cherry control– miR-200c–WM115A and c-cherry Bmi-1–miR-200c–WM115A. The number of colonies formed was counted and averaged in three replicate experiments. *P < 0.05 compared with control. G:BMI-1 overexpression reverses the impact of miR-200c overexpression on E-cadherin, ABCG2, ABCG5, and MDR1. Left panel: Quantitative RT-PCR analysis demonstrate relative levels of E-cadherin, ABCG2, ABCB5, and MDR1 in c-cherry control–miR-200c–WM115A, and c-cherry BMI-1–miR-200c–WM115A cells (n = 3 replicate experiments). *P < 0.05 compared with control. Right panel:Expression of E-cadherin, ABCG2, ABCG5, and MDR1 by Western blot analysis in c-cherry control–miR-200c–WM115A and c-cherry BMI-1–miR-200c–WM115A cells (n = 3 replicate experiments). Data are given as mean ± SD.
miR-200c Inhibits Melanoma Growth and Metastasis in Vivo
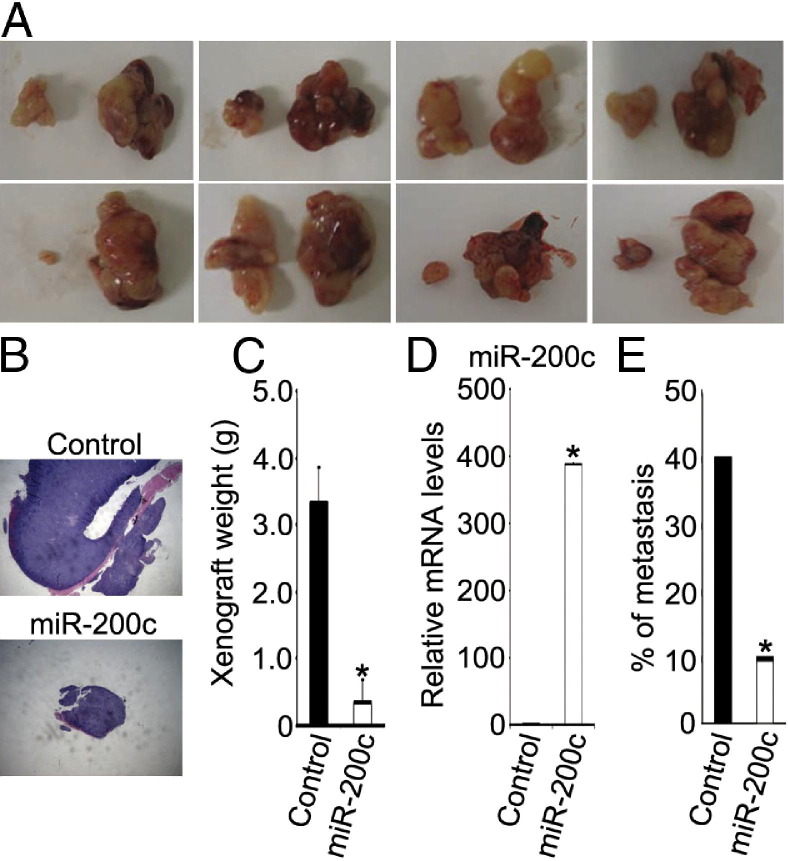
To assess the effect of miR-200c on tumor growth and metastasis in vivo, we injected control or miR-200c–WM115A cells into the flanks of nude mice (eight mice for each group). The mice were observed for 5 weeks, and the resultant xenograft tumors were harvested. The xenografts formed by miR-200c–WM115A cells were significantly smaller than those formed by control cells (Figure 6, A–C), and this correlated with increased levels of miR-200c in these tumors. Necropsies were performed, and organs were examined for the presence of metastases. We found a significantly higher rate of metastasis in tumors derived from WM115A control cells compared with tumors derived from WM115A cells overexpressing miR-200c (∼40% compared with ∼10%; Figure 6E).
Figure 6.
miR-200c inhibits melanoma growth and metastasis in vivo. A: Xenografts are shown (in each panel, the left side shows miR-200c–WM115A and the right side shows control). B: Histopathologic examination of WM115A control cell or miR-200c–WM115A cell tumors. C: Average tumor weights are shown (generated by WM115A control cells and miR-200c–WM115A cells, respectively; n = 8). *P < 0.05 compared with control. D: Quantitative RT-PCR analysis of miR-200c expression in xenograft from WM115A control tumors and miR-200c–WM115A tumors (n = 3 replicate experiments).*P < 0.05 compared with control. E: Frequency of metastases from tumors generated by WM115A control cells and miR-200c–WM115A cells, respectively.). *P < 0.05 compared with control. Data are given as mean ± SD.
