During endocytosis, actin polymerization nucleated by the Arp2/3 complex provides force needed to drive internalization. Las17 is the strongest activator of the Arp2/3 complex in yeast cells. This study shows that Las17 is associated into a stable complex with Sla1, an adaptor that inhibits Las17 activity both in vitro and in vivo.
Abstract
During clathrin-mediated endocytosis, branched actin polymerization nucleated by the Arp2/3 complex provides force needed to drive vesicle internalization. Las17 (yeast WASp) is the strongest activator of the Arp2/3 complex in yeast cells; it is not autoinhibited and arrives to endocytic sites 20 s before actin polymerization begins. It is unclear how Las17 is kept inactive for 20 s at endocytic sites, thus restricting actin polymerization to late stages of endocytosis. In this paper, we demonstrate that Las17 is part of a large and biochemically stable complex with Sla1, a clathrin adaptor that inhibits Las17 activity. The interaction is direct, multivalent, and strong, and was mapped to novel Las17 polyproline motifs that are simultaneously class I and class II. In vitro pyrene-actin polymerization assays established that Sla1 inhibition of Las17 activity depends on the class I/II Las17 polyproline motifs and is based on competition between Sla1 and monomeric actin for binding to Las17. Furthermore, live-cell imaging showed the interaction with Sla1 is important for normal Las17 recruitment to endocytic sites, inhibition during the initial 20 s, and efficient endocytosis. These results advance our understanding of the regulation of actin polymerization in endocytosis.
INTRODUCTION
Endocytosis is essential for a variety of cellular activities, including nutrient uptake, cell surface remodeling, and regulation of signal transduction. Clathrin-mediated endocytosis (CME) is a fundamental endocytic pathway involving numerous proteins that collect cargo into a coated pit, invaginate a vesicle, pinch it off, and transport the vesicle to endosomes (Doherty and McMahon, 2009; Traub, 2009). This process is highly conserved throughout evolution and proceeds through a well-defined sequence of events (Kaksonen et al., 2003, 2005; Galletta and Cooper, 2009; Weinberg and Drubin, 2012). Actin polymerization, a critical component of CME, was first discovered in yeast, in which it forms characteristic cortical actin patches. Mutations in actin cytoskeletal genes, including the Arp2/3 complex, or treatment of yeast cells with drugs that inhibit actin polymerization block endocytic vesicle internalization (Engqvist-Goldstein and Drubin, 2003). The key role of actin in CME has also been established in mammalian cells. A burst of actin polymerization provides force needed for internalization during late stages of CME; however, the mechanisms that control the precise timing of actin polymerization are not well understood (Galletta and Cooper, 2009; Boulant et al., 2011; Taylor et al., 2011).
Budding yeast has been a very fruitful system to dissect CME through a combination of genetics, live-cell fluorescence microscopy, and biochemistry. Several studies have revealed discrete stages for endocytic vesicle generation and a well-choreographed assembly and disassembly pathway of endocytic factors (Kaksonen et al., 2005; Weinberg and Drubin, 2012). First, there is an immobile phase in which clathrin and other components arrive to a defined region of the plasma membrane, form a coated pit, and begin collecting transmembrane cargo, such as receptors. This step is long (1–2 min) and involves proteins such as clathrin, AP-2, and Ede1. Second, another wave of components of the immobile phase, including Las17 and Sla1, assemble ∼20 s before actin polymerization (Sun et al., 2006). Las17 is the yeast Wiskott-Aldrich syndrome protein (WASp) homologue, and the strongest activator of the Arp2/3-mediated actin polymerization, both in vitro and in vivo (Li, 1997; Rodal et al., 2003; Sun et al., 2006; Galletta et al., 2008). Sla1 is both a regulator of the actin cytoskeleton and a clathrin adaptor protein for the internalization of transmembrane protein cargo containing the NPFxD endocytic signal (Holtzman et al., 1993; Howard et al., 2002; Mahadev et al., 2007; Di Pietro et al., 2010). Third, a fast, mobile stage of endocytosis, typically observed by following Abp1, occurs concomitant with Arp2/3-mediated actin polymerization. Components of the coat, such as Sla1, move ∼200 nm into the cell together with the incipient vesicle. This mobile stage of endocytosis is brief (∼10–15 s) and culminates with vesicle scission, facilitated by the BAR-domain proteins Rvs161/167. Fourth, very soon after the scission step, most components of the coat disassemble as the released vesicle moves toward endosomes along actin cables (Toshima et al., 2006). It was initially thought that Las17 dissipates from the plasma membrane rather than moving with the invagination, suggesting actin assembly takes place with barbed ends oriented toward the cell surface (Kaksonen et al., 2003, 2005). However, recent studies indicate Las17 likely moves inward, along with the nascent vesicle, which is consistent with actin polymerization with barbed ends oriented toward the invaginating vesicle (Galletta et al., 2008; Galletta and Cooper, 2009; Idrissi et al., 2008).
The rate-limiting step for de novo actin filament assembly is nucleation (Pollard et al., 2000). The primary cellular factor involved in actin nucleation in endocytosis is the conserved Arp2/3 complex. While isolated Arp2/3 complex is a poor actin nucleator, upon interaction with nucleation-promoting factors (NPFs), Arp2/3 starts nucleating actin assembly with various strengths (Sun et al., 2006; Padrick and Rosen, 2010). Yeast cells express five NPFs: Las17, Myo5, Myo3, Pan1, and Abp1. The strength with which these NPFs stimulate actin polymerization by Arp2/3 ranges greatly, with Las17 reported to be the strongest both in vitro and in vivo, followed by Myo5/3 (Sirotkin et al., 2005; Sun et al., 2006; Galletta et al., 2008). Pan1 and Abp1 display a rather modest NPF activity (Duncan et al., 2001; Goode et al., 2001). Little is known about how the activities of these NPFs are regulated spatially or temporally. Two Arp2/3 NPFs, Las17 and Pan1, arrive to endocytic sites with the same timing as other components of the coat module, ∼20 s before actin assembly is initiated (Sun et al., 2006). Before the regulation of the actin cytoskeleton in endocytosis can be understood, it is essential to determine the mechanism that restricts actin assembly to late stages of the endocytic internalization pathway. This is particularly critical for Las17, because, as stated above, it is the strongest Arp2/3 activator, arrives early to endocytic sites, and it is not autoinhibited, unlike some members of the WASp/WASP family verprolin-homologous protein (WAVE) family (Rodal et al., 2003; Sun et al., 2006; Padrick and Rosen, 2010). Three components of the yeast endocytic machinery, Sla1, Bbc1, and Syp1, have been described to inhibit the Las17 NPF activity in vitro using the well-characterized pyrene-actin assembly reaction (Rodal et al., 2003; Boettner et al., 2009). Therefore Sla1, Bbc1, and Syp1 are candidates to keep Las17 inactive during those initial ∼20 s before actin assembly at endocytic sites. Analysis of the dynamics of these proteins revealed that Las17 and Sla1 arrive at approximately the same time to endocytic sites, Syp1 is recruited much earlier but overlaps partially with Sla1 and Las17, and Bbc1 is recruited ∼20 s later than Las17 (Sun et al., 2006; Boettner et al., 2009; Reider et al., 2009; Stimpson et al., 2009). Taken together, these observations indicate that Sla1 and Syp1 are good candidates for maintaining Las17 in an inactive state during the last 20 s of the immobile phase of endocytosis.
In this paper, we show that Sla1 and Las17 form a large biochemically stable complex in the cytosol and that they are corecruited to sites of endocytosis. Sla1 and Las17 display a direct, multivalent, and high-affinity interaction mediated by specific polyproline-SH3 recognition. The interaction was mapped to distinct Las17 polyproline motifs that are simultaneously class I and class II. Disruption of the Sla1-Las17 interaction uncouples the recruitment of Sla1 and Las17 to endocytic sites, causes a defect in Las17 recruitment, and abrogates the ability of Sla1 to inhibit Las17 NPF activity both in pyrene-actin polymerization assays and in live cells. These data support an important role for Sla1 in negatively regulating the Las17 NPF activity during the initial 20 s of recruitment to endocytic sites.
RESULTS
Las17 and Sla1 display similar spatiotemporal distribution at endocytic sites
Las17–red fluorescent protein (RFP) and Sla1–green fluorescent protein (GFP) display a high level of colocalization in cells carrying a deletion of the SLA2 gene that causes endocytic site arrest (Kaksonen et al., 2003). In addition, live-cell fluorescence microscopy data with wild-type cells suggest Las17 and Sla1 are recruited to endocytic sites at approximately the same time (Sun et al., 2006). However, this was an indirect comparison, rather than a determination made with cells simultaneously expressing both tagged Las17 and Sla1. For more direct assessment of the relative timing of arrival of Las17 and Sla1 at endocytic sites, a strain expressing both Las17-RFP and Sla1-GFP from the corresponding endogenous locus was generated and analyzed by two-color, live-cell confocal fluorescence microscopy (Figure 1A). Kymographs constructed from these movies show Las17 and Sla1 arriving and dissipating at the same time (Figure 1A). We recently reported that Sla1 directly interacts with clathrin and that cells expressing Sla1 with a mutation in its clathrin-binding motif (Sla1AAA-GFP) have a significantly longer patch lifetime compared with wild-type Sla1-GFP (Di Pietro et al., 2010). Interestingly, cells coexpressing the same mutant Sla1 tagged with GFP (Sla1AAA-GFP) and Las17-RFP showed an extended patch lifetime of both proteins and tight colocalization over time (Figure 1B). End3 is a well-established component of the yeast endocytic machinery whose deletion causes a significant endocytic defect and prolongs the patch lifetime of Sla1 and other coat proteins (Raths et al., 1993; Kaksonen et al., 2005). Sla1 and Las17 colocalized over time in cells carrying a deletion of the END3 gene (end3∆; Figure 1C). These results fine-tune the dynamics of Las17 and Sla1 previously reported and suggest the proteins may be corecruited from the cytosol to endocytic sites.
FIGURE 1:
Sla1 and Las17 simultaneously arrive and dissipate at sites of endocytosis and are stably associated in the cytosol. (A) S. cerevisiae cells expressing Las17-RFP and Sla1-GFP from endogenous loci (SDY145) were analyzed by live-cell fluorescence microscopy using a spinning-disk confocal microscope. The areas between white lines on the images (left panels) indicate the region from which kymographs were created (right panels). (B) S. cerevisiae cells expressing Las17-RFP and Sla1AAA-GFP (carrying a mutation in the Sla1 clathrin-binding motif) from endogenous loci (SDY284) were analyzed by live-cell imaging microscopy. (C) S. cerevisiae cells expressing Las17-RFP and Sla1-GFP from endogenous loci and carrying a deletion of the END3 gene (end3∆, SDY474) were analyzed by live-cell imaging microscopy. (D) Cofractionation of Sla1 with Las17 from S. cerevisiae (TVY614) cytosol upon size-exclusion chromatography on a Superose-6 column and immunoblotting analysis (IB) of the fractions. A dotted line is located at the peak (10-ml elution volume) to guide the eye. (E) S. cerevisiae (TVY614) cytosol was fractionated by size-exclusion chromatography followed by immunoprecipitation (IP) of each fraction with the Las-A antibody and immunoblotting analysis with Las-B or anti-Sla1 antibodies. (F) Size-exclusion chromatography and immunoblotting analysis of cytosolic extracts obtained from cells carrying a deletion of the SLA1 gene (sla1∆, GPY3130) or the LAS17 gene (Las17∆, SDY161). Note the shift in Las17 and Sla1 elution profiles compared with cytosolic extracts from wild-type cells in (D). (G) S. cerevisiae (TVY614) cytosol was fractionated in a 5–20% sucrose gradient and fractions were analyzed by immunoblotting, as indicated.
