The retrograde (RTG) pathway transcription factors Rtg1 and Rtg3 are shown to be targets of the Hog1 stress-activated protein kinase (SAPK). Hog1 acts on the RTG complex at multiple levels to mediate gene expression upon stress. The SAPK is required for the nuclear accumulation of the complex, the recruitment of the complex at RTG-responsive promoters, and the regulation of Rtg3 transcriptional activity.
Abstract
Cells modulate expression of nuclear genes in response to alterations in mitochondrial function, a response termed retrograde (RTG) regulation. In budding yeast, the RTG pathway relies on Rtg1 and Rtg3 basic helix-loop-helix leucine Zipper transcription factors. Exposure of yeast to external hyperosmolarity activates the Hog1 stress-activated protein kinase (SAPK), which is a key player in the regulation of gene expression upon stress. Several transcription factors, including Sko1, Hot1, the redundant Msn2 and Msn4, and Smp1, have been shown to be directly controlled by the Hog1 SAPK. The mechanisms by which Hog1 regulates their activity differ from one to another. In this paper, we show that Rtg1 and Rtg3 transcription factors are new targets of the Hog1 SAPK. In response to osmostress, RTG-dependent genes are induced in a Hog1-dependent manner, and Hog1 is required for Rtg1/3 complex nuclear accumulation. In addition, Hog1 activity regulates Rtg1/3 binding to chromatin and transcriptional activity. Therefore Hog1 modulates Rtg1/3 complex activity by multiple mechanisms in response to stress. Overall our data suggest that Hog1, through activation of the RTG pathway, contributes to ensure mitochondrial function as part of the Hog1-mediated osmoadaptive response.
INTRODUCTION
Cells can monitor and respond to changes in the state of mitochondria by inducing changes in the expression of nuclear genes. This response is mediated by the retrograde (RTG) signaling pathway (Parikh et al., 1987; Liao and Butow, 1993; Butow and Avadhani, 2004; Srinivasan et al., 2010). In yeast, the RTG signaling pathway functions as a homeostatic or stress response mechanism to adjust various biosynthetic and metabolic activities when mitochondrial dysfunction occurs (Liao et al., 1991; Small et al., 1995; Liu and Butow, 1999). The key transcriptional activators of the RTG pathway are the Rtg1 and Rtg3 basic helix-loop-helix leucine Zipper transcription factors, which activate transcription by binding as a heterodimer to the consensus sequence GTCAC, which is referred to as the R-box (Liao and Butow, 1993; Jia et al., 1997). Rtg1 and Rtg3 function as a heterodimer, because neither protein alone is able to bind to R-box sites (Jia et al., 1997). Although transcription activation requires both Rtg1 and Rtg3, only Rtg3 has been shown to contain transcriptional activation domains (Rothermel et al., 1997). In respiratory-competent cells, Rtg1 and Rtg3 exist as a complex largely in the cytoplasm, in contrast with cells with mitochondrial dysfunction (e.g., cells without mitochondrial DNA), in which they exist as a complex predominantly localized in the nucleus (Sekito et al., 2000; Ferreira Júnior et al., 2005). Another member of the RTG pathway is Rtg2, a cytoplasmic protein that may act as a proximal sensor of mitochondrial dysfunction, being required for Rtg1/3 complex nuclear accumulation and subsequent activation of gene expression (Rothermel et al., 1997; Liu and Butow, 2006). Remarkably, the RTG pathway is known to be negatively regulated by the target of rapamycin (TOR) pathway (Wullschleger et al., 2006). Nuclear accumulation of Rtg1/3, as well as expression of RTG target genes, is induced by addition of rapamycin (Komeili et al., 2000).
Exposure of yeast cells to increases in external osmolarity activates the Hog1 stress-activated protein kinase (SAPK), which is essential for the induction of diverse osmoadaptive responses (Hohmann, 2002; Hohmann et al., 2007; O'Rourke et al., 2002; Westfall et al., 2004). One of the main functions of activated Hog1 is the regulation of gene expression (Martinez-Montanes et al., 2010; Weake and Workman, 2010; de Nadal and Posas, 2010; de Nadal et al., 2011). Genome-wide transcriptional analyses showed that a large number of genes are regulated by osmostress in a Hog1-dependent manner, including genes that encode proteins implicated in carbohydrate metabolism, general stress protection, protein production, and signal transduction (Posas et al., 2000; Rep et al., 2000; Causton et al., 2001; Capaldi et al., 2008; Miller et al., 2011). One mechanism by which Hog1 modulates gene expression is the direct regulation of transcription factors. For instance, it has been proposed that Sko1, Hot1, the redundant Msn2 and Msn4, and Smp1 are controlled by the Hog1 SAPK (Rep et al., 1999, 2000; Proft et al., 2001; de Nadal et al., 2003). These factors are unrelated, and the mechanisms by which Hog1 regulates their function differ from one to another. Sko1 is an ATF/CREB factor that represses gene expression under nonstress conditions by the recruitment of the general corepressor complex Cyc8-Tup1. In response to osmostress, Hog1 phosphorylates Sko1, switching its activity from a repressing to an activating state, which involves the recruitment of the SWI/SNF and SAGA complexes (Proft et al., 2001; Proft and Struhl, 2002; Guha et al., 2007; Kobayashi et al., 2008). Hot1 physically interacts with Hog1, and its binding to DNA is regulated by Hog1 kinase activity. Moreover, Hog1 mediates RNA polymerase II (Pol II) recruitment at Hot1 target promoters (Rep et al., 1999; Alepuz et al., 2001, 2003). Msn2 and Msn4 are generic stress factors controlled by PKA and Hog1 (Rep et al., 2000; Alepuz et al., 2001). Finally, Smp1 is phosphorylated by Hog1 in response to osmostress, and its transactivation activity is dependent on phosphorylation by the SAPK (de Nadal et al., 2003). However, the transcription factors reported to be under the control of Hog1 cannot account for the regulation of all Hog1-dependent genes (Capaldi et al., 2008; Miller et al., 2011), which suggests that additional transcription factors under the control of the SAPK are required for gene expression in osmostress.
In this paper, we show that Hog1 controls Rtg1/3 transcription factor complex to induce RTG-dependent genes in response to osmostress. Hog1 interacts with the Rtg1/3 transcription complex in vivo. Remarkably, the SAPK is required for the nuclear accumulation of the complex, the recruitment of the complex at Rtg1/3-responsive promoters, and the regulation of Rtg3 transcriptional activity. Therefore the SAPK acts on the RTG complex at multiple levels to mediate gene expression. In addition, our data point out a close communication between mitochondria and nucleus via the RTG pathway to ensure mitochondrial function during adaptation to stress.
RESULTS
Rtg1 and Rtg3 transcription factors are required for cell survival upon osmostress
In an exhaustive genome-wide genetic screen searching for deletions that render cells osmosensitive, we identified several protein complexes involved in transcription, such as SAGA, Mediator RSC, and Ubp3 (Zapater et al., 2007; Mas et al., 2009; Sole et al., 2011). In addition, we found that mutations in RTG1 and RTG3 genes rendered cells unable to grow at high osmolarity. To characterize in more detail the phenotype of osmosensitivity of the rtg1 and rtg3 mutants, we individually spotted these mutant strains onto yeast–peptone–dextrose (YPD) plates with different concentrations of salt and sorbitol. Deletion of RTG1 and RTG3 genes affected cell growth at high osmolarity both in salt and sorbitol media, indicating that these transcription factors are important for adaptation to osmostress (Figure 1A).
FIGURE 1:
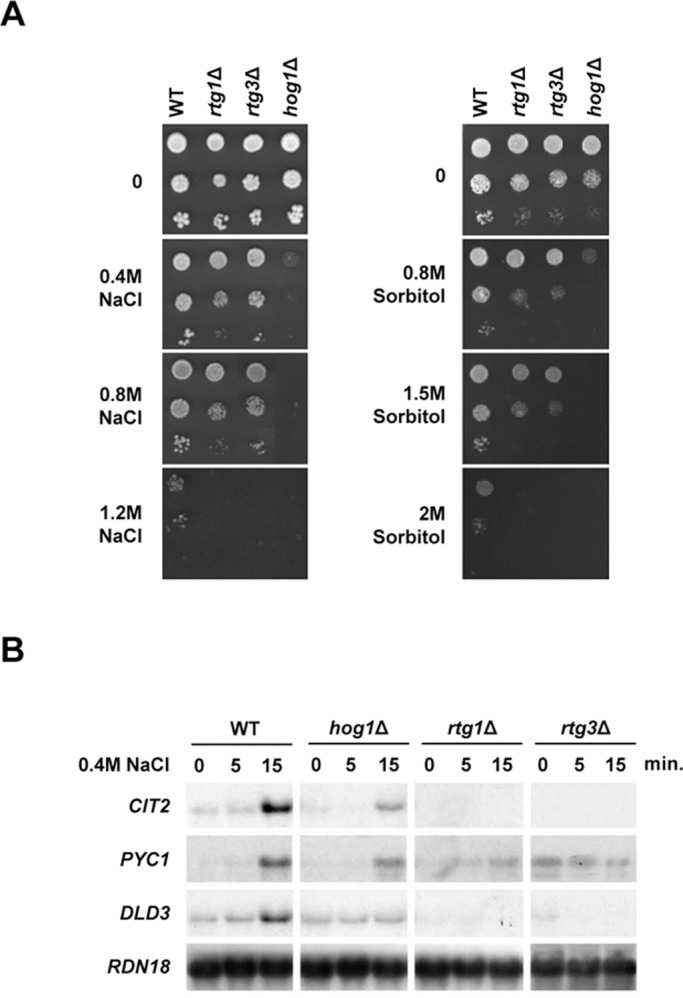
The Rtg1 and Rtg3 transcription factors are essential for adaptation to osmostress. (A) Mutations in RTG1 and RTG3 genes render cells osmosensitive. Wild-type and the indicated mutant strains were grown to logarithmic phase and diluted to 0.05 OD660. Tenfold serial dilution series were spotted on plates with or without NaCl or sorbitol at the indicated concentrations. Growth was scored after 2–5 d. (B) Rtg1, Rtg3, and the Hog1 SAPK are required for the induction of RTG target genes in response to osmostress. Indicated yeast strains were grown on MD-Gln and treated with 0.4 M NaCl at the indicated times. Total RNA was analyzed by Northern blot analysis for transcript levels of CIT2, PYC1, and DLD3, with RDN18 as a loading control.