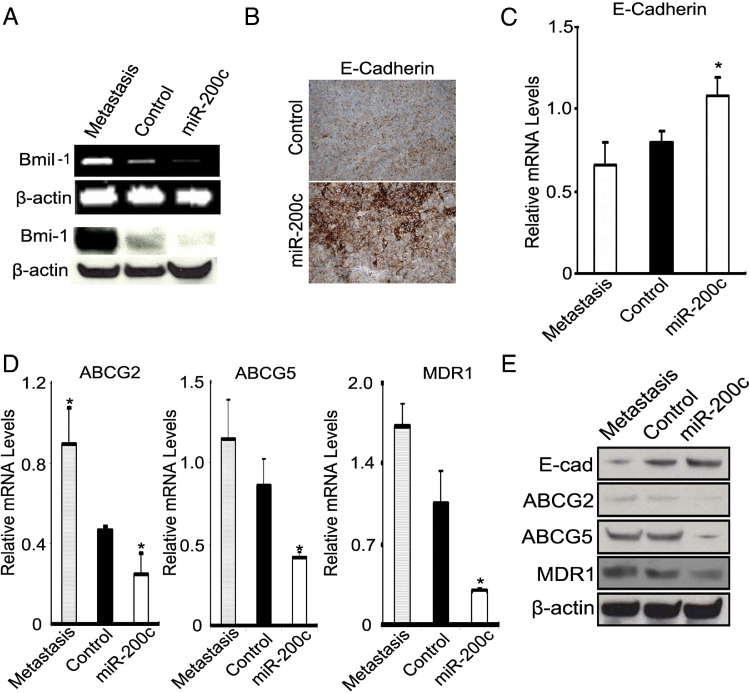
Finally, we examined the expression of Bmi-1 and E-cadherin in the primary xenograft tumors (formed by miR-200c–WM115A and WM115A control cells) and their metastases (derived from control cells). Bmi-1 mRNA and protein (Figure 7A) were reduced in the miR-200c–WM115A tumors compared with the WM115A control tumors and their respective metastases. In addition, E-cadherin was reduced in WM115A control tumors and their respective metastases compared with miR-200c–WM115A tumors (Figure 7, B, C, and E). Finally, there was a progressive, statistically significant decrease in the levels of ABCG2, ABCG5, and MDR1 mRNA and protein expression in the xenografts formed by miR-200–WM115A cells compared with WM115A controls and their metastases, respectively (Figure 7, D and E).
Figure 7.
miR-200c inhibits melanoma growth and metastasis through up-regulation of E-cadherin and down-regulation of Bmi-1. A: Expression of BMI-1 mRNA by quantitative RT-PCR (top panel) and of protein by Western blot analysis (bottom panel) in xenografts generated by WM115A metastasis, WM115A control, and miR-200c–WM115A cells (β-actin used as control). B: Immunohistochemical expression of E-cadherin in xenografts from WM115A control cells (top panel) and miR-200c–WM115A cells (bottom panel). C: RT-PCR analysis of E-cadherin mRNA expression in xenograft generated by metastasis, control, and miR-200c–WM115A cells (n = 3 replicate experiments). *P < 0.05 compared with control. D: Quantitative RT-PCR analysis of relative levels of ABCG2, ABCG5, and MDR1 in xenografts generated by WM115A metastasis, WM115A control, and miR-200c–WM115A cells (n = 3 replicate experiments). *P < 0.05 compared with control. E: Expression of E-cadherin, ABCG2, ABCG5, and MDR1 by Western blot analysis in WM115A metastasis, WM115A control, and miR-200c–WM115A cells (n = 3 replicate experiments). Data are given as mean ± SD.
Discussion
There is a critical need to improve our understanding of the molecular pathogenesis of melanoma. In a previous study, we described a distinct pattern of miRNA expression in nevi compared with melanomas.32 Herein, we describe a progressive diminution of expression of miR-200c in primary and metastatic melanomas compared with melanocytic nevi and in melanoma cell lines derived from melanoma metastases compared with those derived from radial or vertical growth phase–only primary melanoma. In melanoma cells, miR-200c impacts pathways governing cell proliferation, self-renewal, drug sensitivity, and cell migration. Together, the present findings provide evidence of a pivotal role in melanoma progression and resistance to chemotherapeutic agents between the interconnected network of miR-200c, BMI-1, E-cadherin, and ABC transporters (ABCG2, ABCG5, and MDR1).