Las17 is associated with Sla1 into a stable and large complex
To study endogenous Las17, we raised affinity-purified polyclonal antibodies against recombinant Las17 amino- and carboxy-terminal fragments and named them Las-A and Las-B, respectively. As seen by immunoblot analysis, both antibodies recognize a band of the expected molecular weight in Saccharomyces cerevisiae cytosolic extracts (Supplemental Figure S1A). Confirmation that the band corresponds to endogenous Las17 protein was obtained by using cytosolic extracts from cells carrying a deletion of the LAS17 gene (las17∆) in immunoblotting experiments with both the Las-A antibody (Figure S1A, third panel) and the Las-B antibody (unpublished data). Both Las-A and Las-B also cross-react with other bands considered nonspecific, as they are also observed in las17∆ cell extracts, their apparent molecular weight is far from the Las17 molecular mass (67.7 kDa), and they are not detected by both antibodies (Figure S1A). The Las-A antibody works well for immunoprecipitation of endogenous Las17 from yeast cell extracts (Figure S1A, fourth panel).
Epitope-tagged Las17 was reported to coimmunoprecipitate with Sla1 from detergent-containing yeast total extracts, suggesting they interact physically—directly or indirectly—in vivo, presumably at endocytic sites (Li, 1997). The newly generated anti-Las17 antibodies were used in immunoprecipitation–immunoblotting experiments to test whether endogenous Las17 coimmunoprecipitates with endogenous Sla1 from the yeast cytosolic fraction (Figure S1B). For this purpose, a detergent-free extract was prepared and all membrane fractions were removed by ultracentrifugation. Sla1 was detected in Las17 but not in control immunoprecipitates and, conversely, Las17 was detected in Sla1 but not in control immunoprecipitates (Figure S1B). This result indicates Las17 and Sla1 may interact physically—directly or indirectly—both on the plasma membrane and in the cytosol, suggesting they may be more stably associated than previously appreciated.
For testing the possibility of stable association of Las17 with Sla1, yeast cytosolic extracts were fractionated by size-exclusion chromatography, and each fraction was analyzed by immunoblotting (Figure 1D). Las17 cofractionated with Sla1 but not with End3, another endocytic coat protein (Figure 1D). Furthermore, in a separate experiment, each size-exclusion chromatography fraction was subsequently subjected to immunoprecipitation with the Las-A antibody and immunoblotting analysis with both Las-A and Sla1 antibodies (Figure 1E). As expected, Las17 showed the same profile as in Figure 1D. Importantly, Sla1 was also present and peaked in the same fractions as Las17, around an elution volume of 10 ml. This result indicates those fractions contain Las17 associated with Sla1 in a stable complex as opposed to a random cofractionation. For further testing of the possibility of a stable association between Las17 and Sla1, cytosolic extracts were prepared from yeast strains carrying a deletion of the SLA1 (sla1∆) or LAS17 (las17∆) gene and fractionated by size-exclusion chromatography (Figure 1F). Immunoblotting analysis of the fractions showed that in the absence of Sla1, Las17 elution volume changed dramatically (∼10 to ∼17 ml), displaying a significantly smaller Stokes radius (Figure 1F). Conversely, in the absence of Las17, the elution profile of the bulk of Sla1 also changed dramatically (∼10 to ∼16.5 ml) to a considerably smaller Stokes radius (Figure 1F). Together these results show Sla1 and Las17 are associated into a stable complex.
From the experiments with wild-type cells, we estimated the Sla1-Las17 complex has a Stokes radius of 224 ± 6 Å, and from sedimentation velocity experiments (Figure 1G), we estimated a sedimentation coefficient of 8.1 ± 0.5 S. The native molecular mass of the Sla1-Las17 complex was calculated from the Stokes radius and sedimentation coefficient as 822 ± 28 kDa (Siegel and Monty, 1965). A similar analysis for the well-known heterotetrameric AP-2 complex revealed a native molecular mass of 212 ± 5 kDa, while its theoretical molecular mass (one copy each of the Apl1, Apl3, Apm4, and Aps2 subunits) is 286 kDa (unpublished data). Therefore this analysis of the Sla1-Las17 complex may have slightly underestimated its native molecular mass. Because the molecular masses of Sla1 and Las17 are 135.8 and 67.6 kDa, respectively, the complex must contain multiple copies of Sla1 and Las17, include additional proteins, or both. The Stokes radius obtained for Las17 with sla1∆ cells 40 ± 1 Å or for Sla1 with las17∆ cells 47 ± 4 Å (Figure 1F) are compatible with monomeric Las17 and Sla1, respectively, suggesting there may be no additional components to the complex or they dissociate from the complex if Las17 or Sla1 are not present.
For testing the possibility of additional subunits, endogenous Las17 complex was isolated from a strain expressing C-terminus tandem-affinity purification (TAP)-tagged Las17 from the LAS17 locus. Following the TAP purification procedure, proteins associated with Las17 were identified by mass spectrometry. A control experiment was carried out in parallel with an untagged strain subjected to the same TAP purification procedure and mass spectrometry. Proteins detected with the Las17 TAP-tagged strain, but not with the untagged strain, are presented in Table 1. In addition to Sla1, seven proteins were detected. Five of these proteins were found in a previous study using Las17-derived actin networks produced with Las17-functionalized microbeads and yeast extract (Table 1; Michelot et al., 2010). The two additional proteins found here are clathrin heavy chain and Pan1, both known to interact physically with Sla1 (Tang et al., 2000; Di Pietro et al., 2010). These seven proteins are likely transient Las17 or Sla1 interactors, rather than stable components of the Sla1-Las17 complex. For instance, most of these proteins (Vrp1, Bbc1, Myo3, actin, and clathrin) display dynamics at endocytic patches different from those of Sla1 and Las17 and are therefore incompatible as components of a stable complex. Pan1 does have patch dynamics similar to Sla1 and Las17 (Sun et al., 2006), but it has been shown to form a stable complex with End3 and Sla2 (Toshima et al., 2007), and our gel filtration analysis showed End3 is not a component of the Sla1-Las17 stable complex (Figure 1D). Consistently, in similar yeast cytosol gel filtration analysis, the bulk of Pan1 and Sla2 peaked in fractions corresponding to a Stokes radius smaller than Sla1 and Las17, but similar to End3 (unpublished data). Likewise, gel filtration analysis of the eisosome component Pil1 using a cytosolic fraction from a Pil1-GFP strain showed that it does not cofractionate with the Sla1-Las17 complex (unpublished data). These mass spectrometry and gel filtration experiments suggest there may not be additional subunits in the Sla1-Las17 complex. Nevertheless, we cannot rule out putative additional subunits that might have failed mass spectrometry detection for reasons such as poor peptide ionization.
TABLE 1:
Las17-interacting proteins detected by mass spectrometry.
| This study | Michelot et al. (2010) |
|---|---|
| Las17 | + |
| Sla1 | + |
| Vrp1 | + |
| Bbc1 | + |
| Myo3 | + |
| Actin | + |
| Pil1 | + |
| Pan1 | − |
| Clathrin heavy chain | − |
Together these biochemical studies demonstrate all endogenous Las17 in the cytosol is associated into a large and stable complex with Sla1, consistent with the idea of their corecruitment to endocytic sites.
Novel Las17 class I/II polyproline motifs mediate direct and high-affinity binding to Sla1 SH3 domains
From amino- to carboxy-terminus, Las17 is composed of a WH1 domain, a polyproline region, a WH2 domain, and an acidic tail (Figure 2A). While WH2 and the acidic tail (also known as a VCA motif) are the actin- and Arp2/3-binding regions that mediate its conserved NPF activity, the central polyproline region of Las17 can interact with proteins containing SH3 domains (Figure 2A; Tong et al., 2002). Sla1 is composed of three amino-terminal SH3 domains and other domains important for cargo recruitment (SHD1), clathrin binding (LLDLQ), and both Sla1 oligomerization and regulation of binding to clathrin (SHD2) (Figure 2A; Holtzman et al., 1993; Howard et al., 2002; Mahadev et al., 2007; Di Pietro et al., 2010). Importantly, a study by Rodal et al. (2003) showed that the first and second Sla1 SH3 domains, but not the third, can inhibit the Las17 NPF activity in pyrene-actin in vitro polymerization assays. Therefore the first and second Sla1 SH3 domains are good candidates as mediators of a direct interaction with Las17.
FIGURE 2:
Identification of Las17 polyproline motifs involved in Sla1 binding by yeast two-hybrid analysis. (A) Left, cartoon representation of Las17 and Sla1 domains. Gray bars above Las17 indicate the various fragments containing polyproline motifs (P1 through P20) tested for binding to Sla1 SH3 domains 1 and 2 (yellow bar, SH3-1-2). Right, yeast cells (AH109) were cotransformed with expression plasmids containing GAL4 DNA-binding (GAL4 BD) and activation (GAL4 AD) domains fused in frame to the indicated Las17 fragments and Sla1-SH3-1-2, respectively. Double transformants were first selected on minimal media lacking leucine and tryptophan and containing histidine (+His) and then spotted onto plates containing the same medium (as control) or selective medium lacking histidine and containing various concentrations of 3-AT. (B) Yeast two-hybrid analysis comparing the strength of interaction of single and double polyproline motifs within the P8-12 fragment. (C) Yeast two-hybrid analysis showing similar strength of interaction between P1-20, P8-12, or P10-12 with Sla1-SH3-1-2. (D) Various point mutations were introduced that selectively inactivated specific polyproline motifs (class I/II) in the context of the P1-20 construct and analyzed using the yeast two-hybrid system. Mutations consisted of two proline-to-alanine changes per polyproline motif in order to disrupt the PxxP core sequence of each target motif.
