Abstract
5’-R and 5’-S diastereoisomers of 8,5’-cyclo-2’-deoxyadenosine (cdA) and 8,5’-cyclo-2’-deoxyguanosine (cdG) containing a base-sugar covalent bond are formed by hydroxyl radicals. R-cdA and S-cdA are repaired by nucleotide excision repair (NER) in mammalian cellular extracts. Here, we have examined seven purified base excision repair enzymes for their ability to repair S-cdG or S-cdA. We could not detect either excision or binding of these enzymes on duplex oligonucleotide substrates containing these lesions. However, both lesions were repaired by HeLa cell extracts. Dual incisions by human NER on a 136-mer duplex generated 24–32 base-pair fragments. The time course of dual incisions were measured in comparison to cis-anti-B[a]P-N2-dG, an excellent substrate for human NER, which showed that cis-anti-B[a]P-N2-dG was repaired more efficiently than S-cdG, which, in turn, was repaired more efficiently than S-cdA. When NER efficiency of S-cdG with different complementary bases was investigated, the wobble pair S-cdG•dT was excised more efficiently than the S-cdG•dC pair that maintains nearly normal Watson-Crick base pairing. But S-cdG•dA mispair with no hydrogen bonds was excised less efficiently than the S-cdG•dC pair. Similar pattern was noted for S-cdA. The S-cdA•dC mispair was excised much more efficiently than the S-cdA•dT pair, whereas the S-cdA•dA pair was excised less efficiently. This result adds to complexity of human NER, which discriminates the damaged base pairs on the basis of multiple criteria.
Keywords: Cyclopurine deoxynucleosides, NER, Tandem DNA damage, S-cdG, S-cdA, HeLa extract
1. Introduction
The 8,5’-cyclopurine 2’-deoxynucleosides, formed by oxidation and γ-radiation, are unique DNA lesions that exhibit atypical biological effects. They contain damage to both the purine base and the 2’-deoxyribose sugar moiety by forming a covalent bond between C8 of guanine and C5’ of 2’- deoxyribose [1]. These lesions exist as 5’R and 5’S diastereomers and have been detected in vitro and in vivo in DNA derived from various cells and organisms [[2,3,4]. Unlike most other oxidative lesions, instead of base excision repair (BER), 8,5’-cyclo-2’deoxyadenosine (cdA) diastereomers are repaired by nucleotide excision repair (NER) [5,6]. The observation that R-cdA is more efficiently repaired than the S-cdA suggest that the kinetics of NER of the cyclopurine 2’-deoxynucleosides are different [5]. These lesions have been suspected to play a role in neurologic diseases in certain Xeroderma Pigmentosum patients with defects in NER [7]. It was shown that S-cdA accumulates in vivo in genomic DNA of csb−/− mice, suggesting that it may accumulate in Cockayne syndrome patients [8]. Levels of R-cdA and S-cdA increase significantly in neil1−/− mice suggesting involvement of NEIL1 in their repair, although the mechanism is unclear [9]. S-cdA has been reported to be a strong block of gene expression in CHO and human cells [6]. It prevents the binding of TATA binding protein and strongly reduces transcription in vivo [10]. In Escherichia coli S-cdG is significantly mutagenic inducing primarily S-cdG→A mutations [11,12]. Recently, the solution structures of DNA duplexes containing a site-specific S-cdG lesion placed opposite dC, dT, or dA in the complementary strand were obtained by molecular dynamics calculations restrained by distance and dihedral angle restraints obtained from NMR spectroscopy [13,14]. These studies showed that the S-cdG deoxyribose maintains the O4'-exo (west) pseudorotation in each pair. The Watson-Crick base pairing was conserved in the S-cdG•dC pair, whereas S-cdG•dT pair adopts a wobble pairing. In contrast, no hydrogen bonding was observed for the S-cdG•dA pair, which differs in conformation from the dG•dA mismatch pair.
Most of the repair investigations were conducted with cdA, and the repair of cdG is little studied, except that it is repaired inefficiently in vitro by UvrABC, the E. coli NER proteins [11]. Despite the lack of DNA glycosylase activity in mammalian cellular extracts, we found it intriguing that NEIL1 may be involved in the repair of cdA [9]. One possible explanation is that NEIL1 binds to the lesion but is unable to excise the purine base that is linked to the sugar by two covalent bonds. Nevertheless, the protein-bound lesion or the C1’-N9 glycosyl bond cleaved form would likely allow superior recognition by NER. In order to address these issues, we have investigated the repair and binding of S-cdA and S-cdG by several purified DNA glycosylases that repair oxidative DNA damages. Since the NMR data of S-cdG have been reported [13,14], we also compared the repair of these two lesions located opposite different bases by the NER proteins in HeLa cell extract.
