Abstract
We isolated two tomato (Lycopersicon esculentum) cDNA clones, tomPRO1 and tomPRO2, specifying Δ1-pyrroline-5-carboxylate synthetase (P5CS), the first enzyme of proline (Pro) biosynthesis. tomPRO1 is unusual because it resembles prokaryotic polycistronic operons (M.G. García-Ríos, T. Fujita, P.C. LaRosa, R.D. Locy, J.M. Clithero, R.A. Bressan, L.N. Csonka [1997] Proc Natl Acad Sci USA 94: 8249–8254), whereas tomPRO2 encodes a full-length P5CS. We analyzed the accumulation of Pro and the tomPRO1 and tomPRO2 messages in response to NaCl stress and developmental signals. Treatment with 200 mm NaCl resulted in a >60-fold increase in Pro levels in roots and leaves. However, there was a <3-fold increase in the accumulation of the tomPRO2 message and no detectable induction in the level of the tomPRO1 message in response to NaCl stress. Although pollen contained approximately 100-fold higher levels of Pro than other plant tissues, there was no detectable increase in the level of either message in pollen. We conclude that transcriptional regulation of these genes for P5CS is probably not important for the osmotic or pollen-specific regulation of Pro synthesis in tomato. Using restriction fragment-length polymorphism mapping, we determined the locations of tomPRO1 and tomPRO2 loci in the tomato nuclear genome. Sequence comparison suggested that tomPRO1 is similar to prokaryotic P5CS loci, whereas tomPRO2 is closely related to other eukaryotic P5CS genes.
Water stress can be imposed by high salinity, dehydration, or freezing, which are environmental conditions that lead to the loss of water from cells. Water stress triggers the accumulation of Pro in a wide variety of species in all biological kingdoms (Paleg and Aspinall, 1981; Gilles et al., 1987; Csonka and Hanson, 1991). It has been suggested that the accumulation of Pro contributes to the maintenance of proper balance between extracellular and intracellular osmolality under conditions of water stress. Direct evidence supporting this hypothesis was provided by the fact that mutations that resulted in high level Pro overproduction conferred increased osmotic stress tolerance in Salmonella typhimurium (Csonka, 1981). High-level expression of P5CS, a bifunctional enzyme that catalyzes the first and second reactions of Pro biosynthesis, has been reported to result in increased salinity stress tolerance in transgenic tobacco plants (Kishor et al., 1995). However, the significance of Pro accumulation is still controversial (Verma and Hong, 1996; Hare and Cress, 1997), and other functions have been proposed for this response, such as free radical scavenging, nitrogen storage, or pH regulation (Stewart and Hanson, 1980; Delauney and Verma, 1993).
Pro is synthesized by the following four reactions: (a) ATP-dependent phosphorylation of glutamate to γ-glutamyl phosphate, catalyzed by GK; (b) reduction of γ-glutamyl phosphate by NADPH to γ-glutamyl semialdehyde, mediated by GPR; (c) spontaneous cyclization of γ-glutamyl semialdehyde to P5C; and (d) NADPH-dependent reduction of P5C to Pro, carried out by P5C reductase. In addition to this so-called “glutamate pathway” of Pro synthesis, an alternate route to Pro has been suggested, involving the conversion of Orn to P5C by Orn-δ-amino transferase. There are contradictory conclusions in the literature concerning the importance of the latter pathway during salinity stress. Whereas Delauney et al. (1993) found that the level of the Orn-δ-amino transferase mRNA was markedly decreased by high salinity, Roosens et al. (1998) observed that this message was induced by the same stress in Arabidopsis. Isotope-tracing studies suggested that the pathway via Orn is not important for Pro synthesis during osmotic stress in tomato (Lycopersicon esculentum) (Rhodes et al., 1986).
The finding that water stress increases the accumulation of Pro in numerous plant species, together with the demonstration that it is possible to enhance osmotic stress tolerance in bacteria by Pro overproduction provided the motivation for the cloning of genes of the Pro biosynthetic pathway from plants. Genes specifying GK have been cloned from moth bean (Vigna aconitifolia), Arabidopsis, rice (Oryza sativa), and tomato (Hu et al., 1992; Savouré et al., 1995; Yoshiba et al., 1995; Maggio et al., 1996; García-Ríos et al., 1997; Igarashi et al., 1997; Strizhov et al., 1997). The genes that were cloned from moth bean, Arabidopsis, and rice encode a P5CS made up of a hybrid GK and GPR. High salinity or dehydration results in increased accumulation of Pro in Arabidopsis, rice, and moth bean roots, and has been shown to be accompanied by an increase in the P5CS message level (Hu et al., 1992; Savouré et al., 1995; Yoshiba et al., 1995; Igarashi et al., 1997).
The induction in the level of P5CS mRNA has been determined to be 7- to 8-fold in Arabidopsis (Savouré et al., 1995; Yoshiba et al., 1995). Genes for the last enzyme of Pro biosynthesis, P5C reductase, were cloned from soybean, Arabidopsis, and pea (Delauney and Verma, 1990; Williamson and Slocum, 1992; Verbruggen et al., 1993), and it was observed that osmotic stress resulted in a similar increase in the P5C reductase message level as were seen for the P5CS message. The observation that salinity or dehydration stress stimulated the accumulation of the transcripts for the Pro-biosynthetic genes has been interpreted to mean that the transcriptional control of these genes is important for the regulation of Pro synthesis by osmotic stress. However, 50-fold overproduction of P5C reductase in transgenic tobacco plants did not lead to increased Pro accumulation (Szoke et al., 1992), indicating that the much smaller induction of P5C reductase in NaCl-stressed plants is not likely to be of significance for the Pro accumulation.
Plant species exhibit substantial variation both in the relative increases and final levels of Pro attained in response to osmotic stress (Delauney and Verma, 1993). Arabidopsis, pea, and rice, which have been used to probe the importance of transcriptional control for Pro synthesis, are in fact not the best representatives of Pro accumulators. These plants accumulate only approximately 2 to 6 μmol Pro/g fresh weight in response to NaCl stress (Williamson and Slocum, 1992; Savouré et al., 1995; Peng et al., 1996; Igarashi et al., 1997). Thus, unless it is highly concentrated in specific subcellular compartments or organelles, Pro at such low overall concentrations would not be expected to be a substantial determinant of the osmotic potential of the whole cells (Blum et al., 1996; Sharp et al., 1996). However, plants in the family Solanaceae have been found to contain much higher levels of Pro (Treichel et al., 1984; Handa et al., 1986; Rhodes et al., 1986; Delauney and Verma, 1993). The levels of this imino acid can be regulated over 300-fold in tomato tissue-culture cells by osmotic stress (Handa et al., 1986; Rhodes et al., 1986). 15N-isotope-tracing experiments indicated that this increase in the Pro pool in cultured tomato cells upon osmotic stress was primarily due to a 10-fold increase in the rate of Pro synthesis via the glutamate pathway. Therefore, if transcriptional regulation of P5CS is important for the control of Pro synthesis by water stress, as has been suggested for Arabidopsis and rice, then one might expect that the Solanaceae, which accumulate much more robust levels of Pro under osmotic stress, would be more suitable for the study of this effect than the model species studied thus far.
For the above reasons, we cloned the genes that specify the first and second enzymes of Pro biosynthesis in tomato. We obtained two distinct clones, tomPRO1 and tomPRO2. The tomPRO1 clone was isolated from a tomato cDNA library by complementation of GK (proB) and GPR (proA) mutations in Escherichia coli (García-Ríos et al., 1991). Surprisingly, this locus proved to have an unusual structure, in that it contains two open reading frames that encode GK and GPR, arranged as a dicistronic operon (García-Ríos et al., 1997). The tomPRO2 locus was cloned by hybridization to a fragment of the first P5CS gene cloned from Arabidopsis (see below). Like the P5CS genes from Arabidopsis, moth bean, and rice, tomPRO2 specifies a hybrid GK-GPR as a single polypeptide.
Because mitochondria and chloroplasts, like prokaryotes, are able to translate polycistronic messages, we considered the possibility that the tomPRO1 might be present on a plastid genome. To test this, we carried out RFLP mapping of tomPRO1 and tomPRO2 loci and demonstrated that both are present in the tomato nuclear genome. To our knowledge, this is the first example of a polycistronic locus mapped in plant nuclear genome. We also used these clones to probe the transcriptional regulation of the corresponding genes by osmotic stress. Our major finding was that transcriptional induction is not likely to be important for the regulation of Pro synthesis by osmotic stress in tomato, despite the fact that this plant accumulates Pro to much higher levels than Arabidopsis, pea, and rice.