Our analysis of the central polyproline region of Las17 indicates it can be dissected into 20 separate segments, each containing sequences conforming to the SH3-binding PxxP motif (P1 through P20; Figure 2A). Yeast two-hybrid analysis was performed to test for binding of the Las17 polyproline motifs to the first and second Sla1 SH3 domains (Figure 2, A–D). The full-length polyproline region containing all 20 Las17 PxxP motifs (P1-20), and two fragments (P8-11 and P12) interacted with the Sla1 SH3 domains, as evidenced by yeast growth in selective plates lacking histidine (His; Figure 2A). A comparison of the relative interaction strength for the different fragments was obtained by using selective plates with increasing concentrations of 3-amino-1,2,4-triazole (3-AT; Figure 2A). P1-20 displayed the strongest interaction, followed by P8-11 and P12. Sequence analysis of the polyproline motifs in fragments P8-11 and P12 revealed that all five core PxxP sequences are flanked by arginine residues and match class I (RxxPxxP) or class II (PxxPxR) polyproline motifs consensus. This is consistent with a previous study, in which the first and second SH3 domains of Sla1 were predicted to bind class I or class II polyproline motifs (Tonikian et al., 2009). Interestingly, P8, P9, P11, and P12 sequences simultaneously fit the requirement for both polyproline class I and class II motifs, and we refer to these polyproline motifs as class I/II (Figure S2). P10 is an imperfect fit to the class I/II motif (Figure S2). The discovery of the Las17 class I/II polyproline motifs is important, since it implies the binding of Sla1 SH3 domains to Las17 is not restricted to a specific orientation, as was described for other SH3 domains and their ligands (Figure S2; Mayer, 2001).
To further dissect the contribution of these 5 polyproline motifs to complex formation, we examined additional constructs containing one or two motifs (Figure 2B). Analysis of the individual polyproline motifs indicated only P11 and P12 were able to show cell growth in selective plates. In addition, a fragment containing both P10 and P11 displayed cell growth at higher 3-AT concentration than P11 alone. In contrast, P8 and P9 did not show cell growth either individually or together, and contributed only modestly when they were combined with P10 and P11 (P8-11 vs. P10-11; Figure 2B). These findings suggest that the core for the Sla1-binding site spans from P10 to P12 (Figure 2B). To further test the relevance of P10, P11, and P12 and determine their contribution to binding Sla1 SH3 domains, we tested two additional constructs, one containing a Las17 fragment P8 through P12 (P8-12) and another spanning P10 through P12 (P10-12), and compared them with the full central polyproline region (P1-20) (Figure 2C). All three constructs showed a similar level of interaction with cell growth up to 25 mM 3-AT, suggesting that P10, P11, and P12 are the key motifs mediating the Las17-Sla1 interaction (Figure 2C). An alignment between P10, P11, and P12 revealed their sequences are more closely related to one another than to other class I/II (P8 and P9) or non–class I/II motifs (P7) (Figure S2). Features within the P10, P11, and P12 consensus sequence likely explain the preference of Sla1 SH3-1-2 for these three class I/II polyproline motifs.
To confirm the specificity of the interaction, we introduced point mutations that selectively destroyed the class I/II motifs in the context of the P1-20 construct, but maintained all other polyproline motifs intact (Figure 2D). The mutations consisted of two proline-to-alanine changes per polyproline motif in order to disrupt the PxxP core sequence. Consistently, mutation of P10, P11, and P12 (MP10-12) strongly affected the ability of these cells to growth at higher 3-AT concentration, and additional mutations in both P8 and P9 (MP8-12) completely destroyed any residual binding to Sla1 SH3 domains (Figure 2D). Together the yeast two-hybrid experiments have mapped the interaction of the Sla1 SH3 domains 1 and 2 to specific Las17 polyproline motifs that belong to a new class I/II, with a subset composed of polyprolines P10, P11, and P12 likely representing the core binding site.
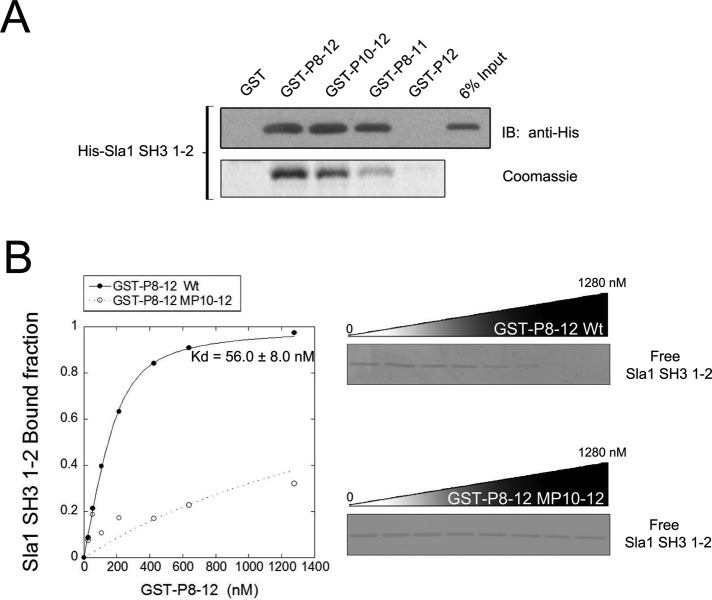
To corroborate a direct association between Las17 and Sla1 specifically mediated by the class I/II polyproline motifs, we carried out glutathione S-transferase (GST)-fusion binding assays. Four GST-fusion proteins, which contained Las17 fragments P8-12, P10-12, P8-11, or P12, were constructed and purified. Binding of these GST-fusion proteins to an hexahistidine-tagged Sla1 fragment spanning the SH3 domains 1 and 2 (His-Sla1-SH3-1-2) was analyzed by Coomassie Blue staining and immunoblotting (Figure 3A). GST-P8-12 and GST-P10-12 showed significant and similar ability to bind His-Sla1-SH3-1-2. GST-P8-11 showed an intermediate level of binding, and GST-P12 displayed a barely detectable binding. The relative level of binding paralleled the data obtained by yeast two-hybrid analyses (Figure 2) and confirmed a direct interaction between Las17 and Sla1. The results also suggest that the presence of multiple polyproline motifs in Las17 may mediate strong binding due to an avidity effect with a divalent Sla1 fragment.
FIGURE 3:
The interaction between Las17 and Sla1 is direct and strong. (A) GST fusion affinity assay was carried out with recombinant GST alone as a control, or GST fused to las17 fragments P8-12 (GST-P8-12), P10-12 (GST-P10-12), P8-11 (GST-P8-11), and P12 (GST-P12). Each GST-fusion protein was bound to glutathione beads and incubated with purified polyhistidine-tagged Sla1 fragment SH3-1-2 (His-Sla1-SH3-1-2). The bound His-Sla1-SH3-1-2 fraction was analyzed by Coomassie staining and immunoblotting with an anti-polyhistidine antibody. (B) A constant concentration of His-Sla1-SH3-1-2 (150 nM) was incubated with increasing concentrations of GST-P8-12 or GST-P8-12-MP10-12 (point mutations in P10, P11, and P12) bound to glutathione beads, as described in (A). The free His-Sla1-SH3-1-2 fraction remaining in the supernatant was determined by densitometry of SDS–PAGE staining. The His-Sla1-SH3-1-2 bound fractions were calculated and fitted to nonlinear regression to estimate the Kd for the interaction.
To estimate the affinity of the interaction between Las17 polyproline motifs and Sla1 SH3 domains, we followed a quantitative ligand-depletion approach (Sirotkin et al., 2005). Using a constant concentration of His-Sla1-SH3-1-2 and increasing concentrations of GST-P8-12 bound to glutathione beads, we determined the free ligand (His-Sla1-SH3-1-2) after incubation. The free His-Sla1-SH3-1-2 fraction present in the supernatant was analyzed by SDS–PAGE and Coomassie staining followed by densitometric quantitation (Figure 3B). Bound fractions were calculated by substraction from the total amount of ligand available for binding (Figure 3B). Binding was saturable, and fitting of the data using nonlinear regression determined a Kd of 56 ± 8 nM (Figure 3B). Binding curves with the same GST-P8-12 protein containing point mutations that substitute alanine for the key proline residues in the P10, P11, and P12 motifs (GST-P8-12 MP10-12) showed minimal residual binding. This high-affinity binding between Sla1 SH3 domains 1 and 2 and specific Las17 class I/II polyproline motifs supports the notion of a stable complex between the full-length proteins.
Class I/II polyproline motifs are necessary for Sla1 inhibition of Las17 NPF activity in vitro
Purified Las17, like purified WAVE, is not autoinhibited and potently activates Arp2/3-mediated actin polymerization (Rodal et al., 2003; Sun et al., 2006). However, Sla1 SH3 domains 1 and 2 were shown to inhibit Las17 NPF activity using the pyrene-actin polymerization assay (Rodal et al., 2003). Similar pyrene-actin polymerization experiments were carried out here using recombinant full-length Las17 purified from bacteria to ensure that no Sla1 or other Las17-interacting protein copurified. Las17 triggered rapid actin polymerization that was inhibited by purified His-Sla1-SH3-1-2 in a concentration-dependent manner, as evidenced by a decrease in the polymerization rate (slope) and an extended lag phase (Figure 4A). Saturating amounts of His-Sla1-SH3-1-2 inhibited Las17 activity by 86%, with a half-maximal concentration (IC50) of 120 nM (Figure 4, A and B). These results are consistent with previously reported data (Rodal et al., 2003).
FIGURE 4:
Class I/II polyproline motifs are critical for Sla1-SH3-1-2 fragment inhibition of Las17 NPF activity in pyrene-actin polymerization assays. (A) Actin (1.5 μM, 99% pyrene-labeled) was polymerized in the presence of Arp2/3 complex (75 nM), full-length, wild-type Las17 (75 nM), and a range of concentrations of His-Sla1-SH3-1-2 fragment. (B) Concentration-dependent effects of His-Sla1-SH3-1-2 inhibition on Las17-Arp2/3 induced actin polymerization. Percent inhibition was calculated from the reduction in polymerization rate, as determined in (A). The ability of (C) full-length, wild-type Las17 (75 nM), (D) Las17-MP10-12 (75 nM), and (E) Las17-MP8-12 (75 nM) to activate Arp2/3-mediated actin polymerization was measured as described in (A) in the absence (black circles) or presence of His-Sla1-SH3-1-2 (150 nM; white circles). The effect of His-Sla1-SH3-1-2 on the Las17 activity is expressed as relative polymerization rate (C–E) or percent inhibition (F). Polymerization rates and inhibition percentages are expressed as the average ± SEM of three independent determinations.
To test the possibility that Sla1 inhibition is dependent on the same Las17 class I/II polyproline motif that mediates the Las17-Sla1 association, we purified from bacteria full-length Las17 containing the same polyproline motif point mutations that abrogated binding, Las17 MP10-12 and Las17-MP8-12 (Figures 2D and 3B). Pyrene-actin polymerization assays showed both Las17-MP10-12 and Las17-MP8-12 retained a strong NPF activity, similar to the wild-type protein assayed under the same conditions, indicating their ability to bind to actin and the Arp2/3 complex was intact (Figure 4, C–E). Addition of 150 nM His-Sla1-SH3-1-2 inhibited the activity of Las17 wild-type by 75% (25% residual NPF activity; Figure 4, C and F). Importantly, addition of 150 nM His-Sla1-SH3-1-2 inhibited Las17-MP10-12 by only 25% (75% residual NPF activity), and did not inhibit Las17-MP8-12 at all (100% residual NPF activity; Figure 4, D–F). These observations indicate that the class I/II polyproline motifs identified here are not only important for Las17-Sla1 strong association but are also key components of Las17 NPF negative regulation by Sla1.