RTG-dependent genes are induced in response to osmostress in a Hog1-dependent manner
The requirement of RTG1 and RTG3 for cell survival at high osmolarity prompted us to analyze the expression of genes known to be under the control of Rtg1 and Rtg3 transcription factors in response to osmostress. Activation of the RTG pathway is dependent on the source of assimilable nitrogen. In the presence of glutamine, Rtg1/3 transcription factors are retained in the cytoplasm and RTG-dependent genes are repressed (Liao and Butow, 1993; Liu and Butow, 1999; Komeili et al., 2000). Growth media for all the experiments was supplemented with glutamine to ensure the RTG pathway remained inactive and Rtg1/3 target genes were repressed (see Materials and Methods). Among the typical Rtg1/3 target genes are CIT2 (encoding a peroxisomal isoform of citrate synthase), PYC1 (encoding pyruvate carboxylase), and DLD3 (encoding a cytoplasmic isoform of d-lactate dehydrogenase). All of them show a strong increase in expression upon respiratory dysfunction (Liao et al., 1991; Chelstowska et al., 1999; Epstein et al., 2001). Notably, these genes were also strongly induced in response to osmostress, and their transcriptional activation was completely dependent on RTG1 and RTG3 transcription factors (Figure 1B). It is worth noting that the expression of CIT2 in YPD medium, in which the RTG pathway is already activated in the absence of stress, still showed an increase upon osmostress (Supplemental Figure S1). When cells suffer mitochondrial dysfunction, the cytoplasmic Rtg2 protein is required for activation of the retrograde response (Liao and Butow, 1993; Ferreira Júnior et al., 2005). Remarkably, under osmostress conditions, cells lacking RTG2 also displayed impaired transcription of the RTG-dependent genes (Figure S2A). These data indicate that the integrity of the RTG pathway is required for gene activation upon osmostress.
We then tested whether induction of the Rtg1/3 target genes in response to osmostress was dependent on Hog1. Expression of CIT2, PYC1, and DLD3 was strongly reduced upon stress in a hog1Δ strain when compared with wild-type (Figure 1B). Therefore Hog1 is required for activation of RTG-dependent genes upon osmostress. To test whether the role of Hog1 on RTG-dependent genes was restricted to osmostress, we analyzed RTG-dependent gene expression in response to rapamycin. Activation of RTG-responsive genes upon addition of rapamycin was not altered in a hog1Δ strain (Figure S2B). Thus the role of Hog1 in RTG-mediated gene expression is restricted to osmostress. Of note, induction of osmoresponsive genes regulated by transcription factors such as CTT1, STL1, and GRE2 was not affected in cells lacking RTG1 or RTG3 (Figure S2C). Therefore Rtg1 and Rtg3 transcription factors are required specifically for activation of a specific subset of osmoresponsive genes.
The Hog1 SAPK interacts with the Rtg1/3 complex in vivo
Hog1 is able to interact with some of the transcription factors regulated by the SAPK (de Nadal and Posas, 2010). To investigate the relationship between Hog1 and the Rtg1/3 transcription complex, we tested whether Hog1 was able to interact with Rtg1 and Rtg3. We performed glutathione S-transferase (GST) pulldown experiments in extracts of cells expressing GST-Hog1 and endogenously hemagglutinin (HA)-tagged RTG1 and RTG3. Hog1 was able to coprecipitate with both Rtg1 and Rtg3 proteins upon osmostress and under nonstressed conditions (Figure 2). Binding of Rtg1/3 to Hog1 was similar when protein extracts were treated with DNase I, suggesting that binding occurs in solution rather than through chromatin (Figure 2). Thus the Hog1 SAPK physically interacts with the Rtg1/3 complex, which provides biochemical evidence for the relationship between the SAPK and the RTG transcription complex.
FIGURE 2:
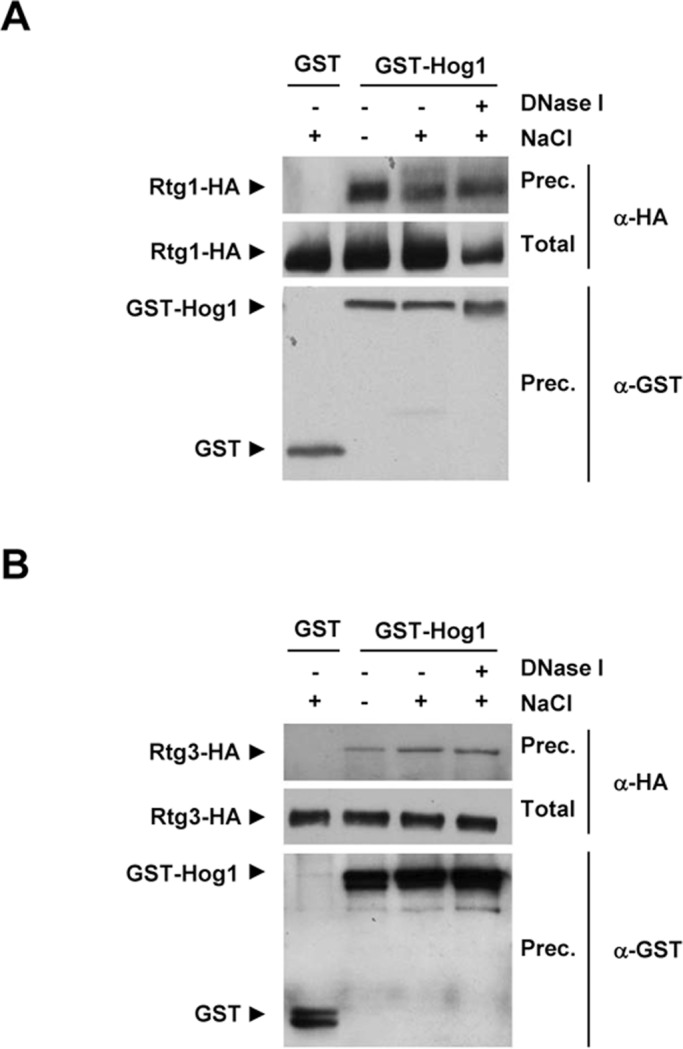
In vivo binding of Hog1 to Rtg1 and Rtg3. Strains expressing Rtg1-HA or Rtg3-HA from the endogenous locus were transformed with a plasmid expressing GST or GST-Hog1 under the PTEF1 promoter. Cells were grown in MD-Gln, and samples were taken before (−) and after (+)treatment with 0.4 M NaCl for 10 min. Protein extracts were treated or not with 200 U/ml of recombinant DNase I for 5 h at 4ºC. GST-Hog1 was pulled-down by glutathione-Sepharose 4B, and the presence of Rtg1-HA (A) and Rtg3-HA (B) was probed by immunoblotting using anti–HA specific monoclonal antibody (top). Total extracts are presented in the middle panel. The amount of precipitated GST proteins was detected using anti–GST specific monoclonal antibody (bottom).
The Rtg1/3 complex and Hog1 bind to RTG-dependent promoters interdependently
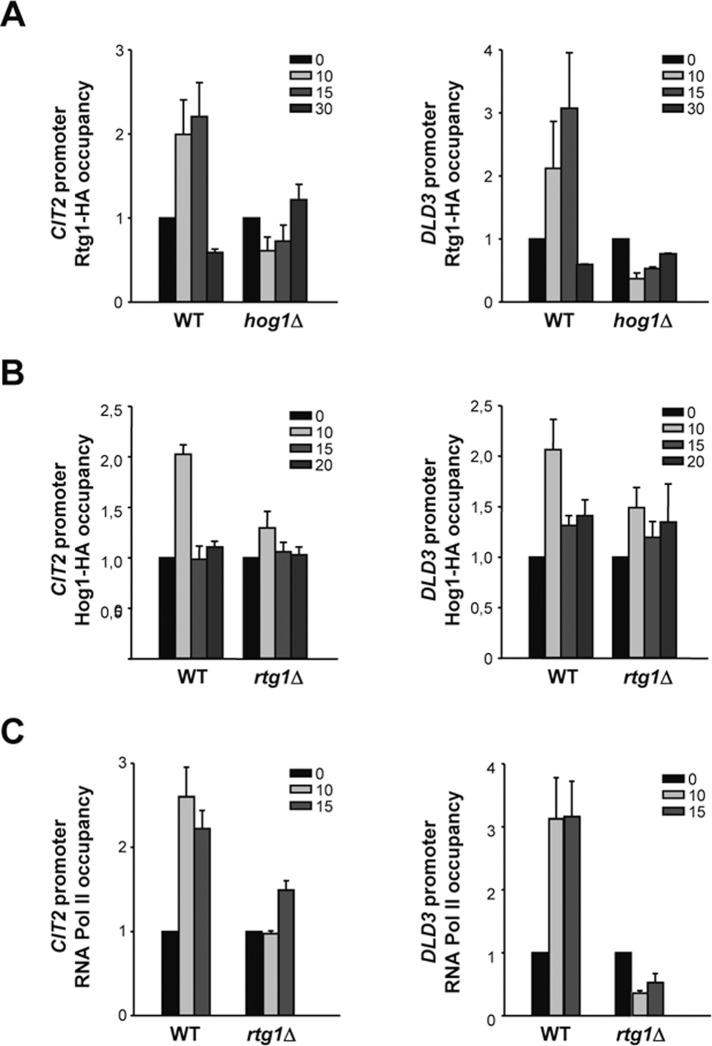
Osmoresponsive transcription factors bind to specific target promoters to regulate gene expression in response to osmostress and, in some cases, Hog1 is required to modulate their association to DNA (Alepuz et al., 2001, 2003). Therefore we asked whether the Rtg1/3 complex was recruited at the corresponding target promoters upon osmostress and whether this recruitment was dependent on Hog1. Rtg1 and Rtg3 form a heterodimer to bind to DNA, since neither protein alone is able to bind to a target R-box site (Jia et al., 1997). We therefore used chromatin immunoprecipitation (ChIP) of Rtg1-HA to follow the binding of the complex to CIT2 and DLD3 promoters upon osmostress. Rtg1 was recruited at CIT2 and DLD3 promoters in response to osmostress. However, binding of Rtg1 was abolished in a hog1Δ strain (Figure 3A). Therefore binding of the transcription complex to chromatin upon osmostress is dependent on the Hog1 SAPK.
