Abstract
Kinases have been implicated in the immunopathological mechanisms of Systemic Lupus Erythematosus (SLE). v-akt murine-thymoma viral-oncogene-homolog 1 (AKT1) and mitogen-activated-protein-kinase 1 (MAPK1) gene expressions in peripheral mononuclear cells from thirteen SLE patients with inactive or mild disease were evaluated using quantitative real-time reverse-transcription polymerase-chain-reaction and analyzed whether there was any correlation with T-helper (Th) transcription factors (TF) gene expression, cytokines, and S100A8/S100A9-(Calprotectin). Age- and gender-matched thirteen healthy controls were examined. AKT1 and MAPK1 expressions were upregulated in SLE patients and correlated with Th17-(Retinoic acid-related orphan receptor (ROR)-C), T-regulatory-(Treg)-(Transforming Growth Factor Beta (TGFB)-2), and Th2-(interleukin (IL)-5)-related genes. MAPK1 expression correlated with Th1-(IL-12A, T-box TF-(T-bet)), Th2-(GATA binding protein-(GATA)-3), and IL-10 expressions. IL-10 expression was increased and correlated with plasma Tumor Necrosis Factor (TNF)-α and Th0-(IL-2), Th1-(IL-12A, T-bet), GATA3, Treg-(Forkhead/winged-helix transcription factor- (FOXP)-3), and IL-6 expressions. FOXP3 expression, FOXP3/RORC, and FOXP3/GATA3 expression ratios were increased. Plasma IL-1β, IL-12(p70), Interferon-(IFN)-γ, and IL-6 cytokines were augmented. Plasma IL-1β, IL-6, IL-2, IFN-γ, TNF-α, IL-10, and IL-13 correlated with C-reactive protein, respectively. Increased Calprotectin correlated with neutrophils. Conclusion, SLE patients presented a systemic immunoinflammatory activity, augmented AKT1 and MAPK1 expressions, proinflammatory cytokines, and Calprotectin, together with increased expression of Treg-related genes, suggesting a regulatory feedback opposing the inflammatory activity.
1. Introduction
Systemic Lupus Erythematosus (SLE) is a chronic inflammatory autoimmune disease most common in women of reproductive age [1], characterized by a relapsing/remitting course and the involvement of multiple organs, including skin, kidneys, and central nervous system. Pathophysiologically is characterized by the dysfunction of T, B, and dendritic cells (DC), skewed cytokine production, breakdown of immunological tolerance, and by the production of antinuclear autoantibodies [2–5].
In SLE bone marrow mononuclear cells, Nakou et al. [6] identified central gene regulators implicated in disease pathogenesis which include activation of multiple kinase pathways (MAPK/extracellular regulated MAP kinase (ERK), Signal Transducer and Activator of Transcription (STAT), AKT, and PI3-kinase (PI3 K)). AKT1 serine/threonine kinase is a key downstream target of PI3 K signaling pathway, and plays a role in the differentiation of peripheral B cells and in T cell homeostasis [7–10]. Upregulated activity of AKT kinases has been documented in B cells from SLE patients [11]. AKT/Mammalian target of rapamycin (mTOR) axis has been successfully targeted with rapamycin for treatment of SLE patients [2]. MAPK/ERK kinases play a significant role in immune-mediated inflammatory responses and are involved in the maintenance of T-cell tolerance that fails in SLE patients [4, 12]. Molad et al. [13] reported increased ERK and JNK activities that correlated with disease activity in SLE patients. Conversely, other studies documented a reduced MAPK activity in human SLE T cells [4]. In mice, a defect in lupus T cell ERK pathway signaling that can cause epigenetic abnormalities in T cells by inhibiting DNA methylation has been reported [12, 14]. In addition, in SLE T cells, alterations in kinases pathways such as the spleen tyrosine kinase (Syk) signaling patterns have been documented by Krishnan et al. [15].
The imbalance in cytokine activities is associated with human autoimmune and autoinflammatory diseases, and it has been reported to play a central role in regulating SLE development [16–20]. There is a key collaboration and regulation between key cytokines in activation/induction of transcription factors during the process of T-helper-(Th)-cell differentiation towards Th1, Th2, Th17, and induced regulatory T (iTreg) cells lineages [21, 22]. Changes in Th-specific transcription factors gene expression ratios have been used as a marker of Th-cytokine balance [20, 23–26].
Moreover, increased inflammation mediators S100A8/S100A9-(Calprotectin) serum and/or plasma levels have been reported in SLE patients [27–29]. Damage-associated molecular pattern molecules (DAMP) S100A8 and S100A9 members of the calcium-binding S100-family make a heterotetrameric form, S100A8/S100A9-(Calprotectin).
The aim of this study is to assess the gene expression levels of intracellular kinases AKT1 and MAPK1 in peripheral blood mononuclear cells (PBMC) from SLE patients with inactive or mild disease and to analyze whether there was any correlation with Th-transcription factors gene expression, with gene expression and plasma levels of a comprehensive panel of cytokines (Th0-, Th1/Th17-, and Th2/Treg-type), with plasma inflammation mediators S100A8/A9-Calprotectin and clinical parameters.
2. Materials and Methods
2.1. Patients and Controls
The study protocol was approved by the SCUH Committee and CSIC Review Board and Ethics Committees. Written informed consent was obtained from all participating patients and controls, according to the Helsinki Declaration. Clinical data and treatments of patients are summarized (Table 1). Thirteen SLE outpatients (eleven women and two men) with inactive or mild disease (SLE disease activity index (SLEDAI) score ≤ 4) (0 (0–3) (median (25% and 75% percentile ranges), see Table 1) that fulfilled at least four of the revised SLE criteria of the American College of Rheumatology (ACR) for the diagnosis of SLE [30, 31] were evaluated. Age- and gender-matched healthy controls (C) included thirteen volunteers subjects, without known autoimmune disease (eight women and five men; 34 (30–51.5) years old; differences versus SLE patients were not statistically significant, Mann-Whitney, P = 0.1435). Likewise, no significant differences in gender distribution were observed between patients and controls (Fisher test: P = 0.3783, for SLE patients versus C). Patients and controls were Caucasians.
Table 1.
Clinical parameters and treatments of SLE patients.
| SLE patients | SLEDAIa | Sexb | Agec | Ld | C3e | C4e | Anti-dsDNAg | RDh | CRPi | Nj | Ddk | Treatmentl |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4 | W | 41 | 1620 | 93 | 17 | − | − | 0.11 | 4670 | 3.67 | HCQ |
| 2 | 0 | W | 27 | 1470 | 108 | 19 | − | − | 0.07 | 9870 | 0.75 | HCQ/PN |
| 3 | 0 | W | 65 | 2530 | 112 | 21 | − | − | 0.17 | 4760 | 4.5 | HCQ/PN |
| 4 | 0 | W | 64 | 2510 | 95 | 25 | − | − | 0.1 | 7130 | 10.5 | HCQ |
| 5 | 4 | W | 43 | 900 | 73 | 6 | + | − | 1.13 | 6870 | 8.67 | HCQ/PN |
| 6 | 2 | M | 59 | 2240 | 88 | 8 | − | − | 4.62 | 8240 | 1.08 | MTX/PN/HCQ |
| 7 | 0 | W | 56 | 1130 | 131 | 20 | − | − | 0.05 | 4070 | 2.58 | NT |
| 8 | 4 | W | 42 | 570 | 66 | 9 | + | + | 0.01 | 3120 | 8.83 | AZA/PN/HCQ |
| 9 | 0 | W | 40 | 750 | 62 | 12 | + | − | 1.16 | 7500 | 8.83 | MTX/HCQ/PN |
| 10 | 0 | W | 55 | 1740 | 110 | 15 | − | − | 0.35 | 8250 | 2.33 | MMF/PN/HCQ |
| 11 | 2 | M | 18 | 3420 | 129 | 11 | + | + | NDf | 12260 | 5.33 | MMF/PN/HCQ |
| 12 | 2 | W | 35 | 2250 | 91 | 23 | − | + | 0.08 | 6190 | 19.25 | AZA/HCQ/PN |
| 13 | 0 | W | 41 | 1150 | NDf | NDf | − | − | 0.7 | 3170 | 6.75 | HCQ |
|
| ||||||||||||
| Medianm | 0 | 42 | 1620 | 94n | 16n | 0.14n | 6870 | 5.33 | ||||
| (0–3) | (37.5–57.5) | (1015–2380) | (76.8 –111.5) |
(9.5– 20.8) | (0.072–1.023) | (4370–8245) | (2.5–8.8) | |||||
aSystemic Lupus Erythematosus Disease Activity index (SLEDAI).
bW: woman; M: man.
cAge (years).
dL: Lymphocytes/mL.
eComplement C3 and C4 (low <90 and/or <10, mg/dL).
fND: no determined.
gSLE patients presented anti-double stranded DNA auto antibodies (31%, three women and one man).
hRD: renal disease, (23% of SLE patients).
iCRP: C-reactive protein (mg/dL).
jN: Neutrophils/microliter.
kDd: Disease duration, at the time of the study, in years (y).
lHCQ: hydroxychloroquine; PN: prednisone; MTX: methotrexate; AZA: azathioprine; MMF: mycophenolate mofetil; NT: no treatment.
mMedian (25th to 75th percentiles).
n n = 12.
Medications taken by the patients at the time the blood was drawn were recorded (Table 1). The majority of SLE patients (n = 12) were treated with hydroxychloroquine (200 mg/day); nine SLE patients were also treated with prednisone (5 mg/day), and six of them were additionally treated with one of methotrexate, azathioprine, or mycophenolate mofetil (Table 1).
2.2. Blood, Plasma, and PBMC Samples
Blood was collected (BD Vacutainer system, K2-EDTA tubes, BD Diagnostics, Franklin Lakes, NJ, USA) and plasma and PBMC were separated using density gradient centrifugation [24].