2. Materials and Methods
2.1. Chemicals, substrates, and enzymes
S-cdG phosphoramidite monomer for DNA synthesis was synthesized and characterized as reported [13,15]. S-cdA phosphoramidite monomer was purchased from Berry and associates (Dexter, MI). Bulk solvents were purchased either from Sigma Aldrich (St. Louis, MO) or Fisher Scientific (Clifton, NJ). The oligonucleotides containing S-cdG or S-cdA in the 12-mer, 5'-GTGCXTGTTTGT-3', and 36-mer, 5’-ACAAACACGCACTCCGGACGAGATGTGCXTGTTTGTATCGCTGCTACC- 3’, where X represents S-cdG or S-cdA, were synthesized, purified, and characterized as reported [13]. cis-anti-B[a]P-N2-dG adducted 11-mer was a gift from Professor Nicholas E. Geacintov (New York University, New York, NY). The purity of the modified oligodeoxynucleotide was assessed by HPLC and MALDI-TOF mass spectrometry. E. coli Fpg, Endo III, Endo V and Endo VIII are routinely prepared in the Kow laboratory, and human OGG1, NEIL1, and NEIL2 were gifts from Dr. Sankar Mitra (UTMB, Galveston, TX).
2.2. Preparation of the 136-mer or 135-mer DNA for NER study
The unmodified and modified 12-mers were 5’-phosphorylated (with 32P radiolabel) and ligated to the 62-mer strands on its 5’- and 3’ ends following a method described in detail in ref. [16]. The sequence is as follows.
5’-GAG CTG AGG ACG TAC GGA ATT CGA TAT CCT CGA GCC AGA TCT GCG CCA GCT GGC CAC CCT GA GTGCXTGTTTGT GAG CGC CAA GCT TGG GCT GCA GCA GGT CGA CTC TAG AGG ATC CCG GGC GAG CTC GAA TTC GC-3’ (where the 12-mer is bold and underlined; X denotes S-cdG, S-cdA, or dG).
Likewise, the cis-anti-B[a]P-N2-dG adducted 11-mer was ligated to two 62-mers to generate the following 135-mer.
5’-GAG CTG AGG ACG TAC GGA ATT CGA TAT CCT CGA GCC AGA TCT GCG CCA GCT GGC CAC CCT GA CCATCG*CTACC GAG CGC CAA GCT TGG GCT GCA GCA GGT CGA CTC TAG AGG ATC CCG GGC GAG CTC GAA TTC GC-3’ (where the 11-mer is bold and underlined; G* denotes cis-anti-B[a]P-N2-dG).
The internally 32P-radiolabeled sequences were annealed with their complementary strands to form the 136-mer or 135-mer duplexes that were PAGE purified and desalted prior to the NER assay.
2.3. Base excision repair (BER) assays
The substrate susceptibility of the BER enzymes was examined on two oligonucleoides (12- mer and 36-mer) containing S-cdG or S-cdG. The sequences of the oligonucleotides used were 5’- GTGCXTGTTTGT-3’ and 5’-CCTGGAAGCGATGTGCXTGTTTGTATCGCTGCTACC (where X denotes S-cdG or S-cdG), which were annealed with the corresponding unmodified complementary strands. Seven BER enzymes were examined: E. coli formamidopyrimidine N-glycosylase (Fpg), endonuclease III (Endo III), endonuclease V (Endo V), endonuclease VIII (Endo VIII) and human OGG1, NEIL1 and NEIL2. The activity of the BER enzymes was confirmed by including in each experiment a 33-mer containing cognate substrates for the BER enzymes. The sequence of the 32-mer substrate is 5’-TTCCAGACTGTCCTTCGTXACTTTCCTCTCAA (for Fpg and OGG1, X=8-OxoG; for Endo III, V and VIII, X=AP site; for Endo III, VIII, NEIL1 and NEIL2, X= 5-hydroxyuracil, 5- OH-U). Each oligonucleotide containing a lesion was 5’ end-labeled with 32P, using T4 polynucleotide kinase. Double stranded oligonucleotide substrates were prepared by adding 1.5 times the amount of complementary oligonucleotide to the labeled single stranded substrate, and the mixture was incubated at 90 °C for 5 min before cooling it down slowly to room temperature. Briefly, reactions were performed in 20 µL reaction buffer (100 mM NaCl, 10 mM Tris, 2 mM MgCl2, pH 8.0) containing 100 fmol of labeled substrate and 40 fmol (10 ng) of protein. The mixture was incubated for 30 min at 37 °C. Equal volume of gel loading buffer (0.05 % bromophenol blue, 0.05 % xylene, and 10 mM EDTA) was added to stop the reaction and immediately heated to 90 °C for 10 min. The product and substrate were separated by electrophoresis on a 12.5% denaturing polyacrylamide gel and the dried gels analyzed for band intensity using a STORM™ phosphoImager (Molecular Dynamics).