MATERIALS AND METHODS
Isolation of tomPRO2 cDNA
A 1.6-kb fragment of the AtP5CS1 gene was generated by García-Ríos (1995) by PCR amplification of Arabidopsis total DNA with primers designed from highly conserved sequences in the tomPRO1 clone (García-Ríos et al., 1997), the moth bean P5CS gene (Hu et al., 1992), and a partial sequence of the AtP5CS2 gene (Strizhov et al., 1997; L. Szabados, personal communication). The two primers were 5′-GATGCTCATTTATGGGCTCC-3′ (specifying amino acids corresponding to residues 283–288 of the tomPRO2 product) and 5′-CCATTCTGCTCCAAATCTTT-3′ (complementary to sequences specifying amino acids corresponding to residues 553–558 of the tomPRO2 product). The amplified fragment was radiolabeled and used to probe a tomato (Lycopersicon esculentum cv Ailsa Craig) cDNA library in λgt10 (kindly provided by Dr. G. Martin; described in Martin et al. [1993]). Hybridization of the plaque blots on Hybond N+ membranes (Amersham) was performed in 6× SSC and 1% SDS at 42°C. The blots were washed with 0.5× SSC and 0.1% SDS at 60°C. Among the positive clones, the one with the longest insert was subcloned into the EcoRI site of pBluescript SKII(−) (Stratagene) and sequenced using an automated fluorescence sequencer (Applied Biosystems).
Plant Materials
Tomato seeds were planted on 3M paper immersed with 0.25× Murashige and Skoog solution (JRH Biosciences, Lenexa, KS) under continuous light at 25°C. About 3 weeks after germination, the seedlings were transplanted to plastic containers filled with one-half-strength Hoagland solution and maintained hydroponically in a greenhouse under natural light. Leaf and root samples were taken at d 2, 6, 16, and 31 after the initiation of NaCl treatment. Various tissues were collected from nonstressed hydroponic plants. Tomato tissue-culture cells were grown in the normal liquid medium (S0 cells) or in the medium containing 15 g/L NaCl (S15 cells), as described by Hasegawa et al. (1980).
Measurement of Pro Content in Tissues
Frozen materials were ground with a mortar and pestle in methanol:chloroform:water (12:5:1, v/v), and Pro content was determined by the acid ninhydrin procedure as described in Troll and Lindsley (1955).
Analyses of RNA
Total RNA was obtained by the LiCl-precipitation method as described in Nagy et al. (1988). For northern-blot analysis, 20 μg of total RNA was electrophoresed on a formaldehyde-agarose gel (1.2% agarose). After electrophoresis, the analysis was carried out by the standard protocol (Sambrook et al., 1989) on Hybond N+ membrane (Amersham). For use as probes of northern blots, fragments containing nucleotides 1 to 899 of the tomPRO1 cDNA clone plus an additional 90 bp derived from the multicloning site in a vector or the full-length EcoRI fragment of tomPRO2 cDNA were amplified with PCR and labeled with [α-32P]dATP and/or dCTP by a random-primer reaction according to the manufacturer's instructions (Amersham). After hybridization, filters were washed three times for 20 min with 0.1× SSC and 0.1% SDS at 42°C. RNase protection analyses were performed as described previously (García-Ríos et al., 1997) with 80 μg of total RNA, and probed with an antisense riboprobe from tomPRO1 cDNA using the HybSpeed RPA kit (Ambion, Austin, TX). Levels of mRNA were quantified by scanning of the autoradioagrams with a densitometer (Molecular Dynamics, Sunnyvale, CA). To correct for loading differences in the northern blots (Figs. 1B and 2B), the blots were also probed with a fragment containing the 18S and 25S rRNA genes of flax (obtained from Dr. Joel Gaffe, Purdue University), and the densitometer readings for the tomPRO2 signal for each sample were normalized to the total 25S and 18S rRNA signal.
Computer Analyses
Analyses of nucleotide and amino acid sequences were carried out with programs in the Genetics Computer Group (GCG) package of the University of Wisconsin, Madison, through a UNIX system. Comparisons against sequences in GenBank and amino acid sequence alignments were performed using the GAP and PILEUP programs, respectively. The codon usage table was derived by the CODONFREQUENCY program, and the codon usage tables for low- and high-expression genes in Escherichia coli and for genes of tomato were also supplied by the GCG package. For constructing phylogenetic trees, the neighbor-joining method was performed on the amino acid-composition data using the SEQBOOT, PROTDIST, NEIGHBOR, and CONSENSE tools from the PHYLIP program (Phylogeny Inference Package, version 3.5c, 1993; J. Felsenstein, Department of Genetics, University of Washington, Seattle). Bootstrapping was performed with 100 replicates. Distances were calculated using the Dayhoff PAM matrix option of PROTDIST. Abbreviations and accession numbers are: tomPRO2, tomato P5CS, U60267; Arabid, Arabidopsis P5CS, D32138; ArabidB, Arabidopsis P5CS2, X86778; Vigna, V. aconitifolia P5CS, M92276; Medicago, Medicago sativa P5CS, X98421; Rice, Oryza sativa P5CS, D49714; Homos, Homo sapiens P5CS, X94453; Cele, Caenorhabditis elegans P5CS, Z50797; Yeast, Saccharomyces cerevisiae GK and GPR, P32264 and X90565; Coryne, Corynebacterium glutamicum GK and GPR, U31230 and X82929; Bacsub, Bacillus subtilis GK and GPR, P39820 and P39821; Tthermo, Thermus thermophilus GK and GPR, D29973; Trepone, Treponema pallidum GK and GPR, U61535; Haein, Hemophilus influenzae GK and GPR, P43763 and U32804; Serma, Serratia marcescens GK and GPR, P17856 and P17857; Ecoli, E. coli GK and GPR, P07005 and P07004; Synecho, Synechocystis sp. GK and GPR, D90903 and D64001; Strept, Streptococcus thermophilus GK and GPR, X92418; tomPRO1, tomato GK and GPR, U27454.
Complementation of Pro Auxotrophy in E. coli
For the construction of a Pro auxotrophic derivative (KC1325) of E. coli strain BL21(DL3)pLysS (Novagen, Madison, WI), the proB1658::Tn10 insertion, which is polar on proA (Mahan and Csonka, 1983), was transduced with P1 phage (Miller, 1972) into BL21(DE3)pLysS, selecting Tetr progeny. Strain KC1325 was not able to grow without Pro, but the parental strain was, confirming that KC1325 is a Pro auxotroph. A tomPRO2 fragment (nucleotides 44–2197) containing a complete open reading frame was amplified by PCR with the primers 5′-TTCCATGGAGACAGTTGATTCAACTCG-3′ and 5′-TTGGATCCATCACCCTTGCTGAGTAAGGT-3′ (which contain NcoI and BamHI restriction enzyme sites, respectively), and the fragment was cloned between the NcoI and BamHI sites of pET32a vector (Novagen) to yield pET32PRO2, resulting in a fusion protein of tomPRO2 with an N-terminal extension from Trx-, His-, and S-tag sequences. Construction of pPRO1, which carries the tomPRO1 cDNA in the EcoRI site of pBluescript KSII(+) (pKS; Stratagene), has been described by García-Ríos et al. (1997). Plasmids pPRO1, pET32PRO2, pKS, and pET32a were electroporated into KC1325, respectively. Complementation tests were carried out at 37°C on solid medium 63 (Cohen and Rickenberg, 1956) containing 10 mm Glc, 0.05 mm thiamine-HCl, and 1 mm IPTG, with or without 1 mm Pro.
Expression of Recombinant Proteins
For tomPRO1 expression, E. coli strain HB101 (ΔproBA leu thi-1) was transformed with pPRO1. The transformants were grown in Luria-Bertani broth with ampicillin (100 μg/mL) at 37°C for 10 h. pET32PRO2 was used for the transformation of the strain KC1325. Production of a recombinant protein for tomPRO2 was induced by 1 mm IPTG at 25°C for 17 h, based on the manufacturer's instructions (Novagen). Cells were collected and resuspended in a 125 mm Tris-HCl (pH 6.8), 4% SDS, 5% β-mercapthoethanol, and 20% glycerol. Total crude extracts were separated by 12% or 10% SDS-PAGE and then visualized with Coomassie brilliant blue R250 as in Sambrook et al. (1989).
RFLP Mapping
RFLP linkage analyses were performed utilizing F2 progeny from the cross between L. esculentum and Lycopersicon pennellii. DNA samples from 67 of the F2 progeny had been digested by various restriction enzymes, separated by electrophoresis, and transferred to Hybond N+ membranes. These membranes, which had been used previously for the mapping of numerous other markers (Tanksley et al., 1992), were generously provided by Dr. G. Martin (Purdue University). RFLP markers were also collected and supplied by Dr. G. Martin. 32P-labeled probe preparation and DNA gel-blot analyses were basically the same as for the RNA gel-blot analyses, except that they were washed with 0.2× SSC, 0.1% SDS at 25°C or with 0.1× SSC and 0.1% SDS at 42°C. Multipoint linkage analyses were performed using the MapMaker program (version 2.0, Lander et al., 1987). Recombination frequencies from multipoint analysis were converted into map distances (in centiMorgans [cM]) using the mapping function of Kosambi (1944).
RESULTS
Isofunctional Enzymes Catalyzing the First Step of Pro Biosynthesis Are Specified by Two Distinct Genes, tomPRO1 and tomPRO2, in Tomato
We isolated a polycistronic tomPRO1 locus in tomato, which specifies GK and GPR divided by an internal stop codon in a single gene (García-Ríos et al., 1997). Because the accumulated sequence information indicated that the first step of Pro biosynthesis is mediated by a bifunctional P5CS in other higher eukaryotes, we screened a tomato cDNA library with a genomic DNA fragment from Arabidopsis encoding P5CS. We identified several inserts that strongly hybridized to this probe, the longest of which was selected for further analysis. Nucleotide sequencing of this clone, which was designated tomPRO2, revealed that it contained a single, long open reading frame, flanked by 43- and 42-bp untranslated regions at the 5′ and 3′ ends, respectively. The predicted amino acid sequence of tomPRO2 indicated that it consists of a GK-GPR hybrid as a monocistron, having an overall 76% identity at the amino acid level to Arabidopsis P5CS. However, tomPRO2 shows only 35% identity to tomPRO1 products, suggesting that tomPRO2 represents a homolog of the Arabidopsis P5CS gene.