The presence of Sla1 SH3 domains disrupts Las17 interaction with G-actin
A second monomeric actin-binding site—in addition to the WH2 domain—was recently discovered in Las17 (Robertson et al., 2009) which may in part explain the very strong Las17 activity compared with other NPFs. This new monomeric actin-binding site was not fully characterized but was located within residues 300 and 422 of Las17. Interestingly, our Las17-P8-12 fragment spans residues 300–404. Thus the ability of the Sla1 SH3 domains to inhibit Las17 NPF activity may depend on competition between monomeric actin and the SH3 domains for binding to this region of Las17. We first tested for the ability of Las17-P8-12 fragment to bind monomeric actin. For this purpose, a pyrene-actin polymerization assay was carried out in conditions under which polymerization of actin (3 μM) by itself—in the absence of Arp2/3 complex—is readily detected (Figure 5). Addition of Las17-P8-12 fragment in excess of the monomeric actin concentration (6 μM) dramatically inhibited actin polymerization, consistent with monomeric actin binding and sequestration (Figure 5). Importantly, further addition of Sla1-SH3-1-2 reverted the Las17-P8-12 fragment inhibition, likely by binding to the Las17-P8-12 fragment, thus competing with monomeric actin and enabling it to return to the free pool available for polymerization. Subsaturating concentrations of Sla1-SH3-1-2 fragment (2 μM) elicited partial rescue, while saturating concentrations (12 μM) caused full rescue (Figure 5). These results indicate that Sla1 inhibits Las17 NPF activity at least in part by disrupting Las17's ability to interact with G-actin through this novel-binding site.
FIGURE 5:

The presence of Sla1 SH3 domains disrupts Las17 interaction with G-actin. (A) Actin (3 μM, 99% pyrene-labeled) was allowed to polymerize alone (in the absence of Arp2/3 complex or any other protein; gray circles), in the presence of Las17-P8-12 fragment (6 μM; black circles), both Las17-P8-12 fragment (6 μM) and His-Sla1-SH3-1-2 (2 μM) (light gray diamonds), or both Las17-P8-12 fragment (6 μM) and His-Sla1-SH3-1-2 (12 μM) (white circles). (B) Relative actin polymerization rates are expressed as the average ± SEM of three independent determinations.
Class I/II polyproline motifs are important for normal Las17 recruitment and inhibition at sites of endocytosis
So far our results indicate that Las17 and Sla1 arrive to endocytic sites at the same time, because Las17 and Sla1 are associated into a stable complex that likely keeps Las17 in an inactive state by competing with G-actin. Moreover, specific Las17 polyproline motifs involved in both the in vitro interaction and inhibition were mapped. We reasoned that disrupting the physical interaction between Las17 and Sla1 in cells may cause a defect in the Las17 recruitment to endocytic sites and/or a lack of inhibition of Las17 NPF activity. To test these possibilities, we tagged the Las17 endogenous gene with GFP and subsequently introduced the same polyproline-motif point mutations that abolished binding to Sla1 (Figures 2D and 3B), and disrupted the inhibition in actin polymerization assays (Figure 4, C–F). We crossed this strain with one expressing Sla1-RFP or Abp1-RFP from the corresponding endogenous locus, sporulated the resulting diploids, and obtained haploid cells expressing the mutant Las17-GFP (Las17-MP8-12-GFP) and Sla1-RFP or Abp1-RFP. Similar strains with wild-type Las17-GFP and Sla1-RFP or Abp1-RFP were also generated for comparison. As observed using confocal fluorescence microscopy analysis, wild-type Las17-GFP and Sla1-RFP displayed an indistinguishable endocytic patch lifetime, and they arrived and dissipated simultaneously (Figures 6A and S3A and Movie S1). Notably, a similar analysis with cells expressing Las17-MP8-12-GFP and Sla1-RFP showed mutant Las17 arrived ∼18 s after Sla1 (Figures 6B and S3B and Movie S2). In addition, the patch lifetime of Las17 decreased and that of Sla1 increased (Figure 6B). During the course of these investigations, it became apparent that while Las17-MP8-12-GFP did localize to endocytic sites, its patch fluorescence intensity was lower and the cytosolic background was higher compared with wild-type Las17-GFP. Quantitation of the fluorescence intensity ratio of Las17-MP8-12-GFP at endocytic patches to cytosol confirmed a statistically significant decrease compared with wild-type Las17-GFP, suggesting fewer mutant Las17 molecules localized to the patch (Figure 7A). This difference is not a result of overall destabilization of mutant Las17, as determined by immunoblotting analysis of total cell extracts (Figure 7B). These results are consistent with a defect in Las17 recruitment to endocytic sites when its association with Sla1 is compromised. We also noticed that in a previous study, sla1∆ cells displayed a higher Las17-GFP background than wild-type cells, although the experiment was not focused on the recruitment aspect, and this defect was not quantified (Kaksonen et al., 2005). Perhaps the known Sla1 interactions with endocytic transmembrane cargo, clathrin, Pan1, and numerous other coat proteins are important for Las17 recruitment in the context of the native complex with Sla1. Las17-MP8-12-GFP residual recruitment to sites of endocytosis may be mediated by some remaining interaction with other Sla1 domains, interactions with other endocytic proteins, or a combination of both.
FIGURE 6:
Uncoupling of the Las17 and Sla1 complex in vivo after mutation of class I/II polyproline motifs. Dynamics of (A) Las17-GFP and (B) Las17-MP8-12-GFP with Sla1-RFP at endocytic sites (SDY382 and SDY384, respectively). Time series (left) showing localization of Las17 and Sla1 at an endocytic patch from two-color movies (1 frame/s). Alignment of Las17 (green) and Sla1 (red) patch intensities as function of time (right panel depicts examples of two distinct patches, solid and dashed lines, respectively). Patch lifetimes ± SEM, delta time (Δtime) ± SEM, and number of patches analyzed (N) are indicated. Differences in Las17 (p = 0.01) and Sla1 (p = 0.0001) patch lifetimes and delta time (p < 0.0001) between Las17 and Las17- MP8-12 cells were statistically significant.
FIGURE 7:

Class I/II polyproline motifs are important for Las17 recruitment to endocytic sites. (A) The patch (membrane) maximal fluorescence intensity and internal (cytosol) fluorescence intensity for Las17-GFP and Abp1-RFP were determined after imaging wild-type Las17 (SDY371) and Las17-MP8-12 (SDY372) cells, and the membrane/cytosol fluorescence intensity ratio was calculated. (B) Immunoblotting (IB) analysis of total extracts from yeast control cells (SDY088) or cells expressing wild-type Las17-GFP (SDY367) or Las17-MP8-12-GFP (SDY363).
Similar to published studies, Abp1-RFP was detected in wild-type cells at endocytic sites 19 s after Las17-GFP, and their patch lifetimes were 14 and 33 s, respectively (Figure 8A and Movie S3; Sun et al., 2006). In contrast, Abp1-RFP was detected only 9 s after Las17-MP8-12-GFP, which can be explained by the lack of inhibition by Sla1 (Figure 8B and Movie S4). This is even more remarkable, considering that fewer molecules of mutant Las17 were concentrated at endocytic sites compared with wild-type Las17 (Figure 7A). In cells expressing Las17-MP8-12-GFP, the patch lifetime of Abp1-RFP was increased from 14 to 20 s (Figure 8), but the maximum Abp1-RFP concentration reached was unchanged (Figure 7A). These results indicate that productive endocytosis likely requires a given amount of Arp2/3-mediated actin polymerization that takes a longer time to achieve in mutant Las17, because there are fewer Las17 molecules.
FIGURE 8:
Class I/II polyproline motifs are critical for Sla1 inhibition of Las17 NPF activity in vivo. Dynamics of (A) wild-type Las17-GFP and (B) Las17-MP8-12-GFP with Abp1-RFP at endocytic sites (SDY371 and SDY372, respectively). Time series (left) showing localization of Las17 and Abp1 from two-color movies (1 frame/s). Alignment of Las17 (green) and Abp1 (red) patch intensities as function of time (right panel depicts examples of two distinct patches, solid and dashed lines, respectively). Patch lifetimes ± SEM, delta time (∆time) ± SEM and number of patches analyzed (N) are indicated. Differences in Las17 (p = 0.0002) and Abp1 (p < 0.0001) patch lifetimes and delta times (p < 0.0001) between wild-type Las17 and Las17- MP8-12 cells were statistically significant.
Overall, live-cell fluorescence microscopy analysis suggests that Las17 molecules that are unable to bind Sla1, and therefore that do not occur in the native complex, are deficiently recruited to sites of endocytosis. However, the Las17 molecules that do get recruited have the NPF activity readily manifested. These results support the idea that Las17 recruitment to sites of endocytosis in wild-type cells is facilitated by being in a preformed complex with Sla1, one in which the NPF activity of Las17 is initially inhibited.
To compare the severity of the Las17 inhibition defect caused by the las17-MP8-12 mutation, we determined the time from Las17-GFP arrival to detection of Abp1-RFP (delta time) in cells carrying deletions of other genes involved in Las17 regulation: SYP1, BBC1, and BZZ1 (Figure 9A). bbc1∆ and bzz1∆ cells did not show a change in early Las17 inhibition, consistent with their later arrival to endocytic patches (Figure 9A). However, syp1∆ cells showed a small but statistically significant defect compared with wild-type cells (15 s vs. 19 s; Figure 9A). This defect is consistent with the known Syp1 ability to inhibit Las17 in vitro and its presence at endocytic sites at the time of Las17 arrival (Boettner et al., 2009; Reider et al., 2009; Stimpson et al., 2009). While this defect is less severe than the one elicited by the Las17-MP8-12 mutation, it suggests Sla1 and Syp1 may cooperate to inhibit las17 during the initial 20 s. Pyrene-actin polymerization assays were used to test the possibility of Sla1 and Syp1 cooperation to inhibit Las17 (Figure 9B). The single presence of the Sla1-SH3-1-2 fragment (1.3 μM) or full-length Syp1 (10 μM) at saturating concentrations inhibited the actin polymerization rate by 87 and 81%, respectively (Figure 9B). Simultaneous addition of Sla1-SH3-1-2 and Syp1, however, caused significantly more—virtually complete—inhibition of Las17 (99% inhibition; Figure 9B). This additive effect between Sla1 and Syp1 inhibition suggests a scenario in which Sla1 and Syp1 cooperate to inhibit Las17 early after its recruitment to endocytic sites.