2.4. Nucleotide excision repair assays
The HeLa cell (ATCC, Camden, NJ) extracts were prepared by cell lysis and ammonium sulfate precipitation following a method described in ref [17]. The modified or control 136-mer (or 135-mer) duplexes were incubated with the HeLa cell extract (50 µg protein in 25 µL aliquot) for a specific time interval. The NER excision products and intact DNA were then precipitated with ethanol (80%) and subjected to electrophoresis on a 12% polyacrylamide gel. The bands were analyzed by autoradiography using a phosphorimager. The NER dual incision products are typically 24–32 base pair in length that contained the 32P label. To determine the yield of NER dual incision products, the densitometry tracings of these DNA fragments in each lane were added and the sum was divided by the total radioactivity in the same lane (which included the intact 136-mer as well as the 24–32 base pair excision products).
3. Results and Discussion
3.1 NEIL1, NEIL2, Fpg, OGG1, Endo III, Endo V, and Endo VIII do not recognize S-cdG and S-cdA
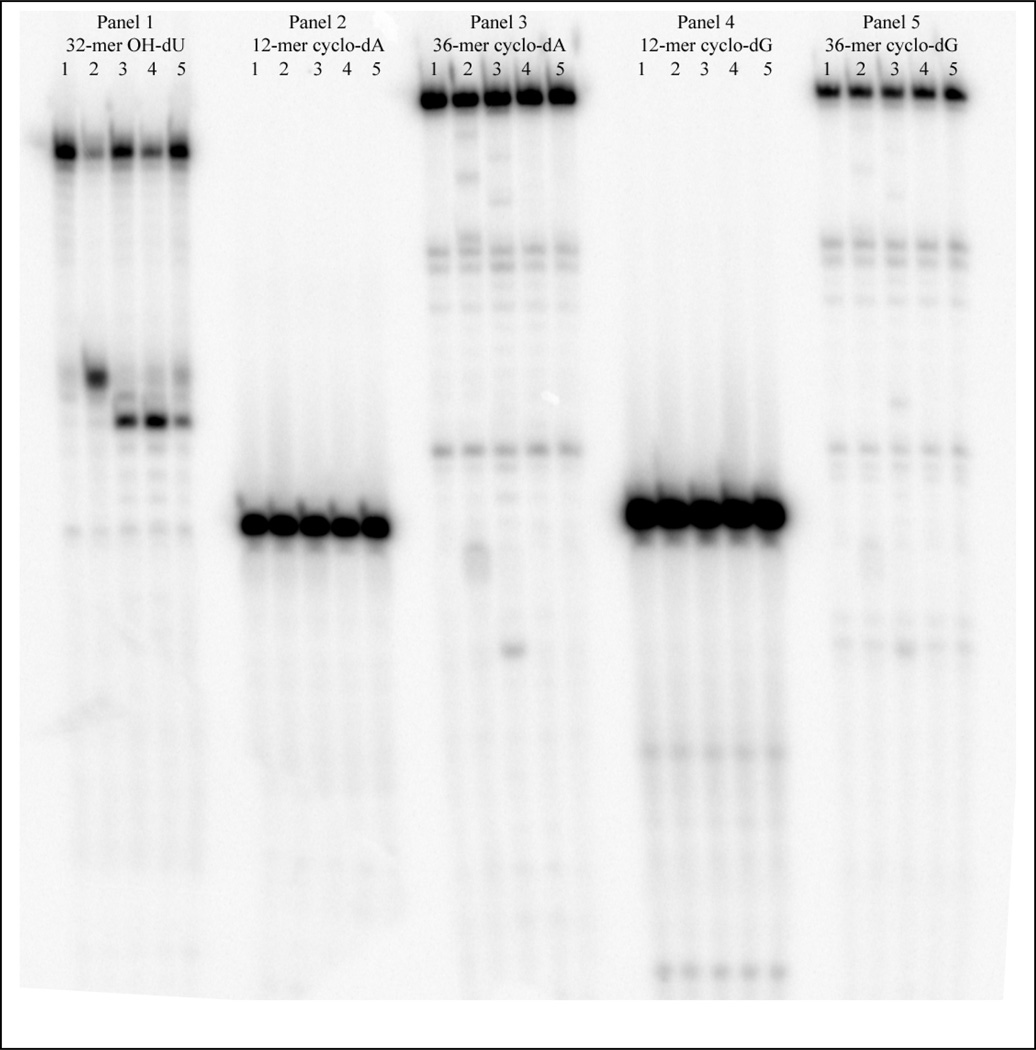
Two prior investigations found no evidence of BER of cdA in mammalian cell extracts [5,6], even though most other hydroxyl radical-induced DNA lesions are subject to repair by various BER glycosylases [18]. Nevertheless, both R-cdA and S-cdA accumulate in neil−/− mice [9], suggesting a possible role of this glycosylase in their repair. Consequently, we investigated if S-cdG and/or S-cdA can be weakly incised by or bound to any of the seven different purified BER proteins. But we did not find any evidence that any of the seven BER enzymes known to be active on a number of oxidative DNA damages either make incisions or bind to a 12- or 36-mer duplex oligonucleotides containing cdG or cdA. As shown in Figure 1, Endo III, Endo V, NEIL1 and NEIL2 made incisions on oligonucleoides containing 5-hydroxy-2’-deoxyuridine (OHdU) (Panel 1), whereas they were not active on oligonucleotides containing either S-cdG or S-cdA. Even with an excess of the enzymes (200 fmol), we found no evidence of cleavage on