The Levels of Pro and of the P5CS Message in Various Tissues in Unstressed Tomato Plants
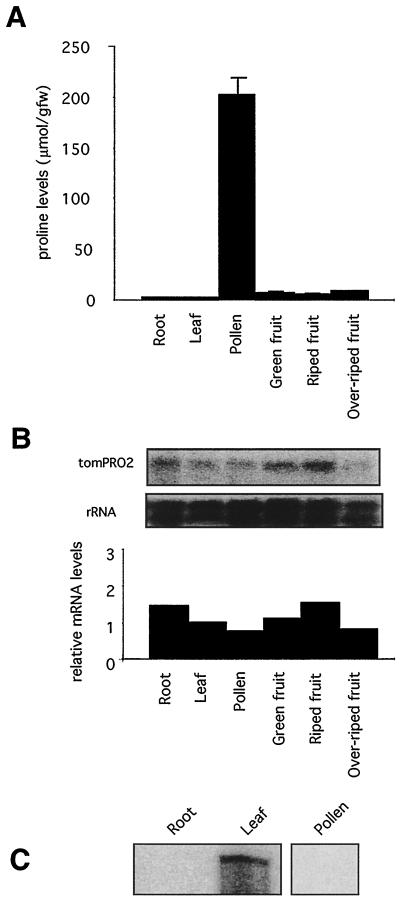
In tomato plants grown under nonstressed conditions, Pro was present in the range of 1 to 7 μmol/g fresh weight in roots, leaves, and fruits of various stages (Fig. 1A). However, in accord with previous reports that pollen are rich in free Pro (Khoo and Stinson, 1957; Hong-qi et al., 1982), we found that the level of this imino acid was 200 μmol/g fresh weight in tomato pollen. We examined the accumulation of tomPRO1 and tomPRO2 mRNAs in the different tissues in the unstressed plants using northern-blot analyses with the tomPRO1 and tomPRO2 cDNA inserts as the probes. The tomPRO2 message was readily observable in all tissues, but the tomPRO1 message was not detectable by northern blots (data not shown), indicating that tomPRO1 is not expressed at all or at a substantially lower level than the tomPRO2 gene in the tissues analyzed. As shown in Figure 1B, the level of the tomPRO2 mRNA, normalized to the rRNA level to correct for loading errors, was nearly the same in all of the tissues, including pollen. Whereas the tomPRO1 message was not observable with northern blots, using the more sensitive RNase-protection assay, we were able to observe it in leaves (Fig. 1C). The tomPRO1 transcript was undetectable in roots and pollen even with this assay. Thus, our data indicate that the high-level accumulation of Pro in pollen was not correlated with a detectable induction in the levels of the tomPRO1 and tomPRO2 transcripts.
Figure 1.
Relationship between Pro accumulation and mRNA levels in unstressed tomato tissues. A, Pro levels in different tissues from unstressed tomato. Pro values are means ± se (n = 3). gfw, Grams fresh weight. B, Relative RNA levels in different tissues from unstressed tomato. Same materials as in A were used to extract total RNA. The top panel shows the result of northern-blot analysis, which was carried out using 20 μg of total RNA and probed with tomPRO2 cDNA, as described in Methods. The rDNA probe was used as a control for sample loading. RNA levels in each sample were quantified by densitometric scanning of the autoradiograms and normalized to the respective rRNA signal. The bottom panel shows normalized levels of the tomPRO2 mRNA in the indicated tissues compared with leaves, where the value for leaves has been set to 1.0. C, RNase-protection assay of tomPRO1 in roots, leaves, and pollen from unstressed tomato. RNase-protection analyses were performed using 80 μg of total RNA with a riboprobe from tomPRO1 cDNA, as described in Methods.
Pro Is Accumulated to High Levels in NaCl-Stressed Plants
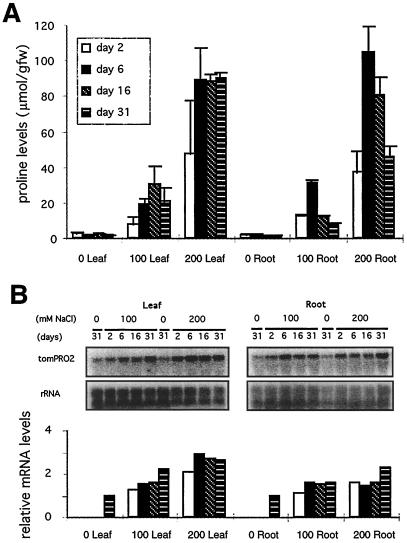
We examined the effect of NaCl stress on Pro levels in hydroponically grown tomato plants (Fig. 2A). Treatment with 100 mm NaCl elicited an approximately 15-fold increase in the level of Pro accumulation in both leaves and roots, and treatment with 200 mm NaCl resulted in 60- and 80-fold increases in these tissues, respectively. In leaves the highest level of Pro was reached after 6 d, and was maintained until 31 d after treatment; in roots, Pro decreased to about one-half of the highest level by this time. The more rapid disappearance of Pro in roots compared with leaves in NaCl-treated plants could reflect a more severe osmotic stress in leaves because of transpiration and/or slower osmotic adjustment than in roots.
Figure 2.
Relationship between Pro accumulation and tomPRO2 mRNA levels in response to NaCl stress in tomato leaves and roots. A, Pro accumulation in leaves and roots of hydroponic tomato after 2, 6, 16, and 31 d of treatment with 0, 100, and 200 mm NaCl. Pro values are means ± se (n = 3). gfw, Grams fresh weight. B, Relative RNA levels in leaves and roots of hydroponic tomato. The conditions for treatment were the same as in A. The top panel represents the result of northern-blot analysis, which was performed using total RNA (20 μg) probed with tomPRO2 cDNA. The rDNA probe was used as a control for sample loading, and the results were measured, as described in the Figure 1B legend. The relative tomPRO2 mRNA levels are shown in the bottom panel, with the message set to 1.0 for the 31-d samples in the untreated leaves and roots, respectively.
The same plants that were used for Pro measurement were also subjected to northern analysis to determine the effect of osmotic stress on the accumulation of the P5CS transcripts. As before, the tomPRO2 mRNA was much more abundant than the tomPRO1 mRNA, which was not detectable with northern analysis in any of the tissues in the NaCl-stressed plants (data not shown). NaCl stress resulted in some increase in the accumulation of the tomPRO2 transcript. However, the tomPRO2 message level, normalized to the rRNA signal, increased only about 2-fold after 2 d of treatment with 100 and 200 mm NaCl in both leaves and roots, and remained at almost the same level for 31 d after the treatment (Fig. 2B). Although Pro accumulation in leaves and roots treated with 200 mm NaCl was 3- to 7-fold higher throughout the entire course of the treatment than in the same tissues treated with 100 mm NaCl (Fig. 2A), we have no evidence that the level of the tomPRO2 mRNA is altered substantially by either the severity or duration of osmotic stress (Fig. 2B). Thus, our results show that the tomPRO2 transcript level was induced much less by NaCl in tomato than in Arabidopsis, rice, and moth bean (Hu et al., 1992; Savouré et al., 1995; Yoshiba et al., 1995; Igarashi et al., 1997; Strizhov et al., 1997), even though tomato accumulated >15-fold higher levels of Pro than those other plants. Thus, our results suggest that control of the accumulation of the tomPRO2 message level is probably not important for the regulation of Pro synthesis by NaCl stress.
Effect of NaCl Stress on the Pro Levels and the Accumulation of the P5CS Transcripts in Tissue-Culture Cells
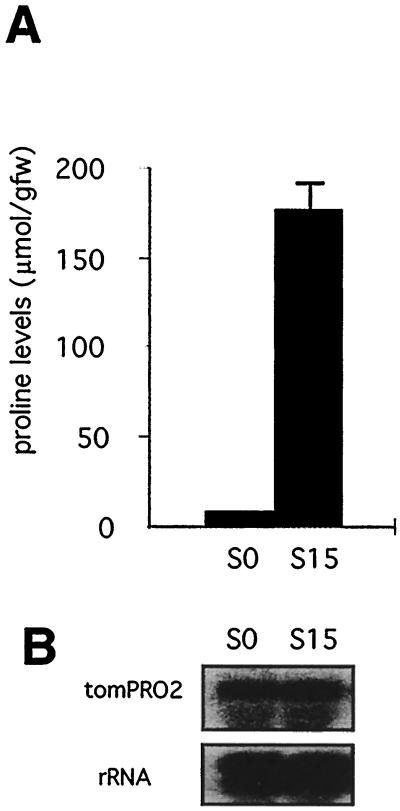
We also measured the Pro levels in normal and NaCl-adapted tissue-culture cells. As shown in Figure 3A, cells grown in normal medium (S0) had a very low level of Pro, whereas cells grown in medium containing 15 g/L NaCl (S15) had an approximately 30-fold higher level of this imino acid. Despite this difference in the Pro content, the level of the tomPRO2 message was essentially the same in the two types of cells, as detected by northern blotting (Fig. 3B), indicating that the 30-fold increase in the Pro levels seen in tissue-culture cells adapted to 15 g/L NaCl occurred without any notable change in the accumulation of the tomPRO2 message. The tomPRO1 mRNA was not detectable by northern-blot analysis in either of the cells, but as reported earlier (García-Ríos et al., 1997), the more sensitive RNase-protection assays demonstrated that the S15 cells had a 4-fold higher level of the tomPRO1 message than the S0 cells. However, because of the low abundance of the tomPRO1 message even in the NaCl-adapted cells, the induction of this message probably is not sufficient to account for the elevation in the Pro pools size in the tissue-culture cells.
