Abstract
BACKGROUND:
Understanding the risk for type 2 diabetes (T2D) early in the life course is important for prevention. Whether genetic information improves prediction models for diabetes from adolescence into adulthood is unknown.
METHODS:
With the use of data from 1030 participants in the Bogalusa Heart Study aged 12 to 18 followed into middle adulthood, we built Cox models for incident T2D with risk factors assessed in adolescence (demographics, family history, physical examination, and routine biomarkers). Models with and without a 38 single-nucleotide polymorphism diabetes genotype score were compared by C statistics and continuous net reclassification improvement indices.
RESULTS:
Participant mean (± SD) age at baseline was 14.4 ± 1.6 years, and 32% were black. Ninety (8.7%) participants developed T2D over a mean 26.9 ± 5.0 years of follow-up. Genotype score significantly predicted T2D in all models. Hazard ratios ranged from 1.09 per risk allele (95% confidence interval 1.03–1.15) in the basic demographic model to 1.06 (95% confidence interval 1.00–1.13) in the full model. The addition of genotype score did not improve the discrimination of the full clinical model (C statistic 0.756 without and 0.760 with genotype score). In the full model, genotype score had weak improvement in reclassification (net reclassification improvement index 0.261).
CONCLUSIONS:
Although a genotype score assessed among white and black adolescents is significantly associated with T2D in adulthood, it does not improve prediction over clinical risk factors. Genetic screening for T2D in its current state is not a useful addition to adolescents’ clinical care.
KEY WORDS: genetic predisposition to disease; diabetes mellitus, type 2; adolescent medicine
What’s Known on This Subject:
Among middle-aged adults, genotype scores predict incident type 2 diabetes but do not improve prediction models based on clinical risk factors including family history and BMI. These clinical factors are more dynamic in adolescence, however.
What This Study Adds:
A genotype score also predicts type 2 diabetes from adolescence over a mean 27 years of follow-up into adulthood but does not improve prediction models based on clinical risk factors assessed in adolescence.
Type 2 diabetes (T2D) is a tremendous source of morbidity, mortality, and health care expenditure.1 As obesity, a major risk factor for T2D, has risen in prevalence across all ages, so has the prevalence of T2D. Because lifestyle modification can delay or prevent the onset of T2D,2–4 the identification of high-risk individuals earlier in the life course might help target prevention efforts. Adolescence may be a key time for such identification, as young people individuate and develop lifelong habits.
Because one’s genetic composition does not change over the life course, genotype information may allow risk prediction in younger age groups. To date, genome-wide association studies based largely on adult data have identified at least 38 independent loci where single-nucleotide polymorphisms (SNPs) are associated with T2D.5–11 Genotype scores using these variants predict incident T2D among middle-aged adults. However, the incorporation of genotype scores into clinical prediction models does not meaningfully improve prediction,12–14 with the exception, perhaps, of young middle age and over a longer follow-up period.13,15 We hypothesized that a 38-SNP genotype score in adolescence predicts incident T2D in adulthood and that this genotype information, unlike in older adults, improves the prediction of T2D in comparison with clinical prediction models based on factors assessed in youth. Because of the great inherent variability present in adolescence in clinical risk factors such as BMI, blood pressure, and lipids, the static nature of a genotype score may be particularly useful in this age group.
Methods
Study Design
The Bogalusa Heart Study is a cohort study of a biracial population in Bogalusa, Louisiana.16,17 Children aged 4 to 18 years between 1973 and 1994 underwent cross-sectional surveys and examinations every 2 to 3 years up to 2010, enabling longitudinal analyses.17 We limited the present analyses to participants with at least 1 examination during adolescence (ages 12–18 years), at least 1 adult follow-up examination (ages 19–51 years), and data available for all baseline predictors, including genotype data. We included unrelated participants, as determined by self-report and genetic analyses. Participants reporting exclusive treatment with insulin were considered to have type 1 diabetes and were excluded. We also excluded participants with diabetes at the baseline examination. Informed consent was obtained from all participants or a parent/guardian, as appropriate. The participating institutions’ institutional review boards approved this study.
Incident T2D
Incident T2D was defined according to the World Health Organization definition18 as a fasting plasma glucose ≥126 mg/dL (≥7.0 mmol/L) or report of receiving oral hypoglycemic agents with or without insulin. Women reporting diagnosis or treatment of diabetes only during pregnancy were considered to have gestational diabetes and were not classified as having T2D.
Clinical Predictors
We chose clinical predictors (demographics, family history of diabetes, physical examination components, and biomarkers of cardiometabolic risk) based on their routine clinical use and their previously demonstrated association with incident T2D among children and adults.19–22
Demographics and Family History
Participant age was calculated at each study examination date. Gender and race (white or black) were self-reported, and gender was confirmed with genetic data. Participants were characterized as having a parental history of diabetes if 1 or both biological parents self-reported having diabetes; the parental history questionnaire did not specify diabetes type.