2.3. RNA Isolation and Quantitative Real-Time Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
Gene expression levels were measured quantitatively by qRT-PCR as previously described [24]. Total RNA was isolated from PBMC from thirteen SLE patients, and four controls, using RNeasy Plus mini kit (Qiagen, Hilden, Germany). RNA quality was assessed by an Experion Automated Electrophoresis Station (Bio-Rad, Hercules, CA, USA). RT reaction, primer sequences, and gene-specific primers used in this studio are indicated (Table 2 and Supplementary Materials). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. To quantify cytokine mRNA expression, a RT2 Profiler PCR Array System (SABiosciences, Qiagen) was used. The PCR Array layout contained gene-specific primers for ten cytokines: Interleukin (IL)-1B, IL-2, IL-5, IL-6, IL-10, IL-12A, Tumor Necrosis Factor (TNF)-α, Interferon (IFN)-γ, Transforming Growth Factor Beta (TGFB)-2, and Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand or TRAIL (TNFSF)-10; two internal loading gene-specific primers controls were included for standardization between samples (GAPDH and β-actin, Table 2). Proportion of transcript present in the samples was calculated using the relative quantification 2−ΔΔCt scheme [32]. Control samples were used as comparative calibrator. Results represented the relative amount of amplicon in patient's sample (fold change) to the mean level of the transcripts in the control samples.
Table 2.
Primers for quantitative RT-PCR.
| Gene1 | Accession no.1 | Description | SABiosciences Cat. no.2 |
|---|---|---|---|
| IL-1B | NM_000576 | Interleukin 1, beta | PPH00171B |
| IFNG | NM_000619 | Interferon, gamma | PPH00380B |
| IL-12A | NM_000882 | Interleukin 12A | PPH00544B |
| IL-6 | NM_000600 | Interleukin 6 | PPH00560B |
| TNF-alpha | NM_000594 | Tumor necrosis factor (TNF superfamily, member 2) | PPH00341E |
| IL-10 | NM_000572 | Interleukin 10 | PPH00572B |
| IL-2 | NM_000586 | Interleukin 2 | PPH00172B |
| TGFB2 | NM_003238 | Transforming growth factor, beta 2 | PPH00524B |
| IL-5 | NM_000879 | Interleukin 5 | PPH00692A |
| TNFSF10 | NM_003810 | Tumor necrosis factor (ligand) superfamily, member 10 | PPH00242E |
| GAPDH | NM_002046 | Glyceraldehyde-3-phosphate dehydrogenase | PPH00150E |
| ACTB | NM_001101 | Actin, beta | PPH00073E |
| TBX21 | NM_013351 | T-box-21 (T-bet) | PPH00396A |
| GATA3 | NM_002051 | GATA-binding protein 3 | PPH02143A |
| FOXP3 | NM_014009 | Forkhead box P3 | PPH00029B |
| STAT4 | NM_003151 | Signal transducer and activator of transcription 4 | PPH00777E |
| RORC | NM_005060 | RAR-related orphan receptor C | PPH05877A |
| MAPK1 | NM_002745 | Mitogen-activated-protein-kinase 1 | PPH00715B |
| AKT1 | NM_005163 | v-akt murine-thymoma viral-oncogene-homolog 1 | PPH00088A |
| CD38 | NM_001775 | CD38 molecule | PPH00856A |
| ZAP70 | NM_001079 | Zeta-chain (TCR) associated protein kinase 70 kDa | PPH00256A |
| CD247 | NM_000734 | CD247 molecule, CD3-zeta. | PPH01484A |
| CCR6 | NM_004367 | Chemokine (C-C motif) receptor 6 | PPH00616E |
|
| |||
| Gene1,3 | Forward primer (5′–3′)4 | Reverse Primer (5′–3′)4 | Ref. |
|
| |||
| GAPDH (NM_002046) | gaaggtgaaggtcggagtc | gaagatggtgatgggatttc | Primer3 software3 |
2SABiosciences, QIAGEN company.
4Primers used for this study based on the literature were computationally checked for sequence specificity. Melt-curve analysis for each primer pair and reaction was also tested to verified specific amplification.
2.4. Plasma Cytokines
Bio-Plex Precision Pro Human Cytokine 10-Plex kit assays were used to simultaneously test 10 cytokines in plasma: IL-1β, IL-2, IL-4, IL-5, IL-6, IL-10, IL-12(p70), IL-13, IFN-γ, and TNF-α, in according with manufacturer's protocol (Bio-Rad).
2.5. Human S100A8/S100A9 Heterodimer Complexes or Calprotectin Enzyme Linked Immunosorbent Assay (ELISA)
Plasma concentration, a noncovalently associated Calprotection, in the presence of calcium, was measured in duplicate using a sandwich ELISA kit (no. HK325; Hycult Biotechnology, Uden, The Netherlands).
2.6. Statistics
Data are given as median and 25% and 75% percentile ranges, or otherwise indicated. Relative mRNA expression levels of the genes in patients' PBMC versus controls were evaluated using the nonparametric Wilcoxon Signed Rank test and comparing the medians against a hypothetical median (value equal 1). Determinations of significant differences between groups were compared using the non-parametric Mann-Whitney U test. Fisher exact test was used for analysis of qualitative variables. Correlations were determined by Spearman's rank correlation test. Differences were considered statistically significant for P values < 0.05 (GarphPad Prism v.5.01, GraphPad, CA, USA).
3. Results
3.1. Intracellular kinases and Immunoreceptors Gene Expressions Levels
AKT1 (7.82 (2.61–11.45)), MAPK1 (19.47 (8.71–33.45)), and ZAP70 (1.98 (1.14–3.66)) relative gene expressions levels were significantly augmented in PBMC from SLE patients (n = 13) as compared with controls (Wilcoxon Signed Rank test: P = 0.0064, P = 0.0012, and P = 0.0024, resp.) (Figure 1(a)). CD38 (1.22 (0.77–1.95)) and CD3zeta (1.15 (0.54–1.83)) immunoreceptors relative gene expression levels were also evaluated; not significantly differences were observed versus controls (Wilcoxon Signed Rank test: P = 0.0805, P = 0.7869, resp.) (Figure 1(a)). In addition, the Th17-associated CC chemokine receptor (CCR)-6 relative gene expression was significantly decreased in SLE's PBMC versus controls (0.50 (0.33–0.95), Wilcoxon Signed Rank test, P = 0.0398) (Figure 1(a)).
Figure 1.
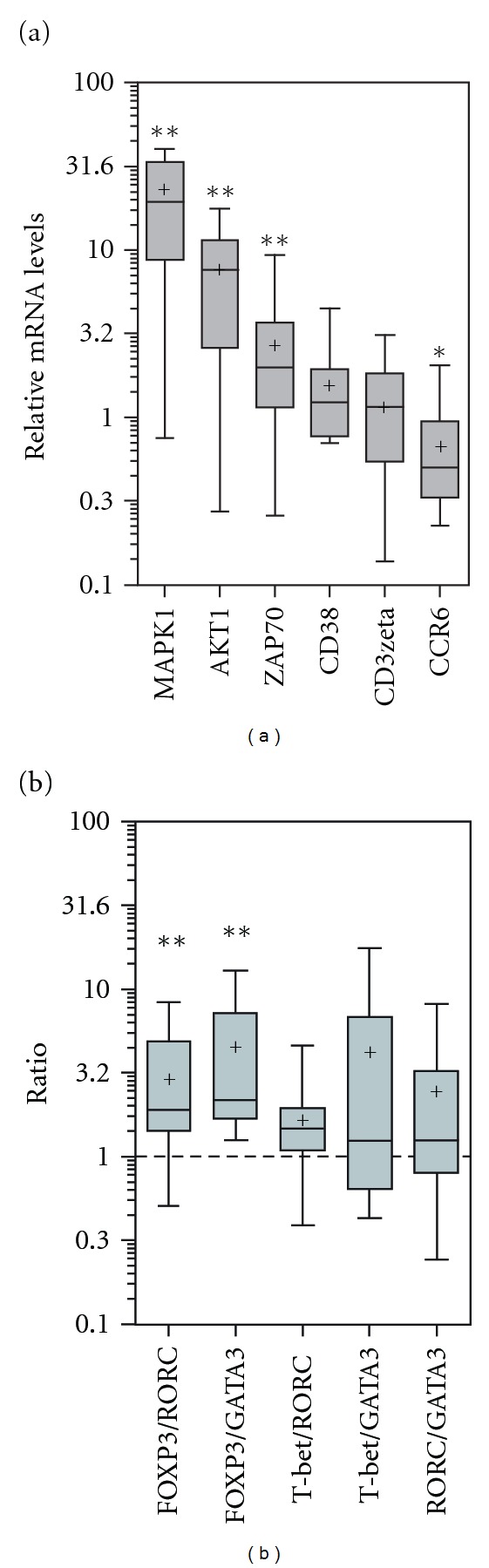
(a) Intracellular kinases and immunoreceptors gene expressions levels. RNAs extracted from PBMC of SLE patients (n = 13) and controls (n = 4) were reverse transcribed, and the gene expression levels were determined by qRT-PCR using gene-specific primers for indicated genes and GAPDH to normalize the mRNA expression (Materials and Methods, Supplementary Materials, and Table 2). Proportion of transcript present in the samples was calculated using the relative quantification 2-ΔΔCt scheme. Control samples were used as comparative calibrator. Final results represent the relative amount of amplicon in patient's sample (fold change) to the mean level of the transcripts in the control samples. In the figure, results are represented in box plots given the median (horizontal bars within boxes), the mean (cross points); box limits correspond to quartiles, and vertical lines indicate the 5–95 percentile range. Results are represented in a log 10 scale, and the indicated numbers are the antilog values. Significance levels set at P values <0.05. AKT1, MAPK1, and ZAP70 gene expressions were increased in SLE patients compared with controls (Wilcoxon Signed Rank test: P = 0.0064, P = 0.0012, P = 0.0024). Gene expression of CCR6 was decreased in SLE patients compared with controls (Wilcoxon Signed Rank test: P = 0.0398). (b) T-helper transcription factors gene expression ratios in SLE patients. Results are expressed as indicated in (a). FOXP3/RORC ratio in SLE patients versus controls (Wilcoxon Signed Rank test, P = 0.0042). FOXP3/GATA3 ratio in SLE versus controls (Wilcoxon test, P = 0.0017).