FIGURE 3:
Occupancy of Rtg1, Hog1, and RNA Pol II at RTG-dependent promoters. (A) Binding of Rtg1/3 complex to CIT2 and DLD3 promoters in response to osmostress is dependent on Hog1. Wild-type and hog1Δ strains carrying HA-tagged Rtg1 were grown in MD-Gln and subjected to osmostress (0.4 M NaCl) for the indicated times. Proteins were immunoprecipitated with anti-HA monoclonal antibody, and binding to the promoter regions of CIT2 and DLD3 loci was analyzed by PCR. Results are shown as the fold induction of treated relative to nontreated (time zero) samples normalized to TEL2. Data represent mean ± SD of three independent experiments. (B) Recruitment of Hog1 to CIT2 and DLD3 promoters in response to osmostress is dependent on Rtg1. Wild-type and rtg1Δ strains were analyzed by ChIP as described for (A). (C) Rtg1 is required for recruitment of RNA Pol II at CIT2 and DLD3 promoters in response to osmostress. Wild-type and rtg1Δ strains were analyzed by ChIP as described for (A), using anti-Rpb1 antibody.
The Hog1 SAPK is targeted to specific osmostress-responsive genes upon stress through association of specific transcription factors (Alepuz et al., 2001; Pascual-Ahuir et al., 2006; Pokholok et al., 2006; Proft et al., 2006). We therefore asked whether Hog1 was recruited at the RTG-dependent promoters in response to stress and whether this recruitment was dependent on the Rtg1/3 transcription factor complex. By performing ChIP experiments, we followed binding of Hog1-HA to CIT2 and DLD3 promoters before and after osmostress in wild-type and rtg1Δ strains. Hog1 associated at these promoters in response to osmostress, and its recruitment was dependent on the presence of Rtg1 (Figure 3B). These data indicate that the Rtg1/3 complex is anchoring the SAPK at the RTG target promoters in response to osmostress.
Moreover, we assessed the presence of RNA Pol II (the largest subunit Rpb1) at the RTG-dependent promoters in wild-type and rtg1Δ cells in response to osmostress. RNA Pol II was recruited at the promoters of CIT2 and DLD3 upon osmostress and, as expected, this recruitment was impaired in rtg1Δ cells (Figure 3C). These results are consistent with the fact that CIT2 and DLD3 genes are not expressed in response to stress in an rtg1Δ strain (Figure 1B).
Hog1 is required for nuclear accumulation of the Rtg1/3 complex upon osmostress
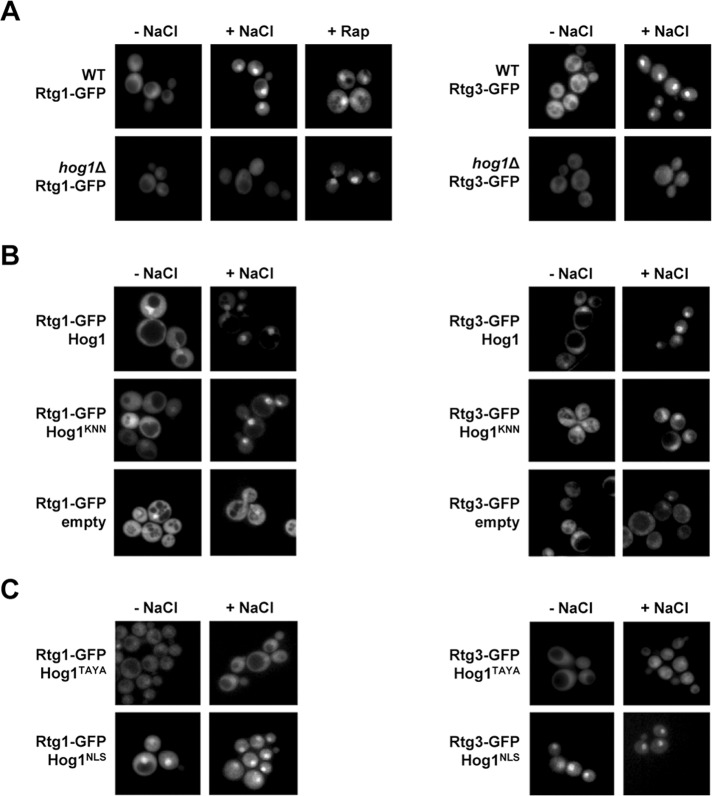
The RTG pathway is regulated by the dynamic localization of the Rtg1/3 heterodimer from the cytoplasm to the nucleus (Sekito et al., 2000). Thus we assessed the subcellular localization of the Rtg1/3 transcription complex upon osmostress, using green fluorescent proteins (GFPs). Rtg1-GFP and Rtg3-GFP fusion proteins expressed under their native promoters were mainly localized in the cytoplasm in the absence of stress, whereas they showed a predominant nuclear localization when cells were subjected to a brief osmotic shock or rapamycin (Figure 4A). We next addressed whether Hog1 regulates the subcellular localization of the Rtg1/3 complex, expressing Rtg1-GFP and Rtg3-GFP in a hog1Δ background. As depicted in Figure 4A, the nuclear accumulation of Rtg1 and Rtg3 was impaired upon osmostress in cells lacking HOG1, indicating that Hog1 is required for the nuclear accumulation of the Rtg1/3 transcription complex in response to osmostress. In contrast, Rtg1 accumulated into the nucleus in hog1Δ-deficient cells when cells were treated with rapamycin (Figure 4A, right). Taken together, these data indicate that Hog1 regulates the subcellular localization of the Rtg1/3 complex specifically upon osmostress.
FIGURE 4:
Role of Hog1 in regulating the subcellular localization of Rtg1/3 complex upon osmostress. (A) Rtg1/3 complex translocates into the nucleus upon osmostress in a Hog1-dependent manner. Wild-type and hog1Δ strains with plasmids carrying GFP-tagged Rtg1 or Rtg3 expressed under their own promoters were grown in MD-Gln to 0.5–0.7 OD660 and stressed or not with 0.4 M NaCl for 5 min before being used for fluorescence microscopy analysis. Cells expressing Rtg1-GFP were treated with 1 μg/ml of rapamycin (Rap) for 15 min. (B) Rtg1/3 nuclear translocation is not dependent on Hog1 catalytic activity. hog1Δ strains expressing Rtg1-GFP or Rtg3-GFP with an empty monocopy vector or carrying wild-type Hog1 or Hog1KNN were treated as in (A). (C) Subcellular localization of the Rtg1/3 complex depends on the subcellular localization of Hog1. hog1Δ strains expressing Rtg1-GFP or Rtg3-GFP with a monocopy vector carrying wild-type Hog1, Hog1TAYA, or a vector carrying NLS-containing Hog1 (Hog1NLS) were treated as in (A).
Nuclear accumulation of the Rtg1/3 complex upon osmostress is independent on Hog1 catalytic activity
The above results prompted us to investigate whether Hog1 catalytic activity was required for Rtg1/3 accumulation into the nucleus upon osmostress. For this purpose, hog1Δ cells expressing Rtg1-GFP or Rtg3-GFP were transformed with an empty vector, a vector containing wild-type HOG1, or a catalytically inactive Hog1 mutant (Hog1KNN; Wurgler-Murphy et al., 1997). Cells lacking HOG1 failed to accumulate Rtg1 and Rtg3 into the nucleus upon osmostress. Expression of wild-type Hog1 restored the ability of cells to accumulate Rtg1 and Rtg3 into the nucleus upon stress. Interestingly, cells expressing Hog1KNN concentrated Rtg1 and Rtg3 into the nucleus upon osmostress, similar to wild-type (Figure 4B). Therefore our results indicated that it is not the catalytic activity of Hog1 but the presence of Hog1 that is required for Rtg1/3 complex accumulation into the nucleus upon osmostress.