Physical Examination
Weight, height, and systolic and diastolic blood pressures were measured with standard protocols.16 BMI was calculated as weight in kilograms divided by height in meters squared. Age- and gender-standardized BMI z scores were derived from the Centers for Disease Control and Prevention growth reference year 2000.23 Overweight was defined as a BMI z score of at least the 85th percentile for age and gender. Mean arterial pressure was calculated as 1/3(systolic blood pressure) + 2/3(diastolic blood pressure).
Biomarkers of Cardiometabolic Risk
Three biomarkers of cardiometabolic risk were used in prediction models: glucose, high-density lipoprotein (HDL) cholesterol, and triglyceride levels. Fasting blood samples used for these assays were measured as described previously.24
Genotyping
DNA was extracted from banked blood samples in the Bogalusa archive and was available for genotyping from 1202 Bogalusa participants from the 2001–2002 examinations as described previously.25 Genotyping was performed with the Illumina Human610 Genotyping BeadChip and Human CVD BeadChip with >99.99% concordance on duplicate samples. SNP imputation was performed by using MACH v.1.0.16 (http://www.sph.umich.edu/csg/yli/mach/)7 with HapMap CEU and YRI (phase II, release 22) as the reference populations. Genotyping quality control and SNP filtering in Bogalusa data have been discussed previously,25 and no SNP of interest violated Hardy-Weinberg equilibrium at a P value <1×10−6.
Genotype Score
We based our genotype score on a published score consisting of 40 SNPs associated with T2D in adults.15 Of the 40 SNPs used in this score, we had genotyped or imputed data for 38 of the SNPs in both whites and blacks (not rs4457053 near ZBED3 or rs11634397 near ZFAND6) (see Supplemental Table 4). In general, the effect direction of individual SNPs was similar in Bogalusa compared with the published effect direction, despite that we had low power to show significant associations for individual SNPs. Similar to previous reports,13,14,26 we calculated a score as the unweighted sum of the number of risk alleles (0, 1, or 2) at each of the 38 SNPs. Instead of weighting the risk alleles by their effect sizes from genome-wide association studies in predominantly European ancestral groups, we used an unweighted score because of the biracial composition of the Bogalusa cohort and the lack of published SNP effect size estimates in populations of African ancestry. No genotyped participants had missing data for >3 SNPs. The 83 participants with missing data at ≤3 SNPs were given a score of 1 for each missing value, equivalent to being heterozygous (the most common genotype) at that locus.
Statistical Analyses
We used Cox regression to build nested prediction models for time to incident T2D. For each eligible participant, we identified the baseline adolescent examination for the present analyses as the first study examination occurring between 12 and 18 years of age. Time-to-event was calculated from the date of this baseline adolescent examination to the date of the first follow-up examination meeting our criteria for incident T2D or to the date of the last examination for each censored participant. Models sequentially included (1) demographics (age, gender, and race), (2) parental history of diabetes, (3) physical examination (BMI z score and mean arterial pressure), and (4) routine laboratory predictors (fasting glucose, HDL cholesterol, and triglycerides), all as assessed at the baseline adolescent examination. Triglyceride and HDL cholesterol levels were log-transformed to improved model fit. In separate analyses, we also calculated a within-sample z score for each continuous variable (age, BMI z score, mean arterial pressure, fasting glucose, log-transformed HDL cholesterol and triglycerides, and the genotype risk score) to use in determining standardized hazard ratios, each corresponding to the risk associated with a 1-SD increase in that variable. In ancillary analyses, models included the first axis of African ancestry (based on the HapMap YRI population, phase II, release 22) obtained from the Local Ancestry in adMixed Populations method27 as a continuous variable in place of dichotomized race. Because the results were unchanged, here, we report only results with reported race. Inclusion of a time-dependent interaction term with each variable in the full prediction model confirmed the validity of the proportional hazards assumption.
At each stage of model building (1 through 4 above), model improvement was assessed after the addition of the 38-SNP genotype score. We assessed model calibration with a Hosmer-Lemeshow χ2 statistic comparing observed and expected event counts in deciles of event probability.28 The fit of nested models was compared with likelihood ratio tests. To assess model performance, we calculated C statistics and continuous net reclassification improvement indices (NRIs) for survival data and their 95% confidence intervals (CIs) by using the methods described by Pencina et al.29,30 When discrete risk categories are used, a category-based NRI compares 2 prediction models by summing the difference in proportion of cases correctly placed in a higher category minus the proportion of cases incorrectly placed in a lower category and the difference in the proportion of noncases correctly placed in a lower category minus the proportion of noncases incorrectly placed in a higher category.12,15 The continuous NRI does not require such categories and relies, rather, on the proportions of cases correctly assigned a higher model probability and noncases correctly assigned a lower model probability.30 Continuous NRI values of 0.2 correspond to a low predictive effect of a variable added to a model, whereas values of 0.4 correspond to an intermediate effect.31 Hosmer-Lemeshow statistics, C statistics, and NRI were calculated at 30 years of follow-up. We estimated 95% CIs around C statistics and NRI by using 999 bootstrap replications.30,32 In secondary analyses, age-, gender-, and race-adjusted models included interaction terms between genotype score and gender, race, parental history of diabetes, and overweight (BMI ≥85th percentile for age and gender). All analyses were performed with SAS v. 9.3 software (SAS Institute Inc, Cary, NC).