3.2. Increased Treg/Th17 and Treg/Th2 Transcription Factors Gene Expression Ratios
In PBMC from patients and controls, relative gene expression of T-box transcription factor (T-bet or TBX21) and signal transducer and activator of transcription (STAT)-4, both for Th1 cells; GATA3, a member of the GATA family of zinc finger proteins for Th2; retinoic acid-related orphan receptor (ROR)-C, for Th17 cells and forkhead/winged helix transcription factor (FOXP)-3 for Treg cells [33, 34] were assessed and Th-transcription factors gene expression ratios calculated. FOXP3 relative gene expression was significantly increased in SLE patients versus controls (18.3 (1.39–44.14), n = 13, Wilcoxon test, P = 0.0012). However, there were no statistically significant differences between medians of the T-bet (5.94 (0.44–63.98)), STAT4 (1.09 (0.60–2.07)), RORC (4.49 (0.60–30.56)) and GATA3 (2.80 (0.79–8.19)) relative gene expressions levels, respectively, versus controls (Wilcoxon Signed Rank test, P = 0.0681; P = 0.3054, P = 0.0681, P = 0.0574, for T-bet, STAT4, RORC, and GATA3, resp.).
In SLE patients, gene expression ratios of both FOXP3/RORC (1.90 (1.41–4.85)) and FOXP3/GATA3 (2.17 (1.67–7.22)) were significantly increased versus controls (Wilcoxon Signed Rank test, P = 0.0042 and P = 0.0017, resp.) (Figure 1(b)). While, ratios of T-bet/RORC (1.48 (1.09–1.95)), T-bet/GATA3 (1.24 (0.64–6.77)), and RORC/GATA3 (1.25 (0.79–3.23)) were evaluated and no significantly differences were observed between SLE versus controls (Wilcoxon Signed Rank test, P = 0.0503, P = 0.1099, and P = 0.0942, resp., Figure 1(b)).
3.3. Cytokines Gene Expression, Plasma Levels, and Correlations
In SLE patients, cytokines mRNA expressions were evaluated using a RT2 Profiler PCR Array System. Results indicated that IL-10 relative gene expression was significantly increased versus controls (Table 3). In addition, ten cytokines were simultaneously assessed in each plasma sample using a Bio-Plex (Table 4). Results indicated that plasma levels of proinflammatory cytokines IL-1β, IL-6 (both Th17), IL-12(p70), IFN-γ (both Th1) together with IL-5 (Th2) were significantly augmented in SLE patients versus controls (Table 4).
Table 3.
Cytokines relative mRNA levels in PBMC.
| Genesa | SLE (n = 13)b |
|---|---|
| IL-1B | 1.36 (0.75–1.88) |
| IL-2 | 1.20 (0.26–6.03) |
| IL-5 | 1.41 (0.60–2.70) |
| IL-6 | 1.41 (0.58–6.08) |
| IL-10 | 1.98c (1.21–5.37) |
| IL-12A | 1.67 (0.58–2.81) |
| IFN-γ | 1.24 (0.74–2.32) |
| TNF-α | 0.20 (0.16–2.20) |
| TGFB2 | 1.17 (0.25–2.24) |
| TNFSF10 | 1.37 (0.95–1.92) |
aRNAs extracted from PBMC of SLE patients (n = 13) and controls (n = 4) were reverse transcribed, and the gene expression levels were determined by qRT-PCR using gene-specific primers for indicated genes to quantify cytokine mRNA expression, by means of a RT2 Profiler PCR Array System (Table 2). qRT-PCR methods (Materials and Methods and Supplementary material). Two internal loading gene-specific primers controls were included for standardization between samples (GAPDH and β-actin, Table 2). Proportion of transcript present in the samples was calculated using the relative quantification 2−ΔΔCt scheme. Control samples were used as comparative calibrator. Final results represent the relative amount of amplicon in patient's sample (fold change) to the mean level of the transcripts in the control samples.
bValues of the relative mRNA levels are presented as Median (25th to 75th percentile).
cIL-10 gene expression was increased in SLE patients compared with controls (Wilcoxon Signed Rank test, P = 0.0046).
Analysis of the data was done using the GraphPad Prism v.5.01 software.
Table 4.
Cytokines plasma levels.
| Cytokinesa, b | SLE (n = 13) | Controls (n = 13) | P valuec |
|---|---|---|---|
| IL-1β | 0.24 (0.10–0.96) | 0.10 (0.10–0.13) | 0.0112 |
| IL-2 | 0.61 (0.60–20.24) | 0.60 (0.60–2.62) | 0.1893 |
| IL-4 | 0.10 (0.10–0.68) | 0.10 (0.10–0.19) | 0.6607 |
| IL-5 | 1.00 (1.00–1.00) | 0.90 (0.80–1.00) | 0.0077 |
| IL-6 | 1.00 (1.00–22.35) | 1.00 (0.85–1.00) | 0.0117 |
| IL-10 | 2.82 (0.75–9.64) | 0.63 (0.60–3.22) | 0.1368 |
| IL-12(p70) | 1.02 (0.60–1.68) | 0.18 (0.06–0.50) | 0.0009 |
| IL-13 | 0.20 (0.20–3.87) | 0.20 (0.19–0.21) | 0.0685 |
| IFN-γ | 0.64 (0.20–3.76) | 0.20 (0.20–0.33) | 0.0445 |
| TNF-α | 0.34 (0.10–2.10) | 0.10 (0.10–0.30) | 0.1193 |
aA multiplex bead array (Bio-Plex, BioRad) was used to simultaneously measure 10 different cytokines in each plasma sample. Samples from patients and controls were analyzed in parallel. Values are in pg/mL. Limits of detection (pg/mL) for the indicated cytokines were: 0.1, 0.64, 0.17, 0.88, 0.29, 0.17, 0.37, 0.19, 0.35, and 0.2, respectively. Analyses of data were performed using five-parameter logistic curve fitting to standard analyte values. Intra-assay and interassay CV were ≤8% and ≤10%, respectively.
bValues are presented as Median (25th to 75th percentiles).
cMann-Whitney U test for comparison between SLE and controls.
Significance levels set at P values <0.05. The analysis of the data was done using the GraphPad Prism v.5.01 software.
In SLE (n = 13) patients, IL-1β and IL-6 plasma levels were positively correlated to IL-1B and IL-6 gene expression levels, respectively (Table 5). IL-6 gene expression also positively correlated with Th1 (IL-12(p70), IFN-γ, and TNF-α), Th0 (IL-2), Th17 (IL-1β), and IL-10 plasma cytokines, respectively. There was a positive correlation between IL-10 gene expression and plasma TNF-α (Table 5). While there was a negative correlation between IL-10 and TNF-α gene expressions (r = −0.6905, P = 0.009). There were significant positive correlations in the IL-2, IL-6, IL-10, IL-12A, and TGFB2 gene expressions, which were correlated to one another; except for IL-6 and TGFB2 (Table 5). In addition, IFN-γ and IL-12A gene expressions were positively correlated (Table 5).
Table 5.
Spearman's rank correlations obtained between cytokines gene expressions in PBMCs and cytokines plasma levels in SLE patients.
| Gene | SLE (n = 13) | ||||||
|---|---|---|---|---|---|---|---|
| Plasma cytokines | |||||||
| IL-1β | IL-2 | IL-6 | IL-10 | IL-12(p70) | IFN-γ | TNF-α | |
| IL-1B | |||||||
| r | 0.6082 | ||||||
| P | 0.0274 | ||||||
| IL-6 | |||||||
| r | 0.6110 | 0.7427 | 0.7204 | 0.8253 | 0.6877 | 0.7856 | 0.8970 |
| P | 0.0265 | 0.0036 | 0.0055 | 0.0005 | 0.0094 | 0.0015 | <0.0001 |
| IL-10 | |||||||
| r | 0.6101 | ||||||
| P | 0.0268 |
||||||
|
| |||||||
| Gene | Gene | ||||||
| IL-2 | IL-6 | IL-10 | IL-12A | IFN-γ | TGFB2 | ||
|
| |||||||
| IL-2 | |||||||
| r | 1 | 0.6538 | 0.6319 | 0.9106 | 0.7868 | ||
| P | 0.0153 | 0.0205 | <0.0001 | 0.0014 | |||
| IL-6 | |||||||
| r | 1 | 0.7582 | 0.7180 | ||||
| P | 0.0027 | 0.0057 | |||||
| IL-10 | |||||||
| r | 1 | 0.7813 | 0.6190 | ||||
| P | 0.0016 | 0.0241 | |||||
| IL-12A | |||||||
| r | 1 | 0.6905 | 0.7796 | ||||
| P | 0.0090 | 0.0017 | |||||
Significant Spearman's rank correlations coefficients are indicated.
Significance levels set at P values <0.05.