Figure 3.
Relationship between Pro accumulation and mRNA levels in tomato tissue-culture cells. A, Comparison of Pro levels in tissue-culture cells grown in normal medium (S0) and in medium containing 15 g/L NaCl (S15). Pro values are means ± se (n = 3). gfw, Grams fresh weight. B, Relative RNA levels in the tissue-culture cells S0 and S15. Same materials as in A were used to extract total RNA. The analysis was carried out using 20 μg of total RNA and probed with tomPRO2 cDNA. The rDNA probe was used as a control for sample loading.
The tomPRO1 and tomPRO2 Loci Are Structurally Different and May Have Evolved from Separate Ancestral Genes
In addition to the different levels of expression of tomPRO1 and tomPRO2 described above, the two cDNAs have remarkable structural differences. The tomPRO1 has a dicistronic structure (García-Ríos et al., 1997), which is unusual in eukaryotes (Kozak, 1986). Thus far, all genes encoding GK and GPR in bacteria and yeast have been reported as separate genes, organized as an operon in most bacteria. In contrast, tomPRO2 encodes a bifunctional enzyme formed by a hybrid of GK and GPR without an intervening stop codon, similar to genes encoding a hybrid GK and GPR, or P5CS, that have been cloned from several higher eukaryotes, such as C. elegans, H. sapiens, and higher plants (Hu et al., 1992; Savouré et al., 1995; Yoshiba et al., 1995; Liu et al., 1996; Igarashi et al., 1997).
Table I shows that the predicted amino acid sequence of tomPRO2 has greater than 76% identity with other plant P5CS, but shows a much lower identity (35%–44%) with the tomPRO1 product and the bacterial and yeast GK and GPR enzymes. In contrast, the predicted tomPRO1 product has a low amino acid identity (32%–47%) with GK and GPR from yeast and prokaryotes and with P5CS from all other eukaryotes. Although the tomPRO1 product has a similar homology to the corresponding proteins from different organisms, sequence alignment shown in Figure 4 revealed that tomPRO1 is closer to bacterial GK and GPR than to P5CS of plants. Some regions that are highly conserved in P5CS proteins from plants are either missing, divergent, or carry insertions in tomPRO1 and in bacterial GK and GPR (e.g. amino acids 71–83, 151–161, and 194–219 in GK, and amino acids 426–451, 476–497, and 571–577 in GPR, where the numbers of amino acid residues are based on tomPRO1 sequences). However, the GK part of tomPRO1 also shows a striking difference from most bacterial GKs, because it lacks an approximately 100-amino acid C-terminal tail that is conserved in most other prokaryotic GKs (represented by E. coli GK, amino acids 259–367, and B. subtilis GK, amino acids 256–354 [García-Ríos et al., 1997]). At present, S. thermophilus is the only exception among bacteria that also lacks this C-terminal tail in GK. The tomPRO1 product has the closest sequence similarity to the GK and GPR from the latter organism (Table I).
Table I.
Comparison of predicted amino acids from various GKs, GPRs, and P5CSs
| Namea | tomPRO1 | tomPRO2 |
|---|---|---|
| % | ||
| tomPRO2 | 35 /57b | 100 |
| Arabid | 34 /56 | 76 /87 |
| ArabidB | 34 /56 | 76 /88 |
| Vigna | 32 /54 | 76 /88 |
| Medicago | 33 /56 | 77 /87 |
| Rice | 34 /55 | 76 /85 |
| Homos | 34 /56 | 48 /66 |
| Cele | 32 /56 | 47 /66 |
| Yeast | 37 /59 | 39 /59 |
| Coryne | 40 /61 | 34 /57 |
| Bacsub | 39 /56 | 35 /56 |
| Tthermo | 39 /61 | 35 /56 |
| Trepone | 41 /60 | 35 /55 |
| Haein | 39 /60 | 37 /59 |
| Serma | 40 /60 | 38 /59 |
| Ecoli | 40 /61 | 39 /60 |
| Synecho | 41 /62 | 44 /62 |
| Strept | 47 /65 | 37 /59 |
| tomPRO1 | 100 | 35 /57 |
Comparison was done by the GAP program of the GCG. Abbreviations and accession numbers for these proteins are as in Methods.
Each number is depicted as identity/similarity.
Figure 4.
Amino acid sequence comparison of tomPRO1, tomPRO2, and other related genes. Predicted amino acid sequences of proB, proA, and P5CS genes were aligned using the multiple alignment program PILEUP, and the results were highlighted with the BOXSHADE program. Letters in the black and gray backgrounds indicate identical and similar residues, respectively. Representative regions that are highly conserved in P5CS proteins in plants but are either missing, divergent, or carry insertions in GK and GPR for tomPRO1 and in bacterial GK and GPR are underlined. Extended C-terminal tails of GK, which are conserved in most of prokaryotic GK, are shown by a dashed line. Abbreviations and accession numbers are provided in Methods.
We compared codon usage of tomPRO1 and tomPRO2 with the average codon usage of tomato genes and of genes expressed at low or high levels in E. coli. The tomPRO2 codon usage agrees well with the preference of average codon usage from tomato genes, whereas tomPRO1 codon usage deviates from the usage of tomato genes (Table II), and in fact, appears to be between the preference in genes in tomato and in E. coli (results not shown).
Table II.
Comparison of codon frequency usage in tomPRO1, tomPRO2, and general tomato genes
| Amino Acid | Codon | tomPRO1 | tomPRO2 | Tomato |
|---|---|---|---|---|
| % | ||||
| Arg | CGA | 14 | 12 | 9 |
| CGC | 29′ a | 5 | 7 | |
| CGG | 43" b | 5 | 4 | |
| CGU | 14 | 29′ | 18 | |
| AGA | 0 | 36" | 37" | |
| AGG | 0 | 14 | 24′ | |
| Leu | CUA | 7 | 10 | 9 |
| CUC | 4 | 11 | 13 | |
| CUG | 10 | 14 | 7 | |
| CUU | 7 | 35" | 30" | |
| UUA | 28′ | 12 | 12 | |
| UUG | 42" | 19′ | 30" | |
| Ser | UCA | 28′ | 21 | 26" |
| UCC | 0 | 7 | 13 | |
| UCG | 10 | 5 | 7 | |
| UCU | 14 | 28′ | 25′ | |
| AGC | 3 | 7 | 13 | |
| AGU | 45" | 33" | 16 | |
| Thr | ACA | 24 | 35′ | 32′ |
| ACC | 27′ | 9 | 22 | |
| ACG | 41" | 6 | 8 | |
| ACU | 8 | 50" | 38" | |
| Pro | CCA | 56" | 40′ | 47" |
| CCC | 7 | 15 | 13 | |
| CCG | 22′ | 0 | 6 | |
| CCU | 15 | 45" | 35′ | |
| Ala | GCA | 13 | 32′ | 30′ |
| GCC | 31′ | 13 | 18 | |
| GCG | 34" | 4 | 5 | |
| GCU | 21 | 51" | 46" | |
| Gly | GGA | 10 | 44" | 38" |
| GGC | 27 | 22 | 13 | |
| GGG | 29′ | 11 | 11 | |
| GGU | 34" | 24′ | 38" | |
| Val | GUA | 7 | 17 | 14 |
| GUC | 24′ | 7 | 16 | |
| GUG | 48" | 33′ | 25′ | |
| GUU | 20 | 43" | 45" | |
| Lys | AAA | 58 | 55 | 46 |
| AAG | 42 | 45 | 54 | |
| Asn | AAC | 52 | 33 | 45 |
| AAU | 48 | 67 | 55 | |
| Gln | CAA | 76 | 44 | 64 |
| CAG | 24 | 56 | 36 | |
| His | CAC | 20 | 38 | 39 |
| CAU | 80 | 63 | 61 | |
| Glu | GAA | 76 | 51 | 52 |
| GAG | 24 | 49 | 48 | |
| Asp | GAC | 21 | 22 | 33 |
| GAU | 79 | 78 | 67 | |
| Tyr | UAC | 13 | 19 | 53 |
| UAU | 87 | 81 | 47 | |
| Cys | UGC | 0 | 43 | 46 |
| UGU | 100 | 57 | 54 | |
| Phe | UUC | 15 | 22 | 44 |
| UUU | 85 | 78 | 56 | |
| Ile | AUA | 4 | 25 | 18 |
| AUC | 16 | 23 | 29 | |
| AUU | 80 | 52 | 52 | |
Values are given of each codon in each amino acid. Trp, Met, and stop codons are not included.