Results
Baseline Participant Characteristics
The mean (± SD) age of the 1030 eligible adolescents was 14.4 ± 1.6 years (range, 12.0–18.5 years) (Table 1). Slightly more than half (55.3%) were girls, and approximately one-third (32.2%) were black. At the baseline adolescent examination, 214 (20.8%) were overweight, including 19.5% of whites and 23.5% of blacks. On average, participants had 41.8 ± 4.1 risk alleles at the 38 loci, with blacks having a higher number of risk alleles than whites (44.5 ± 3.3 vs 40.5 ± 3.8, P < .001).
TABLE 1.
Baseline Characteristics of Adolescents in the Bogalusa Heart Study, in Overall Cohort and Stratified by Race
| Characteristic | Overall (N = 1030) | Whites (n = 698) | Blacks (n = 332) |
|---|---|---|---|
| Age, y | 14.4 ± 1.6 | 14.5 ± 1.6 | 14.2 ± 1.6 |
| Female gender, n (%) | 570 (55.3) | 368 (52.7) | 202 (60.8) |
| Parental history of diabetes, n (%) | 84 (8.2) | 47 (6.7) | 37 (11.1) |
| BMI z score | 0.11 ± 1.13 | 0.04 ± 1.15 | 0.25 ± 1.1 |
| Mean arterial pressure, mm Hg | 81.1 ± 7.8 | 81.1 ± 7.6 | 81.0 ± 8.2 |
| Glucose, mg/dL | 86.6 ± 8.5 | 87.0 ± 7.9 | 85.6 ± 9.5 |
| HDL cholesterol, mg/dL | 56.6 ± 19.0 | 54.6 ± 19.3 | 60.8 ± 17.5 |
| Triglycerides, mg/dL | 73.9 ± 38.8 | 78.3 ± 41.4 | 64.7 ± 30.9 |
| 38-variant genotype score | 41.8 ± 4.1 | 40.5 ± 3.8 | 44.5 ± 3.3 |
Plus-minus values are means ± SD.
Incident T2D
Participants were followed to a mean age of 41.4 ± 5.5 years. During a mean 26.9 ± 5.0 years of observation (range, 9.0–33.8 years), 90 participants (8.7%) developed incident T2D. Study detection of incident T2D occurred at a mean age of 40.5 ± 5.2 years. Over the total 27 526 person-years in the cohort, the incidence of T2D was 3.3 cases/1000 person-years.
Predictive Value of Genotype Risk Score
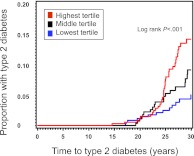
In a model adjusted for age, gender, and race, the 38-variant genotype score was significantly associated with time to incident T2D (hazard ratio per 1-allele increase 1.09, 95% CI 1.03–1.15; P < .001) (Table 2). Figure 1 demonstrates the different cumulative incidence of T2D (log rank P < .001) by tertile of genotype risk. After adjustment for age and gender, each 1-SD increase in the standardized BMI z score, glucose, and genotype score had a hazard ratio of 2.15 (95% CI 1.70–2.73), 1.20 (95% CI 0.97–1.49), and 1.49 (95% CI 1.20–1.85), respectively. Table 3 shows the results of the nested models with the use of unstandardized variables. Genotype score remained significantly associated with time to incident T2D in all 4 models. In the fully adjusted model, genotype risk score, parental history of diabetes, BMI z score, and log-transformed HDL cholesterol were all independent significant predictors. We observed no interaction between genotype risk score and gender, race, overweight, or parental history of diabetes in age- and gender-adjusted models (P > .05 for all interaction terms).
TABLE 2.
Age- and Gender-Adjusted Hazard Ratios From Cox Proportional Hazards Models for Incident T2D
| Hazard Ratio (95% CI) | P | |
|---|---|---|
| Demographics | ||
| Age, per increase of 1 ya | 1.00 (0.89–1.14) | .95 |
| Gender, male versus femalea | 1.05 (0.70–1.60) | .80 |
| Self-reported race, black versus white | 1.76 (1.15–2.67) | .009 |
| Parental history of diabetes, yes versus no | 4.09 (2.48–6.76) | <.001 |
| Physical examination | ||
| BMI z score, per increase of 1 unit | 1.97 (1.60–2.43) | <.001 |
| Mean arterial pressure, per increase of 1 mm Hg | 1.04 (1.01–1.07) | .005 |
| Laboratory analyses | ||
| Glucose, per increase of 1 mg/dL | 1.02 (1.00–1.05) | .10 |
| HDL cholesterol,b per increase of 1 mg/dL | 0.68 (0.54–0.85) | <.001 |
| Triglycerides,b per increase of 1 mg/dL | 1.76 (1.11–2.78) | .02 |
| Genotype score, per increase of 1 allele | 1.10 (1.05–1.16) | <.001 |
Hazard ratio for age is gender-adjusted and hazard ratio for gender is age-adjusted.