3.4. AKT1, MAPK1, and Th-Transcription Factors Gene Expressions Correlations
In SLE patients, there were significant correlations between MAPK1, AKT1, T-bet, RORC, and GATA3 relative gene expressions, respectively (Table 6). Additionally, AKT1 gene expression was correlated to both RORC (Table 6) and CD38 expressions (Table 8), respectively. Moreover, there were significant correlations in the T-bet, STAT4, GATA3, RORC, and FOXP3 gene expressions, which were correlated to one another (Table 6). Both AKT1 and MAPK1 gene expressions were correlated with gene expression levels of Treg-(TGFB2) and Th2-(IL-5) cytokine-related genes, respectively (Table 7). There was also a correlation between AKT1 and TNFSF10 (TRAIL) gene expressions (Table 7). Additionally, MAPK1 gene expression was correlated to IL-10 and IL-12A gene expressions (Table 7). Furthermore, significant correlations between IL10 relative gene expression and FOXP3, RORC, GATA3, T-bet, (Table 7), and CD38 gene expressions (Table 8), respectively, were observed. Correlations between FOXP3 gene expression and Th1 (IL-12A, IFN-γ), Th0 (IL-2), Treg-(TGFB2), Th2-(IL-5), and IL-6 gene expressions, respectively, were observed (Table 7).
Table 6.
Spearman's rank correlations found between MAPK1, AKT1, and Th-transcription factors gene expressions in SLE patients.
| SLE (n = 13) | ||||||
|---|---|---|---|---|---|---|
| Gene | AKT1 | T-bet | STAT4 | GATA3 | RORC | FOXP3 |
| MAPK1 | ||||||
| r | 0.9188 | 0.5659 | 0.5609 | 0.6154 | ||
| P | <0.0001 | 0.0438 | 0.0463 | 0.0252 | ||
| AKT1 | ||||||
| r | 1 | 0.6300 | ||||
| P | 0.0210 | |||||
| T-bet | ||||||
| r | 1 | 0.7912 | 0.9231 | 0.8958 | 0.9066 | |
| P | 0.0013 | <0.0001 | <0.0001 | 0.0004 | ||
| STAT4 | ||||||
| r | 1 | 0.7308 | 0.7253 | 0.5659 | ||
| P | 0.0045 | 0.005 | 0.0438 | |||
| GATA3 | ||||||
| r | 1 | 0.9066 | 0.9066 | |||
| P | <0.0001 | <0.0001 | ||||
| RORC | ||||||
| r | 1 | 0.7802 | ||||
| P | 0.0017 | |||||
Significant Spearman's rank correlations coefficients are indicated.
Significance levels set at P values <0.05.
Table 8.
Spearman's rank correlation test.
| SLE (n = 13) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| CRPc | Nd | Age | C4c | Lf | CD38e | ZAP70e | CCR6e | ||
| Cytokinesa | Genee | ||||||||
| IL-1β | IL-2 | ||||||||
| r | 0.5944 | r | 0.6658 | ||||||
| P | 0.0415 | P | 0.0130 | ||||||
| IL-2 | IL-10 | ||||||||
| r | 0.6671 | r | 0.6107 | 0.5915 | |||||
| P | 0.0178 | P | 0.0266 | 0.0332 | |||||
| IL-6 | IL-12A | ||||||||
| r | 0.6160 | r | 0.6419 | ||||||
| P | 0.0329 | P | 0.0180 | ||||||
| IL-10 | TGFB2 | ||||||||
| r | 0.6690 | r | 0.5909 | ||||||
| P | 0.0174 | P | 0.0335 | ||||||
| IL-13 | TNFSF10 | ||||||||
| r | 0.8629 | r | −0.5750 | ||||||
| P | 0.0003 | P | 0.0398 | ||||||
| IFN-γ | TNF-α | ||||||||
| r | 0.7261 | r | 0.6740 | ||||||
| P | 0.0075 | P | 0.0115 | ||||||
| TNF-α | AKT1 | ||||||||
| r | 0.6834 | r | 0.6061 | ||||||
| P | 0.0143 | P | 0.0281 | ||||||
| GATA-3 | |||||||||
| r | 0.6190 | ||||||||
| P | 0.0241 | ||||||||
| STAT4 | |||||||||
| r | 0.7836 | 0.5659 | |||||||
| P | 0.0026 | 0.0438 | |||||||
| Calprotectinb | CCR6 | ||||||||
| r | 0.6593 | r | 0.7343 | ||||||
| P | 0.0142 | P | 0.0065 | ||||||
aPlasma Cytokines (pg/mL).
bCalprotectin plasma levels (ng/mL).
c n = 12.
dN: Neutrophils.
eGene: Gene expression, as indicated in legend of Figure 1.
fL: Lymphocytes.
Significant Spearman's rank correlations coefficients are indicated.
Significance levels set at P values <0.05.
Table 7.
Spearman's rank correlations found between gene expressions of AKT1, MAPK1, FOXP3, cytokines, IL-10, and Th-transcription factors.
| SLE (n=13) | ||||||
|---|---|---|---|---|---|---|
| Gene | AKT1 | MAPK1 | FOXP3 | RORC | GATA3 | T-bet |
| IL-2 | ||||||
| r | 0.7692 | |||||
| P | 0.0021 | |||||
| IL-5 | ||||||
| r | 0.6970 | 0.7895 | 0.6575 | |||
| P | 0.0081 | 0.0013 | 0.0106 | |||
| IL-6 | ||||||
| r | 0.6484 | |||||
| P | 0.0165 | |||||
| IL-10 | ||||||
| r | 0.5549 | 0.8022 | 0.6923 | 0.7143 | 0.5604 | |
| P | 0.0490 | 0.0010 | 0.0087 | 0.0061 | 0.0463 | |
| IL-12A | ||||||
| r | 0.5942 | 0.9051 | ||||
| P | 0.0322 | <0.0001 | ||||
| IFN-γ | ||||||
| r | 0.6703 | |||||
| P | 0.0122 | |||||
| TNFSF10 | ||||||
| r | 0.6556 | |||||
| P | 0.0150 | |||||
| TGFB2 | ||||||
| r | 0.6584 | 0.6960 | 0.6990 | |||
| P | 0.0144 | 0.0082 | 0.0082 | |||
Significant Spearman's rank correlations coefficients are indicated.
Significance levels set at P values <0.05.
3.5. Plasma S100A8/A9 Calprotectin, Clinical Parameters, and Correlations
Plasma levels of inflammation mediator Calprotectin were augmented in SLE patients (175.6 (138.9–229.2) ng/mL, n = 13), as compared with controls (143.3 (133.4–151.9) ng/mL, n = 11) although differences were not statistical significant (Mann-Whitney, P = 0.0637). Our data are consistent with previous reports [27–29, 35]. In SLE patients, Calprotectin was correlated with neutrophils numbers (Table 8). In addition, significant correlations were obtained between disease-marker C-reactive protein (CRP) and plasma cytokines type Th1-(IFN-γ, TNF-α), Th0-(IL-2), Th2-(IL-13), Th17-(IL-1β, IL-6), and IL-10, respectively (Table 8). Patients' age was correlated with gene expression levels of IL-2, IL-10, IL-12A, TGFB2, and GATA3 relative gene expressions, respectively (Table 8). Moreover, in SLE patients, C4-complement levels were correlated to CCR6 (lymphocyte trafficking) and STAT4 gene expressions levels, respectively, which were correlated to one another (Table 8). In mice, CCR6 was recently identified by Wei et al. [36] among the STAT4-occupied genes. Lastly, lymphocytes numbers negatively correlated with TNFSF10 (TRAIL) gene expression (Table 8).
4. Discussion
In this study, we observed that AKT1 and MAPK1 gene expressions were upregulated in SLE patients with mild or inactive disease and correlate with gene expressions levels of Th17-(RORC), Treg-(TGFB2), and Th2-(IL-5)-related genes, respectively. Additionally, AKT1 gene expression also correlates with MAPK1 expression in SLE patients. PI3 K/AKT and MAPK pathways are regulated by extensive crosstalk at different levels [9, 37]. AKT effect on the generation and function of Treg and Th17 cells is not well understood. Wan et al. [38] reported that RORC was necessary, but not sufficient, for IL-17 production by human memory T cells, and they proposed that RORC synergized with common γ chain-cytokines mediated PI3 K and AKT signaling to transactivate IL-17 expression. Recently, Kurebayashi et al. [39] demonstrated in mice that the suppression of PI3 K-AKT-mammalian target of Rapamycin complex (mTORC)-1 axis impaired Th17 differentiation in vitro and in vivo in S6 kinases 1 and 2-dependent fashion. In mice, Sauer et al. [40] documented that T cell receptor (TCR) signaling via PI3Kp110α, p110δ, AKT, and mTOR controls FOXP3 expression in activated CD4+ thymocytes and peripheral T cells. PI3 K-AKT axis has also been reported augments clonal expansion of Th1 and Th2 cells [41, 42]. In mature T cells, constitutively active AKT could induce growth and survival of CD4+ [8]; and a direct role of AKT in CD8+ T cells has been reported, regulating whether their differentiation leads them to either memory or effector fate [10]. In SLE patients, Suarez-Fueyo et al. [43] reported that PI3 K pathway activation paralleled activated/memory T cell accumulation. Our results show in SLE patients that AKT1 expression positively correlates with TNFSF10 (TRAIL) expression, and TRAIL expression negatively correlates with lymphocytes numbers. MAPK/ERK and PI3 K/AKT signaling pathways are activated by TRAIL [44]. Up-regulation of TRAIL mRNA expression in PBMC from active SLE patients was reported [45]. TRAIL-induced apoptosis by binding to death receptors but also can enhance T cell proliferation after TCR engagement via transducing a costimulation signal [46].