′, Second-most frequently used codon.
", Most frequently used codon.
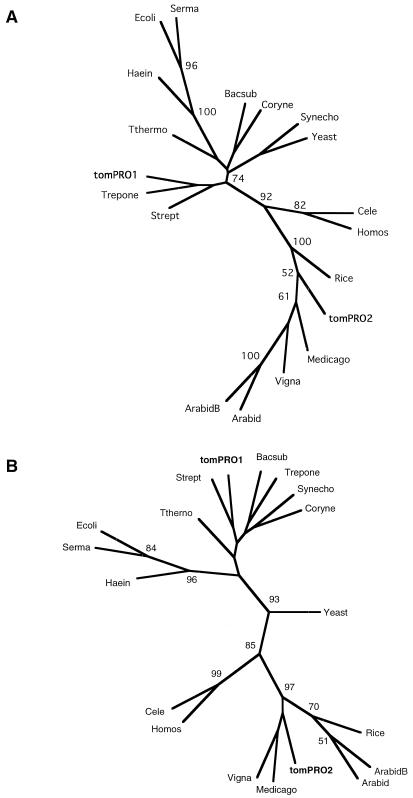
Because of the intriguing differences in nucleotide and predicted amino acid sequences of tomPRO1 and tomPRO2, the relationship of these two loci to other GK, GPR, and P5CS from various species was examined further by a phylogenetic analysis. A phylogenetic tree was constructed by the neighbor-joining method from a highly conserved region of GK (amino acids 84–149 of tomPRO1). Figure 5A revealed that P5CS proteins from higher eukaryotes are clearly monophyletic, and that tomPRO2 was tightly clustered as a member of plant P5CS within this group. On the other hand, tomPRO1 appeared together with T. pallidum and S. thermophilus within a bacterial group. This trend is also true for the phylogenetic tree constructed from a highly conserved region of GPR (361–436 amino acids of tomPRO1) as shown in Figure 5B. Whereas the GPR domain of tomPRO2 grouped with the corresponding regions of other eukaryotes, the tomPRO1-encoded GPR clustered with bacterial ones. These results suggest that tomPRO1 and tomPRO2 were probably incorporated into the tomato genome separately during evolution.
Figure 5.
Possible evolutionary relationship among the GK (A) and the GPR (B) proteins. The phylogenetic tree was generated using the PHYLIP program (Felsenstein, 1993). Numbers are bootstrap values given as percentages, and only 50% or greater values are indicated at a node. Abbreviations and accession numbers are as described in Methods.
tomPRO1 and tomPRO2 Encode Functional Enzymes Catalyzing GK and GPR Activities
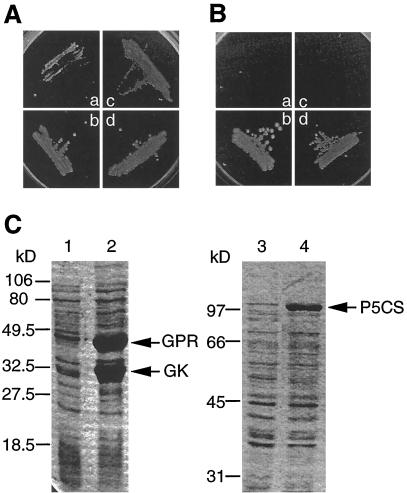
We demonstrated that the tomPRO2 gene encodes a functional P5CS by the fact that it can complement both a GK and a GPR defect in E. coli. As shown in Figure 6B, the plasmid carrying the tomPRO2 fragment could complement the polar proB1658::Tn10 mutation, whereas the vector by itself was unable to do so. In accord with our earlier observations (García-Ríos et al., 1997), the tomPRO1 cDNA clone inserted pBluescript KSII(+) could likewise complement the Pro auxotrophic mutation in KC1325 (Fig. 6B). All strains could grow on the medium containing Pro (Fig. 6A). These results demonstrate that although both tomPRO1 and tomPRO2 show only 35% amino acid sequence identity, they both specify functional GK and GPR.
Figure 6.
Complementation of a proBA mutation by tomPRO1 and tomPRO2, and their products expressed in E. coli. A and B, Expression vectors containing the tomPRO1 and tomPRO2 cDNA clones were introduced into strain KC1325 (a derivative of BL21[DL3]pLysS carrying the proB1658::Tn10 insertion, which is polar on proA). a, KC1325 harboring the vector, pKS only. b, KC1325 harboring pPRO1. c, KC1325 harboring pET32a only. d, KC1325 harboring pET32PRO2. Strain KC1325 containing each plasmid was streaked on minimal M63 medium containing Glc, thiamine, and IPTG with (A) and without (B) Pro, and incubated for 2 d at 37°C. All strains could grow on the media supplemented with Pro (A). C, Total cell extracts from either E. coli strain HB101, containing pKS (lane 1) and pPRO1 (lane 2), or strain KC1325, containing pET32a (lane 3) and pET32PRO2 (lane 4), were analyzed by SDS-PAGE. The gels were stained with Coomasie brilliant blue. tomPRO1 products are indicated as GK and GPR, and the tomPRO2 product as P5CS. Numbers at left refer to size standards (in kD).
We verified with SDS-PAGE analysis that the tomPRO2 gene directs the synthesis of a single polypeptide of the expected mass of 98 kD in E. coli (Fig. 6C), whereas tomPRO1 directs the synthesis of two polypeptides: the approximately 33-kD GK and the approximately 44-kD GPR, in accord with our previous report (García-Ríos et al., 1997) that tomPRO1 is recognized as a polycistronic locus in E. coli.
tomPRO1 and tomPRO2 Are Located at Different Loci within the Tomato Nuclear Genome
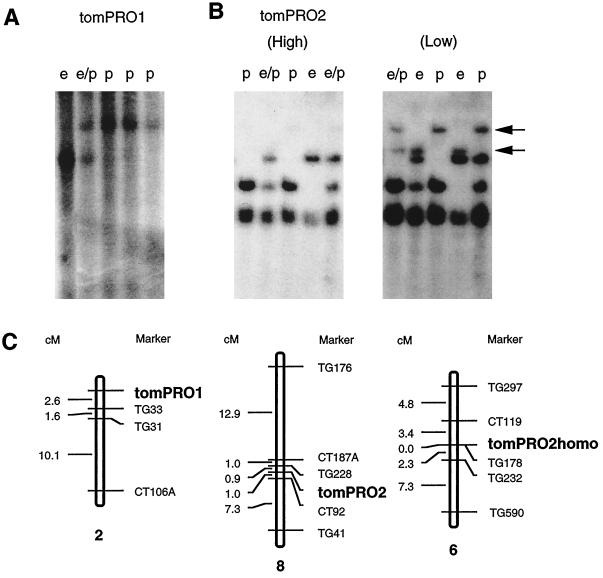
Restriction fragments of total tomato DNA were probed with sequences from the GK region of tomPRO1 and with full-length tomPRO2. Probes made from both clones hybridized with an efficiency of less than 10 copies per haploid genome (data not shown). This result suggested that each locus is present in the nuclear genome but not in an organelle genome, which is present at a much higher copy number (>50 copies).
We mapped both genes using a segregating population of progeny from the cross L. esculentum × L. pennellii. As shown in Figure 7, the tomPRO1 locus was mapped to a region of 2.6 cM adjacent to the TG33 locus on chromosome 2. Mapping of the tomPRO1 locus to chromosome 2 supports unambiguously our conclusion that a polycistronic locus is present in the tomato nuclear genome, as opposed to being in a chloroplast or mitochondrial genome, which would not only be in a much higher copy, but would also be maternally inherited. The tomPRO2 locus proved to be present in a region of approximately 1 cM between the TG228 and CT92 markers on chromosome 8 (Fig. 7B, left panel). Southern analysis with a tomPRO2 probe at a milder stringency (Fig. 7B, right panel) revealed additional bands that appeared homologous to tomPRO2, which did not hybridize to a tomPRO1 probe under these conditions. These tomPRO2-related bands were mapped to chromosome 6.
Figure 7.
Mapping of tomPRO1, tomPRO2, and tomPRO2 homolog in tomato nuclear genome. A and B, Southern-blot analyses of total DNA from the F2 population of crosses between L. esculentum and L. pennellii. Hybridizations were performed with the GK part of the tomPRO1 cDNA fragment (A) and the full length of tomPRO2 cDNA as the probes (B) at a high-stringency wash condition, 0.1× SSC, 0.1% SDS, at 42°C (High), and at a low-stringency wash condition, 0.2× SSC, 0.1% SDS, at 25°C (Low). Two bands that appeared at a low-stringency condition are depicted by arrows. This figure shows a representative portion of the blots, in which a total of 67 of the F2 populations were used for the RFLP mapping. Shown at the top of each lane is the RFLP pattern representative for the L. esculentum homozygote (e), the L. pennellii homozygote (p), or their heterozygote (e/p). C, Map position of tomPRO1, tomPRO2, and tomPRO2 homologs on the tomato chromosome. The maps were drawn by segregation analysis of RFLPs based on data by Tanksley et al. (1992). The map distances (in cM) are indicated on the left. Maps are not drawn to scale. tomPRO1, tomPRO2, and tomPRO2 homologs (tomPRO2homo) were located on chromosomes 2, 8, and 6, respectively.