FIGURE 9:
Syp1 deletion has a subtle in vivo effect on the delta time between Las17 and Abp1 recruitment and cooperates with Sla1 to inhibit Las17 in vitro. (A) The time from Las17-GFP arrival to detection of Abp1-RFP (delta time) was determined in cells carrying deletions of genes involved in Las17 regulation, SYP1 (syp1∆, SDY471), BBC1 (bbc1∆, SDY472), or BZZ1 (bzz1∆, SDY473). A strain carrying the Las17 mutation, Las17-MP8-12-GFP (SDY372), was also included to facilitate comparison. Differences in delta time between wild-type cells and syp1∆ cells were statistically significant (p < 0.01; number of patches analyzed = 20–45). (B) His-Sla1-SH3-1-2 and His-Syp1 have additive inhibitory effects on Las17. Actin (1.5 μM, 99% pyrene-labeled) was polymerized in the presence of Arp2/3 complex (75 nM), full-length Las17 (75 nM), and the indicated concentrations of His-Sla1-SH3-1-2 fragment and/or full-length His-Syp1. Relative polymerization rates are expressed as the average ± SEM of three independent determinations.
Class I/II polyproline motifs are important for normal overall endocytic patch dynamics and for fluid phase and CME
The defects in Las17, Sla1, and Abp1 dynamics described above in live cells expressing Las17-MP8-12 suggest that overall endocytic patch dynamics may be abnormal, and endocytosis may be less efficient in these mutant cells. To investigate these possibilities, we first examined the patch lifetime of early (Syp1), intermediate (Pan1), and late (Myo5) coat components in cells expressing Las17-MP8-12 or wild-type Las17 from the endogenous LAS17 locus (Figure 10). Early (Syp1) and intermediate (Pan1) proteins showed a statistically significant increase in their patch lifetime in mutant Las17 cells compared with wild-type Las17 cells (Figure 10, A and B). Interestingly, the number of patches counted at steady state also increased in these mutant cells (Figure 10C). The simplest explanation for these results is that initial formation of the endocytic site is not affected, but the endocytic event takes a longer time to complete, and the number of patches counted at any given time is therefore higher. The difference in patch lifetime and number of patches appeared to be more marked in early and intermediate markers (Syp1, Pan1) than in late markers (Myo5).
FIGURE 10:
Class I/II polyproline motifs are important for normal overall endocytic patch dynamics. Patch lifetime and number of patches was analyzed for endocytic patch components Syp1-GFP, Pan1-GFP, and Myo5-GFP in cells expressing wild-type Las17 (SDY465, SDY466, and SDY467, respectively) or Las17-MP8-12 (SDY468, SDY469, and SDY470, respectively) from the corresponding endogenous loci. (A) Kymographs (right) were constructed from time- lapse movies of Syp1-GFP (1 frame/5 s), Pan1-GFP (1 frame/s) and Myo5-GFP (1 frame/s). A frame from each movie is shown (left). (B) Patch lifetimes are expressed as the average ± SEM (n = 25). Patch lifetime differences between Syp1-GFP (p = 0.04) and Pan1-GFP (p = 0.008) in wild-type Las17 and Las17-MP8-12 cells were statistically significant. (C) Number of patches is expressed as the average ± SEM (n = 25). The differences in patch number of Syp1-GFP (p < 0.0001) and Pan1-GFP (p < 0.0001) between wild-type Las17 and Las17-MP8-12 cells was statistically significant.
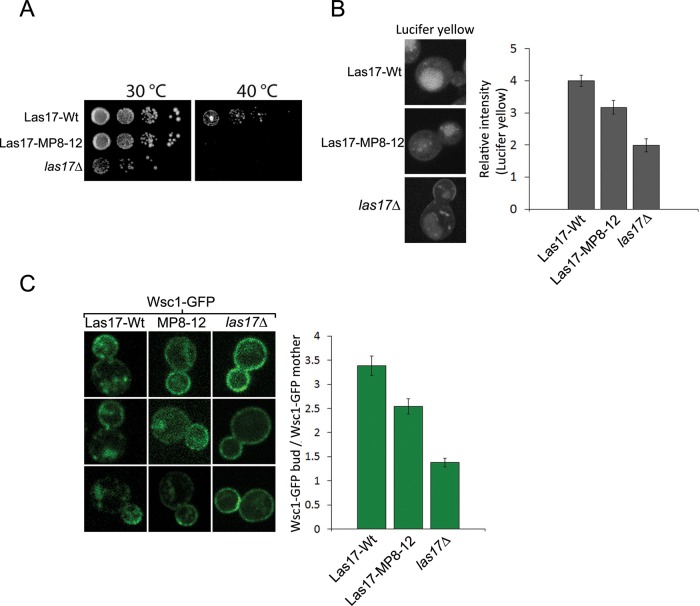
To evaluate the effect of the Las17-MP8-12 mutation on cell physiology, we examined growth at 30ºC and did not observe a significant defect compared with wild-type cells (Figure 11A). However, growth at 40ºC was severely impaired in Las17-MP8-12 cells compared with wild-type cells (Figure 11A). As previously reported (Li, 1997), cells carrying a deletion of the LAS17 gene (las17∆) did show a significant growth defect at all temperatures (Figure 11A). Bulk endocytosis, assayed by lucifer yellow uptake, was less efficient in Las17-MP8-12 cells than in wild-type cells, although it was not as severely affected as in las17∆ cells (Figure 11B). The impact of the Las17-MP8-12 mutation in CME was addressed by examining localization of Wsc1, a native clathrin-dependent endocytic cargo (Di Pietro et al., 2010). Wsc1 is a cell wall stress sensor that normally displays a polarized localization with high concentration at sites of new cell surface growth through clathrin- and Sla1-mediated endocytosis and recycling from endosomes (Piao et al., 2007; Di Pietro et al., 2010). In wild-type cells, polarized localization of Wsc1 is manifested as an enrichment in the plasma membrane of the bud compared with the mother cell, which can be detected in living cells expressing Wsc1-GFP. Endocytosis defects lead to uniform distribution of Wsc1-GFP in both the mother cell and the bud. Consequently, polarized localization provides a sensitive measure of Wsc1-GFP endocytosis (Mahadev et al., 2007; Piao et al., 2007). Accordingly, we determined Wsc1-GFP localization in wild-type cells, Las17-MP8-12 cells, and las17∆ cells (Figure 11C). In wild-type cells, Wsc1-GFP was primarily located at the cell surface of buds. In both Las17-MP8-12 cells and las17∆ cells, there was a significantly higher level of Wsc1-GFP at the surface of mother cells, indicative of a partial defect in endocytosis (Figure 11C). Fluorescence intensities of bud and mother cell surfaces were determined, and a ratio was calculated in order to quantify the defect (Figure 11C). The analysis indicates a statistically significant defect in Wsc1 endocytosis in Las17-MP8-12 cells and more so in las17∆ cells (Figure 11C). Overall, characterization of the Las17-MP8-12 mutant cells indicates that this region of interaction with Sla1 is important for normal progression of the endocytic patch and efficient endocytosis.
FIGURE 11:
Class I/II polyproline motifs are important for normal fluid-phase endocytosis and CME. (A) Wild-type, Las17MP8-12, and las17∆ (SDY367, SDY363, and SDY161) cells were grown to early log phase (OD600 = 0.2), plated in 10-fold serial dilutions on yeast–peptone–dextrose plates, and incubated for 72 h at 30°C or 40°C. (B) Wild-type, Las17MP8-12, and las17∆ cells were grown to early log phase, incubated for 2 h at 24°C with lucifer yellow, and imaged by confocal fluorescence microscopy. The fluorescence intensity of the vacuole was measured, normalized by the intensity of the background, and expressed as the average ± SEM (n = 15). Differences between wild-type and Las17MP8-12 (p = 0.007) or las17∆ (p = 0.0006) cells, and between Las17MP8-12 and las17∆ (p = 0.02) cells were statistically significant. An example of each cell type is presented in the left panels. (C) A pRS313-WSC1-GFP plasmid was introduced in wild-type, Las17MP8-12, and las17∆ cells to generate strains SDP478, SDP479, and SDP480, respectively. The Wsc1-GFP fluorescence intensities in the cortex of the bud and mother cells was determined, and the intensity ratio between the bud and mother was calculated for each bud–mother pair. The bud-to-mother Wsc1-GFP intensity ratios were expressed as the average ± SEM for wild-type (n = 40), Las17MP8-12, (n = 30) and las17∆ (n = 25). Both Las17MP8-12 and las17∆ cells displayed differences with wild-type cells that were statistically significant (p = 0.002 and p < 0.0001, respectively). las17∆ was also significantly different from Las17MP8-12 (p < 0.0001). Three examples of each cell type are presented in the left panels.
DISCUSSION
Arp2/3-mediated actin assembly plays a central role in CME, providing force needed for the internalization step (Engqvist-Goldstein and Drubin, 2003; Galletta and Cooper, 2009). Importantly, Arp2/3 activity must be tightly regulated to enable the endocytic patch to properly mature before the burst of actin polymerization that initiates coated vesicle invagination. Las17, the most potent yeast Arp2/3 nucleation-promoting factor, is recruited to endocytic sites well before actin polymerization is initiated and is constitutively active in purified form and therefore predicted to be inhibited by direct binding partners, in a manner similar to WAVE family proteins (Rodal et al., 2003; Sun et al., 2006; Boettner et al., 2009). Thus an important outstanding question has been how Las17 is inhibited during the initial ∼20 s after its arrival to sites of endocytosis. Sla1 is encoded by a gene that is synthetic lethal with ABP1, and has long been recognized as a central factor for regulation of the actin cytoskeleton in endocytosis. Moreover, the two most amino-terminal SH3 domains of Sla1 were shown to inhibit Las17 NPF activity in vitro (Rodal et al., 2003). Our findings that Las17 exists in a stable complex with Sla1 and that Las17 and Sla1 are corecruited from the cytosol to sites of endocytosis provide an excellent explanation to this long-standing problem. In addition to Sla1, two other components of the endocytic machinery, Bbc1 and Syp1, reduce the Las17 NPF activity in vitro. Because Bbc1 arrives to endocytic sites ∼20 s after Las17, it cannot be the inhibitor during the initial ∼20 s of Las17 recruitment, although it may modulate Las17's activity during late stages of endocytosis (Sun et al., 2006). Cooperation between Bbc1 and Sla1 to modulate actin polymerization in later stages of endocytosis is consistent with their additive inhibitory effect in pyrene-actin polymerization assays and the exaggerated endocytic actin structures found in live cells carrying deletions of both genes (Rodal et al., 2003; Kaksonen et al., 2005). Syp1 arrives to endoytic sites much earlier than Las17 and could therefore be an inhibitor in vivo during the initial ∼20 s of Las17 recruitment (Boettner et al., 2009; Reider et al., 2009; Stimpson et al., 2009). Previous studies found syp1∆ cells, much like bbc1∆ cells, display relatively mild defects in growth or endocytic dynamics (Kaksonen et al., 2005; Boettner et al., 2009; Reider et al., 2009; Stimpson et al., 2009). Results in Figure 9A indicate that syp1∆ cells display a small but statistically significant reduction in the time from Las17 arrival to detection of actin polymerization, suggesting a role in early Las17 inhibition. It is therefore possible that Sla1 and Syp1 cooperate to inhibit Las17 during its initial recruitment. This possibility is supported by the pyrene-actin polymerization assays showing additive effect between Sla1-SH3-1-2 fragment and Syp1 to inhibit Las17 (Figure 9B). Importantly, the mechanism mediating the Syp1 inhibitory effect is not fully understood, but since Syp1 does not contain SH3 domains, it is not expected to involve the Las17 polyproline motifs bound by Sla1. Therefore the fact that the time from Las17 recruitment to actin polymerization is significantly shorter in Las17-MP8-12 (9 s) than in wild-type Las17 (19 s) strains is best explained by the lack of Las17 inhibition by Sla1 (Figure 8). The emerging picture is one in which Sla1 is an important inhibitor of Las17 NPF activity in vivo during initial recruitment to the endocytic patch, likely in cooperation with Syp1. In the future, it will be important to better delineate the molecular mechanism of Syp1 inhibition of Las17 NPF activity and cooperation with Sla1. The physical interaction reported between Sla1 and Syp1 argues for a potentially more intertwined relationship between these two Las17 inhibitors (Tonikian et al., 2009).