BMI-1 expression is up-regulated in numerous human cancers,10,11,40–55 including melanoma.16 Increased BMI-1 seems to correlate with a more aggressive phenotype. Primary melanomas with metastases demonstrated increased BMI-1 expression compared with primary melanomas without metastases (83% compared with 52%).16 Similar findings have been described in breast carcinoma, where there is a statistically significant relationship between BMI-1 expression and the presence of axillary lymph node metastases.46 Increased BMI-1 expression also correlated with either a worse outcome or a more aggressive cancer phenotype in nasopharyngeal carcinoma,40 colonic adenocarcinoma,41 gastric carcinoma,42 and hepatocellular carcinoma.44 Although the mechanism by which cancer cells acquire BMI-1 up-regulation is unclear, studies in different cancer cell types demonstrate a central role for miRNAs in this process. In ovarian carcinoma cells, BMI-1 is targeted by miR-15a and miR1656; in endometrial carcinoma cells, miR-194 represses a BMI-1–mediated epithelial to mesenchymal transition57; and in breast carcinoma and glioblastoma, miR-128 represses BMI-158,59 Finally, miR-200c targets BMI-1 in breast and pancreatic cancer, and this functional relationship provided an important mechanistic association between miRNAs, epithelial to mesenchymal transition, and a stem cell–like phenotype.23,60 In breast cancer stem cells, miR-200c expression was decreased compared with nontumorigenic breast cancer cells; enforced expression of miR-200c not only repressed BMI-1 expression but also compromised the ability of breast cancer stem cells and normal mammary stem cells to form tumors and normal mammary ducts in vivo, respectively.23 Similarly, in pancreatic cancer, miR-200c targets ZEB1 and BMI-1, both of which are required to preserve stem cell–like properties (self-renewal, characteristic marker expression, drug resistance, and tumorigenicity) in pancreatic cancer cells.60 Together with the finding of an inverse relationship between ZEB1 expression in pancreatic carcinoma tissue samples and long-term survival in those patients, these findings established an important relationship between miR-200c, ZEB1/E-cadherin (epithelial to mesenchymal transition), and BMI-1. Namely, miR-200c directly represses ZEB1 and BMI-1; in turn, diminished expression of miR-200c correlates with acquisition of a stem cell–like phenotype during the course of tumor progression.60
The present findings in clinical specimens and cell lines provide additional support for such a model in melanoma progression. miR-200c exhibits progressively diminished expression from benign melanocytic nevi to primary melanomas in human tissue and in cell culture, and the expression of Bmi-1 correlates inversely with miR-200 expression. Similar to a recent report describing diminished expression of miR-200c in melanoma and a reduced capacity for colony formation when miR-200c is overexpressed,61 we demonstrate that enforced expression of miR-200c in melanoma cells impaired cell proliferation and self-renewal, enhanced drug sensitivity, and compromised cell migration. These phenotypic alterations were accompanied by a decrease in the expression of BMI-1, ABCG2, ABCB5, and MDR1 and a concomitant increase in E-cadherin expression. Enforced expression of Bmi-1 in these same cells reversed the phenotypic effects of miR-200c, whereas knockdown of Bmi-1 seems to phenocopy miR-200c overexpression. The expression of miR-200 family members in melanoma and their effect on cell migration in melanoma cells is somewhat controversial. Elson-Schwab et al62 demonstrated that miR-200a and miR-200c seem to be overexpressed in melanoma cell lines compared with normal human melanocytes, whereas the present results and others characterizing miRNA expression patterns in tissue samples of melanomas and nevi63 demonstrated a progressive diminution in their expression. In addition, Elson-Schwab et al62 found that overexpression of miR-200c and miR-200a primarily affects cell morphology, and miR-200c seems to promote cell migration in certain melanoma cell lines using a three-dimensional collagen I matrix. The discrepancy between these findings is, in part, attributable to the differences in cell lines and different assays used to define the role of miR-200c in melanoma cells.62 Nevertheless, we showed that melanoma cells overexpressing miR-200c developed significantly smaller tumors with a reduced propensity for metastasis compared with controls, and the more aggressive control tumors and their metastases exhibited diminished expression of E-cadherin and increased expression of Bmi-1. These findings demonstrate that perturbations in the miR200c/Bmi-1/Zeb1/E-cadherin axis correlate with a more aggressive melanoma phenotype.