HDL cholesterol and triglyceride level log-transformed to improve model fit.
FIGURE 1.
Cumulative incidence of T2D from adolescence by tertile of genotype score in the Bogalusa Heart Study.
TABLE 3.
Nested Cox Proportional-Hazards Models for Incident T2D in 1030 Bogalusa Heart Study Participants Followed From Adolescence to Adulthood
| Models Without Genotype Score, HR (95% CI) or C Statistic (95% CI) | P | Models With Genotype Score, HR (95% CI), C Statistic (95% CI), or NRI (95% CI) | P | |
|---|---|---|---|---|
| Model 1: Demographics | ||||
| Age, per increase of 1 y | 1.02 (0.90 to 1.15) | .78 | 1.02 (0.90 to 1.15) | .76 |
| Gender, male versus female | 1.11 (0.73 to 1.68) | .64 | 1.09 (0.72 to 1.66) | .68 |
| Race, black versus white | 1.75 (1.15 to 2.67) | .01 | 1.26 (0.79 to 2.03) | .33 |
| Genotype score, per increase of 1 allele | — | — | 1.09 (1.03 to 1.15) | .01 |
| C statistic | 0.562 (0.492 to 0.632) | 0.613 (0.541 to 0.684) | ||
| Continuous NRI | — | 0.408 (0.165 to 0.646) | ||
| Hosmer-Lemeshow χ2 | 30.39 | 5.36 | ||
| Model 2: Model 1 plus parental history | ||||
| Age, per increase of 1 y | 0.99 (0.88 to 1.12) | .90 | 1.00 (0.88 to 1.13) | .95 |
| Gender, male versus female | 1.09 (0.72 to 1.65) | .69 | 1.07 (0.71 to 1.63) | .74 |
| Race, black versus white | 1.57 (1.03 to 2.40) | .04 | 1.24 (0.77 to 1.99) | .37 |
| Parental history of diabetes, yes versus no | 3.78 (2.28 to 6.27) | <.001 | 3.40 (2.03 to 5.69) | <.001 |
| Genotype score, per increase of 1 allele | — | — | 1.07 (1.01 to 1.13) | .03 |
| C statistic | 0.637 (0.562 to 0.712) | 0.674 (0.604 to 0.744) | ||
| Continuous NRI | — | 0.246 (−0.022 to 0.507) | ||
| Hosmer-Lemeshow χ2 | 18.76 | 13.66 | ||
| Model 3: Model 2 plus physical examination | ||||
| Age, per increase of 1 y | 1.00 (0.87 to 1.14) | .94 | 1.00 (0.87 to 1.14) | .96 |
| Gender, male versus female | 1.13 (0.74 to 1.73) | .57 | 1.10 (0.72 to 1.68) | .66 |
| Race, black versus white | 1.34 (0.87 to 2.06) | .18 | 1.07 (0.67 to 1.72) | .78 |
| Parental history of diabetes, yes versus no | 2.96 (1.77 to 4.97) | <.001 | 2.61 (1.53 to 4.44) | <.001 |
| BMI z score, per increase of 1 SD | 1.75 (1.41 to 2.17) | <.001 | 1.73 (1.40 to 2.15) | <.001 |
| Mean arterial pressure, per increase of 1 mm Hg | 1.02 (0.99 to 1.05) | .14 | 1.02 (0.99 to 1.05) | .13 |
| Genotype score, per increase of 1 allele | — | — | 1.06 (1.00 to 1.13) | .04 |
| C statistic | 0.757 (0.695 to 0.819) | 0.760 (0.699 to 0.820) | ||
| Continuous NRI | — | 0.281 (0.020 to 0.547) | ||
| Hosmer-Lemeshow χ2 | 21.90 | 16.35 | ||
| Model 4: Model 3 plus laboratory analyses | ||||
| Age, per increase of 1 y | 0.98 (0.86 to 1.12) | .81 | 0.98 (0.86 to 1.12) | .81 |
| Gender, male versus female | 1.02 (0.66 to 1.58) | .92 | 1.00 (0.65 to 1.54) | >.99 |
| Race, black versus white | 1.53 (0.96 to 2.42) | .07 | 1.22 (0.73 to 2.02) | .45 |
| Parental history of diabetes, yes versus no | 2.96 (1.77 to 4.95) | <.001 | 2.67 (1.58 to 4.53) | <.001 |
| BMI z score, per increase of 1 unit | 1.68 (1.35 to 2.09) | <.001 | 1.67 (1.34 to 2.08) | <.001 |
| Mean arterial pressure, per increase of 1 mm Hg | 1.02 (0.99 to 1.05) | .15 | 1.02 (0.99 to 1.05) | .13 |
| Glucose, per increase of 1 mg/dL | 1.02 (0.99 to 1.05) | .15 | 1.02 (0.99 to 1.04) | .20 |
| HDL cholesterol,a per increase of 1 mg/dL | 0.66 (0.48 to 0.90) | .01 | 0.66 (0.48 to 0.90) | .01 |
| Triglycerides,a per increase of 1 mg/dL | 0.93 (0.51 to 1.69) | .81 | 0.92 (0.51 to 1.65) | .77 |
| Genotype score, per increase of 1 allele | — | — | 1.06 (1.00 to 1.13) | .05 |
| C statistic | 0.756 (0.692 to 0.821) | 0.760 (0.697 to 0.823) | ||
| Continuous NRI | — | 0.261 (−0.007 to 0.529) | ||
| Hosmer-Lemeshow χ2 | 10.23 | 7.88 | ||
C statistics, continuous NRI, and Hosmer-Lemeshow χ2 values calculated at 30 y. HR, hazard ratio; —, corresponds to nonapplicable results.