DISCUSSION
Pro Accumulation Is Not Correlated with the tomPRO1 and tomPRO2 Message Levels in Tomato
It has been proposed that transcriptional control of the P5CS gene, which encodes a bifunctional enzyme catalyzing the first and second reactions of Pro synthesis, is important for the regulation of accumulation of this imino acid during osmotic stress in plants. This conclusion was based on the observation that NaCl stress increased the P5CS transcript level in moth bean, Arabidopsis, and rice (Hu et al., 1992; Savouré et al., 1995; Yoshiba et al., 1995; Igarashi et al., 1997). In rice and Arabidopsis, the increases in the Pro levels were accompanied by coordinate increases in the P5CS transcript levels. (The accumulation of Pro was not monitored in moth bean during the course of induction of the P5CS message [Hu et al., 1992].) Arabidopsis has two P5CS isoenzymes, encoded in the AtP5CS1 and AtP5CS2 genes (Savouré et al., 1995; Yoshiba et al., 1995; Strizhov et al., 1997; Zhang et al., 1997). AtP5CS1, which was estimated to synthesize about 60% to 80% of the total P5CS mRNA (Strizhov et al., 1997), exhibited up to an 8-fold induction upon osmotic stress (Savouré et al., 1995; Yoshiba et al., 1995; Strizhov et al., 1997), whereas AtP5CS2 exhibited a <4-fold regulation (Strizhov et al., 1997; Zhang et al., 1997). Because tomato accumulates much more Pro than Arabidopsis or rice, our initial hypothesis had been that tomato might show an even more sensitive regulation of P5CS transcript accumulation than the other two plants. To test whether this is the case, we determined the effect of NaCl stress on the accumulation of the tomPRO1 and tomPRO2 transcripts. We also determined whether there is a special transcriptional regulation of these two loci in pollen, which contain very high levels of Pro.
Although NaCl stress was shown to cause some Pro accumulation in rice and Arabidopsis, this metabolite reached only a maximum of 2 and 6 μmol Pro/g fresh weight in these two species (Savouré et al., 1995; Igarashi et al., 1997; Zhang et al., 1997). In contrast, we found that tomato accumulated to 90 and 105 μmol Pro/g fresh weight in leaves and roots, respectively, after 6 d of treatment with 200 mm NaCl (Fig. 2), representing a 60- to 80-fold increase over the level in unstressed plants. Surprisingly, in view of the results reported for Arabidopsis and rice, there was only about a 2- to 3-fold change in the tomPRO2 transcript level throughout the entire time course of NaCl treatment. In roots the accumulation of Pro was maximal at d 6 of NaCl treatment, after which it declined gradually, but this was not reflected by a decrease in the tomPRO2 message level (Fig. 2). The Pro pool size was 30-fold higher in the NaCl-adapted S15 tomato tissue-culture cells than in the control, unadapted S0 cells (Fig. 3). Despite this large difference in Pro content, the tomPRO2 message was present at similar levels in the two types of cells.
Previously, we showed that the level of the tomPRO1 message in the S15 cells was approximately 4-fold higher than in the S0 cells (García-Ríos et al., 1997). However, because of the low level of this transcript even in the NaCl-stressed cell line, this induction of the transcript probably is not sufficient to account for the increase in the Pro content. The highest level of Pro in all tissues tested was found in pollen of unstressed plants. (We did not measure the Pro content in pollen of NaCl-stressed plants.) The tomPRO2 message level, however, was unchanged compared with other tissues, and the tomPRO1 message was undetectable in pollen. These results indicate that in tomato, the large increases in Pro levels in response to NaCl stress or pollen-specific developmental signals are brought about without substantial increases in the levels of the tomPRO1 and tomPRO2 messages.
Because we only determined the steady-state accumulation of these messages, we cannot infer that tomPRO1 and tomPRO2 are transcribed constitutively. In principle, it is conceivable that changes in the rates of synthesis of these transcripts could be compensated for by comparable changes in their turnover. Zhang et al. (1997) demonstrated with a GUS reporter fusion that the 2- to 4-fold increase in the level of the AtP5CS2 message after dehydration or NaCl stress in transgenic Arabidopsis and tobacco plants was the result of transcriptional induction. However, all of the other studies on the regulation of the accumulation of the P5CS messages in Arabidopsis and rice (Savouré et al., 1995; Yoshiba et al., 1995; Igarashi et al., 1997; Strizhov et al., 1997) involved only measurements of the steady-state levels of these messages, and, therefore, direct evidence is lacking that the increases in the accumulation of these transcripts upon water stress are necessarily brought about by induction of transcription initiation.
Aside from control of the accumulation of P5CS message (at the level of synthesis or turnover), there are several other possible mechanisms for the control of Pro biosynthesis. The enzyme for the first step, GK, is sensitive to feedback regulation by Pro (Hu et al., 1992; García-Ríos et al., 1997). However, there may be important differences in the allosteric properties of the enzymes in tomato and other plants, as indicated by the observations that the activity of the tomPRO1-encoded P5CS was inhibited 50% by 0.07 mm Pro (García-Ríos et al., 1997), whereas 5 mm Pro was required to elicit 50% inhibition of the GK activity of moth bean P5CS (Zhang et al., 1995). We have not been successful in measuring the kinetic properties of tomPRO2 product because of difficulties in obtaining this enzyme in a soluble form. However, we have preliminary evidence that this enzyme, which is more similar in its amino acid sequence to the moth bean P5CS than to the tomPRO1 product, is also sensitive to feedback inhibition by Pro. It is possible that the regulation of synthesis of Pro in tomato is effected by relief of allosteric inhibition of the activities of the tomPRO1 and tomPRO2 products under NaCl or dehydration stress. Tomato may have an additional gene related to tomPRO2 (Fig. 7), for which we have no sequence information. If it proves to be related to P5CS, it could participate in the regulation of Pro synthesis.
The accumulation of Pro could also be regulated by changes in the rate of its catabolism to glutamate by the combined action of Pro dehydrogenase and P5C dehydrogenase. However, because Pro dehydrogenase is a mitochondrial enzyme (Kiyosue et al., 1996), effective catabolism of Pro would presumably require transport of Pro from the cytosol to the mitochondria. In Arabidopsis NaCl stress or dehydration down-regulates the accumulation of the message for Pro dehydrogenase (Kiyosue et al., 1996; Peng et al., 1996; Verbruggen et al., 1996). Although repression of the synthesis of Pro dehydrogenase could have a role in the long-term regulation of Pro accumulation in response to water stress, the effect of water stress on the activity or stability of Pro dehydrogenase itself has not been determined in Arabidopsis. Repression of transcription of the gene for Pro dehydrogenase would be an efficient mechanism for increasing the Pro pools size only if this response is accompanied by a simultaneous inactivation or turnover of preexisting Pro dehydrogenase molecules. Direct evidence on the relative contributions of the biosynthetic and catabolic pathways for the regulation of Pro pool size was provided in cultured tomato cells by the 15N-isotope-tracing experiments of Rhodes et al. (1986). These studies indicated that the 300-fold increase in the Pro accumulation resulting from 25% PEG stress was mainly due to a 10-fold increase in the rate of biosynthesis and provided no evidence that the rate of Pro catabolism was inhibited under these conditions.
Changes in the intracellular Pro levels could also be accomplished by translocation of this metabolite between different tissues or cell compartments. Two genes, ProT1 and ProT2, which encode closely related Pro-transport proteins, have been cloned from Arabidopsis (Rentsch et al., 1996). Accumulation of the ProT2 message was strongly elevated by NaCl stress, indicating that control of the synthesis of Pro-transport proteins also could be involved in the regulation of the cellular Pro pool sizes.
Two Evolutionarily Distinct Genes Are Present in the Tomato Nuclear Genome
We showed that the tomPRO1 and tomPRO2 loci are present in the tomato nuclear genome. Comparison of protein sequence, codon usage, and phylogenetic analysis suggested that tomPRO2 is in a tight group containing the P5CS proteins from plants and other higher eukaryotes. In contrast, tomPRO1 has several unique features that distinguish it from the eukaryotic P5CS group. First, tomPRO1 did not show a high identity to eukaryotic P5CS (32%–35% at the amino acid level). A comparison of codon usage of tomPRO1 with genes from E. coli or tomato suggested that the codon usage of tomPRO1 is not typical for either E. coli or tomato genes, but something in between. Second, tomPRO1 has a dicistronic structure, similar to polycistronic operons found in bacteria, and in fact, tomPRO1 is recognized as a dicistronic operon in E. coli (Fig. 6C). Third, phylogenetic analysis placed tomPRO1 within the same clade of prokaryotes, separate from the other eukaryotic P5CS genes. These characteristics suggest that tomPRO1 and tomPRO2, which are both nuclear loci, might have different origins. We present alternative hypotheses for the possible origin of the tomPRO1 locus, but we would like to emphasize that at this stage we do not have sufficient data to distinguish among these speculative alternatives.