Our results show that MAPK1 gene expression also correlates with gene expression levels of Th1-(IL-12A, T-bet), Th2-(GATA3), and IL-10 genes. MAPK/ERK signaling pathways integrate cytoplasmic signals to produce changes in transcription associated with differentiation, proliferation, and survival in multiple stages of T cell development [47–49]. The results show a clear increase of IL-10 relative gene expression in SLE PBMC, although IL-10 plasma levels are not significantly augmented. Meta-analysis of microarray data indicated that one of the biological pathways consistently enhanced in SLE patients was the IL-10 signaling [50]. IL-10 has been identified as one of the risk loci for SLE in a recent genome-wide association study [51]. Despite that no correlation of IL-10 gene expression with SLEDAI was observed, IL-10 expression positively correlates with plasma TNF-α (Table 8), and with expression levels of Th0-(IL-2), Th1-(IL-12A, T-bet), Th2-(GATA3), Treg-(TGFB2, FOXP3), pro-inflammatory IL-6, and Th17-(RORC)-cytokine related genes, respectively. IL-10 is a key cytokine regulating immune responses, and the absence results in spontaneous inflammatory disorders as well as exacerbated inflammation in T cell responses during host defense or autoimmune inflammation (see reviews [52, 53]). IL-10 is expressed by cells of the adaptive immune system (Th1, Th2, Treg cells, and regulatory B cells (Bregs) and by cells of the innate immune system (DCs, macrophages, mast cells, NK, eosinophils, and neutrophils) [54, 55]. IL-10 provides a highly regulated feedback loop to avoid the extremes of excessive inflammation or chronic infections and also allow a protective response to pathogens [54]. In mice, Saraiva et al. [56] reported that development of IL-10-producing Th1 cells required high TCR ligation, sustained ERK1 and ERK2 MAPKs phosphorylation, and IL-12-induced STAT4 activation, indicating that ERK1 and ERK2 activation was a common pathway required for the production of IL-10 by Th1, Th2, and Th17 cell subsets. Regulation of IL-10 expression in mouse NK cells reported by Grant et al. [57] indicated that like IFN-γ, IL-10 expression was induced by IL-2 and IL-12 stimulation. Cytokines like TGF-β and IL-6 are also important regulators of IL10-producing cells [58]. It has been reported by Avagyan et al. [59] that TGF-β2 specifically enhanced signaling by the hematopoietic growth factor fms-related tyrosine kinase (FLT)-3 in hematopoietic stem cells. In activated human T cells, Astier et al. [60] documented that FLT3 growth factor was acting as a negative regulator of IL-10 levels. Furthermore, a relevant role of IL-10 and TNF-α genotypes in SLE has been reported [61], and IL-10 cytokine inhibits transcription elongation of the human TNF gene in primary macrophages [62]. Our results also show that IL-10 gene expression in SLE patients correlates with CD38 (activation, maturation, and trafficking marker) gene expression; CD38 expression correlates with AKT1 expression. In human T and B cells, upon receptor stimulation CD38 leads to PKB/AKT and ERK activation [63, 64]. Moreover, in a kinase-dead mutant of PI3Kp110δ mice, Patton et al. [65] reported that PI3Kp110δ kinase regulates expression of CD38 on Treg. Furthermore, Blair et al. [66] documented that CD19+CD24hiCD38hi-B cells exhibit regulatory capacity in healthy individuals while the same B cells from SLE patients produced less IL-10 and lacked the suppressive capacity.
Our data show a clear increase in FOXP3-(Treg) gene expression levels in SLE patients and positively correlate with IL-10 gene expression. Recent reports provided new data about the mechanisms linking IL-10 and Treg cells by demonstrating that IL-10 directly signals in Th17 and Treg cells to maintain control of Th17 cell-mediated inflammation [67, 68]. In addition, IL-10-producing B cells, also known as regulatory B cells (Bregs), play a key role in controlling autoimmunity. Carter et al. [55] reported that chimeric mice specifically lacking IL-10-producing B cells developed an exacerbated arthritis compared with chimeric wild-type B cell mice. Moreover, a significant decrease in the absolute numbers of FOXP3 regulatory T cells (Tregs), in their expression level of FOXP3 and a marked increase in inflammatory Th1 and Th17 cells were detected in chimeric mice lacking IL-10-producing B cells compared with WT B-cell mice [55].
How Treg cell function and FOXP3 expression are regulated is an important question under intensive investigation [69]. Barreto et al. [70] reported a promoter polymorphism in FOXP3 gene associated with SLE. In SLE patients, increased gene expression of FOXP3 compared to healthy controls has been reported [71]. In SLE patients, several reports indicated that increased proportions of CD4+CD25−Foxp3+ T cells were correlated with the clinical disease activity and the daily cortisone dose [72, 73]. In our study, we cannot exclude that treatments received by SLE patients could have an effect on the results. Although, a recent report from Sbiera et al. [74] documented that short-term glucocorticoid therapy in vivo either in immunologically uncompromised humans or in mice did not induce an increased in the frequency of circulating Treg cells.
Balance between FOXP3 and RORC function has been postulated that determines CD4+T cell fate and the type of immune response that will be generated [75]. Th17 and Treg seem to be closely linked [33, 76]. Several studies reported in active SLE patients an imbalance between Treg and Th17 cells [77–79]. Moreover new information has been reported about the interplay between Tregs and Th2 cells [80]. Wang et al. [81] demonstrated an essential role for GATA3, in controlling Treg cell homeostasis by stabilizing expression of FOXP3. Our results show an increased balance of both FOXP3 (Treg)/RORC (Th17) and FOXP3 (Treg)/GATA3 (Th2) gene expression ratios in PBMC from SLE patients with inactive or middle disease, suggesting a trend towards Treg polarization. In addition, our results show no significant increases compared with controls in the T-bet (Th1)/RORC (Th17), T-bet (Th1)/GATA3 (Th2), and RORC (Th17)/GATA3 (Th2) gene expression ratios in PBMC from SLE patients. Our data also show a low relative gene expression levels of Th17-associated CCR6 (is a trait of tissue-homing effector T cells lymphocyte trafficking) in PBMC from the SLE patients with inactive or middle disease. CCR6 expression on CD4+T cells has been considered as a marker of disease activity in SLE patients [82]. In addition, Sallusto et al. [83] reported that following TCR stimulation, human memory/effector T cells down-regulate receptors for constitutive chemokines (including CCR6). They suggested that following TCR stimulation effector/memory T cells transiently acquire responsiveness to constitutive chemokines. As a result, T cells that are activated in tissues may either recirculate to draining lymph nodes or migrate to nearby sites of organized ectopic lymphoid tissues [83].
Several studies have shown a trend towards increased Th17 cells and Th17/Th1 ratio in SLE patients with active disease [82, 84]. Furthermore, reports on function and numbers and Treg cells in SLE have been contradictory (reviews [75, 85]), suggesting that not all FOXP3+T cells in SLE have protective suppressive activity. Plasticity of CD4+FOXP3+T cells has been documented [34, 86], and it has been reported that conversion of human FOXP3+ Tregs into IL-17-producing cells can be enhanced in the context of an inflammatory cytokine milieu (IL-1β and IL-6) [75, 76, 86, 87].
Tregs presented core suppressive mechanisms driven by FOXP3. Nevertheless they are also able to adapt in response to stimuli by homing to sites of inflammation and exerting suppressive functions. It has been reported that Treg are able to express other transcription factors normally associated with other Th cell subtypes in order to better control immunopathology (see review [34]). In our study, plasma levels of proinflammatory cytokines IL-1β, IL-12(p70), IFN-γ, and IL-6 are significantly augmented in SLE patients with inactive or middle disease compared with controls. In SLE it has been suggested that aberrant control of immune cell responses to inflammatory cytokines may disrupt the delicate balance between immunity and self-reactivity [2, 3, 88]. IL-6 has been studied in human SLE, and IL-6 was found to reflect disease activity [19, 89–93]. IL-6 contributes to host defence against acute environmental stress, and continuous deregulated IL-6 production plays a significant pathological role in systemic autoimmune diseases [94]. IL-6 has been known to have a key role in the maturation of B cells, as well to be one of the members of cytokines that drives the acute inflammatory response together with TNF-alpha and IL-1 [94]. Additionally, plasma IL-5 (Th2) was significantly augmented in SLE patients versus controls. IL-5 is produced by natural helper cells and is involved in antibody production [95, 96]. IL-5 is a critical growth factor for B1 cells present in the peritoneal cavity and plays an important role in innate-type immune responses by producing natural antibodies [97–99]. Wen et al. [100] reported that deregulated, continuous high expression of IL-5 in SLE-prone mice may directly or indirectly mediate a skewed signaling of proliferation/differentiation of self-antigen-activated B1 cells, leading to suppression of autoimmune disease, but instead of aberrant expansion of B1 cells, giving rise to B-cell chronic lymphocytic leukaemia (B-CLL). Moreover, despite no correlation with SLEDAI, our results indicate that Th17-(IL-1β, IL-6), Th0-(IL-2), Th1-(IFN-γ, TNF-α), Th2-(IL-13), and IL-10 plasma cytokines are positively correlated with the inflammation mediator CRP. During inflammation altered cytokine levels are produced by different cell populations in an interdependent mode [101]. These results are consistent with those of other studies [102, 103].
Moreover, our results show a moderate increase of ZAP70 tyrosine kinase gene expression in SLE's PBMC and ZAP70 correlate with gene expressions levels of TNF-α (Table 8). Syk and ZAP-70 kinases also associated differently with key molecules involved in cytoskeletal and calcium signaling in SLE T cells [15]. In human mature T cells, Meinl et al. [104] demonstrated a key role of ZAP70 in CD2-mediated phosphorylation of CD3zeta chain of TCR, in Ca2+ influx, proliferation, and production of IFN-γ and TNF-α.
Furthermore our data show that increased inflammation mediator S100A8/S100A9-Calprotectin plasma levels in SLE patients positively correlate with neutrophils numbers. S100A8/S100A9-Calprotectin is expressed by neutrophils, monocytes, activated macrophages [105] and released in Neutrophil Extracellular Traps [106]. These results also accord with our earlier observations [29], which showed in SLE PBMCs that increased S100A9 levels, correlated positively with the abnormal presence of low-density granulocytes detected by flow cytometry in the mononuclear cells.
Despite the fact that the study has been performed in a small number of patients, the results and the positive correlations found suggest biological functions and associations that need to be further confirmed in a larger cohort of patients.