Log-transformed to improved model fit.
Comparison of Models With and Without Genotype Score
As assessed by likelihood ratio tests, the fit of each of the 4 nested models was improved by the addition of the genotype score (all P < .05). When added to the basic demographic model (model 1), genotype score had a moderate effect on reclassification (continuous NRI 0.408) and improved model performance (C statistics 0.562 without and 0.613 with genotype score). Genotype score did improve the performance of a prediction model including only demographics and parental history of diabetes (C statistics 0.637 and 0.674 for model 2 without and with genotype score, respectively). However, it did not improve more complex models that added physical examination and laboratory measures (C statistics 0.756 and 0.760 for model 4 without and with genotype risk score, respectively). Genotype score had a relatively weak effect on reclassification in the full clinical model (continuous NRI 0.261 for model 4).
Discussion
The present analyses demonstrate that a genotype score predicts incident T2D in a biracial population of adolescents followed on average 26 years into adulthood. The majority of the 38 variants included in the genotype score were identified in cohort studies of adult participants of European descent. Our results suggest that such genotype information can also predict T2D in a younger and more ancestrally diverse population than those in the discovery studies. The genotype score remained a significant predictor of incident T2D in models adjusted for clinical risk predictors in adolescence but did not improve model performance. This suggests that common SNP genotype information available today, when measured in adolescence, is unlikely to improve the detection of risk for adult T2D.
Several large prospective studies have developed clinical prediction models for incident T2D in black and white adults.21,26,33,34 In general, these models include age, gender, race, family history, adiposity, blood pressure, and blood glucose and lipids levels. Because few prospective studies in children have sufficient follow-up to have accrued an adequate number of cases of T2D, risk prediction models from this age are less well developed but contain similar risk factors.19,22,24 Guided by these models, we built prediction models by sequentially layering components of the routine clinical encounter: basic demographics, family history, examination, and laboratory analyses. The addition of a genotype score did not improve the model discrimination for T2D from adolescence, compared with a prediction model based on these predictors. Similar analyses in adults have shown that the addition of genotype information to T2D prediction models generally does not yield a significant categorical NRI.12,15,35
The strengths of the present analyses include the use of a large prospective study from adolescence with excellent clinical measures and sufficient accrual of cases of T2D over a long follow-up period. The inclusion of a large number of black participants improves the generalizability of the analysis to diverse populations. Although most of the variants comprising our genotype score were discovered in populations of European ancestry, recent work has demonstrated an association between such scores and T2D in black adult populations.36,37 However, the majority of these variants are in noncoding regions of the genome and likely are proxies in linkage disequilibrium with the true causal variants. The discovery of these causal variants through targeted sequencing may strengthen the predictive value of updated genotype scores in diverse ancestral groups.
The present analysis has a few limitations. First, some participants classified as noncases in these analyses probably went on to develop T2D after the observation period and were thus misclassified as without diabetes in the current study. It is possible that a study with follow-up >30 years would detect a stronger association between genotype score and incident T2D, as a greater number of cases accrued as the study population aged.13 Our diagnostic criteria for T2D may have also misclassified individuals with latent autoimmune diabetes or on oral diabetes medications for a reason other than T2D. That a genotype score consisting of known T2D loci significantly predicted diabetes in this cohort suggests that such misclassification was minimized. Second, censoring from nonrandom loss to follow-up may have resulted in a bias if loss to follow-up was also associated with genotype risk score, which seems unlikely. There is the additional possibility of bias because genotype data were only available on a subset of all Bogalusa participants, but no evidence for such bias has been identified in the context of incident T2D.25 Third, we were not able to determine the diabetes type for participants reporting a parental history of diabetes. Despite this potential misclassification, we observed a strong effect size of parental history of diabetes in our models that is consistent with previous reports for T2D.21,38 The relatively small decrease in the effect size of parental history after the addition of genotype score to the model is consistent with the low percentage of T2D heritability explained by currently identified SNPs.39
Our findings in adolescence suggest that genotype scores based on known common variants might predict adult T2D among younger children, perhaps even when tested at birth. However, despite the variability and change in clinical risk factors from childhood through adolescence, it seems unlikely that genotype information at even younger ages would outperform routine clinical information. Thus, the utility of using common variant genotype scores in early life to screen for T2D risk much later in life is questionable. Some have suggested that parents may be interested in having their children tested for genetic susceptibility for such conditions.40,41 However, several professional societies agree that timely medical benefit is the main justification for testing of minors for adult-onset diseases.42–47 Several large studies have demonstrated that T2D is preventable through lifestyle modification among high-risk adults.2–4 Such data in children and adolescents are lacking.48–51 In addition, whether children would be motivated to change behaviors based on their genotype risk score is unknown, as are the ramifications of genotype testing for a child’s self-concept and for parent-child interactions. The benefits of testing would need to outweigh these and other potential sequelae for genotype screening to have clinical utility. Future research is needed to understand whether the knowledge of genotypic susceptibility for T2D would lead to the adoption of improved health-related behaviors and, if so, at which ages such risk information is effective. Moreover, unintended consequences of genotype screening in childhood need further exploration. As the state of the science moves forward and the allele spectrum of diabetes-causing variants is pushed to rare and rarer variants presumably of greater individual effect, genotype scores may be developed that do lead to improved predictive ability and, therefore, suggest clinical utility.