the S-cdG and S-cdA oligonucleoides. Fpg, Endo V and human OGG1 were also found to be inactive against these oligonucleotide duplexes (data not shown). In parallel binding experiments, none of the BER enzymes examined were able to form detectable amount of high molecular weight DNA-protein complex with either 12-mer or 36-mers containing S-cdG and S-cdA (data not shown). It is noteworthy that NEIL1, at very high concentrations (e.g., 10–20 pmol), binds to DNA containing OHdU, S-cdA, or S-cdG, but it also binds to undamaged DNA, suggesting that the DNA-protein complexes formed were non-specific (Figure S1 in Supporting Information). This is not surprising, because NEIL1 has been reported to exhibit high affinity for undamaged DNA[19]. However, the fact that at a much lower concentration, NEIL1 can incise DNA containing OHdU but not S-cdA or S-cdG suggests that the interaction of NEIL1 with OHdU was not stable enough to be resolved by a gel-shift assay. We conclude that these seven repair proteins do not directly interact with S-cdG and S-cdA.
Figure 1.
BER enzymes do not cleave oligonucleotides containing S-cdA or S-cdG. Oligonucleoides (12-mers and 36-mers) containing S-cdA or S-cdG were examined to determine if they are recognized by E. coli Fpg, Endo III, Endo V, Endo VIII, human OGG1, human NEIL1 and NEIL2. A typical experiment is shown here with Endo III, Endo V, NEIL1 and NEIL2. Panel 1 shows the reactions with a positive control, 5-OH-dU. For each panel, lane 1 =control, no enzyme; lane 2 =Endo III; lane 3 =Endo VIII; lane 4 = NEIL1; lane 5 = NEIL2. Similar experiments were performed with Fpg, Endo V, and OGG1 (data not shown).
3.2. Both S-cdG and S-cdA are repaired by HeLa cell extracts
We have constructed 136-mer double-stranded DNA containing a single S-cdG or S-cdA near the center of the duplex, which also contained a 32P label between the 4th and 5th base 5’ to the lesion. A 135-mer DNA duplex containing cis-anti-B[a]P-N2-dG, an excellent substrate for the human NER, was used as a positive control, whereas an unmodified 136-mer served as a negative control.