We also demonstrate a functional relationship among miR-200c, Bmi-1, and the expression of ABC transporters (ABCG2, ABCG5, and MDR1), and this relationship affects sensitivity to various chemotherapeutic agents. The role of ABC transporters in mediating chemoresistance during the course of melanoma progression has been proposed to arise by exploiting pathways and molecules that melanocytes normally express during melanogenesis.3,8 In benign pigment-synthesizing melanocytes, ABC transporters function to prevent cellular injury by sequestering potentially cytotoxic melanin intermediates into various subcellular organelles, enabling these intermediates to be safely exported from the cell. During the course of melanoma progression, it has been hypothesized that melanoma cells hijack these intrinsic pathways of melanosome biosynthesis to impart a drug resistance phenotype.3,8 Indeed, aberrant ABC transporter expression and function in melanoma is well described: ABCG2 exhibits increased expression in so-called melanoma-initiating cells, which function as melanoma stemlike cells capable of self-renewal and differentiation.9 Other ABC transporters (ABCB5) are expressed at high levels in melanoma64 and mediate chemoresistance.39 Similar to ABCG2, ABCB5 also exhibits increased expression in melanoma-initiating cells, conferring a stem cell–like phenotype on these cells.65
However, the mechanisms by which the expression and activity of these endogenously used transporters are hijacked to effect drug resistance in melanoma cells are not well defined. The present results implicate alterations in miRNA expression in this process. Namely, enforced expression of miR-200c coincides with reduced expression of Bmi-1, ABCG2, ABCG5, and MDR1 and increased sensitivity to various chemotherapeutic agents. Enforced expression of Bmi-1 seems to reverse these effects, consistent with a model in which BMI-1, itself a known direct target of miR-200c in other cancers,23 directly or indirectly activates ABC transporter expression. Thus, loss of miR-200c in melanoma progression would result in increased levels of BMI-1 and ABC transporters and a resultant chemotherapeutic resistance phenotype. Consistent with this model is the observation of coincidently increased BMI-1 and ABC transporter expression in hepatocellular carcinomas66 and ameloblastic tumors.67 Future studies are needed to define more completely the precise mechanisms whereby miR-200c and BMI-1 leverage ABC transporter expression and activity to affect chemoresistance in melanomas.
Herein, we defined additional pivotal components in the molecular pathogenesis of melanoma and highlighted important new opportunities for the rational design of targeted therapies. Relying first on patient specimens, we identified a progressive diminution of miR-200c expression in primary and metastatic melanomas compared with nevi. We demonstrate further that miR-200c governs the expression of an important axis in melanoma progression, including BMI-1, E-cadherin, and ABC transporters, which conspire together to regulate cell proliferation, cell motility, and resistance to chemotherapeutic agents. Together, these results support the utility of miRNA expression profiling as a powerful tool for interrogating clinical samples of melanomas as a window to clinically and biologically important pathways central to the aggressive behavior of this disease.
Acknowledgments
We thank George Daley for providing Bmi-1 knockdown constructs and James Martin for helping with the FACS analysis.
Footnotes
Supported by NIH grants CA-116103, CA-093372, and AR-054593 (X.X.).
S.L and M.T.T. contributed equally to this work.
References
- 1.Balch C.M., Buzaid A.C., Soong S.J., Atkins M.B., Cascinelli N., Coit D.G., Fleming I.D., Gershenwald J.E., Houghton A., Jr, Kirkwood J.M., McMasters K.M., Mihm M.F., Morton D.L., Reintgen D.S., Ross M.I., Sober A., Thompson J.A., Thompson J.F. Final version of the American Joint Committee on Cancer staging system for cutaneous melanoma. J Clin Oncol. 2001;19:3635–3648. doi: 10.1200/JCO.2001.19.16.3635. [DOI] [PubMed] [Google Scholar]