Conclusions
We have demonstrated that a T2D genotype score predicts incident adult T2D from adolescence and that this prediction persists after inclusion of common clinical risk factors. However, the inclusion of this genotype score does not improve the performance of the full clinical prediction model. The ability to distinguish those at highest risk for T2D early in life would increase both the efficiency and efficacy of prevention efforts, but at this time, common variant genotype information in adolescence does not provide such distinguishing information and currently is not recommended as a primary care tool for screening for T2D risk in youth.
Supplementary Material
Glossary
- CI
confidence interval
- HDL
high-density lipoprotein
- NRI
net reclassification improvement index
- SNP
single-nucleotide polymorphism
- T2D
type 2 diabetes
Footnotes
Dr Vassy contributed to the intellectual content of this article including conception and design, statistical analysis and interpretation of the data, and the drafting of and critical revision of the manuscript; Dr DasMahapatra contributed to the design of the study, statistical analysis of data, and revision of manuscript for intellectual content; Drs Meigs and Goodman contributed to the conception and design of the study, the analysis and interpretation of the data, and the drafting and critical revision of the manuscript; Drs Schork, Chen, and Berenson contributed to the conception of this study, the acquisition and interpretation of data, and critical revision of the article; Drs Magnussen and Raitakari and Seema Jamal contributed to the interpretation of data and revision of the manuscript; Dr Pencina contributed to the analysis and interpretation of data and critical revision of the article; and all authors gave final approval of the manuscript.
FINANCIAL DISCLOSURE: The authors have indicated they have no financial relationships relevant to this article to disclose.
FUNDING: Dr Vassy is supported by NIH National Research Service Award grant T32 HP12706 from the Health Resources and Services Administration and the NIH Loan Repayment Program (NIDDK); Dr Meigs is supported by NIH grants K24 DK080140 and R01 DK078616; Dr Schork is funded in part by NIH/NCRR grant UL1 RR025774; Dr Magnussen holds an NHMRC Early Career Fellowship (Public Health Fellowship, APP1037559); Dr Goodman is supported by NIH grant DK046200; Drs DasMahapatra, Chen, and Berenson are supported by grants HD-061437 and HD-062783 from the National Institute of Child Health and Human Development and AG-16592 from the National Institute on Aging. Funded by the National Institutes of Health (NIH).