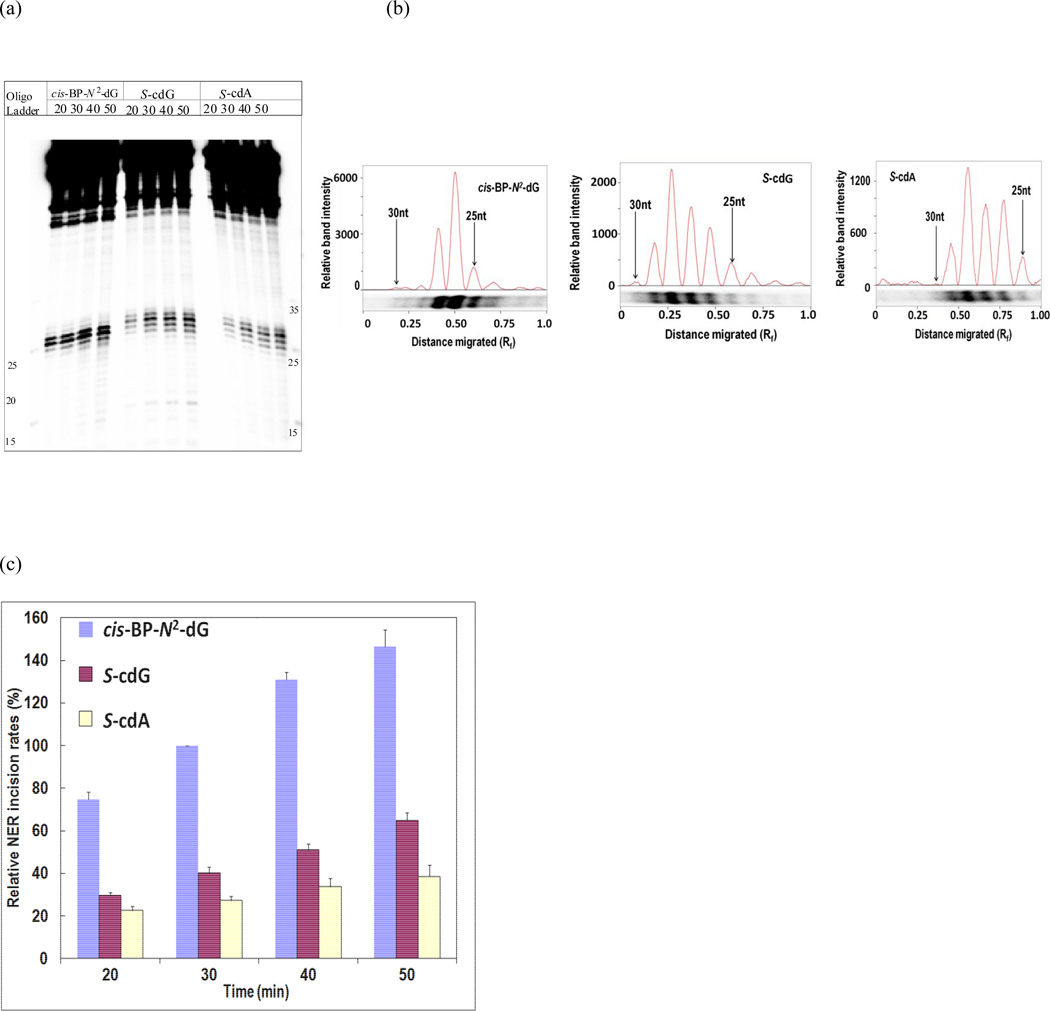
The duplex DNA substrates were incubated with HeLa cell nuclear extracts for 0, 20, 30, 40, and 50 min. Dual incisions by the human NER generated 24–32 nucleotides in length products with the 32P label. These fragments were resolved by denaturing PAGE and visualized by autoradiography. A typical experiment is shown in Figure 2 (a) that excluded the 0 min lane, for which earlier experiments showed no detectable cleavage products. Densitometry tracing of the products (as shown in Figure 2 (b)) were combined to determine the efficiency of dual incisions. Three separate sets of experiments were conducted, which showed that the NER efficiencies followed the order: cis-anti- B[a]P-N2-dG ≫ S-cdG > S-cdA (Figure 2 (c)). So, S-cdG, like cdA, was repaired by human cell extracts. It is noteworthy that S-cdA was incised at a faster rate than S-cdG by the purified NER proteins from E. coli, suggesting important differences between the E. coli and human NER proteins. Similar observation of different specificity between the human and E. coli NER system was reported for a benzo[a]pyrene DNA adduct [20].