5. Conclusions
SLE patients with inactive or mild disease presented a systemic immunoinflammatory activity with augmented AKT1 and MAPK1 gene expressions, plasma proinflammatory cytokines, and Calprotectin, together with increased gene expression of Treg-related genes, suggesting an ongoing regulatory feedback opposing the inflammatory activity.
Supplementary Material
qRT-PCR procedures: Reverse transcription (RT) reaction was performed on 1 µg total RNA, 20-µl reaction, containing 200 U SuperScript III reverse transcriptase (Invitrogen), 5 mM random hexamer primers, 0.5mM each deoxyribonucleotide triphosphate (dNTP), 20 U RNase inhibitor, and 1X-RT buffer. Primer sequences and gene-specific primers used are available in Table 2. Gene expressions assays were performed on iQ-Cycler (Bio-Rad), with 3 replicates with iQ-SYBR Green Supermix, 21-microliter reaction (200nM each primer and 10 ng of cDNA). PCR program: 10 min incubation at 95°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C. Specificity of PCR amplification procedure was checked with a heat dissociation protocol (from 65°C to 100°C), after the final cycle of the PCR.
Authors' Contribution
S. Garcia-Rodriguez, J.-L. Callejas-Rubio, N. Ortego-Centeno, J. Sancho, and M. Zubiaur participated in acquisition, analysis, and interpretation of data; E. Zumaquero and P. Navarro participated in data acquisition; J.-L. Callejas-Rubio, N. Ortego-Centeno, R. Ríos-Fernandez, and S. Arias-Santiago performed clinical evaluation of patients and controls. M. Zubiaur wrote the paper revised by S. Garcia-Rodriguez, J.-L. Callejas-Rubio, N. Ortego-Centeno, R. Ríos-Fernandez, S. Arias-Santiago, and J. Sancho. Authors approved the final paper.
Conflict of Interests
The authors declare no financial conflict of interests.
Acknowledgments
Thanks are due to patients, controls, and staff of Systemic Autoimmune Diseases Unit, Departments of Internal Medicine and Department of Dermatology (SCUH-Granada). This paper is supported by European Commission-European Regional Development Fund (ERDF/FEDER), in collaboration with: ISCIII-FIS06-1502 (M. Zubiaur); CSIC-PI 200820I216 (M. Zubiaur); Junta de Andalucía (JA), Consejerías Innovación-Ciencia-Empresa y Educación-Ciencia (CVI 226, CVI 908-2006 and PC08-CTS-04046) (J. Sancho and M. Zubiaur); ME-MICINN (SAF-2005-060656-C02-01 and SAF-2008-03685) (J. Sancho and M. Zubiaur) and SAF-2011- 27261 (J. Sancho); JA-FEDER-Fellowship (E. Zumaquero) and JAE-Doc-CSIC-FEDER (S. Garcia-Rodriguez).
References
- 1.Cervera R. Systemic lupus erythematosus in Europe at the change of the millennium: lessons from the ‘Euro-Lupus Project’. Autoimmunity Reviews. 2006;5(3):180–186. doi: 10.1016/j.autrev.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Perl A. Pathogenic mechanisms in systemic lupus erythematosus. Autoimmunity. 2010;43(1):1–6. doi: 10.3109/08916930903374741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenks SA, Sanz I. Altered B cell receptor signaling in human systemic lupus erythematosus. Autoimmunity Reviews. 2009;8(3):209–213. doi: 10.1016/j.autrev.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crispín JC, Kyttaris VC, Terhorst C, Tsokos GC. T cells as therapeutic targets in SLE. Nature Reviews Rheumatology. 2010;6(6):317–325. doi: 10.1038/nrrheum.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsokos GC. Systemic lupus erythematosus. The New England Journal of Medicine. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 6.Nakou M, Bertsias G, Stagakis I, et al. Gene network analysis of bone marrow mononuclear cells reveals activation of multiple kinase pathways in human systemic lupus erythematosus. PLoS ONE. 2010;5(10) doi: 10.1371/journal.pone.0013351.e13351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juntilla MM, Koretzky GA. Critical roles of the PI3K/Akt signaling pathway in T cell development. Immunology Letters. 2008;116(2):104–110. doi: 10.1016/j.imlet.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finlay D, Cantrell DA. Metabolism, migration and memory in cytotoxic T cells. Nature Reviews Immunology. 2011;11(2):109–117. doi: 10.1038/nri2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bononi A, Agnoletto C, De Marchi E, et al. Protein kinases and phosphatases in the control of cell fate. Enzyme Research. 2011;2011:26 pages. doi: 10.4061/2011/329098.329098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macintyre AN, Finlay D, Preston G, et al. Protein kinase B controls transcriptional programs that direct cytotoxic T cell fate but is dispensable for T cell metabolism. Immunity. 2011;34(2):224–236. doi: 10.1016/j.immuni.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taher TE, Parikh K, Flores-Borja F, et al. Protein phosphorylation and kinome profiling reveal altered regulation of multiple signaling pathways in B lymphocytes from patients with systemic lupus erythematosus. Arthritis and Rheumatism. 2010;62(8):2412–2423. doi: 10.1002/art.27505. [DOI] [PubMed] [Google Scholar]
- 12.Gorelik G, Richardson B. Key role of ERK pathway signaling in lupus. Autoimmunity. 2010;43(1):17–22. doi: 10.3109/08916930903374832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molad Y, Amit-Vasina M, Bloch O, Yona E, Rapoport MJ. Increased ERK and JNK activities correlate with disease activity in patients with systemic lupus erythematosus. Annals of the Rheumatic Diseases. 2010;69(1):175–180. doi: 10.1136/ard.2008.102780. [DOI] [PubMed] [Google Scholar]
- 14.Sawalha AH, Jeffries M, Webb R, et al. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes and Immunity. 2008;9(4):368–378. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnan S, Juang YT, Chowdhury B, et al. Differential expression and molecular associations of Syk in systemic lupus erythematosus T cells. Journal of Immunology. 2008;181(11):8145–8152. doi: 10.4049/jimmunol.181.11.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Apostolidis SA, Lieberman LA, Kis-Toth K, et al. The dysregulation of cytokine networks in systemic lupus erythematosus. Journal of Interferon & Cytokine Research. 2011;31:769–779. doi: 10.1089/jir.2011.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob N, Stohl W. Cytokine disturbances in systemic lupus erythematosus. Arthritis Research & Therapy. 2011;13, article 228 doi: 10.1186/ar3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kariuki SN, Niewold TB. Genetic regulation of serum cytokines in systemic lupus erythematosus. Translational Research. 2010;155(3):109–117. doi: 10.1016/j.trsl.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su DL, Lu ZM, Shen MN, et al. Roles of pro- and anti-inflammatory cytokines in the pathogenesis of SLE. Journal of Biomedicine and Biotechnology. 2012;2012:15 pages. doi: 10.1155/2012/347141.347141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu SL, Kuan WP, Wong CK, et al. Immunopathological roles of cytokines, chemokines, signaling molecules, and pattern-recognition receptors in systemic lupus erythematosus. Clinical and Developmental Immunology. 2012;2012:14 pages. doi: 10.1155/2012/715190.715190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J, Paul WE. Heterogeneity and plasticity of T helper cells. Cell Research. 2010;20(1):4–12. doi: 10.1038/cr.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakayamada S, Takahashi H, Kanno Y, et al. Helper T cell diversity and plasticity. Current Opinion in Immunology. 2012;24:297–302. doi: 10.1016/j.coi.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan RWY, Lai FMM, Li EKM, et al. Imbalance of Th1/Th2 transcription factors in patients with lupus nephritis. Rheumatology. 2006;45(8):951–957. doi: 10.1093/rheumatology/kel029. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Rodriguez S, Arias-Santiago S, Perandres-Lopez R, et al. Increased gene expression of Toll-like receptor 4 on peripheral blood mononuclear cells in patients with psoriasis. doi: 10.1111/j.1468-3083.2011.04372.x. Journal of the European Academy of Dermatology and Venereology. In press. [DOI] [PubMed] [Google Scholar]
- 25.Chung BH, Oh HJ, Piao SG, et al. Clinical significance of the ratio between FOXP3 positive regulatory T cell and interleukin-17 secreting cell in renal allograft biopsies with acute T-cell-mediated rejection. Immunology. 2012;136(3):344–351. doi: 10.1111/j.1365-2567.2012.03588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyake K, Akahoshi M, Nakashima H. Th subset balance in lupus nephritis. Journal of Biomedicine and Biotechnology. 2011;2011:7 pages. doi: 10.1155/2011/980286.980286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soyfoo MS, Roth J, Vogl T, Pochet R, Decaux G. Phagocyte-specific S100A8/A9 protein levels during disease exacerbations and infections in systemic lupus erythematosus. Journal of Rheumatology. 2009;36(10):2190–2194. doi: 10.3899/jrheum.081302. [DOI] [PubMed] [Google Scholar]
- 28.Lood C, Stenström M, Tydén H, et al. Protein synthesis of the pro-inflammatory S100A8/A9 complex in plasmacytoid dendritic cells and cell surface S100A8/A9 on leukocyte subpopulations in systemic lupus erythematosus. Arthritis Research and Therapy. 2011;13(2, article R60) doi: 10.1186/ar3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavon EJ, Garcia-Rodriguez S, Zumaquero E, et al. Increased expression and phosphorylation of the two S100A9 isoforms in mononuclear cells from patients with systemic lupus erythematosus: a proteomic signature for circulating low-density granulocytes. Journal of Proteomics. 2012;75(6):1778–91. doi: 10.1016/j.jprot.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 30.Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH. Derivation of the SLEDAI: a disease activity index for lupus patients. Arthritis and Rheumatism. 1992;35(6):630–640. doi: 10.1002/art.1780350606. [DOI] [PubMed] [Google Scholar]