References
- 1.Centers for Disease Control and Prevention National Diabetes Fact Sheet: National Estimates and General Information of Diabetes and Prediabetes in the United States, 2011. Atlanta, GA: US Department of Health and Human Services, Centers for Disease Control and Prevention; 2011 [Google Scholar]
- 2.Knowler WC, Barrett-Connor E, Fowler SE, et al. Diabetes Prevention Program Research Group . Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346(6):393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuomilehto J, Lindström J, Eriksson JG, et al. Finnish Diabetes Prevention Study Group . Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med. 2001;344(18):1343–1350 [DOI] [PubMed] [Google Scholar]
- 4.Pan XR, Li GW, Hu YH, et al. Effects of diet and exercise in preventing NIDDM in people with impaired glucose tolerance. The Da Qing IGT and Diabetes Study. Diabetes Care. 1997;20(4):537–544 [DOI] [PubMed] [Google Scholar]
- 5.Zeggini E, Scott LJ, Saxena R, et al. Wellcome Trust Case Control Consortium . Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40(5):638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voight BF, Scott LJ, Steinthorsdottir V, et al. MAGIC investigators. GIANT Consortium . Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42(7):579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rung J, Cauchi S, Albrechtsen A, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41(10):1110–1115 [DOI] [PubMed] [Google Scholar]
- 9.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–885 [DOI] [PubMed] [Google Scholar]
- 10.Saxena R, Voight BF, Lyssenko V, et al. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research . Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336 [DOI] [PubMed] [Google Scholar]
- 11.Zeggini E, Weedon MN, Lindgren CM, et al. Wellcome Trust Case Control Consortium (WTCCC) . Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meigs JB, Shrader P, Sullivan LM, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359(21):2208–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyssenko V, Jonsson A, Almgren P, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359(21):2220–2232 [DOI] [PubMed] [Google Scholar]
- 14.van Hoek M, Dehghan A, Witteman JC, et al. Predicting type 2 diabetes based on polymorphisms from genome-wide association studies: a population-based study. Diabetes. 2008;57(11):3122–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Miguel-Yanes JM, Shrader P, Pencina MJ, et al. Genetic risk reclassification for type 2 diabetes by age below or above 50 years using 40 type 2 diabetes risk single nucleotide polymorphisms. Diabetes Care. 2011;34(1):121–125 [DOI] [PMC free article] [PubMed]
- 16.Berenson GS, McMahan CA, Voors AW, et al. Cardiovascular Risk Factors in Children: The Early Natural History of Atherosclerosis and Essential Hypertension. New York, NY: Oxford University Press; 1980 [Google Scholar]
- 17.Pickoff AS, Berenson GS, Schlant RC. Introduction to the symposium celebrating the Bogalusa Heart Study. Am J Med Sci. 1995;310(suppl 1):S1–S2 [DOI] [PubMed] [Google Scholar]
- 18.World Health Organization. Part 1: Diagnosis and Classification of Diabetes Mellitus. Geneva, Switzerland: World Health Organization; 1999. WHO/NCD/NCS/99.2
- 19.Franks PW, Hanson RL, Knowler WC, et al. Childhood predictors of young-onset type 2 diabetes. Diabetes. 2007;56(12):2964–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen QM, Srinivasan SR, Xu JH, Chen W, Berenson GS. Fasting plasma glucose levels within the normoglycemic range in childhood as a predictor of prediabetes and type 2 diabetes in adulthood: the Bogalusa Heart Study. Arch Pediatr Adolesc Med. 2010;164(2):124–128 [DOI] [PubMed] [Google Scholar]
- 21.Wilson PWF, Meigs JB, Sullivan L, Fox CS, Nathan DM, D’Agostino RB Sr. Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Arch Internal Med. 2007;167(10):1068–1074 [DOI] [PubMed]
- 22.Morrison JA, Glueck CJ, Horn PS, Wang P. Childhood predictors of adult type 2 diabetes at 9- and 26-year follow-ups. Arch Pediatr Adolesc Med. 2010;164(1):53–60 [DOI] [PubMed] [Google Scholar]
- 23.Kuczmarski RJ, Ogden CL, Guo SS, et al. 2000 CDC Growth Charts for the United States: Methods and Development. Hyattsville, MD: National Center for Health Statistics; 2002 [PubMed] [Google Scholar]
- 24.Nguyen QM, Srinivasan SR, Xu JH, Chen W, Kieltyka L, Berenson GS. Utility of childhood glucose homeostasis variables in predicting adult diabetes and related cardiometabolic risk factors: the Bogalusa Heart Study. Diabetes Care. 2010;33(3):670–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith EN, Chen W, Kähönen M, et al. Longitudinal genome-wide association of cardiovascular disease risk factors in the Bogalusa heart study. PLoS Genet. 2010;6(9):e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balkau B, Lange C, Fezeu L, et al. Predicting diabetes: clinical, biological, and genetic approaches: data from the Epidemiological Study on the Insulin Resistance Syndrome (DESIR). Diabetes Care. 2008;31(10):2056–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sankararaman S, Sridhar S, Kimmel G, Halperin E. Estimating local ancestry in admixed populations. Am J Hum Genet. 2008;82(2):290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosmer DW, Lemeshow S. A goodness-of-fit test for the multiple logistic regression model. Comm Stat. 1980;9(10):1043–1069 [Google Scholar]
- 29.Pencina MJ, D’Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23(13):2109–2123 [DOI] [PubMed] [Google Scholar]