- 2.Balch C.M., Soong S.J., Gershenwald J.E., Thompson J.F., Reintgen D.S., Cascinelli N., Urist M., McMasters K.M., Ross M.I., Kirkwood J.M., Atkins M.B., Thompson J.A., Coit D.G., Byrd D., Desmond R., Zhang Y., Liu P.Y., Lyman G.H., Morabito A. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. J Clin Oncol. 2001;19:3622–3634. doi: 10.1200/JCO.2001.19.16.3622. [DOI] [PubMed] [Google Scholar]
- 3.La Porta C.A. Mechanism of drug sensitivity and resistance in melanoma. Curr Cancer Drug Targets. 2009;9:391–397. doi: 10.2174/156800909788166574. [DOI] [PubMed] [Google Scholar]
- 4.Davies M.A., Gershenwald J.E. Targeted therapy for melanoma: a primer. Surg Oncol Clin N Am. 2011;20:165–180. doi: 10.1016/j.soc.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flaherty K.T., Puzanov I., Kim K.B., Ribas A., McArthur G.A., Sosman J.A., O'Dwyer P.J., Lee R.J., Grippo J.F., Nolop K., Chapman P.B. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim K.B., Eton O., Davis D.W., Frazier M.L., McConkey D.J., Diwan A.H., Papadopoulos N.E., Bedikian A.Y., Camacho L.H., Ross M.I., Cormier J.N., Gershenwald J.E., Lee J.E., Mansfield P.F., Billings L.A., Ng C.S., Charnsangavej C., Bar-Eli M., Johnson M.M., Murgo A.J., Prieto V.G. Phase II trial of imatinib mesylate in patients with metastatic melanoma. Br J Cancer. 2008;99:734–740. doi: 10.1038/sj.bjc.6604482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottesman M.M., Fojo T., Bates S.E. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 8.Chen K.G., Valencia J.C., Gillet J.P., Hearing V.J., Gottesman M.M. Involvement of ABC transporters in melanogenesis and the development of multidrug resistance of melanoma. Pigment Cell Melanoma Res. 2009;22:740–749. doi: 10.1111/j.1755-148X.2009.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monzani E., Facchetti F., Galmozzi E., Corsini E., Benetti A., Cavazzin C., Gritti A., Piccinini A., Porro D., Santinami M., Invernici G., Parati E., Alessandri G., La Porta C.A. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur J Cancer. 2007;43:935–946. doi: 10.1016/j.ejca.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs J.J., van Lohuizen M. Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim Biophys Acta. 2002;1602:151–161. doi: 10.1016/s0304-419x(02)00052-5. [DOI] [PubMed] [Google Scholar]
- 11.Piunti A., Pasini D. Epigenetic factors in cancer development: polycomb group proteins. Future Oncol. 2011;7:57–75. doi: 10.2217/fon.10.157. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs J.J., Kieboom K., Marino S., DePinho R.A., van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs J.J., Scheijen B., Voncken J.W., Kieboom K., Berns A., van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherr C.J. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 15.Molofsky A.V., Pardal R., Iwashita T., Park I.K., Clarke M.F., Morrison S.J. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mihic-Probst D., Kuster A., Kilgus S., Bode-Lesniewska B., Ingold-Heppner B., Leung C., Storz M., Seifert B., Marino S., Schraml P., Dummer R., Moch H. Consistent expression of the stem cell renewal factor BMI-1 in primary and metastatic melanoma. Int J Cancer. 2007;121:1764–1770. doi: 10.1002/ijc.22891. [DOI] [PubMed] [Google Scholar]
- 17.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 18.Lee E.J., Gusev Y., Jiang J., Nuovo G.J., Lerner M.R., Frankel W.L., Morgan D.L., Postier R.G., Brackett D.J., Schmittgen T.D. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortez M.A., Ivan C., Zhou P., Wu X., Ivan M., Calin G.A. microRNAs in cancer: from bench to bedside. Adv Cancer Res. 2010;108:113–157. doi: 10.1016/B978-0-12-380888-2.00004-2. [DOI] [PubMed] [Google Scholar]
- 20.Fabbri M., Calin G.A. Epigenetics and miRNAs in human cancer. Adv Genet. 2010;70:87–99. doi: 10.1016/B978-0-12-380866-0.60004-6. [DOI] [PubMed] [Google Scholar]
- 21.Ferdin J., Kunej T., Calin G.A. Non-coding RNAs: identification of cancer-associated microRNAs by gene profiling. Technol Cancer Res Treat. 2010;9:123–138. doi: 10.1177/153303461000900202. [DOI] [PubMed] [Google Scholar]