- 31.Tan EM, Cohen AS, Fries JF. The 1982 revised criteria for the classification of systemic lupus erythrematosus. Arthritis and Rheumatism. 1982;25(11):1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Zhu J, Paul WE. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunological Reviews. 2010;238(1):247–262. doi: 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wing JB, Sakaguchi S. Multiple treg suppressive modules and their adaptability. Frontiers in Immunology. 2012;3:p. 178. doi: 10.3389/fimmu.2012.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haga HJ, Brun JG, Berntzen HB, Cervera R, Khamashta M, Hughes GRV. Calprotection in patients with systemic lupus erythematosus: relation to clinical and laboratory parameters of disease activity. Lupus. 1993;2(1):47–50. doi: 10.1177/096120339300200108. [DOI] [PubMed] [Google Scholar]
- 36.Wei L, Vahedi G, Sun HW, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32(6):840–851. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salmond RJ, Emery J, Okkenhaug K, Zamoyska R. MAPK, phosphatidylinositol 3-kinase, and mammalian target of rapamycin pathways converge at the level of ribosomal protein S6 phosphorylation to control metabolic signaling in CD8 T cells. Journal of Immunology. 2009;183(11):7388–7397. doi: 10.4049/jimmunol.0902294. [DOI] [PubMed] [Google Scholar]
- 38.Wan Q, Kozhaya L, ElHed A, et al. Cytokine signals through PI-3 kinase pathway modulate Th17 cytokine production by CCR6+ human memory T cells. The Journal of Experimental Medicine. 2011;208:1875–1887. doi: 10.1084/jem.20102516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurebayashi Y, Nagai S, Ikejiri A, et al. PI3K-Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORgamma. Cell Reports. 2012;1:360–373. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Sauer S, Bruno L, Hertweck A, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(22):7797–7802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arimura Y, Shiroki F, Kuwahara S, et al. Akt is a neutral amplifier for Th cell differentiation. The Journal of Biological Chemistry. 2004;279(12):11408–11416. doi: 10.1074/jbc.M309063200. [DOI] [PubMed] [Google Scholar]
- 42.Okkenhaug K, Patton DT, Bilancio A, Garçon F, Rowan WC, Vanhaesebroeck B. The p110δ isoform of phosphoinositide 3-kinase controls clonal expansion and differentiation of Th cells. Journal of Immunology. 2006;177(8):5122–5128. doi: 10.4049/jimmunol.177.8.5122. [DOI] [PubMed] [Google Scholar]
- 43.Suarez-Fueyo A, Barber DF, Martinez-Ara J, et al. Enhanced phosphoinositide 3-kinase delta activity is a frequent event in systemic lupus erythematosus that confers resistance to activation-induced T cell death. The Journal of Immunology. 2011;187:2376–2385. doi: 10.4049/jimmunol.1101602. [DOI] [PubMed] [Google Scholar]
- 44.Morel J, Audo R, Hahne M, Combe B. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces rheumatoid arthritis synovial fibroblast proliferation through mitogen-activated protein kinases and phosphatidylinositol 3-kinase/Akt. The Journal of Biological Chemistry. 2005;280(16):15709–15718. doi: 10.1074/jbc.M414469200. [DOI] [PubMed] [Google Scholar]
- 45.Komatsuda A, Wakui H, Iwamoto K, et al. Up-regulation of TRAIL mRNA expression in peripheral blood mononuclear cells from patients with active systemic lupus erythematosus. Clinical Immunology. 2007;125(1):26–29. doi: 10.1016/j.clim.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 46.Huang SC, Tsai HF, Tzeng HT, Liao HJ, Hsu PN. Lipid raft assembly and LcK recruitment in TRAIL costimulation mediates NF-κB activation and T cell proliferation. Journal of Immunology. 2011;186(2):931–939. doi: 10.4049/jimmunol.1001092. [DOI] [PubMed] [Google Scholar]
- 47.Fischer AM, Katayama CD, Pagès G, Pouysségur J, Hedrick SM. The role of Erk1 and Erk2 in multiple stages of T cell development. Immunity. 2005;23(4):431–443. doi: 10.1016/j.immuni.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 48.von Kriegsheim A, Baiocchi D, Birtwistle M, et al. Cell fate decisions are specified by the dynamic ERK interactome. Nature Cell Biology. 2009;11(12):1458–1464. doi: 10.1038/ncb1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu Y, Bertics PJ. Chemoattractant-induced signaling via the Ras-ERK and PI3K-Akt networks, along with leukotriene C4 release, is dependent on the tyrosine kinase Lyn in IL-5- and IL-3-primed human blood eosinophils. Journal of Immunology. 2011;186(1):516–526. doi: 10.4049/jimmunol.1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arasappan D, Tong W, Mummaneni P, Fang H, Amur S. Meta-analysis of microarray data using a pathway-based approach identifies a 37-gene expression signature for systemic lupus erythematosus in human peripheral blood mononuclear cells. BMC Medicine. 2011;9, article 65 doi: 10.1186/1741-7015-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gateva V, Sandling JK, Hom G, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nature Genetics. 2009;41(11):1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li MO, Flavell RA. Contextual regulation of inflammation: a duet by transforming growth factor-β and interleukin-10. Immunity. 2008;28(4):468–476. doi: 10.1016/j.immuni.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 53.Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annual Review of Immunology. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- 54.Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nature Reviews Immunology. 2010;10(3):170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 55.Carter NA, Vasconcellos R, Rosser EC, et al. Mice lacking endogenous IL-10-producing regulatory B cells develop exacerbated disease and present with an increased frequency of Th1/Th17 but a decrease in regulatory T cells. Journal of Immunology. 2011;186(10):5569–5579. doi: 10.4049/jimmunol.1100284. [DOI] [PubMed] [Google Scholar]
- 56.Saraiva M, Christensen JR, Veldhoen M, Murphy TL, Murphy KM, O'Garra A. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity. 2009;31(2):209–219. doi: 10.1016/j.immuni.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grant LR, Yao ZJ, Hedrich CM, et al. Stat4-dependent, T-bet-independent regulation of IL-10 in NK cells. Genes and Immunity. 2008;9(4):316–327. doi: 10.1038/gene.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGeachy MJ, Bak-Jensen KS, Chen Y, et al. TGF-β and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain TH-17 cell-mediated pathology. Nature Immunology. 2007;8(12):1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 59.Avagyan S, Aguilo-Payeras F, Kamezaki K, et al. Quantitative trait mapping reveals a regulatory axis involving peroxisome proliferator-activated receptors, PRDM16, transforming growth factor-beta2 and FLT3 in hematopoiesis. Blood. 2011;118:6078–6086. doi: 10.1182/blood-2011-07-365080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Astier AL, Beriou G, Eisenhaure TM, Anderton SM, Hafler DA, Hacohen N. RNA interference screen in primary human T cells reveals FLT3 as a modulator of IL-10 levels. Journal of Immunology. 2010;184(2):685–693. doi: 10.4049/jimmunol.0902443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lopez P, Gutierrez C, Suarez A. IL-10 and TNFalpha genotypes in SLE. Journal of Biomedicine and Biotechnology. 2010;2010 doi: 10.1155/2010/838390.838390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smallie T, Ricchetti G, Horwood NJ, Feldmann M, Clark AR, Williams LM. IL-10 inhibits transcription elongation of the human TNF gene in primary macrophages. Journal of Experimental Medicine. 2010;207(10):2081–2088. doi: 10.1084/jem.20100414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zubiaur M, Fernández O, Ferrero E, et al. CD38 is associated with lipid rafts and upon receptor stimulation leads to Akt/protein kinase B and Erk activation in the absence of the CD3-ζ immune receptor tyrosine-based activation motifs. Journal of Biological Chemistry. 2002;277(1):13–22. doi: 10.1074/jbc.M107474200. [DOI] [PubMed] [Google Scholar]
- 64.Zumaquero E, Muñoz P, Cobo M, et al. Exosomes from human lymphoblastoid B cells express enzymatically active CD38 that is associated with signaling complexes containing CD81, Hsc-70 and Lyn. Experimental Cell Research. 2010;316(16):2692–2706. doi: 10.1016/j.yexcr.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 65.Patton DT, Wilson MD, Rowan WC, Soond DR, Okkenhaug K. The PI3K p110δ regulates expression of CD38 on regulatory T cells. PLoS ONE. 2011;6(3) doi: 10.1371/journal.pone.0017359.e17359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blair PA, Noreña LY, Flores-Borja F, et al. CD19+CD24hiCD38hi B cells exhibit regulatory capacity in healthy individuals but are functionally impaired in systemic lupus erythematosus patients. Immunity. 2010;32(1):129–140. doi: 10.1016/j.immuni.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 67.Chaudhry A, Samstein RM, Treuting P, et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34(4):566–578. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huber S, Gagliani N, Esplugues E, et al. Th17 cells express interleukin-10 receptor and are controlled by Foxp3− and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity. 2011;34(4):554–565. doi: 10.1016/j.immuni.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rudensky AY. Regulatory T cells and Foxp3. Immunological Reviews. 2011;241(1):260–268. doi: 10.1111/j.1600-065X.2011.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barreto M, Ferreira RC, Lourenço L, et al. Low frequency of CD4+CD25+ Treg in SLE patients: a heritable trait associated with CTLA4 and TGFβ gene variants. BMC Immunology. 2009;10, article 5 doi: 10.1186/1471-2172-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suggs MJL, Majithia V, Lewis RE, Cruse JM. HLA DRB1*1503 allelic haplotype predominance and associated immunodysregulation in systemic lupus erythematosus. Experimental and Molecular Pathology. 2011;91(2):548–562. doi: 10.1016/j.yexmp.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 72.Azab NA, Bassyouni IH, Emad Y, Abd El-Wahab GA, Hamdy G, Mashahit MA. CD4+CD25+ regulatory T cells (TREG) in Systemic Lupus Erythematosus (SLE) patients: the possible influence of treatment with corticosteroids. Clinical Immunology. 2008;127(2):151–157. doi: 10.1016/j.clim.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 73.Prado C, Gómez J, López P, de Paz B, Gutiérrez C, Suárez A. Dexamethasone upregulates FOXP3 expression without increasing regulatory activity. Immunobiology. 2011;216(3):386–392. doi: 10.1016/j.imbio.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 74.Sbiera S, Dexneit T, Reichardt SD, et al. Influence of short-term glucocorticoid therapy on regulatory T cells in vivo. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0024345.e24345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abdulahad WH, Boots AMH, Kallenberg CGM. FoxP3+ CD4+ T cells in systemic autoimmune diseases: the delicate balance between true regulatory T cells and effector Th-17 cells. Rheumatology. 2011;50(4):646–656. doi: 10.1093/rheumatology/keq328. [DOI] [PubMed] [Google Scholar]