There are several reports in which isozymes are encoded by multigene families in nuclear genomes, as exemplified by the existence of dual genes for P5CS in Arabidopsis (Strizhov et al., 1997). Multigene families may be derived by gene duplication or by gene conversion from a single gene. It is, however, unlikely that the tomPRO1 and tomPRO2 loci arose in tomato by such mechanisms, because of the difference in their coding regions. The prokaryotic features of tomPRO1 are consistent with the notion that it may have been acquired by organelle-to-nucleus gene transfer, or by uptake of DNA of prokaryotic origin into the nuclear genome. According to the theory of endosymbiosis, mitochondria and chloroplasts originated from once free-living eubacteria (Gray, 1989), followed by the loss of genes from the organellar genomes or transfer to the nucleus (Weeden, 1981; Palmer, 1985). The tufA gene, encoding the chloroplast protein synthesis elongation factor Tu in Arabidopsis, and the rpl22 gene, encoding chloroplast ribosomal protein CL22, are examples of genes that were transferred from the chloroplast genome to the nucleus (Baldauf and Palmer, 1990; Gantt et al., 1991). There are two isoenzymes of glyceraldehyde-3-phosphate dehydrogenase in tobacco and maize, one found in the chloroplasts and the other in the cytosol. Although both of these isoenzymes are encoded in nuclear genome, they display sequence divergence corresponding to the prokaryotic/eukaryotic separation (Shih et al., 1986; Brinkmann et al., 1987). These examples support the endosymbiotic theory of chloroplast evolution, with subsequent transfer of genes from the endosymbiont to the host nucleus.
An alternative explanation for the origin of tomPRO1 is that it may have been acquired from a bacterium or virus by a horizontal gene transfer. This mechanism has been invoked to explain the close homology between vertebrate hemoglobin genes and the leghaemoglobin gene from legume (Lewin, 1981). Some soil bacteria (e.g. the genus Rhizobium) have two forms of Gln synthetase, a prokaryotic type and a eukaryotic type. It has been proposed that the eukaryotic-type genes may have been incorporated by a horizontal transfer from a host plant to symbiont bacteria (Carlson and Chelm, 1986; Smith et al., 1992).
It is unlikely that tomPRO1 has been translocated from an organelle to the nucleus, because if tomPRO1 originated from organelles, then there should be tomPRO1 homologs in other plants. However, we did not find any homologs in organellar genomes using a computer search. This is also supported by the results of Southern-blot analysis that sequences homologous to tomPRO1 are present in species of the Solanaceae family, such as tobacco, potato, and two wild species of tomato (L. pennellii and Lycopersicon cheesmanii), but could not be detected in rice and maize (García-Ríos, 1995). These results suggest that horizontal gene transfer may have been responsible for the integration of the tomPRO1 gene into the nuclear genome after the divergence of dicots and monocots, but before divergence of the family Solanaceae. An examination of the subcellular localization of the tomPRO1 product and a more detailed search of tomPRO1 homologs in other plants may lead to clues as to the origin of tomPRO1, as well as to the mechanism of its transfer. There is little evidence about the possibility that bacteria or viruses could be responsible for the introduction of the tomPRO1 gene into the tomato genome. However, because of its close sequence similarity to the S. thermophilus proBA and the common lack of the C-terminal 100-amino acid tail in the GK region, the tomPRO1 locus may have been derived from a bacterium related to S. thermophilus.
Although we found evidence for tomPRO1-like genes in some other Solanaceae (see above), it is not clear whether this locus is present in other plants. The tomPRO1 clone and the P5CS clone from moth bean (Hu et al., 1992) were isolated by complementation of a proB point mutation in E. coli, but all subsequent plant P5CS clones, including tomPRO2, were isolated on the basis of sequence homology with the P5CS gene family. It is possible that homologs of tomPRO1 might be present in other plants, but because of the sequence divergence between tomPRO1 and the other plant P5CS clones, it is unlikely that the former type of gene could be cloned by sequence hybridization with P5CS clones.
There are few reports of the coexistence of prokaryotic and eukaryotic forms of a gene in a single genome. The coexistence of dual genes specifying isoforms of enzymes in one organism may serve two functions. Multiple copies of genes could satisfy a need for high amounts of a particular gene product, or they could provide an efficient means for differential regulation of gene-expression development in response to different factors (Long and Dawid, 1980). It seems likely that tomPRO1 and tomPRO2 will fit into the latter type of gene family, because of their distinct pattern of expression. Because the tomPRO2 message was much more abundant than the tomPRO1 in all tissues under the conditions we tested, it is likely that tomPRO2 may have the predominant responsibility for Pro production in these situations, and it is possible that the expression of the tomPRO1 gene might be restricted to very specific cell types or developmental stages. The significance of the existence of tomPRO1 and the coexistence of tomPRO1 and tomPRO2 at this time remains elusive.
ACKNOWLEDGMENTS
We thank S. Fletcher for technical support, Dr. M. Hasebe for help with the construction of the phylogenetic tree, Dr. G. Martin for materials and assistance with RFLP mapping, Dr. D. Rhodes for helpful discussions, and Dr. L. Szabados for the sequence of portions of the ATP5CS2 gene prior to its publication.
Abbreviations:
- GK
γ-glutamyl kinase
- GPR
γ-glutamyl phosphate reductase
- IPTG
isopropyl-β-d-thiogalactopyranoside
- P5C
Δ1-pyrroline-5-carboxylate
- P5CS
Δ1-pyrroline-5-carboxylate synthetase
- RFLP
restriction fragment-length polymorphism
Footnotes
This work was funded by the U.S. Department of Agriculture (grant no. 93-37100-8871).
LITERATURE CITED
- Baldauf SL, Palmer JD. Evolutionary transfer of the chloroplast tufA gene to the nucleus. Nature. 1990;344:262–265. doi: 10.1038/344262a0. [DOI] [PubMed] [Google Scholar]
- Blum A, Munns R, Passioura J, Turner N. Genetically engineered plants resistant to soil drying and salt stress: how to interpret osmotic relations? Plant Physiol. 1996;110:1051. doi: 10.1104/pp.110.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann H, Martinez P, Quigley F, Martin W, Cerff R. Endosymbiotic origin and codon bias of the nuclear gene for chloroplast glyceraldehyde-3-phosphate dehydrogenase from maize. J Mol Evol. 1987;26:320–328. doi: 10.1007/BF02101150. [DOI] [PubMed] [Google Scholar]
- Carlson TA, Chelm BK. Nature. 1986;322:568–570. [Google Scholar]
- Cohen GN, Rickenberg RH. Concentration specifique reversible des amino acides chez E. coli. Ann Inst Pasteur Paris. 1956;91:693–720. [PubMed] [Google Scholar]
- Csonka LN. Proline over-production results in enhanced osmotolerance in Salmonella typhimurium. Mol Gen Genet. 1981;182:82–86. doi: 10.1007/BF00422771. [DOI] [PubMed] [Google Scholar]
- Csonka LN, Hanson AD. Prokaryotic osmoregulation: genetics and physiology. Annu Rev Microbiol. 1991;45:569–606. doi: 10.1146/annurev.mi.45.100191.003033. [DOI] [PubMed] [Google Scholar]
- Delauney AJ, Hu C, Kishor K, Verma DPS. Ornithine aminotransferase and proline biosynthesis in Vigna aconitifoiia. J Biol Chem. 1993;268:18673–18678. [PubMed] [Google Scholar]
- Delauney AJ, Verma DPS. A soybean gene encoding Δ1-pyrroline-5-carboxylate reductase was isolated by functional complementation in Escherichia coliand is found to be osmoregulated. Mol Gen Genet. 1990;221:299–305. doi: 10.1007/BF00259392. [DOI] [PubMed] [Google Scholar]
- Delauney AJ, Verma DPS. Proline biosynthesis and osmoregulation in plants. Plant J. 1993;4:215–223. [Google Scholar]
- Gantt JS, Baldauf SL, Calie PJ, Weeden NF, Palmer JD. Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 1991;10:3073–3078. doi: 10.1002/j.1460-2075.1991.tb07859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ríos MG (1995) Cloning and characterization of PRO1, a tomato cDNA encoding the first two enzymes of proline biosynthesis. PhD thesis. Purdue University, West Lafayette, IN
- García-Ríos MG, Fujita T, Christopher LaRosa P, Locy RD, Clithero JM, Bressan RA, Csonka LN. Cloning of a polycistronic cDNA from tomato encoding γ-glutamyl kinase and γ-glutamyl phosphate reductase. Proc Natl Acad Sci USA. 1997;94:8249–8254. doi: 10.1073/pnas.94.15.8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ríos MG, LaRosa PC, Bressan RA, Csonka LN, Hanquier JM (1991) Cloning by complementation of the gamma-glutamyl kinase gene from a tomato expression library. InR. B. Hallick ed, Molecular Biology of Plant Growth and Development. October 6–11, 1991, Third International Congress of Plant Molecular Biology, Tucson, AZ, abstract no. 1507
- Gilles R, Kleinzeller A, Bolis L (1987) Cell volume control: fundamental and comparative aspects in animal cells. In Current Topics in Membranes and Transport, Vol 30. Academic Press, San Diego, CA