To confirm that only the presence of Hog1 is required for the localization of the Rtg1/3 complex, we followed the localization of Rtg1-GFP and Rtg3-GFP in cells expressing mutant versions of Hog1 with altered subcellular localization. We tested a Hog1 mutant containing point mutations at the Pbs2 mitogen-activated protein kinase kinase (MAPKK) phosphorylation sites, which is unable to translocate into the nucleus upon stress (Hog1TAYA; Ferrigno et al., 1998), and a Hog1 fused to a nuclear localization site (NLS), which is retained in the nucleus even in the absence of stress (Ferrigno et al., 1998). As shown in Figure 4C, cells expressing Hog1TAYA showed Rtg1 and Rtg3 localized throughout the cell under nonstressed conditions; this did not change upon stress. Correspondingly, cells expressing the nuclear-targeted Hog1 (Hog1NLS) showed Rtg1 and Rtg3 concentrated into the nucleus, even in absence of stress (Figure 4C). Therefore the localization of the Rtg1/3 complex depends on the localization of Hog1. Overall, these results suggest that it is not the catalytic activity of Hog1 but only the interaction of Hog1 with the Rtg1/3 transcription complex that is required for nuclear accumulation of the complex in response to osmostress.
Hog1 catalytic activity is required for binding of Rtg1/3 at promoters and proper RTG-dependent gene expression
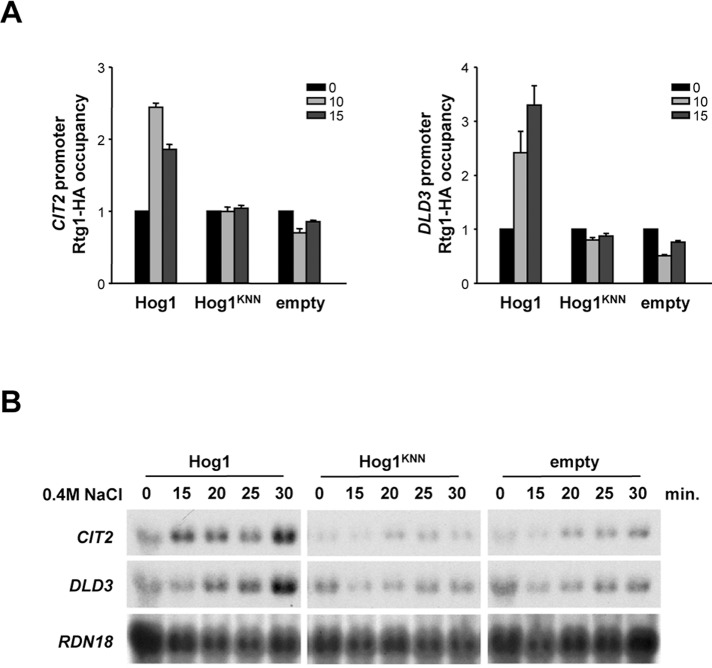
We then asked whether the catalytic activity of Hog1 could play a role in the regulation of the Rtg1/3 complex. We initially analyzed the role of Hog1 catalytic activity in the binding of Rtg1 to chromatin. hog1Δ cells with HA-tagged Rtg1 were transformed with a vector expressing wild-type Hog1, Hog1KNN, and an empty vector. As shown previously, deletion of HOG1 resulted in impaired recruitment of Rtg1/3 complex to chromatin (i.e., CIT2 and DLD3 promoters) upon osmostress, which is consistent with a deficient nuclear accumulation of Rtg1 in the nucleus upon stress. However, in cells expressing Hog1KNN, Rtg1 accumulated in the nucleus upon stress, but its recruitment to chromatin upon osmostress was clearly impaired (Figure 5A). Correspondingly, cells carrying the Hog1KNN mutant were not able to induce transcription of the RTG-dependent genes upon osmostress (Figure 5B). Taken together, our data indicate that whereas the presence of the SAPK permits nuclear accumulation of Rtg1 upon stress, and its activity is dispensable for this phenomenon, catalytic activity of Hog1 is absolutely required for chromatin association of the Rtg1/3 complex and proper transcriptional activation upon osmostress.
FIGURE 5:
Hog1 catalytic activity is required for Rtg1/3 chromatin binding and RTG-dependent gene expression. (A) Association of Rtg1/3 complex to CIT2 and DLD3 promoters in response to osmostress is dependent on Hog1 catalytic activity. hog1Δ cells expressing HA-tagged Rtg1 with an empty monocopy vector or carrying full-length Hog1 or Hog1KNN were grown in MD-Gln and treated with 0.4 M NaCl at the indicated times. Proteins were immunoprecipitated with anti-HA monoclonal antibody, and binding to the promoter regions of CIT2 and DLD3 loci was analyzed by PCR. Results are shown as the fold induction of treated relative to nontreated (time zero) samples normalized to TEL2. Data represent mean ± SD of three independent experiments. (B) The Hog1 SAPK activity is required for the induction of RTG-dependent genes in response to osmostress. The same strains and growth conditions as in (A) were used. Total RNA was analyzed by Northern blot analysis for transcript levels of CIT2 and DLD3, with RDN18 as a loading control.
Phosphorylation of Rtg1 does not alter RTG-mediated transcription upon stress
We next assessed whether Rtg1 was a direct substrate of Hog1 by using Escherichia coli–purified proteins in an in vitro kinase assay in which Hog1 was activated in the presence of a constitutive MAPKK allele (Pbs2EE; Proft et al., 2001). Initially, the in vitro kinase assay revealed that Rtg1 was directly phosphorylated by Hog1 (Figure S3A). Rtg1 contains a unique putative phosphorylation site for SAPKs (Ser-Pro or Thr-Pro), namely Thr-60. We created a point mutation version of Rtg1 in which Thr-60 was replaced with Ala (Rtg1T60A). Notably, phosphorylation of Rtg1T60A by Hog1 was completely abolished (Figure S3A), indicating that Hog1 phosphorylates Rtg1 in vitro specifically at the Thr-60 residue.
We next tested whether Rtg1 was phosphorylated in vivo in response to osmostress. Wild-type and hog1Δ cells expressing HA-tagged Rtg1 were subjected to a brief osmotic shock, and proteins were probed using a specific monoclonal antibody against the HA epitope. The mobility pattern of Rtg1 in SDS–PAGE was altered in wild-type cells subjected to an osmotic shock, but this change in mobility was not observed in hog1Δ cells (Figure S3B). When extracts from osmostressed cells were treated with alkaline phosphatase (AP), the mobility pattern of Rtg1 could be reversed, confirming that the mobility change was induced by phosphorylation (Figure S3B). Therefore Rtg1 is rapidly phosphorylated upon osmostress, and this modification depends on the Hog1 SAPK.
To assess the role of Rtg1 phosphorylation by Hog1, we studied the effect of the mutation of the Thr-60 phosphorylation site in gene expression. Vectors containing full-length RTG1 and RTG1T60A allele were transformed in rtg1Δ cells, and the effect on transcription was measured by Northern blot. Induction of CIT2 and DLD3 genes in response to osmostress was not affected in cells containing the mutant allele compared with wild-type (Figure S3C). These results indicated that Rtg1 phosphorylation by Hog1 is not a key determinant for Rtg1/3-mediated gene expression in response to osmostress.
Rtg3 is phosphorylated by Hog1 in vitro
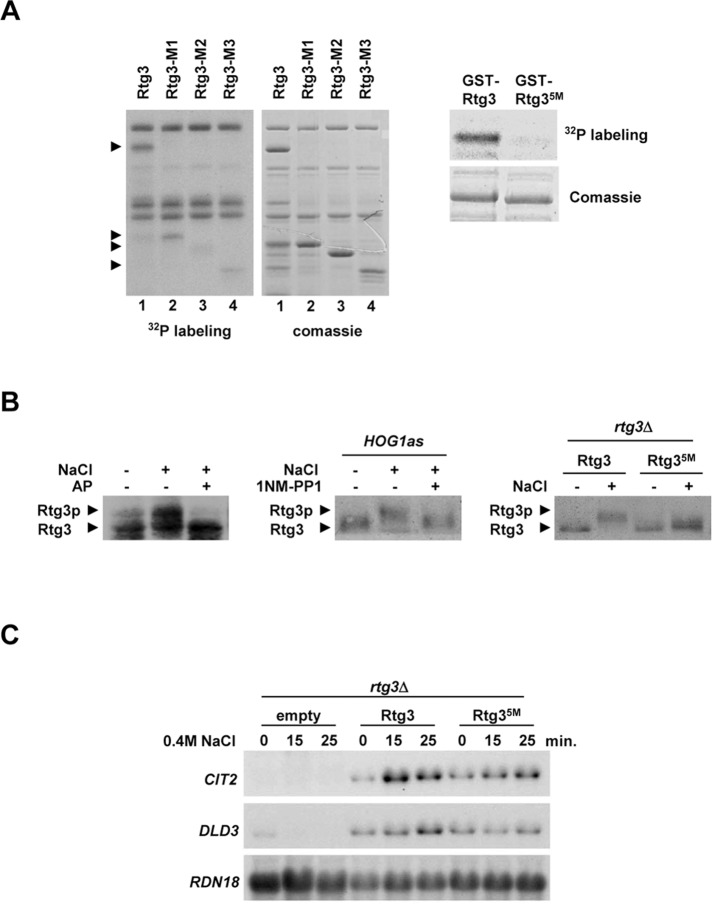
We then tested whether Rtg3 was directly phosphorylated by Hog1 in an in vitro kinase assay (Figure S3A). As shown in Figure 6A, full-length Rtg3 was phosphorylated by Hog1. Rtg3 contains 11 putative phosphorylation sites for SAPKs. To map the phosphorylation site(s) for Hog1 in Rtg3, we created several truncated RTG3 alleles, expressed as GST-tagged proteins in E. coli and subjected to in vitro phosphorylation. Whereas a truncated form of Rtg3 containing amino acids 1–210 (Rtg3-M1) was phosphorylated by Hog1, phosphorylation of a region containing amino acids 1–183 (Rtg3-M2) was strongly reduced (Figure 6A, lanes 2 and 3). Rtg3-M1 contains a putative phosphorylation site (Thr-197) for SAPKs that is not present in the Rtg3-M2 fragment, suggesting that this residue could be a Hog1 phosphorylation site in Rtg3. Moreover, a C-terminal region of Rtg3 containing amino acids 211–486 (Rtg3-M3) was also phosphorylated by Hog1, although to a lesser extent than Rtg3-M1 (Figure 6A, lane 4). Four consensus phosphorylation sites for SAPKs are present in this region (Ser-222, Ser-227, Thr-249, and Ser-376). Notably, the combination of point mutations that replace Thr-197 and these four candidate residues (Rtg35M) completely abolished the phosphorylation in vitro by Hog1 (Figure 6A, right).