The complex between Las17 and Sla1 serves other roles in addition to negatively regulate Las17 NPF activity:
First, mutant Las17 impaired for Sla1 binding displayed a significant recruitment defect, both in terms of timing and total amount of protein localized to sites of endocytosis (Figures 6, 7, and S3). The multiple interactions Sla1 makes with transmembrane protein cargo and coat components, such as clathrin, Pan1, Sla2, End3, and Syp1, are likely important for proper Las17 recruitment in the context of the complex with Sla1 (Ayscough et al., 1999; Costa and Ayscough, 2005; Tang et al., 2000; Howard et al., 2002; Warren et al., 2002; Mahadev et al., 2007; Tonikian et al., 2009; Di Pietro et al., 2010). Therefore in vivo Sla1 has both positive effects (recruitment to endocytic sites) and negative effects (inhibition of actin polymerization) on Las17 NPF functions. Mutant Las17 impaired for Sla1 binding displayed defects in overall coat dynamics (Figure 10), growth at high temperature (Figure 11A), and both fluid-phase endocytosis and CME (Figure 11, B and C). These defects may be the result of both defective recruitment of mutant Las17 to endocytic sites (fewer molecules and delayed arrival; Figures 6, 7, and S3) and misregulation of actin polymerization (Figures 8 and 9).
Second, given that Sla1 is a clathrin adaptor for cargo containing the NPFxD endocytic signal, it is possible that Sla1 senses the maturation state of the coated pit and transmits that information to Las17 to initiate actin polymerization when the incipient vesicle is ready for internalization. Therefore this complex has the potential to be a key regulator of the transition from early to late stages of the endocytic pathway.
Third, the Sla1-Las17 complex is large and likely consists of several copies of both Sla1 and Las17 (Figure 1). Supporting this possibility, mass spectrometry analysis of the complex purified under conditions mild enough to detect transient interactors failed to uncover additional subunits of the stable Sla1-Las17 complex (Table 1). This is also consistent with a multivalent interaction between Las17 and Sla1: three polyproline motifs (P10, P11, and P12) are the core binding site for the Sla1 first and second SH3 domains (Figures 2 and 3). This raises the possibility of several Sla1 molecules bridging two or more Las17 molecules within the complex. Additionally, we have recently shown that Sla1 can self-oligomerize through SHD2-SHD2 interactions and that homooligomerization occurs in vivo and appears to be mechanistically linked to clathrin binding at the membrane (Di Pietro et al., 2010). Such Sla1 oligomers may represent the small residual peak around 10 ml elution volume in gel filtration analysis of cytosol from the las17∆ strain (Figure 1F). Therefore Sla1 has the ability to bring multiple Las17 molecules together at endocytic sites and, at least in part, link multimerization to association with the clathrin lattice. This is important in light of the new finding that the activity of NPFs increases ∼100 times when they are in dimeric form compared with monomeric molecules (Padrick et al., 2008; Padrick and Rosen, 2010). This has been elegantly explained at the mechanistic level by the very recent discovery that the Arp2/3 complex has two different binding sites for the VCA motif (Padrick et al., 2011; Ti et al., 2011). Consequently a dimer or multimer of Las17 within the complex with Sla1, when activated (see below), would have significantly more potency with respect to the Arp2/3 complex, due to a cooperativity effect. In fact, it has been predicted that binding of two or more WASp molecules to an SH3 protein scaffold/adaptor could bring them together in cells to mediate such stronger activation (Padrick et al., 2008; Padrick and Rosen, 2010). The Las17-Sla1 complex uncovered here is likely a very good example of such proposed mechanism.
This work constitutes an important step forward in understanding the mechanism of actin polymerization regulation in endocytosis but also raises several important questions. For instance, how is Las17 inhibition accomplished at the molecular level by the Sla1 SH3 domains? There are at least two conceivable scenarios to explain these results. One possibility is that the carboxy-terminal WH2 and acidic tail (or VCA motif) may fold back and interact with a surface of the Sla1 SH3 domains 1 and/or 2 that is not involved in binding to Las17 polyproline motifs P10, P11, and/or P12. In this way, the WH2/acidic tail would not be available to bind monomeric actin and/or the Arp2/3 complex and would therefore be inhibited. Another possibility involves a second monomeric actin-binding site—in addition to the WH2 domain—recently described in Las17 that may in part explain the very strong Las17 activity compared with other yeast NPFs (Robertson et al., 2009). This new monomeric actin-binding site was not fully mapped but is located within residues 300 and 422 of Las17. This region includes the Sla1-binding site, P10-12 (Figures 2 and 3), thus opening up the possibility of mutually exclusive binding between one or both Sla1 SH3 domains and monomeric actin. Results presented here indicate monomeric actin and the Sla1 SH3 domains indeed compete for binding to a region of Las17 spanning residues 300–404 (P8-12; Figure 5). Although it is unlikely that monomeric actin binds a PxxP motif as the Sla1 SH3 domains do, it could bind partially overlapping regions of Las17. For better delineation of this mechanism, it will be important to define which residues within the 300–404 fragment are involved in monomeric actin binding. These two possible mechanisms for Sla1 inhibition of Las17 NPF activity are not mutually exclusive, and there may be additional regulatory contacts with full-length Sla1 in the context of the native complex. This result also raises the interesting possibility that other Las17 inhibitors, such as Syp1, could engage the WH2–acidic tail motif, while Sla1 operates on this new actin-binding site to produce more complete inhibition. The fact that Syp1 and Sla1 have an additive inhibitory effect on Las17 to produce nearly complete inhibition, at least in vitro, supports this possibility (Figure 9B).
Results presented here demonstrate Las17 arrives together with Sla1 in a complex that keeps its NPF activity inhibited during the initial ∼20 s. However, Las17 NPF activity must be released later to allow internalization to proceed. This could be achieved in two possible ways: by disassembly of the complex or by rearrangement without disassembly, such that the newly discovered actin-binding site (and the WH2-acidic tail) are free to exert their NPF activity. The first possibility indicates Las17 would cycle between Sla1-bound and Sla1-free states. Our gel filtration analysis of endogenous (untagged) Las17 in wild-type cells found only the large species that is in complex with Sla1, but no free Las17 (Figure 1). The most likely explanation for this result is that Las17 is always in complex with Sla1. In this respect, Las17 would be similar to mammalian and Drosophila WAVE, which is part of a constitutive WAVE regulatory complex that keeps WAVE inhibited and, upon activation by Rac1, releases its VCA motif without complex dissociation (Ismail et al., 2009). We cannot, however, exclude the possibility that activation of Las17 involves complex dissociation, but reassociation is so quick that the free Las17 fraction at steady state is too small to be detected.
One unresolved aspect of Las17 function has been its localization during the motile phase of endocytosis. Does it stay at the plasma membrane or does it move with the invaginating coat? This detail is of fundamental importance because the former possibility indicates that Las17 mediates actin polymerization with barbed ends oriented toward the plasma membrane, but the latter implies polymerization with barbed ends pointing to the invaginating vesicle (Takenawa and Suetsugu, 2007; Galletta and Cooper, 2009). In early studies, GFP-tagged Las17 appeared to stay at the plasma membrane (Kaksonen et al., 2003, 2005). More recent studies found that GFP-tagged Las17 does indeed move inward and internalizes with the incipient vesicle (Galletta et al., 2008; Galletta and Cooper, 2009). Immunoelectron microscopy data of epitope-tagged Las17 were also interpreted as indicating Las17 moves from the plasma membrane, although not quite at the tip of the invagination (Idrissi et al., 2008). The emerging picture from these later studies supports an Arp2/3-actin meshwork with barbed ends oriented toward the vesicle. Such an arrangement of actin filaments around clathrin-coated endocytic structures was recently observed in mouse cells, using platinum replica electron microscopy in combination with electron tomography (Collins et al., 2011). Given the high similarity of the endocytic machinery among eukaryotes, this orientation is expected to be conserved in yeast cells. Because Sla1 is a coat component that internalizes with the invagination, our data of a biochemically stable Sla1-Las17 complex are more in line with the idea that Las17 internalizes along with the incipient vesicle. However, as described above, we cannot exclude a very rapid dissociation and reassociation between Las17 and Sla1 that would be compatible with Las17 remaining at the plasma membrane.
In summary, this work demonstrates that Las17 is associated in a large and biochemically stable complex with Sla1 that is important for normal Las17 recruitment and regulation of endocytosis. Because this complex between Sla1 and Las17 regulates actin polymerization during clathrin-mediated endocytosis, we propose to name it “SLAC.” Specific SH3-polyproline interactions hold SLAC together and are responsible for inhibition of Las17 NPF activity during initial recruitment to sites of endocytosis. Las17 inhibition is caused, at least in part, by Sla1 SH3 domains masking a novel actin-binding site in Las17. A new type of polyproline motif within Las17 that simultaneously conforms to class I and class II was uncovered and mapped. The multivalent, direct, and high-affinity binding supports both a stable and large SLAC complex in which multiple copies of Las17 and Sla1 are likely present. Activation is not expected to involve complex dissociation and thus likely maintains multiple Las17 molecules together, providing strong Arp2/3 activation through cooperative binding. Release of Las17 NPF activity is essential for endocytosis to proceed, and the data presented here suggest activation occurs through SLAC rearrangement, rather than through dissociation. In the future, it will be interesting to investigate whether SH3 proteins arriving later to the patch, such as Bzz1, are responsible for the release of Sla1 inhibition in vivo, as was observed in in vitro experiments (Sun et al., 2006).