- 22.Le XF, Merchant O, Bast RC, Calin GA: The roles of microRNAs in the cancer invasion-metastasis cascade. Cancer Microenviron 3:137–147 [DOI] [PMC free article] [PubMed]
- 23.Shimono Y., Zabala M., Cho R.W., Lobo N., Dalerba P., Qian D., Diehn M., Liu H., Panula S.P., Chiao E., Dirbas F.M., Somlo G., Pera R.A., Lao K., Clarke M.F. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leskela S., Leandro-Garcia L.J., Mendiola M., Barriuso J., Inglada-Perez L., Munoz I., Martinez-Delgado B., Redondo A., de Santiago J., Robledo M., Hardisson D., Rodriguez-Antona C. The miR-200 family controls β-tubulin III expression and is associated with paclitaxel-based treatment response and progression-free survival in ovarian cancer patients. Endocr Relat Cancer. 2010;18:85–95. doi: 10.1677/ERC-10-0148. [DOI] [PubMed] [Google Scholar]
- 25.Vallejo D.M., Caparros E., Dominguez M. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. EMBO J. 2011;30:756–769. doi: 10.1038/emboj.2010.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J.W., Park Y.A., Choi J.J., Lee Y.Y., Kim C.J., Choi C., Kim T.J., Lee N.W., Kim B.G., Bae D.S. The expression of the miRNA-200 family in endometrial endometrioid carcinoma. Gynecol Oncol. 2011;120:56–62. doi: 10.1016/j.ygyno.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 27.Dykxhoorn D.M. MicroRNAs and metastasis: little RNAs go a long way. Cancer Res. 2010;70:6401–6406. doi: 10.1158/0008-5472.CAN-10-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., Goodall G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 29.Park S.M., Gaur A.B., Lengyel E., Peter M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu M.Y., Meier F.E., Nesbit M., Hsu J.Y., Van Belle P., Elder D.E., Herlyn M. E-cadherin expression in melanoma cells restores keratinocyte-mediated growth control and down-regulates expression of invasion-related adhesion receptors. Am J Pathol. 2000;156:1515–1525. doi: 10.1016/S0002-9440(10)65023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tucci M.G., Lucarini G., Brancorsini D., Zizzi A., Pugnaloni A., Giacchetti A., Ricotti G., Biagini G. Involvement of E-cadherin, β-catenin: Cdc42 and CXCR4 in the progression and prognosis of cutaneous melanoma. Br J Dermatol. 2007;157:1212–1216. doi: 10.1111/j.1365-2133.2007.08246.x. [DOI] [PubMed] [Google Scholar]
- 32.Liu S., Tetzlaff M.T., Liu A., Liegl-Atzwanger B., Guo J., Xu X. Loss of microRNA-205 expression is associated with melanoma progression. Lab Invest. 2012;92:1084–1096. doi: 10.1038/labinvest.2012.62. [DOI] [PubMed] [Google Scholar]
- 33.Tetzlaff M.T., Liu A., Xu X., Master S.R., Baldwin D.A., Tobias J.W., Livolsi V.A., Baloch Z.W. Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocr Pathol. 2007;18:163–173. doi: 10.1007/s12022-007-0023-7. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S.M., Yu H., Edwards R., Chen L., Kazianis S., Brafford P., Acs G., Herlyn M., Xu X. Mutant V600E BRAF increases hypoxia inducible factor-1α expression in melanoma. Cancer Res. 2007;67:3177–3184. doi: 10.1158/0008-5472.CAN-06-3312. [DOI] [PubMed] [Google Scholar]
- 35.Park I.K., Qian D., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 36.Onder T.T., Kara N., Cherry A., Sinha A.U., Zhu N., Bernt K.M., Cahan P., Marcarci B.O., Unternaehrer J., Gupta P.B., Lander E.S., Armstrong S.A., Daley G.Q. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S., Kumar S.M., Lu H., Liu A., Yang R., Pushparajan A., Guo W., Xu X. MicroRNA-9 up-regulates E-cadherin through inhibition of NF-κB1-Snail1 pathway in melanoma. J Pathol. 2012;226:61–72. doi: 10.1002/path.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu A., Tetzlaff M.T., Vanbelle P., Elder D., Feldman M., Tobias J.W., Sepulveda A.R., Xu X. MicroRNA expression profiling outperforms mRNA expression profiling in formalin-fixed paraffin-embedded tissues. Int J Clin Exp Pathol. 2009;2:519–527. [PMC free article] [PubMed] [Google Scholar]
- 39.Frank N.Y., Margaryan A., Huang Y., Schatton T., Waaga-Gasser A.M., Gasser M., Sayegh M.H., Sadee W., Frank M.H. ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma. Cancer Res. 2005;65:4320–4333. doi: 10.1158/0008-5472.CAN-04-3327. [DOI] [PubMed] [Google Scholar]