FIGURE 6:
Rtg3 is phosphorylated by the Hog1 SAPK. (A) Hog1 phosphorylates Rtg3 in vitro. Left, GST-tagged full-length Rtg3 and fragments Rtg3-M1 (aa 1–210), Rtg3-M2 (aa 1–183), and Rtg3-M3 (aa 211–486) were purified from E. coli and assayed with in vitro activated Hog1. Phosphorylated proteins were detected by autoradiography, and proteins were detected by Coomassie Blue staining. Arrows indicate the positions of Rtg3 fragments. Right, mutations of Rtg3 Thr-197, Ser-222, Ser-227, Thr-249, and Ser-376 to Ala (Rtg35M) abolish in vitro phosphorylation by Hog1. (B) Rtg3 is phosphorylated upon osmostress. Left, rtg1Δ cells expressing Rtg3-HA were grown to 0.6–1 OD660, and samples were taken before (−) or 5 min after (+) addition of NaCl to a final concentration of 0.4 M. Protein extracts were treated (+) or not (−) with 10 U of AP. Rtg3-HA mobility was assessed by immunoblotting using anti-HA monoclonal antibody. Middle, rtg1Δhog1Δ cells expressing Rtg3-HA and carrying a mutant allele of HOG1 (HOG1as) were preincubated or not for 10 min with 5 μM of 1NM-PP1 prior to osmostress at the same conditions as in the left panel. Right, mutations of Rtg3 Thr-197, Ser-222, Ser-227, Thr-249, and Ser-376 to Ala (Rtg35M) abolish phosphorylation by osmostress. rtg1Δrtg3Δ strains were transformed with HA-tagged Rtg3 wild-type and the Rtg3 nonphosphorylatable mutant (Rtg35M) and processed as in the left panel. (C) Hog1 phosphorylation sites in Rtg3 are required for transcriptional activity. rtg3Δ strain was transformed with plasmids expressing wild-type Rtg3 or the Rtg3 nonphosphorylatable mutant (Rtg35M), grown in MD-Gln to 0.5–0.7 OD660, and subjected to osmotic stress (0.4 M NaCl) for the indicated length of time. Total RNA was assayed by Northern blot analysis for transcript levels of CIT2 and DLD3, with RDN18 as a loading control.
Rtg3 is phosphorylated in vivo by Hog1 upon osmostress
We next attempted to determine whether Rtg3 was phosphorylated in vivo upon osmostress. It has been reported that Rtg3 is phosphorylated under nonstressing conditions and that deletion of RTG1 abolishes this phosphorylation (Komeili et al., 2000). We therefore tested whether Rtg3 was phosphorylated upon stress in RTG1 mutant cells. HA-tagged Rtg3 was subjected to a brief osmotic shock and found to undergo a mobility shift that was reversed when samples were treated with AP (Figure 6B, left). Thus Rtg3 is phosphorylated upon osmostress. To determine whether this phosphorylation is Hog1-dependent, we analyzed the mobility of the Rtg3 protein in a yeast strain carrying a mutant allele of HOG1 (hog1as) that is specifically inhibited by the small molecule inhibitor 1NM-PP1 (Macia et al., 2009). We used this approach to inhibit Hog1 specifically, due to a basal change in mobility of Rtg3 in a hog1 mutant. The addition of the inhibitor 1NM-PP1 to cells inhibited Hog1 effectively and prevented the change in mobility of Rtg3 upon stress (Figure 6B, middle). Therefore the Rtg3 phosphorylation in response to stress is Hog1-dependent. We then analyzed the phosphorylation of the nonphosphorylatable mutant Rtg35M. As shown in Figure 6B (right), Rtg3 phosphorylation was abolished in the RTG3 allele that contains the mutant Hog1-phosphorylated sites (Rtg35M). Taken together, these results show that Rtg3 is phosphorylated by Hog1 upon osmostress.
Phosphorylation at multiple sites of Rtg3 by Hog1 affects Rtg3 function
We then aimed to determine whether phosphorylation of Rtg3 by Hog1 was important for gene expression upon osmostress. We assessed transcriptional activation in rtg3Δ cells carrying wild-type RTG3 or Rtg35M. Cells expressing the nonphosphorylatable mutant Rtg35M showed a partial impairment of induction of transcription compared with wild-type, indicating that Hog1-dependent phosphorylation of Rtg3 is important for Rtg3-mediated gene expression (Figure 6C). Of note, the combination of Rtg1T60A with Rtg35M does not synergize the defect in transcription of the Rtg35M mutant upon stress (Figure S5). Because Hog1 catalytic activity is required for the recruitment of the Rtg1/3 transcription complex at the responsive promoters (Figure 5A), we asked whether Rtg3 phosphorylation by Hog1 was also required for the association of Rtg1/3 to chromatin. Recruitment of the Rtg1/3 complex to DNA in cells expressing the nonphosphorylatable Rtg3 (Rtg35M) was almost identical to wild-type (Figure S4). It is worth noting that the reduction in gene expression observed in the Rtg35M mutant is not as dramatic as in a hog1-deficient strain. This might be because Rtg3 cannot concentrate in the nucleus, associate to chromatin, nor be phosphorylated by Hog1 in a hog1 strain, whereas the Rtg35M is only deficient in the last regulatory step. Overall the results suggest that Rtg3 phosphorylation by Hog1 is important for regulation of its transcription factor activity.
DISCUSSION
Yeast cells respond to increases in external osmolarity by activating the stress-activated Hog1 SAPK. A major outcome of the activation of Hog1 is the regulation of gene expression, and several transcription factors have been proposed as being controlled by the SAPK (de Nadal and Posas, 2010; de Nadal et al., 2011; Martinez-Montanes et al., 2010; Weake and Workman, 2010). However, data suggest that additional transcription factors may be required for the osmostress-induced gene expression controlled by Hog1 (Miller et al., 2011). In this paper, we show that the Rtg1/3 transcriptional complex is a new target for Hog1, linking the SAPK to mitochondrial activity during adaptation to stress. Deletion of RTG1 or RTG3 results in a deficient osmostress-induced expression of key genes in the trichloroacetic acid (TCA) cycle, such as CIT2, DLD3, and PYC1, known to be activated by the RTG pathway upon respiratory dysfunction (Liao et al., 1991; Chelstowska et al., 1999; Epstein et al., 2001). In addition deletion of RTG1 or RTG3 displays osmosensitivity. Of note, the osmosensitivity observed in rtg1 and rtg3 mutants is stronger than that observed in deletions of other known transcription factors targeted by Hog1, but less sensitive than observed in hog1 cells.
Osmostress alters mitochondrial function by reducing the mitochondrial electron transport in plants (Hamilton and Heckathorn, 2001). Recently it was reported that constitutive activation of Hog1 provokes a decrease in respiration levels, which leads to cell death caused by an increase in reactive oxygen species (Vendrell et al., 2011). On the other hand, the Rtg proteins are essential for respiration under limiting respiratory conditions, when several TCA cycle and respiratory chain enzymes are significantly up-regulated (Fendt and Sauer, 2010). Therefore it is likely that RTG-mediated transcription increases to compensate the down-regulation of respiration that rapidly occurs upon stress. Correspondingly, mutants in different mitochondrial components showed hypersensitivity to increases in NaCl, suggesting that mitochondrial biogenesis and function are needed for efficient adaptation to osmostress (Martinez-Pastor et al., 2010). Taken together, the data suggest that cells exposed to osmostress have compromised mitochondrial respiration, and Rtg1 and Rtg3 transcription factors might be important determinants for maintaining or restoring mitochondrial function.
Production of glycerol is critical for adaptation in response to osmostress. Glycerol synthesis requires the action of NADH-dependent enzymes, and mitochondria are designed to oxidize NADH back to NAD+. Thus it could seem contradictory that osmostress can up-regulate mitochondrial function. However, the pool of NADH/NAD+ in the cytosol and mitochondria are different, although NADH can diffuse from the cytosol into the mitochondria in certain conditions (Rigoulet et al., 2004). Therefore the function of inducing the retrograde function by osmostress might be related to the ability of the cells to reactivate respiration once cells have started to adapt rather than to maintenance of the balance between of NADH/NAD+. Alternatively, due to the delay required for the Rtg response when compared with glycerol accumulation, it is possible that an elevation in mitochondrial respiration provides more ATP to improve long-term survival under osmostress.