- 30.Pencina MJ, D’Agostino RB, Sr, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30(1):11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pencina MJ, D’Agostino RB, Sr, Demler OV. Novel metrics for evaluating improvement in discrimination: net reclassification and integrated discrimination improvement for normal variables and nested models. Stat Med. 2012;31(2):101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hosmer DW, Lemeshow S. Confidence interval estimates of an index of quality performance based on logistic regression models. Stat Med. 1995;14(19):2161–2172 [DOI] [PubMed] [Google Scholar]
- 33.Stern MP, Williams K, Haffner SM. Identification of persons at high risk for type 2 diabetes mellitus: do we need the oral glucose tolerance test? Ann Intern Med. 2002;136(8):575–581 [DOI] [PubMed] [Google Scholar]
- 34.Schmidt MI, Duncan BB, Bang H, et al. Atherosclerosis Risk in Communities Investigators . Identifying individuals at high risk for diabetes: The Atherosclerosis Risk in Communities study. Diabetes Care. 2005;28(8):2013–2018 [DOI] [PubMed] [Google Scholar]
- 35.Talmud PJ, Hingorani AD, Cooper JA, et al. Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. BMJ. 2010;340:b4838 [DOI] [PMC free article] [PubMed]
- 36.Cooke JN, Ng MC, Palmer ND, et al. Genetic risk assessment of type 2 diabetes-associated polymorphisms in African Americans. Diabetes Care. 2012;35(2):287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waters KM, Stram DO, Hassanein MT, et al. Consistent association of type 2 diabetes risk variants found in Europeans in diverse racial and ethnic groups. PLoS Genet. 2010;6(8):e1001078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meigs JB, Cupples LA, Wilson PW. Parental transmission of type 2 diabetes: the Framingham Offspring Study. Diabetes. 2000;49(12):2201–2207 [DOI] [PubMed]
- 39.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGuire AL, Diaz CM, Wang T, Hilsenbeck SG. Social networkers’ attitudes toward direct-to-consumer personal genome testing. Am J Bioeth. 2009;9(6-7):3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tercyak KP, Hensley Alford S, Emmons KM, Lipkus IM, Wilfond BS, McBride CM. Parents’ attitudes toward pediatric genetic testing for common disease risk. Pediatrics 2011;127(5). Available at: www.pediatrics.org/cgi/content/full/127/5/e1288 [DOI] [PMC free article] [PubMed]
- 42.National Society of Genetic Counselors. Position Statement: Prenatal and Childhood Testing for Adult-Onset Disorders. Chicago, IL: National Society of Genetic Counselors; 1995
- 43.Canadian Paediatric Society and Canadian College of Medical Geneticists. Guidelines for genetic testing of healthy children. Paediatr Child Health. 2003;8(1):42–45 [DOI] [PMC free article] [PubMed]
- 44.European Society of Human Genetics . Statement of the ESHG on direct-to-consumer genetic testing for health-related purposes. Eur J Hum Genet. 2010;18(12):1271–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Points to consider: ethical, legal, and psychosocial implications of genetic testing in children and adolescents. American Society of Human Genetics Board of Directors, American College of Medical Genetics Board of Directors. Am J Hum Genet. 1995;57(5):1233–1241 [PMC free article] [PubMed] [Google Scholar]
- 46.Committee on Bioethics . Ethical issues with genetic testing in pediatrics. Pediatrics. 2001;107(6):1451–1455 [DOI] [PubMed] [Google Scholar]
- 47.European Society of Human Genetics . Genetic testing in asymptomatic minors: recommendations of the European Society of Human Genetics. Eur J Hum Genet. 2009;17(6):720–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foster GD, Linder B, Baranowski T, et al. HEALTHY Study Group . A school-based intervention for diabetes risk reduction. N Engl J Med. 2010;363(5):443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katz DL, O’Connell M, Njike VY, Yeh MC, Nawaz H. Strategies for the prevention and control of obesity in the school setting: systematic review and meta-analysis. Int J Obes (Lond). 2008;32(12):1780–1789 [DOI] [PubMed] [Google Scholar]
- 50.Rosenbaum M, Nonas C, Weil R, et al. Camino Diabetes Prevention Group . School-based intervention acutely improves insulin sensitivity and decreases inflammatory markers and body fatness in junior high school students. J Clin Endocrinol Metab. 2007;92(2):504–508 [DOI] [PubMed] [Google Scholar]
- 51.Whitlock EA, O’Connor EP, Williams SB, Beil TL, Lutz KW. Effectiveness of weight management programs in children and adolescents. Evid Rep Technol Assess (Full Rep). 2008;(170):1–308 [PMC free article] [PubMed] [Google Scholar]
- 52.Yasuda K, Miyake K, Horikawa Y, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40(9):1092–1097 [DOI] [PubMed] [Google Scholar]
- 53.Grant SFA, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38(3):320–323 [DOI] [PubMed] [Google Scholar]
- 54.Gloyn AL, Weedon MN, Owen KR, et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52(2):568–572 [DOI] [PubMed] [Google Scholar]
- 55.Altshuler D, Hirschhorn JN, Klannemark M, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26(1):76–80 [DOI] [PubMed] [Google Scholar]
- 56.Dupuis J, Langenberg C, Prokopenko I, et al. DIAGRAM Consortium. GIANT Consortium. Global BPgen Consortium. Hamsten A on behalf of Procardis Consortium. MAGIC investigators . New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39(8):977–983 [DOI] [PubMed] [Google Scholar]
- 58.Winckler W, Weedon MN, Graham RR, et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007;56(3):685–693 [DOI] [PubMed] [Google Scholar]
- 59.Qi L, Cornelis MC, Kraft P, et al. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum Mol Genet. 2010;19(13):2706–2715 [DOI] [PMC free article] [PubMed]
- 60.Kong A, Steinthorsdottir V, Masson G, et al. DIAGRAM Consortium . Parental origin of sequence variants associated with complex diseases. Nature. 2009;462(7275):868–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.