- 40.Song L.B., Zeng M.S., Liao W.T., Zhang L., Mo H.Y., Liu W.L., Shao J.Y., Wu Q.L., Li M.Z., Xia Y.F., Fu L.W., Huang W.L., Dimri G.P., Band V., Zeng Y.X. Bmi-1 is a novel molecular marker of nasopharyngeal carcinoma progression and immortalizes primary human nasopharyngeal epithelial cells. Cancer Res. 2006;66:6225–6232. doi: 10.1158/0008-5472.CAN-06-0094. [DOI] [PubMed] [Google Scholar]
- 41.Li D.W., Tang H.M., Fan J.W., Yan D.W., Zhou C.Z., Li S.X., Wang X.L., Peng Z.H. Expression level of Bmi-1 oncoprotein is associated with progression and prognosis in colon cancer. J Cancer Res Clin Oncol. 2010;136:997–1006. doi: 10.1007/s00432-009-0745-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J.H., Song L.B., Zhang X., Guo B.H., Feng Y., Li X.X., Liao W.T., Zeng M.S., Huang K.H. Bmi-1 expression predicts prognosis for patients with gastric carcinoma. J Surg Oncol. 2008;97:267–272. doi: 10.1002/jso.20934. [DOI] [PubMed] [Google Scholar]
- 43.Li W., Li Y., Tan Y., Ma K., Cui J. Bmi-1 is critical for the proliferation and invasiveness of gastric carcinoma cells. J Gastroenterol Hepatol. 2010;25:568–575. doi: 10.1111/j.1440-1746.2009.06045.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang H., Pan K., Zhang H.K., Weng D.S., Zhou J., Li J.J., Huang W., Song H.F., Chen M.S., Xia J.C. Increased polycomb-group oncogene Bmi-1 expression correlates with poor prognosis in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2008;134:535–541. doi: 10.1007/s00432-007-0316-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sasaki M., Ikeda H., Itatsu K., Yamaguchi J., Sawada S., Minato H., Ohta T., Nakanuma Y. The overexpression of polycomb group proteins Bmi1 and EZH2 is associated with the progression and aggressive biological behavior of hepatocellular carcinoma. Lab Invest. 2008;88:873–882. doi: 10.1038/labinvest.2008.52. [DOI] [PubMed] [Google Scholar]
- 46.Kim J.H., Yoon S.Y., Jeong S.H., Kim S.Y., Moon S.K., Joo J.H., Lee Y., Choe I.S., Kim J.W. Overexpression of Bmi-1 oncoprotein correlates with axillary lymph node metastases in invasive ductal breast cancer. Breast. 2004;13:383–388. doi: 10.1016/j.breast.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 47.van Leenders G.J., Dukers D., Hessels D., van den Kieboom S.W., Hulsbergen C.A., Witjes J.A., Otte A.P., Meijer C.J., Raaphorst F.M. Polycomb-group oncogenes EZH2: BMI1, and RING1 are overexpressed in prostate cancer with adverse pathologic and clinical features. Eur Urol. 2007;52:455–463. doi: 10.1016/j.eururo.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 48.Abdouh M., Facchino S., Chatoo W., Balasingam V., Ferreira J., Bernier G. BMI1 sustains human glioblastoma multiforme stem cell renewal. J Neurosci. 2009;29:8884–8896. doi: 10.1523/JNEUROSCI.0968-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Godlewski J., Nowicki M.O., Bronisz A., Williams S., Otsuki A., Nuovo G., Raychaudhury A., Newton H.B., Chiocca E.A., Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 50.Bea S., Tort F., Pinyol M., Puig X., Hernandez L., Hernandez S., Fernandez P.L., van Lohuizen M., Colomer D., Campo E. BMI-1 gene amplification and overexpression in hematological malignancies occur mainly in mantle cell lymphomas. Cancer Res. 2001;61:2409–2412. [PubMed] [Google Scholar]
- 51.Raaphorst F.M., van Kemenade F.J., Blokzijl T., Fieret E., Hamer K.M., Satijn D.P., Otte A.P., Meijer C.J. Coexpression of BMI-1 and EZH2 polycomb group genes in Reed-Sternberg cells of Hodgkin's disease. Am J Pathol. 2000;157:709–715. doi: 10.1016/S0002-9440(10)64583-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Breuer R.H., Snijders P.J., Sutedja G.T., Sewalt R.G., Otte A.P., Postmus P.E., Meijer C.J., Raaphorst F.M., Smit E.F. Expression of the p16(INK4a) gene product, methylation of the p16(INK4a) promoter region and expression of the polycomb-group gene BMI-1 in squamous cell lung carcinoma and premalignant endobronchial lesions. Lung Cancer. 2005;48:299–306. doi: 10.1016/j.lungcan.