Gene expression analysis showed that, specifically upon osmostress, the HOG pathway is required for proper induction of Rtg1/3 target genes. The retrograde response is linked to other pathways, such as TOR signaling (Komeili et al., 2000; Butow and Avadhani, 2004). However, activation of the retrograde pathway by addition of rapamycin does not require the SAPK, indicating that RTG transcription can be activated by different signaling pathways, depending on the extracellular stimuli. A key regulatory step in the regulation of the RTG pathway is the translocation of Rtg1/3 proteins from the cytoplasm to the nucleus. When the RTG pathway is inactive, Rtg1 and Rtg3 are sequestered together in the cytoplasm. On activation, Rtg3 undergoes changes in its phosphorylation state and translocates into the nucleus together with Rtg1 (Sekito et al., 2000). Hog1 interacts with the Rtg1/3 complex in vivo, and this interaction seems to be critical for the nuclear accumulation of the complex in response to osmostress. Indeed, localization of Rtg1 and Rtg3 in cells expressing Hog1 with abolished catalytic activity and altered subcellular localization correlates with the localization of Hog1 independently upon its catalytic activity. Thus the sole interaction of the transcription factor with the SAPK is sufficient for Rtg1 and Rtg3 nuclear accumulation. There are other stress-responsive transcription factors for which translocation is the key regulatory event in stress induction of transcription. For instance, Msn2 and Msn4 activators accumulate in the nucleus under stress conditions. However, in this case, the nuclear localization seems to occur independent of Hog1 (Gorner et al., 1998).
In addition to its role in the regulation of the Rtg1/3 subcellular localization, Hog1 is further required for Rtg1/3 chromatin binding and transcriptional activity. It is known that Hog1 becomes intimately linked with stress-responsive loci upon osmostress, and this binding is dependent on the presence of stress-mediating transcriptional activators (Alepuz et al., 2001, 2003; Pascual-Ahuir et al., 2006; Pokholok et al., 2006; Proft et al., 2006). Similarly, Hog1 is recruited by Rtg1 to the RTG-dependent promoters in response to osmostress. Thus the Rtg1/3 transcription complex targets the SAPK to specific promoters. On the other hand, our results also showed that recruitment of the Rtg1/3 complex to RTG-dependent promoters during osmostress is strongly reduced in a hog1Δ strain. These results are in agreement with the subcellular localization experiments indicating that Hog1 regulates the localization of the complex upon stress. However, the association of RTG at promoters not only requires the presence of Hog1 but also its catalytic activity. For instance, in cells containing the catalytic-inactive Hog1KNN, Rtg1 and Rtg3 are nuclear upon stress but cannot associate to chromatin. Similarly, it has been described that nuclear accumulation of the MAPK or the transcription activator Hot1 are not sufficient for the association of the transcription factor to chromatin, which is also dependent on Hog1 activity (Alepuz et al., 2003). Overall the interdependence of binding of Hog1 and Rtg1 to promoters of target genes reveals, as in the case of other stress-mediating transcription factors, a functional connection between both proteins for making a stable complex at stress-responsive promoters.
Hog1 also directly phosphorylates Rtg1 and Rtg3 proteins. We have not been able to unravel the role of Rtg1 phosphorylation by Hog1, but Rtg3 phosphorylation seems to be important for proper gene activation. Gene induction is reduced in an Rtg3 nonphosphorylatable mutant, whereas Rtg1/3 recruitment remains unaltered. Moreover, combining Rtg1 and Rtg3 nonphosphorylatable mutants is not synergistic for proper gene activation. Thus Hog1 phosphorylation plays an important role in the regulation of Rtg3 transcriptional activity. A similar scenario has been described for other transcription factors under the control of the SAPK. This is the case of the MEF2-like activator Smp1, the transcriptional activity of which is regulated by Hog1-dependent phosphorylation upon stress (de Nadal et al., 2003).
Taken together, our results show that, in contrast with what is known for the regulation of other transcription factors targeted by Hog1, the SAPK regulates the Rtg1/3 heterodimeric transcription complex by multiple mechanisms (see schematic diagram in Figure 7), which are: 1) the regulation of the nuclear accumulation of the complex, 2) the stimulation of the association of the complex at RTG-responsive promoters, and 3) the control of Rtg3 transcriptional activity. Notably, whereas the presence of Hog1 is essential for Rtg1/3 nuclear localization upon stress, the activity of the SAPK is required for Rtg1/3 chromatin binding and Rtg3 transcriptional activity regulation. Our data also provide a direct link between Hog1 and the regulation of genes important for mitochondrial activity during stress adaptation.
FIGURE 7:
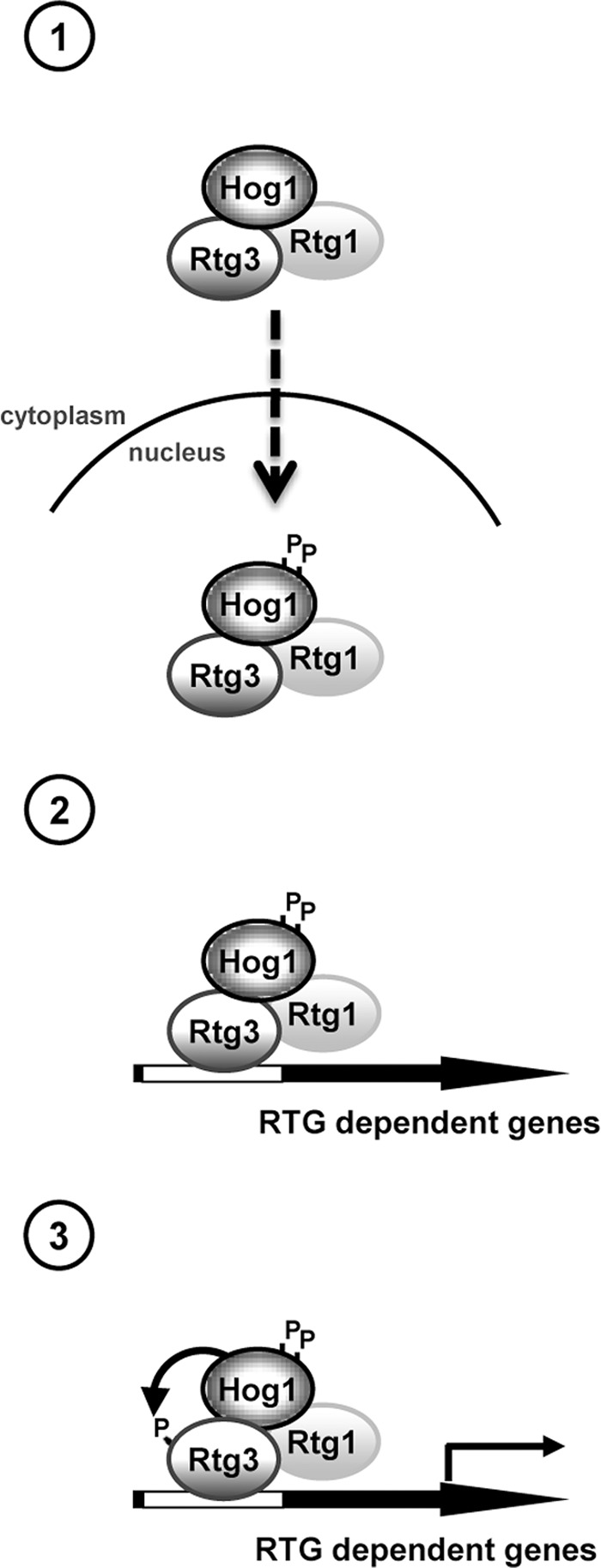
The Hog1 SAPK directly modulates the Rtg1/3 complex upon osmostress by a triple mechanism: (1) regulating the nuclear accumulation of the heterodimeric complex (no catalytic activity of Hog1 is required); (2) stimulating the association of the complex to the responsive promoters (Hog1 catalytic activity is required, but not Rtg3 phosphorylation); and (3) controlling Rtg3 transcriptional activity (Hog1 catalytic activity and Rtg3 phosphorylation are required).
MATERIALS AND METHODS
Yeast strains
Saccharomyces cerevisiae strain K699 (MATa leu2-3112 ura3-1 his3-11 trp1-1 can1-100) and its derivatives YNN15 (RTG1::kanMX4), YNN20 (RTG3::kanMX4), YNN41 (RTG2::kanMX4), YNN17 (HOG1::kanMX4), and YNN51 (RTG1::KanMX KAN::NAT RTG3::KanMX) were used in this work. Genomic disruptions were made by long flanking-homology PCR-based gene disruption. C-terminal genomic tagging of Rtg1 in wild-type and hog1Δ strains yielded YNN21 (RTG1-6HA::HIS3) and YNN23 (HOG1::kanMX4 RTG1-6HA::HIS3). C-terminal genomic tagging of Rtg3 yielded YNN44 (RTG3-6HA::HIS3), YNN56 (RTG1::KanMX RTG3-HA6::HIS3), and YAD165 (RTG1::KanMX HOG1::URA3 RTG3-HA6::HIS3). YAD189 (RTG1::KanMX RTG3-HA6::HIS3 HOG1::URA3::HOG1AS) was obtained from YAD165 by introducing the HOG1AS mutated gene in the HOG1 genomic locus. For Hog1 ChIP assays, YCR109 (HOG1-6HA::HIS3) and YCR110 (RTG1::kanMX4 HOG1-6HA::HIS3) were used. Tagging of genomic open reading frames (ORFs) with HA epitopes was done with a PCR-based strategy.