- 76.Murphy KM, Stockinger B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nature Immunology. 2010;11(8):674–680. doi: 10.1038/ni.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Valencia X, Yarboro C, Illei G, Lipsky PE. Deficient CD4+CD25high T regulatory cell function in patients with active systemic lupus erythematosus. Journal of Immunology. 2007;178(4):2579–2588. doi: 10.4049/jimmunol.178.4.2579. [DOI] [PubMed] [Google Scholar]
- 78.Ma J, Yu J, Tao X, Cai L, Wang J, Zheng SG. The imbalance between regulatory and IL-17-secreting CD4+ T cells in lupus patients. Clinical Rheumatology. 2010;29(11):1251–1258. doi: 10.1007/s10067-010-1510-7. [DOI] [PubMed] [Google Scholar]
- 79.Xing Q, Wang B, Su H, Cui J, Li J. Elevated Th17 cells are accompanied by FoxP3+ Treg cells decrease in patients with lupus nephritis. Rheumatology International. 2011;32(4):949–958. doi: 10.1007/s00296-010-1771-0. [DOI] [PubMed] [Google Scholar]
- 80.Chapoval S, Dasgupta P, Dorsey NJ, Keegan AD. Regulation of the T helper cell type 2 (Th2)/T regulatory cell (Treg) balance by IL-4 and STAT6. Journal of Leukocyte Biology. 2010;87(6):1011–1018. doi: 10.1189/jlb.1209772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35:337–348. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shah K, Lee WW, Lee SH, et al. Dysregulated balance of Th17 and Th1 cells in systemic lupus erythematosus. Arthritis Research and Therapy. 2010;12(2, article R53) doi: 10.1186/ar2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sallusto F, Kremmer E, Palermo B, et al. Switch in chemokine receptor expression upon TCR stimulation reveals novel homing potential for recently activated T cells. European Journal of Immunology. 1999;29:2037–2045. doi: 10.1002/(SICI)1521-4141(199906)29:06<2037::AID-IMMU2037>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 84.Prado C, de Paz B, Gomez J, et al. Glucocorticoids enhance Th17/Th1 imbalance and signal transducer and activator of transcription 3 expression in systemic lupus erythematosus patients. Rheumatology. 2011;50:1794–1801. doi: 10.1093/rheumatology/ker227. [DOI] [PubMed] [Google Scholar]
- 85.Horwitz DA. Identity of mysterious CD4+CD25−Foxp3+ cells in SLE. Arthritis Research & Therapy. 2010;12(1):p. 101. doi: 10.1186/ar2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou X, Bailey-Bucktrout S, Jeker LT, Bluestone JA. Plasticity of CD4+ FoxP3+ T cells. Current Opinion in Immunology. 2009;21(3):281–285. doi: 10.1016/j.coi.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu L, Zhou X, Wang J, Zheng SG, Horwitz DA. Characterization of protective human CD4CD25 FOXP3 regulatory T cells generated with IL-2, TGF-β and retinoic acid. PloS ONE. 2010;5(12):p. e15150. doi: 10.1371/journal.pone.0015150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hale MB, Krutzik PO, Samra SS, Crane JM, Nolan GP. Stage dependent aberrant regulation of cytokine-STAT signaling in murine systemic lupus erythematosus. PLoS ONE. 2009;4(8) doi: 10.1371/journal.pone.0006756.e6756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cash H, Relle M, Menke J, et al. Interleukin 6 (IL-6) deficiency delays lupus nephritis in MRL-Fas lpr mice: the IL-6 pathway as a new therapeutic target in treatment of autoimmune kidney disease in systemic lupus erythematosus. Journal of Rheumatology. 2010;37(1):60–70. doi: 10.3899/jrheum.090194. [DOI] [PubMed] [Google Scholar]
- 90.Chun HY, Chung JW, Kim HA, et al. Cytokine IL-6 and IL-10 as biomarkers in systemic lupus erythematosus. Journal of Clinical Immunology. 2007;27(5):461–466. doi: 10.1007/s10875-007-9104-0. [DOI] [PubMed] [Google Scholar]
- 91.Hu S, Xiao W, Kong F, Ke D, Qin R, Su M. Regulatory T cells and their molecular markers in peripheral blood of the patients with systemic lupus erythematosus. Journal of Huazhong University of Science and Technology. 2008;28(5):549–552. doi: 10.1007/s11596-008-0513-y. [DOI] [PubMed] [Google Scholar]
- 92.Ohl K, Tenbrock K. Inflammatory cytokines in systemic lupus erythematosus. Journal of Biomedicine and Biotechnology. 2011;2011:14 pages. doi: 10.1155/2011/432595.432595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sabry A, sheashaa H, El-husseini A, et al. Proinflammatory cytokines (TNF-α and IL-6) in Egyptian patients with SLE: its correlation with disease activity. Cytokine. 2006;35(3-4):148–153. doi: 10.1016/j.cyto.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 94.Naugler WE, Karin M. The wolf in sheep's clothing: the role of interleukin-6 in immunity, inflammation and cancer. Trends in Molecular Medicine. 2008;14(3):109–119. doi: 10.1016/j.molmed.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 95.Sonoda E, Matsumoto R, Hitoshi Y, et al. Transforming growth factor β induces IgA production and acts additively with interleukin 5 for IgA production. The Journal of Experimental Medicine. 1989;170(4):1415–1420. doi: 10.1084/jem.170.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hayashi T. Therapeutic strategies for SLE involving cytokines: mechanism-oriented therapies especially IFN-λ targeting gene therapy. Journal of Biomedicine and Biotechnology. 2010;2010:19 pages. doi: 10.1155/2010/461641.461641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Erickson LD, Foy TM, Waldschmidt TJ. Murine B1 B cells require IL-5 for optimal T cell-dependent activation. Journal of Immunology. 2001;166(3):1531–1539. doi: 10.4049/jimmunol.166.3.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martin F, Kearney JF. B-cell subsets and the mature preimmune repertoire. Marginal zone and B1 B cells as part of a ″natural immune memory″ . Immunological Reviews. 2000;175:70–79. [PubMed] [Google Scholar]
- 99.Koyasu S, Moro K. Type 2 innate immune responses and the natural helper cell. Immunology. 2011;132(4):475–481. doi: 10.1111/j.1365-2567.2011.03413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wen X, Zhang D, Kikuchi Y, et al. Transgene-mediated hyper-expression of IL-5 inhibits autoimmune disease but increases the risk of B cell chronic lymphocytic leukemia in a model of murine lupus. European Journal of Immunology. 2004;34(10):2740–2749. doi: 10.1002/eji.200425267. [DOI] [PubMed] [Google Scholar]
- 101.Valeyev NV, Hundhausen C, Umezawa Y, et al. A systems model for immune cell interactions unravels the mechanism of inflammation in human skin. PLoS Computational Biology. 2010;6(12) doi: 10.1371/journal.pcbi.1001024.e1001024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Capper ER, Maskill JK, Gordon C, Blakemore AIF. Interleukin (IL)-10, IL-1ra and IL-12 profiles in active and quiescent systemic lupus erythematosus: could longitudinal studies reveal patient subgroups of differing pathology? Clinical and Experimental Immunology. 2004;138(2):348–356. doi: 10.1111/j.1365-2249.2004.02607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rhodes B, Meek J, Whittaker JC, Vyse TJ. Quantification of the genetic component of basal C-reactive protein expression in SLE nuclear families. Annals of Human Genetics. 2008;72(5):611–620. doi: 10.1111/j.1469-1809.2008.00444.x. [DOI] [PubMed] [Google Scholar]
- 104.Meinl E, Lengenfelder D, Blank N, Pirzer R, Barata L, Hivroz C. Differential requirement of ZAP-70 for CD2-mediated activation pathways of mature T cells. Journal of Immunology. 2000;165(7):3578–3583. doi: 10.4049/jimmunol.165.7.3578. [DOI] [PubMed] [Google Scholar]
- 105.Foell D, Wittkowski H, Vogl T, Roth J. S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. Journal of Leukocyte Biology. 2007;81(1):28–37. doi: 10.1189/jlb.0306170. [DOI] [PubMed] [Google Scholar]
- 106.Urban CF, Ermert D, Schmid M, et al. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathogens. 2009;5(10) doi: 10.1371/journal.ppat.1000639.e1000639 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qRT-PCR procedures: Reverse transcription (RT) reaction was performed on 1 µg total RNA, 20-µl reaction, containing 200 U SuperScript III reverse transcriptase (Invitrogen), 5 mM random hexamer primers, 0.5mM each deoxyribonucleotide triphosphate (dNTP), 20 U RNase inhibitor, and 1X-RT buffer. Primer sequences and gene-specific primers used are available in Table 2. Gene expressions assays were performed on iQ-Cycler (Bio-Rad), with 3 replicates with iQ-SYBR Green Supermix, 21-microliter reaction (200nM each primer and 10 ng of cDNA). PCR program: 10 min incubation at 95°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C. Specificity of PCR amplification procedure was checked with a heat dissociation protocol (from 65°C to 100°C), after the final cycle of the PCR.