- Gray MW. The evolutionary origins of organelles. Trends Genet. 1989;5:294–299. doi: 10.1016/0168-9525(89)90111-x. [DOI] [PubMed] [Google Scholar]
- Handa S, Handa A, Hasegawa PM, Bressan RA. Proline accumulation and the adaptation of cultured plant cells to water stress. Plant Physiol. 1986;80:938–945. doi: 10.1104/pp.80.4.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare PD, Cress WA. Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Reg. 1997;21:79–102. [Google Scholar]
- Hasegawa PM, Bressan RA, Handa AK. Growth characteristics of NaCl-selected and nonselected cells of Nicotiana tabacumL. Plant Cell Physiol. 1980;21:1347–1355. doi: 10.1093/pcp/21.8.1347. [DOI] [PubMed] [Google Scholar]
- Hong-qi Z, Croes AF, Liskens HF. Protein synthesis in germinating pollen of Petunia: role of proline. Plant. 1982;154:99–203. doi: 10.1007/BF00387864. [DOI] [PubMed] [Google Scholar]
- Hu C-AA, Delauney AJ, Verma DPS. A bifunctional enzyme (Δ1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proc Natl Acad Sci USA. 1992;89:9354–9358. doi: 10.1073/pnas.89.19.9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y, Yoshiba Y, Sanada Y, Yamaguchi-Shinozaki K, Wada K, Shinozaki K. Characterization of the gene for Δ1-pyrroline-5-carboxylate synthetase and correlation between the expression of the gene and salt tolerance in Oryza sativaL. Plant Mol Biol. 1997;33:857–865. doi: 10.1023/a:1005702408601. [DOI] [PubMed] [Google Scholar]
- Khoo U, Stinson HT., Jr Free amino acid differences between cytoplasmic male sterile and normal fertile anthers. Proc Natl Acad Sci USA. 1957;43:603–607. doi: 10.1073/pnas.43.7.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishor PBK, Hong Z, Miao G-H, Hu C-AA, Verma DPS. Overexpression of Δ1-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol. 1995;108:1387–1394. doi: 10.1104/pp.108.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Yoshiba Y, Yamaguchi-Shinozaki K, Shinozaki K. A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. Plant Cell. 1996;8:1323–1335. doi: 10.1105/tpc.8.8.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Annals of Eugenics. 1944;12:172–175. [Google Scholar]
- Kozak M. Bifunctional messenger RNAs in eukaryotes. Cell. 1986;74:481–483. doi: 10.1016/0092-8674(86)90609-4. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lewin R. Evolutionary history written in globin genes. Science. 1981;214:426–429. doi: 10.1126/science.7291984. [DOI] [PubMed] [Google Scholar]
- Liu G, Maunoury C, Kamoun P, Aral B. Assignment of the human gene encoding the Δ1-pyrroline-5-carboxylate synthetase (P5CS) to 10q24.3 by in situhybridization. Genomics. 1996;37:145–146. doi: 10.1006/geno.1996.0535. [DOI] [PubMed] [Google Scholar]
- Long EO, Dawid IB. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Maggio A, García-Ríos M, Fujita T, Bressan RA, Joly RJ, Hasegawa PM, Csonka LN. Cloning of tomPRO1 (accession no. U27454) and tomPRO2 (accession no. U60267) from Lycopersicon esculentum L. Coexistence of polycistronic and monocistronic genes which encode the enzymes catalyzing the first two steps of proline biosynthesis (PGR 96-077) Plant Physiol. 1996;112:862. [Google Scholar]
- Mahan MJ, Csonka LN. Genetic analysis of the proBA genes of Salmonella typhimurium: physical and genetic analyses of the cloned proB+A+ genes of Escherichia coliand of a mutant allele that confers proline overproduction and increased osmotolerance. J Bacteriol. 1983;156:1249–1262. doi: 10.1128/jb.156.3.1249-1262.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Nagy F, Kay SA, Chua NH. Analysis of gene expression in transgenic plants. In: Gelvin SB, Schilperoort RA, Verma DPS, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. pp. 1–29. [Google Scholar]
- Paleg LG, Aspinall D. Physiology and Biochemistry of Drought Resistance in Plants. Sydney, Australia: Academic Press; 1981. [Google Scholar]
- Palmer JD. Comparative organization of chloroplast genomes. Annu Rev Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- Peng Z, Lu Q, Verma DPS. Reciprocal regulation of Δ1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol Gen Genet. 1996;253:334–341. doi: 10.1007/pl00008600. [DOI] [PubMed] [Google Scholar]
- Rentsch D, Hirner B, Schmelzer E, Frommer WB. Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. Plant Cell. 1996;8:1437–1446. doi: 10.1105/tpc.8.8.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D, Handa S, Bressan RA. Metabolic changes associated with adaptation of plant cells to water stress. Plant Physiol. 1986;82:890–903. doi: 10.1104/pp.82.4.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosens NHCJ, Thu TT, Iskandar HM, Jacobs M. Isolation of the ornithine-δ-aminotrasnferase cDNA and effect of salt stress on its expression in Arabidopsis thaliana. Plant Physiol. 1998;117:263–271. doi: 10.1104/pp.117.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Savouré A, Jaoua S, Hua XJ, Ardiles W, Van Montagu M, Verbruggen N. Isolation, characterization, and chromosomal location of a gene encoding the Δ1-pyrroline-5-carboxylate synthetase in Arabidopsis thaliana. FEBS Lett. 1995;372:13–19. doi: 10.1016/0014-5793(95)00935-3. [DOI] [PubMed] [Google Scholar]
- Sharp R, Boyer JS, Nguyen H, Hsiao T. Genetically engineered plants resistant to soil drying and salt stress. How to interpret osmotic relations? Plant Physiol. 1996;110:1051–1052. doi: 10.1104/pp.110.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih M-C, Lazar G, Goodman HM. Evidence in favor of the symbiotic origin of chloroplasts: primary structure and evolution of tobacco glyceraldehyde-3-phosphate dehydrogenases. Cell. 1986;47:73–80. doi: 10.1016/0092-8674(86)90367-3. [DOI] [PubMed] [Google Scholar]
- Smith MW, Feng D-F, Doolittle RF. Evolution by acquisition: the case for horizontal gene transfers. Trends Biochem Sci. 1992;17:489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- Stewart CR, Hanson AD (1980) Proline accumulation as a metabolic response to water stress. In NC Turner, PJ Kramer, eds, Adaptation to Water and High Temperature Stress. John Wiley and Sons, New York, pp 173–189
- Strizhov N, Ábrahám E, Ökrész L, Blickling S, Zilberstein A, Schell J, Koncz C, Szabados L. Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. 1997;12:557–569. doi: 10.1046/j.1365-313x.1997.00557.x. [DOI] [PubMed] [Google Scholar]
- Szoke A, Miao G-H, Hong Z, Verma DPS. Subcellular location of Δ1-pyrroline-5-carboxylate reductase in root/nodule and leaf of soybean. Plant Physiol. 1992;99:1642–1649. doi: 10.1104/pp.99.4.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB and others. High density molecular linkage maps of the tomato and potato genomes. Genetics. 1992;132:1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treichel S, Brinckmann E, Scheitler B, von Willert DJ. Occurrence and changes of proline content in plants in the southern Namib Desert in relations to increasing and decreasing drought. Planta. 1984;162:236–242. doi: 10.1007/BF00397445. [DOI] [PubMed] [Google Scholar]
- Troll W, Lindsley J. A photometric method for the determination of proline. J Biol Chem. 1955;215:655–660. [PubMed] [Google Scholar]
- Verbruggen N, Hua X-J, May M, Van Montagu M. Environmental and developmental signals modulate proline homeostasis: evidence for a negative transcriptional regulator. Proc Natl Acad Sci USA. 1996;93:8787–8791. doi: 10.1073/pnas.93.16.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Villarroel R, Van Montagu M. Osmoregulation of a pyrroline-5-carboxylate reductase gene in Arabidopsis thaliana. Plant Physiol. 1993;103:771–781. doi: 10.1104/pp.103.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma DPS, Hong Z. Genetically engineered plants resistant to soil drying and salt stress: how to interpret osmotic relations? Plant Physiol. 1996;110:1053. doi: 10.1104/pp.110.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeden NF. Genetic and biological implications of the endosymbiotic origin of the chloroplast. J Mol Evol. 1981;17:133–139. doi: 10.1007/BF01733906. [DOI] [PubMed] [Google Scholar]
- Williamson CL, Slocum RD. Molecular cloning and evidence for osmoregulation of the Δ1-pyrroline-5-carboxylate reductase (proC) gene in pea (Pisum sativumL.) Plant Physiol. 1992;100:1464–1470. doi: 10.1104/pp.100.3.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, Wada K, Harada Y, Shinozaki K. Correlation between the induction of a gene for Δ1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thalianaunder osmotic stress. Plant J. 1995;7:751–760. doi: 10.1046/j.1365-313x.1995.07050751.x. [DOI] [PubMed] [Google Scholar]
- Zhang C-s, Lu Q, Verma DPS. Removal of feedback inhibition of Δ1-pyrroline-5-carboxylate synthetase, a bifunctional enzyme catalyzing the first two steps of proline biosynthesis in plants. J Biol Chem. 1995;270:20491–20496. doi: 10.1074/jbc.270.35.20491. [DOI] [PubMed] [Google Scholar]
- Zhang C-s, Lu Q, Verma DPS. Characterization of Δ1-pyrroline-5-carboxylate synthetase gene promoter in transgenic Arabidopsis thalianasubjected to water stress. Plant Sci. 1997;129:81–89. [Google Scholar]