MATERIALS AND METHODS
Plasmids and yeast strains
For generation of recombinant GST- or polyhistidine-tagged (His) fusion proteins, the corresponding DNA sequence fragments of LAS17, SLA1, or SYP1 were amplified by PCR and cloned into pGEX-5X-1 (Amersham Biosciences, Piscataway, NJ) and/or pET-30a+ (Novagen, Madison, WI) bacterial expression vectors, respectively. For yeast two-hybrid experiments the corresponding LAS17 and SLA1 DNA fragments were amplified by PCR and cloned into pGBT9 or pGAD424 vector, respectively (Clontech–BD Biosciences, San Jose, CA). A DNA fragment containing 200 base pairs of the LAS17 5′ untranslated region followed by full-length LAS17 open reading frame was amplified by PCR and cloned into NotI/SalI sites of pRS-315 (Sikorski and Hieter, 1989; pSDP320; Supplemental Table S1). pSDP320 was used as a template for various proline-to-alanine site-directed mutageneses using the QuikChange system (Stratagene, La|Jolla, CA). For actin polymerization assays, full-length, wild-type LAS17 was amplified from pSDP320 by PCR and cloned into pGEX-5X-1, and a sequence encoding amino acids AAAHHHHHH was added to the reverse primer used in the PCR so that the protein contained a carboxy-terminal His tag for purification purposes (pSDP438). Similar full-length LAS17 constructs containing mutations in polyproline motifs 10, 11, and 12 (MP10-12-LAS17; pSDP439) and polyproline motifs 8, 9, 10, 11, and 12 (MP8-12-LAS17; pSDP440) were generated. Table S1 shows the plasmids generated in this study.
AH109 cells (Clontech–BD Biosciences) were cotransformed with the respective pGBT9 and pGAD424 construct combinations and were analyzed as previously described (Starcevic and Dell'Angelica, 2004; Mahadev et al., 2007).
SDY088 (MATa ura3-52 leu2-3112 his3-∆200 trp1-∆901 lys2-801 suc2-∆9) was crossed with SDY169 (MATα ura3-52 leu2-3112 his3-∆200 trp1-∆901 lys2-801 suc2-∆9 LAS17-GFP::HIS3) and the resulting heterozygous diploid Las17-GFP strain, SDY392 (MATa/MATα ura3-52 leu2-3112 his3-∆200 trp1-∆901 lys2-801 suc2-∆9 LAS17-GFP::HIS3) was used to introduce mutations in the LAS17-GFP gene while maintaining an intact LAS17 gene. A two-step approach similar to the one described previously was followed (Di Pietro et al., 2010; Feliciano et al., 2011). For the first step, a sequence upstream of the central Las17 polyproline stretch (−200 to + 570) was amplified by PCR (the reverse primer contained XbaI and BamHI restriction sites in series) and cloned into the NotI/BamHI sites of pRS-315. Subsequently, a PCR fragment containing URA3 was amplified using a reverse primer containing BglII and BamHI restriction sites in series and was subcloned into the XbaI/BamHI sites. An additional fragment containing the LAS17 sequence downstream of the central polyproline stretch (nucleotides 1441–1899) followed by the GFP sequence was amplified by PCR using genomic DNA from SDY169 and subcloned into BglII/SalI sites of the above construct. The resulting construct was cleaved with NotI/SalI, and the URA3 fragment was introduced by lithium acetate transformation (Ito et al., 1983) into SDY392 to generate SDY362, in which the central Las17 polyproline stretch was replaced by URA3 in the LAS17-GFP gene, leaving the rest of the LAS17 gene intact. In the second step, NotI/SalI fragments from pSDP320 and pSDP375 were cotransformed with pRS-314 (TRP1; Sikorski and Hieter, 1989) into SDY362. Colonies that grew in plates lacking tryptophan were replica-plated onto agar medium containing 5-fluorotic acid to identify cells in which the wild-type or mutant sequences replaced URA3. These diploid cells were then subjected to sporulation and tetrad dissection, thus generating strains SDY367 (LAS17-GFP) and SDY363 (Las17MP8-12-GFP), respectively.
All other yeast strains generated are described in the Supplemental Material.
Antibodies
The generation and characterization of antibodies against Las17 were carried out as described previously (Di Pietro et al., 2004). The polyclonal antibody Las-A was raised in rabbit by immunizing with recombinant GST-Las17 (433–633) protein, and the polyclonal antibody Las-B was raised in rat by immunizing with recombinant GST-Las17 (1–151) protein. Subsequently, these antibodies were affinity-purified using as a ligand recombinant His-Las17 (433–633; Las-A) and GST-Las17 (1–151; Las-B) that had been covalently coupled to Affi-Gel 15 beads (Bio-Rad, Hercules, CA). Las-B was adsorbed using immobilized GST protein.
Biochemical methods
Cytosolic extracts were obtained as follows: liquid nitrogen frozen yeast pellets were ground in a blender, resuspended in phosphate-buffered saline (PBS: 12 mM phosphate, 147 mM NaCl, 3 mM KCl, pH 7.35) or buffer A (20 mM HEPES, 150 mM NaCl, 1 mM ethylene glycol tetraacetic acid (EGTA), 1 mM dithiothreitol, pH 7.4) supplemented with protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO), and subjected to ultracentrifugation at 4°C for 15 min at 400,000 × g, as previously described (Feliciano et al., 2011). The following yeast strains were used: TVY614 (MATa ura3-52 leu2-3112 his3-∆200 trp1-∆901 lys2-801 suc2-∆9 pep4::LEU2 prb1::HISG prc1::HIS3; Vida and Emr, 1995), GPY3130 (MATa ura3-52 leu2-3112 his3-∆200 trp1-∆901 suc2-∆9 sla1∆::URA3; Di Pietro et al., 2010), SDY159 (MATa his3-∆1 leu2-∆0 ura3-∆0), and SDY161 (MATa his3-∆1 leu2-∆0 ura3-∆0 Las17∆::KanMX4).
Coimmunoprecipitation analysis was performed as previously described using cytosolic extracts prepared in PBS (Di Pietro et al., 2010; Feliciano et al., 2011). GST-fusion protein affinity assays were performed in PBS containing 0.5% TX100, as previously described (Mahadev et al., 2007; Di Pietro et al., 2010; Feliciano et al., 2011). Quantitative GST-fusion protein affinity assays were performed as previously described (Sirotkin et al., 2005). Briefly, recombinant His-Sla1-SH3-1-2 (150 nM) was incubated with increasing concentrations of wild-type GST-Las17-P8-12 or GST-Las17-P8-12-MP10-12 bound to glutathione-Sepharose beads for 1 h at 4°C. Unbound His-Sla1-SH3-1-2 was recovered in the supernatant, and protein concentration was determined by gel densitometry.
Size-exclusion chromatography fractionation of cytosolic extracts was carried out with a Superose 6 column (10 × 300 mm; Amersham Biosciences) connected to a fast protein liquid chromatography system (Amersham Biosciences) and equilibrated with buffer A. Elution was performed at a flow rate of 0.4 ml/min at 4°C. Fractions (0.750 ml) were collected and analyzed by immunoblotting or immunoprecipitation–immunoblotting using antibodies to Las17, Sla1, and End3. The column was calibrated using blue dextran and the following standard proteins of known Stokes radii: bovine thyroglobulin (85 Å), γ-globulin (55 Å), ovalbumin (28 Å), myoglobin (19 Å), vitamin B12 (Å) (Bio-Rad). Sedimentation velocity analysis of cytosolic extracts (0.2 ml) was performed using linear 5–20% (wt/vol) sucrose gradients prepared in a Gradient Master (BioComp Instruments, Fredericton, New Brunswick, Canada) in buffer A (total volume: 12 ml). The samples were layered on top of the gradients and centrifuged in a SW41 rotor (Beckman Coulter, Fullerton, CA) at 39,000 rpm (261,000 × g) for 18 h at 4°C. Fractions (0.75 ml) were collected from the top of the tube using a density gradient fractionator and analyzed by immunoblotting. The following protein standards of known sedimentation coefficient were analyzed in parallel: bovine catalase (11.3 S), bovine serum albumin (4.6 S), ovalbumin (3.6 S), and cytochrome C (1.5 S) (Sigma-Aldrich).
Protein purification
GST- and His-fusion proteins were expressed in Escherichia coli (BL21) and affinity purified using glutathione–Sepharose-4B or the TALON cobalt affinity resin (Clontech–BD Biosciences), as previously described (Falcon-Perez et al., 2002). Full-length, wild-type Las17, Las17-MP10-12, and Las17-MP8-12 were expressed in E. coli (BL21) and subjected to a sequential affinity purification using glutathione–Sepharose-4B (GST-tag at amino-terminus), which was followed by TALON cobalt affinity resin (His-tag at carboxy-terminus).
Actin polymerization assay
Actin polymerization experiments were performed with 1.5 μM rabbit actin (99% pyrene-labeled) and 75 nM bovine Arp2/3 complex (Cytoskeleton, Denver, CO). Pyrene-actin (0.4 mg/ml) was incubated in G buffer (10 mM Tris, pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP) on ice for 1.5 h to promote depolymerization and subsequently centrifuged at 100,000 rpm in a TLA100.3 rotor (Beckman Coulter) at 4°C for 1.5 h. Pyrene-actin was incubated in exchange buffer (1 mM EGTA, 0.1 mM MgCl2) for 10 min and added to a 96-well plate already containing the proteins of interest (75 nM wild-type or mutant Las17, and variable concentrations of His-Sla1-SH3-1-2 in Figure 4A or 150 nM in Figure 4, C–E). In Figure 9B, His-Sla1-SH3-1-2 was used at 1.3 μM and His-Syp1 at was used at 10 μM. For G-actin sequestering studies (Figure 5), 3 μM rabbit actin (99% pyrene-labeled) was used in all experiments, along with 6 μM Las17-P8-12 fragment and 2 or 12 μM Sla1 SH3-1-2, and no Arp2/3 complex was added to the reactions. Polymerization buffer (50 mM KCl, 2 mM MgCl2, 1 mM ATP) was added to the reaction (t = 0) and actin polymerization was measured over time using a Victor3 V microplate reader (Perkin Elmer-Cetus, Waltham, MA) with excitation and emission wavelengths of 365 nm and 406 nm, respectively. Actin polymerization rates were calculated from the slope of the linear portion of assembly curves (25–50% polymerization).
Fluorescence microscopy
Fluorescence microscopy was performed as previously described (Feliciano et al., 2011) using an Olympus IX81 spinning-disk confocal microscope with Photometrics Cascade II camera (Tucson, AZ) and a 100×/1.40 numerical aperture objective. Time-lapse movies were generated by collecting 120 or 150 images at 1 frame/s at room temperature using cells grown until early log phase. For quantifying the number of patches, patch lifetimes, and patch intensities, Slidebook 5 software (Intelligent Imaging Innovations, Denver, CO) was used to generate a mask, covering endocytic spots or regions inside the cell. KaleidaGraph (Synergy Sofware, Reading, PA) was used for photobleaching corrections and patch-lifetime determination for each GFP- and RFP-tagged protein by integrating the intensities of each consecutive time point and establishing the minimum and maximum value of the integral. Delta times (time that takes for Abp1-RFP to appear after Las17-GFP detection) were calculated by subtraction of Abp1-RFP first recruitment time point (minimum value of the Abp1-RFP integral) to Las17-GFP first recruitment time point (minimum value of the Las17-GFP integral) within the same time lapse. Lucifer yellow uptake experiments were performed as previously described (Duncan et al., 2001). Cells were incubated for 2 h at 24°C with lucifer yellow (Invitrogen, Carlsbad, CA), and endocytic uptake was analyzed by fluorescence microscopy. The fluorescence intensity of the vacuole was measured and normalized by the intensity of the background. Quantification of Wsc1-GFP localization was done as previously described (Di Pietro et al., 2010). Student's t test was used to determine statistical significance.