Plasmids
GFP-fused constructs were generated by amplifying RTG1 and RTG3 coding regions plus promoter regions (∼800) from genomic DNA and cloning them into the XhoI site of the pRS416-GFP construct (Raitt et al., 2000). To analyze the role of Hog1 catalytic activity, we used YCpLac111-Hog1-3HA and YCpLac111-Hog1KNN-3HA vectors (Wurgler-Murphy et al., 1997). pGBT9 (Gal4DBD-HOG1) and pRS426TEG2-Hog1TAYA were also used in this study (Ferrigno et al., 1998). Full-length RTG1 and RTG3 coding regions were PCR-amplified from genomic DNA and cloned into the BamHI site of the bacterial expression vector pGEX-4T (GE Healthcare, Waukesha, WI), which allowed the expression of GST-tagged proteins in E. coli. pGEX-4T plasmids containing mutated alleles (described in Results) were generated by PCR amplification with the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, Agilent, Santa Clara, CA) from the pGEX-4T plasmids containing the wild-type RTG1 and RTG3 ORFs. Each mutation was verified by DNA sequencing. pGEX4T-Hog1 and pGEX4T-Pbs2EE (PBS2 with Ser-514-Glu and Thr-518-Glu mutations) are described in Bilsland-Marchesan et al. (2000). For obtaining HA fusion proteins, the promoter (∼800) and coding regions of wild-type RTG1 and RTG3 were PCR-amplified and cloned into the BamHI site of the pRS415 vector. Mutated alleles (described in Results) were generated and verified as before. The 6xHA tags were inserted before the stop codon by recombination, using a specific cassette containing the HA epitopes.
Buffers and media
Buffer A consisted of 50 mM Tris-HCl (pH 7.5), 15 mM EDTA, 15 mM ethylene glycol tetraacetic acid, 2 mM dithiothreitol (DTT), 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 5 μg/ml of pepstatin, and 5 μg/ml of leupeptin. AP buffer consisted of 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 10 mM MgCl2. STET buffer consisted of 10 mM Tris-HCl (pH 8.0), 0.1M NaCl, 1 mM EDTA, and 5% Triton X-100. Kinase buffer consisted of 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 2 mM DTT. Phosphatase inhibitor cocktail contained 10 mM NaF, 1 mM sodium pyrophosphate, and 10 mM β-glycerophosphate. For all experiments, cell cultures were grown in minimal dextrose (MD) media that contained 0.7% yeast nitrogen base and 2% dextrose (pH 5.5). Glutamine was added to MD media to a final concentration of 0.2%. To supplement the auxotrophic requirements of strains, we added required amino acids (0.006% histidine, 0.008% leucine, 0.006% adenine, 0.005% tryptophan, 20 μg/ml uracil, and 20 μg/ml methionine), lacking for the specific ones to select for plasmid maintenance. Cells were grown in YPD (1% yeast extract, 2% peptone, and 2% dextrose) as indicated in Results.
Cell viability assays
Yeast cultures were grown to early log phase (0.5–0.8 OD660) and diluted to a 0.05 OD660. Serial dilutions of cultures were spotted directly onto YPD plates containing glutamine and the indicated concentrations of salt and sorbitol (Figure 1A) and were incubated at 30ºC. Growth was scored after 2–5 d.
Northern blot analysis
Yeast cultures were grown to early log phase (0.5–0.8 OD660) and either subjected to stress (0.4 M NaCl, times indicated in the figure legends) or untreated. Total RNA was extracted from 15 ml of culture by acid phenol treatment. Twenty micrograms of total RNA per sample was run in 1% agarose gels using electrophoresis. RNAs were transferred to a nylon membrane (Roche, Indianapolis, IN) by a vacuum blotter (model 785; Bio-Rad, Hercules, CA). Total RNA and expression of specific genes were probed by using radiolabeled PCR fragments containing the ORF region of CIT2, DLD3, PYC1, CTT1, STL1, or GRE2, or the noncoding exon of RDN18 as a loading control. Signals were quantified by using a storage phosphor screen in a Typhoon 8600 phosphorimager.
In vivo coprecipitation assays
In vivo interaction of Rtg1-HA and Rtg3-HA fusion proteins with GST-Hog1 was determined by GST pulldown experiments. Exponentially growing cells (0.5–0.8 OD660) were subjected to a brief osmotic shock (0.4 M NaCl for 10 min). Two milligrams of yeast extract in a mixture of buffer A plus 150 mM NaCl and phosphatase inhibitors was prepared and incubated with glutathione-Sepharose beads overnight at 4ºC. Beads were washed extensively with buffer A, resuspended in loading buffer, and separated on SDS–15% PAGE (Rtg1-HA) or SDS–10% PAGE (Rtg3-HA). The lysis buffer used in the coimmunoprecipitation, treated or not with DNase I, was 50 mM Tris (pH 7.5), 150 mM NaCl, 0.1% Triton X-100, 1 mM EDTA, 2 mM DTT, and the entire cocktail of antiproteases and phosphatase inhibitors. Protein extracts were treated with 200 U/ml of recombinant DNase I RNase-free for 5 h at 4ºC (Roche) before being incubated with glutathione-Sepharose beads as above. Immunoblotting was done by using anti-HA monoclonal antibody 12CA5 (Roche) and anti-GST monoclonal antibody (Pharmacia) together with ECL reagent (GE Healthcare, Waukesha, WI).
ChIP
ChIP experiments were performed as described before (Zapater et al., 2007). Yeast cultures were grown to early log phase (0.6–0.8 OD660) before samples of the culture were exposed to osmostress (0.4 M NaCl; times indicated in the figure legends). For cross-linking, yeast cells were treated with 1% formaldehyde for 20 min at room temperature. Antibodies used were mouse polyclonal anti-Rpb1 (8WG16; Covance, Princeton, NJ), and monoclonal anti-HA 12CA5. For quantitative PCR analysis of osmoresponsive genes and constitutively expressed genes, the following primers, with locations indicated by the distance from the respect ATG initiation codon, were used: CIT2, −460/−141; DLD3, −253/+46; and TEL2 (telomeric region on the right arm of chromosome VI).
GFP fluorescence microscopy
Exponentially growing cells (0.6–0.8 OD660) were observed without fixation using a Nikon Eclipse 901 microscope (Nikon, Tokyo, Japan) with an ORCA II charge-coupled device camera (Hamamatsu, Hamamatsu City, Japan). Images were taken at 100× magnification and converted to Photoshop 7.0.1 (Adobe Systems, San Jose, CA).
Purification of GST proteins in E. coli and in vitro kinase assays
GST fusion proteins encoding Pbs2EE, Hog1, and wild-type or mutant Rtg1 and Rgt3 were constructed by using pGEX-4T (Pharmacia), expressed in E. coli, and purified using glutathione-Sepharose beads (Pharmacia) in STET buffer, as described previously (Posas et al., 1996). Phosphorylation by Hog1 of Rtg1, Rtg3, and mutant versions were monitored as follows: 1 μg of recombinant GST-Hog1 was activated by phosphorylation by using 0.5 μg of GST-Pbs2EE in the presence of kinase buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 2 mM DTT) and 50 μM ATP. After 15 min at 30°C, 5 μg of GST-Rtg1, GST-Rtg2, or GST-Rtg3 was added to the previous mixture together with [γ-32P]ATP (0.2 μCi/μl). The mixture was then incubated for 10 min at 30°C, and the reactions were stopped by the addition of 2X SDS loading buffer. The labeled proteins were resolved by SDS–PAGE, stained with Coomassie Blue, and detected by autoradiography.
In vivo phosphorylation assays
Cells containing the RTG1 and RTG3 ORF fused to HA (6xHA) epitope were grown to mid–log phase (0.6–0.8 OD660), subjected to osmostress treatment (0.4 M NaCl for 5 min), and harvested by centrifugation. Yeast extracts were obtained in a mixture of buffer A plus 150 mM NaCl and phosphatase inhibitors. Protein concentration was determined by Bradford analysis (Bio-Rad Protein Assay). When necessary, 1 mg of total yeast extract was treated for 2 h at 37ºC with 0.5 μl phosphatase alkaline (20 U/μl; Roche). In experiments that did not require the use of AP, protein extraction was performed using the TCA protocol, as described before (Macia et al., 2009). 1NM-PP1 was used at 5 μM to inhibit analogue-sensitive mutants (Macia et al., 2009). Total crude extracts were loaded onto SDS–15% PAGE, and the Rtg1-HA and Rtg3-HA fusion proteins were detected by immunoblotting using an anti-HA monoclonal antibody (12CA5; Roche).
Supplementary Material
Acknowledgments
We are grateful for technical assistance from L. Subirana and S. Obejas. We thank Markus Proft for helpful comments and discussion. F.P. and E.d.N. are the recipients of an ICREA Acadèmia (Generalitat de Catalunya). This work was supported by the Botin Foundation and grants from the Spanish Ministry of Science and Innovation (BFU2011-26722 to E.d.N. and BIO2009-07762 to F.P.) and the FP7 (UNICELLSYS) framework program.
Abbreviations used:
- AP
alkaline phosphatase
- ChIP
chromatin immunoprecipitation
- DTT
dithiothreitol
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- HA
hemagglutinin
- MAPKK
mitogen-activated protein kinase kinase
- MD
minimal dextrose
- NLS
nuclear localization site
- ORF
open reading frame
- Pol II
polymerase II
- RTG
retrograde
- SAPK
stress-activated protein kinase
- TCA
trichloroacetic acid
- TOR
target of rapamycin
- YPD
yeast–peptone–dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-04-0289) on September 5, 2012.
*These authors contributed equally to this work.