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 53.Kang M.K., Kim R.H., Kim S.J., Yip F.K., Shin K.H., Dimri G.P., Christensen R., Han T., Park N.H. Elevated Bmi-1 expression is associated with dysplastic cell transformation during oral carcinogenesis and is required for cancer cell replication and survival. Br J Cancer. 2007;96:126–133. doi: 10.1038/sj.bjc.6603529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shafaroudi A.M., Mowla S.J., Ziaee S.A., Bahrami A.R., Atlasi Y., Malakootian M. Overexpression of BMI1, a polycomb group repressor protein, in bladder tumors: a preliminary report. Urol J. 2008;5:99–105. [PubMed] [Google Scholar]
- 55.Zhang X., Wang C.X., Zhu C.B., Zhang J., Kan S.F., Du L.T., Li W., Wang L.L., Wang S. Overexpression of Bmi-1 in uterine cervical cancer: correlation with clinicopathology and prognosis. Int J Gynecol Cancer. 2010;20:1597–1603. [PubMed] [Google Scholar]
- 56.Bhattacharya R., Nicoloso M., Arvizo R., Wang E., Cortez A., Rossi S., Calin G.A., Mukherjee P. MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res. 2009;69:9090–9095. doi: 10.1158/0008-5472.CAN-09-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong P., Kaneuchi M., Watari H., Hamada J., Sudo S., Ju J., Sakuragi N. MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. Mol Cancer. 2011;10:99. doi: 10.1186/1476-4598-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu Y., Yu F., Jiao Y., Feng J., Tang W., Yao H., Gong C., Chen J., Su F., Zhang Y., Song E. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin Cancer Res. 2011;17:7105–7115. doi: 10.1158/1078-0432.CCR-11-0071. [DOI] [PubMed] [Google Scholar]
- 59.Cui J.G., Zhao Y., Sethi P., Li Y.Y., Mahta A., Culicchia F., Lukiw W.J. Micro-RNA-128 (miRNA-128) down-regulation in glioblastoma targets ARP5 (ANGPTL6), Bmi-1 and E2F-3a, key regulators of brain cell proliferation. J Neurooncol. 2010;98:297–304. doi: 10.1007/s11060-009-0077-0. [DOI] [PubMed] [Google Scholar]
- 60.Wellner U., Schubert J., Burk U.C., Schmalhofer O., Zhu F., Sonntag A., Waldvogel B., Vannier C., Darling D., zur Hausen A., Brunton V.G., Morton J., Sansom O., Schuler J., Stemmler M.P., Herzberger C., Hopt U., Keck T., Brabletz S., Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 61.Xu Y., Brenn T., Brown E.R., Doherty V., Melton D.W. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer. 2012;106:553–561. doi: 10.1038/bjc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elson-Schwab I., Lorentzen A., Marshall C.J. MicroRNA-200 family members differentially regulate morphological plasticity and mode of melanoma cell invasion. PLoS One. 2010;5:e13176. doi: 10.1371/journal.pone.0013176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen J., Feilotter H.E., Pare G.C., Zhang X., Pemberton J.G., Garady C., Lai D., Yang X., Tron V.A. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol. 2010;176:2520–2529. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen K.G., Szakacs G., Annereau J.P., Rouzaud F., Liang X.J., Valencia J.C., Nagineni C.N., Hooks J.J., Hearing V.J., Gottesman M.M. Principal expression of two mRNA isoforms (ABCB 5α and ABCB 5β ) of the ATP-binding cassette transporter gene ABCB 5 in melanoma cells and melanocytes. Pigment Cell Res. 2005;18:102–112. doi: 10.1111/j.1600-0749.2005.00214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schatton T., Murphy G.F., Frank N.Y., Yamaura K., Waaga-Gasser A.M., Gasser M., Zhan Q., Jordan S., Duncan L.M., Weishaupt C., Fuhlbrigge R.C., Kupper T.S., Sayegh M.H., Frank M.H. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Effendi K., Mori T., Komuta M., Masugi Y., Du W., Sakamoto M. Bmi-1 gene is upregulated in early-stage hepatocellular carcinoma and correlates with ATP-binding cassette transporter B1 expression. Cancer Sci. 2010;101:666–672. doi: 10.1111/j.1349-7006.2009.01431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumamoto H., Ohki K. Detection of CD133, Bmi-1, and ABCG2 in ameloblastic tumors. J Oral Pathol Med. 2010;39:87–93. doi: 10.1111/j.1600-0714.2009.00807.x. [DOI] [PubMed] [Google Scholar]