Mass spectrometry analysis
TAP of Las17 was performed using cells expressing C-terminally TAP-tagged Las17 (Open Biosystems, Huntsville, AL) and untagged control cells, as previously described (Rigaut et al., 1999). The samples were concentrated in a preparative polyacrylamide gel and digested in-gel with trypsin. Mass spectrometry was performed at the Colorado State University Proteomics and Metabolomics facility. Peptides were purified and concentrated using an on-line enrichment column (Zorbax C18, 5 μm, 5 mm × 0.3 mm; Agilent, Santa Clara, CA). Subsequent chromatographic separation was performed on a reverse-phase nanospray column (1100 nanoHPLC, Zorbax C18, 5 μm, 75-μm inner diameter × 150-mm column; Agilent) using a 42-min linear gradient from 25 to 55% buffer B (90% acetonitrile, 0.1% formic acid) at a flow rate of 300 nl/min. Peptides were eluted directly into the mass spectrometer (LTQ linear ion trap; Thermo Fisher Scientific, Lafayette, CO), and spectra were collected over an m/z range of 200–2000 Da using a dynamic exclusion limit of two tandem mass spectrometry spectra of a given peptide mass for 30 s (exclusion duration of 90 s). Compound lists of the resulting spectra were generated using BioWorks version 3.0 software (Thermo Fisher Scientific) with an intensity threshold of 5000 and 1 scan/group. Tandem mass spectrometry spectra were searched against the appropriate protein database (baker's yeast; National Center for Biotechnology Information) using the Mascot database search engine (version 2.3). Peptides were validated using Scaffold 3 file (version 3.3.1) with minimum 99% protein, 95% peptide, and minimum 2% peptide thresholds.
Supplementary Material
Acknowledgments
We thank Olve Peersen for help with the microplate reader and KaleidaGraph analysis of actin polymerization curves and patch-lifetime determinations. We are grateful to Michael Hughes for helping with protein purification and to other members of the Di Pietro lab for helpful discussions. D. F. acknowledges an NSF Bridges to the Doctorate fellowship and an American Heart Association predoctoral fellowship. The microscope used in this work is supported in part by the Microscope Imaging Network core infrastructure grant from Colorado State University. This work was supported by NSF grant 1052188 to S.M.D.P.
Abbreviations used:
- 3-AT
3-amino-1,2,4-triazole
- CME
clathrin-mediated endocytosis
- EGTA
ethylene glycol tetraacetic acid
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- NPF
nucleation-promoting factor
- PBS
phosphate-buffered saline
- RFP
red fluorescent protein
- TAP
tandem-affinity purification
- WASp
Wiskott-Aldrich syndrome protein
- WAVE
WASP family verprolin-homologous protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-12-1022) on September 12, 2012.
REFERENCES
- Ayscough KR, Eby JJ, Lila T, Dewar H, Kozminski KG, Drubin DG. Sla1p is a functionally modular component of the yeast cortical actin cytoskeleton required for correct localization of both Rho1p-GTPase and Sla2p, a protein with talin homology. Mol Biol Cell. 1999;10:1061–1075. doi: 10.1091/mbc.10.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner DR, D'Agostino JL, Torres OT, Daugherty-Clarke K, Uygur A, Reider A, Wendland B, Lemmon SK, Goode BL. The F-BAR protein Syp1 negatively regulates WASp-Arp2/3 complex activity during endocytic patch formation. Curr Biol. 2009;19:1979–1987. doi: 10.1016/j.cub.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant S, Kural C, Zeeh JC, Ubelmann F, Kirchhausen T. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat Cell Biol. 2011;13:1124–1131. doi: 10.1038/ncb2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, Warrington A, Taylor KA, Svitkina T. Structural organization of the actin cytoskeleton at sites of clathrin-mediated endocytosis. Curr Biol. 2011;21:1167–1175. doi: 10.1016/j.cub.2011.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R, Ayscough KR. Interactions between Sla1p, Lsb5p and Arf3p in yeast endocytosis. Biochem Soc Trans. 2005;33:1273–1275. doi: 10.1042/BST0331273. [DOI] [PubMed] [Google Scholar]
- Di Pietro SM, Cascio D, Feliciano D, Bowie JU, Payne GS. Regulation of clathrin adaptor function in endocytosis: novel role for the SAM domain. EMBO J. 2010;29:1033–1044. doi: 10.1038/emboj.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro SM, Falcon-Perez JM, Dell'Angelica EC. Characterization of BLOC-2, a complex containing the Hermansky-Pudlak syndrome proteins HPS3, HPS5 and HPS6. Traffic. 2004;5:276–283. doi: 10.1111/j.1600-0854.2004.0171.x. [DOI] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Duncan MC, Cope MJ, Goode BL, Wendland B, Drubin DG. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- Falcon-Perez JM, Starcevic M, Gautam R, Dell'Angelica EC. BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J Biol Chem. 2002;277:28191–28199. doi: 10.1074/jbc.M204011200. [DOI] [PubMed] [Google Scholar]
- Feliciano D, Bultema JJ, Ambrosio AL, Di Pietro SM. In vivo and in vitro studies of adaptor-clathrin interaction. J Vis Exp. 2011;47:e2352. doi: 10.3791/2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta BJ, Chuang DY, Cooper JA. Distinct roles for Arp2/3 regulators in actin assembly and endocytosis. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.0060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta BJ, Cooper JA. Actin and endocytosis: mechanisms and phylogeny. Curr Opin Cell Biol. 2009;21:20–27. doi: 10.1016/j.ceb.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Rodal AA, Barnes G, Drubin DG. Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J Cell Biol. 2001;153:627–634. doi: 10.1083/jcb.153.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DA, Yang S, Drubin DG. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J Cell Biol. 1993;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard JP, Hutton JL, Olson JM, Payne GS. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J Cell Biol. 2002;157:315–326. doi: 10.1083/jcb.200110027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrissi FZ, Grotsch H, Fernandez-Golbano IM, Presciatto-Baschong C, Riezman H, Geli MI. Distinct acto/myosin-I structures associate with endocytic profiles at the plasma membrane. J Cell Biol. 2008;180:1219–1232. doi: 10.1083/jcb.200708060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AM, Padrick SB, Chen B, Umetani J, Rosen MK. The WAVE regulatory complex is inhibited. Nat Struct Mol Biol. 2009;16:561–563. doi: 10.1038/nsmb.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Li R. Bee1, a yeast protein with homology to Wiscott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J Cell Biol. 1997;136:649–658. doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadev RK, Di Pietro SM, Olson JM, Piao HL, Payne GS, Overduin M. Structure of Sla1p homology domain 1 and interaction with the NPFxD endocytic internalization motif. EMBO J. 2007;26:1963–1971. doi: 10.1038/sj.emboj.7601646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- Michelot A, Costanzo M, Sarkeshik A, Boone C, Yates JR, III, Drubin DG. Reconstitution and protein composition analysis of endocytic actin patches. Curr Biol. 2010;20:1890–1899. doi: 10.1016/j.cub.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, et al. Hierarchical regulation of WASP/WAVE proteins. Mol Cell. 2008;32:426–438. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA. 2011;108:E472–479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Rosen MK. Physical mechanisms of signal integration by WASP family proteins. Annu Rev Biochem. 2010;79:707–735. doi: 10.1146/annurev.biochem.77.060407.135452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao HL, Machado IM, Payne GS. NPFXD-mediated endocytosis is required for polarity and function of a yeast cell wall stress sensor. Mol Biol Cell. 2007;18:57–65. doi: 10.1091/mbc.E06-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Baez L, Hurley JH, Traub LM, Wendland B. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 2009;28:3103–3116. doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Robertson AS, Allwood EG, Smith AP, Gardiner FC, Costa R, Winder SJ, Ayscough KR. The WASP homologue Las17 activates the novel actin-regulatory activity of Ysc84 to promote endocytosis in yeast. Mol Biol Cell. 2009;20:1618–1628. doi: 10.1091/mbc.E08-09-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Manning AL, Goode BL, Drubin DG. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr Biol. 2003;13:1000–1008. doi: 10.1016/s0960-9822(03)00383-x. [DOI] [PubMed] [Google Scholar]
- Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of macromolecules in impure systems: aggregation of urease. Biochem Biophys Res Commun. 1965;19:494–499. doi: 10.1016/0006-291x(65)90152-x. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotkin V, Beltzner CC, Marchand JB, Pollard TD. Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast. J Cell Biol. 2005;170:637–648. doi: 10.1083/jcb.200502053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic M, Dell'Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- Stimpson HE, Toret CP, Cheng AT, Pauly BS, Drubin DG. Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast. Mol Biol Cell. 2009;20:4640–4651. doi: 10.1091/mbc.E09-05-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Martin AC, Drubin DG. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Tang HY, Xu J, Cai M. Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol Cell Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti SC, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci USA. 2011;108:E463–E471. doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, et al. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 2002;295:321–324. doi: 10.1126/science.1064987. [DOI] [PubMed] [Google Scholar]
- Tonikian R, et al. Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 2009;7:e1000218. doi: 10.1371/journal.pbio.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshima JY, Toshima J, Kaksonen M, Martin AC, King DS, Drubin DG. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent α-factor derivatives. Proc Natl Acad Sci USA. 2006;103:5793–5798. doi: 10.1073/pnas.0601042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshima J, Toshima JY, Duncan MC, Cope MJ, Sun Y, Martin AC, Anderson S, Yates JR, III, Mizuno K, Drubin DG. Negative regulation of yeast Eps15-like Arp2/3 complex activator, Pan1p, by the Hip1R-related protein, Sla2p, during endocytosis. Mol Biol Cell. 2007;18:658–668. doi: 10.1091/mbc.E06-09-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat Rev Mol Cell Biol. 2009;10:583–596. doi: 10.1038/nrm2751. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren DT, Andrews PD, Gourlay CW, Ayscough KR. Sla1p couples the yeast endocytic machinery to proteins regulating actin dynamics. J Cell Sci. 2002;115:1703–1715. doi: 10.1242/jcs.115.8.1703. [DOI] [PubMed] [Google Scholar]
- Weinberg J, Drubin DG. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 2012;22:1–13. doi: 10.1016/j.tcb.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.