REFERENCES
- Alepuz PM, de Nadal E, Zapater M, Ammerer G, Posas F. Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. EMBO J. 2003;22:2433–2442. doi: 10.1093/emboj/cdg243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alepuz PM, Jovanovic A, Reiser V, Ammerer G. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol Cell. 2001;7:767–777. doi: 10.1016/s1097-2765(01)00221-0. [DOI] [PubMed] [Google Scholar]
- Bilsland-Marchesan E, Arino J, Saito H, Sunnerhagen P, Posas F. Rck2 kinase is a substrate for the osmotic stress-activated mitogen-activated protein kinase Hog1. Mol Cell Biol. 2000;20:3887–3895. doi: 10.1128/mcb.20.11.3887-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- Capaldi AP, Kaplan T, Liu Y, Habib N, Regev A, Friedman N, O'Shea EK. Structure and function of a transcriptional network activated by the MAPK Hog1. Nat Genet. 2008;40:1300–1306. doi: 10.1038/ng.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelstowska A, Liu Z, Jia Y, Amberg D, Butow RA. Signalling between mitochondria and the nucleus regulates the expression of a new D-lactate dehydrogenase activity in yeast. Yeast. 1999;15:1377–1391. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1377::AID-YEA473>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- de Nadal E, Ammerer G, Posas F. Controlling gene expression in response to stress. Nat Rev Genet. 2011;12:833–845. doi: 10.1038/nrg3055. [DOI] [PubMed] [Google Scholar]
- de Nadal E, Casadome L, Posas F. Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol Cell Biol. 2003;23:229–237. doi: 10.1128/MCB.23.1.229-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E, Posas F. Multilayered control of gene expression by stress-activated protein kinases. EMBO J. 2010;29:4–13. doi: 10.1038/emboj.2009.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CB, Waddle JA, Hale W, Dave V, Thornton J, Macatee TL, Garner HR, Butow RA. Genome-wide responses to mitochondrial dysfunction. Mol Biol Cell. 2001;12:297–308. doi: 10.1091/mbc.12.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt SM, Sauer U. Transcriptional regulation of respiration in yeast metabolizing differently repressive carbon substrates. BMC Syst Biol. 2010;4:12. doi: 10.1186/1752-0509-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira Júnior JR, Spirek M, Liu Z, Butow RA. Interaction between Rtg2p and Mks1p in the regulation of the RTG pathway of Saccharomyces cerevisiae. Gene. 2005;354:2–8. doi: 10.1016/j.gene.2005.03.048. [DOI] [PubMed] [Google Scholar]
- Ferrigno P, Posas F, Koepp D, Saito H, Silver PA. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin β homologs NMD5 and XPO1. EMBO J. 1998;17:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha N, Desai P, Vancura A. Plc1p is required for SAGA recruitment and derepression of Sko1p-regulated genes. Mol Biol Cell. 2007;18:2419–2428. doi: 10.1091/mbc.E06-10-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton EW, Heckathorn SA. Mitochondrial adaptations to NaCl. Complex I is protected by anti-oxidants and small heat shock proteins, whereas complex II is protected by proline and betaine. Plant Physiol. 2001;126:1266–1274. doi: 10.1104/pp.126.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S, Krantz M, Nordlander B. Yeast osmoregulation. Methods Enzymol. 2007;428:29–45. doi: 10.1016/S0076-6879(07)28002-4. [DOI] [PubMed] [Google Scholar]
- Jia Y, Rothermel B, Thornton J, Butow RA. A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol. 1997;17:1110–1117. doi: 10.1128/mcb.17.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Inai T, Mizunuma M, Okada I, Shitamukai A, Hirata D, Miyakawa T. Identification of Tup1 and Cyc8 mutations defective in the responses to osmotic stress. Biochem Biophys Res Commun. 2008;368:50–55. doi: 10.1016/j.bbrc.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Komeili A, Wedaman KP, O'Shea EK, Powers T. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J Cell Biol. 2000;151:863–878. doi: 10.1083/jcb.151.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- Liao XS, Small WC, Srere PA, Butow RA. Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:38–46. doi: 10.1128/mcb.11.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol Cell Biol. 1999;19:6720–6728. doi: 10.1128/mcb.19.10.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- Macia J, Regot S, Peeters T, Conde N, Sole R, Posas F. Dynamic signaling in the Hog1 MAPK pathway relies on high basal signal transduction. Sci Signal. 2009;2:ra13. doi: 10.1126/scisignal.2000056. [DOI] [PubMed] [Google Scholar]
- Martinez-Montanes F, Pascual-Ahuir A, Proft M. Toward a genomic view of the gene expression program regulated by osmostress in yeast. OMICS. 2010;14:619–627. doi: 10.1089/omi.2010.0046. [DOI] [PubMed] [Google Scholar]
- Martinez-Pastor M, Proft M, Pascual-Ahuir A. Adaptive changes of the yeast mitochondrial proteome in response to salt stress. OMICS. 2010;14:541–552. doi: 10.1089/omi.2010.0020. [DOI] [PubMed] [Google Scholar]
- Mas G, de Nadal E, Dechant R, Rodriguez de la Concepcion ML, Logie C, Jimeno-Gonzalez S, Chavez S, Ammerer G, Posas F. Recruitment of a chromatin remodelling complex by the Hog1 MAP kinase to stress genes. EMBO J. 2009;28:326–336. doi: 10.1038/emboj.2008.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol. 2011;7:458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke SM, Herskowitz I, O'Shea EK. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 2002;18:405–412. doi: 10.1016/s0168-9525(02)02723-3. [DOI] [PubMed] [Google Scholar]
- Parikh VS, Morgan MM, Scott R, Clements LS, Butow RA. The mitochondrial genotype can influence nuclear gene expression in yeast. Science. 1987;235:576–580. doi: 10.1126/science.3027892. [DOI] [PubMed] [Google Scholar]
- Pascual-Ahuir A, Struhl K, Proft M. Genome-wide location analysis of the stress-activated MAP kinase Hog1 in yeast. Methods. 2006;40:272–278. doi: 10.1016/j.ymeth.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- Posas F, Chambers JR, Heyman JA, Hoeffler JP, de Nadal E, Arino J. The transcriptional response of yeast to saline stress. J Biol Chem. 2000;275:17249–17255. doi: 10.1074/jbc.M910016199. [DOI] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Proft M, Mas G, de Nadal E, Vendrell A, Noriega N, Struhl K, Posas F. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell. 2006;23:241–250. doi: 10.1016/j.molcel.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Proft M, Pascual-Ahuir A, de Nadal E, Arino J, Serrano R, Posas F. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 2001;20:1123–1133. doi: 10.1093/emboj/20.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Struhl K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell. 2002;9:1307–1317. doi: 10.1016/s1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- Raitt DC, Posas F, Saito H. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 2000;19:4623–4631. doi: 10.1093/emboj/19.17.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Krantz M, Thevelein JM, Hohmann S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem. 2000;275:8290–8300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]
- Rep M, Reiser V, Gartner U, Thevelein JM, Hohmann S, Ammerer G, Ruis H. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol. 1999;19:5474–5485. doi: 10.1128/mcb.19.8.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigoulet M, Aguilaniu H, Avéret N, Bunoust O, Camougrand N, Grandier-Vazeille X, Larsson C, Pahlman IL, Manon S, Gustafsson L. Organization and regulation of the cytosolic NADH metabolism in the yeast Saccharomyces cerevisiae. Mol Cell Biochem. 2004;256–257:73–81. doi: 10.1023/b:mcbi.0000009888.79484.fd. [DOI] [PubMed] [Google Scholar]
- Rothermel BA, Thornton JL, Butow RA. Rtg3p, a basic helix-loop-helix/leucine zipper protein that functions in mitochondrial-induced changes in gene expression, contains independent activation domains. J Biol Chem. 1997;272:19801–19807. doi: 10.1074/jbc.272.32.19801. [DOI] [PubMed] [Google Scholar]
- Sekito T, Thornton J, Butow RA. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol Biol Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small WC, Brodeur RD, Sandor A, Fedorova N, Li G, Butow RA, Srere PA. Enzymatic and metabolic studies on retrograde regulation mutants of yeast. Biochemistry. 1995;34:5569–5576. doi: 10.1021/bi00016a031. [DOI] [PubMed] [Google Scholar]
- Sole C, Nadal-Ribelles M, Kraft C, Peter M, Posas F, de Nadal E. Control of Ubp3 ubiquitin protease activity by the Hog1 SAPK modulates transcription upon osmostress. EMBO J. 2011;30:3274–3284. doi: 10.1038/emboj.2011.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V, Kriete A, Sacan A, Jazwinski SM. Comparing the yeast retrograde response and NF-κB stress responses: implications for aging. Aging Cell. 2010;9:933–941. doi: 10.1111/j.1474-9726.2010.00622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrell A, Martinez-Pastor M, Gonzalez-Novo A, Pascual-Ahuir A, Sinclair DA, Proft M, Posas F. Sir2 histone deacetylase prevents programmed cell death caused by sustained activation of the Hog1 stress-activated protein kinase. EMBO Rep. 2011;12:1062–1068. doi: 10.1038/embor.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- Westfall PJ, Ballon DR, Thorner J. When the stress of your environment makes you go HOG wild. Science. 2004;306:1511–1512. doi: 10.1126/science.1104879. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Wurgler-Murphy SM, Maeda T, Witten EA, Saito H. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol. 1997;17:1289–1297. doi: 10.1128/mcb.17.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapater M, Sohrmann M, Peter M, Posas F, de Nadal E. Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol Cell Biol. 2007;27:3900–3910. doi: 10.1128/MCB.00089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.