Figure 2.
Comparison of excision of 24–32 base-pair fragments from 136-mer S-cdG and S-cdA duplexes and 135-mer cis-anti-B[a]P-N2-dG duplex in HeLa cell extracts. A typical experiment is shown in panel (a), which includes incubation times of 20, 30, 40, and 50 min for each lesion. Panel (b) shows the densitometry tracings of the dual incision products in the 30 min lane of each lesion. The densitometry tracings of these bands in each lane were combined to obtain the cleavage products at each time point. Panel (c) shows the time course of formation of the dual incision products from three separate experiments. The data points were normalized to the 30-min excision products from the cis-anti-B[a]P-N2-dG containing duplex.
3.3. NER efficiency of S-cdG and S-cdA depends on the complementary base
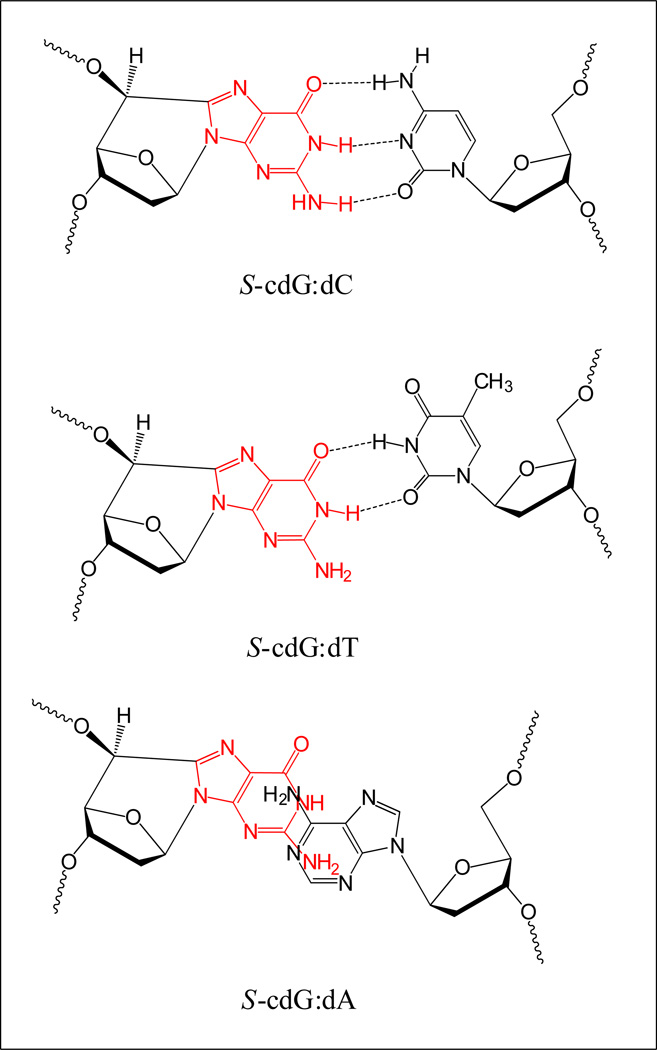
Since the NMR solution structure of S-cdG•dC, S-cdG•dT, and S-cdG•dA in DNA duplexes have been reported, we availed the opportunity to investigate the relationship of the solution structure and thermodynamic stability of these pairs with the human NER efficiency. The NMR investigation of S-cdG•dC pair showed that although the Watson-Crick base pairing was conserved throughout the sequence including the S-cdG•dC pair (Chart 1), the O4’-exo puckering of the S-cdG deoxyribose caused substantial perturbations on the helical twist and base pair shift at the lesion site [13]. The base stacking perturbations at the S-cdG•dC pair decreased the stability of the duplex. Compared with the unmodified DNA (Tm: 55 °C), the S-cdG•dC 12-mer duplex exhibited a melting temperature (Tm) of 46 °C, which accounts for a destabilization of the duplex by 9 °C. It is conceivable that these perturbations are detected by the human NER system. NMR investigation of the structure of the S-cdG• dT mismatch pair showed that it adopts a wobble base pairing (Chart 1) [14]. The duplex containing the S-cdG•dT base pair exhibited a Tm of 38 °C, whereas the Tm of the unmodified dG•dT mismatch was 43 °C under the same conditions. Thus, the incorporation of S-cdG reduced the Tm by 5 °C. When S-cdG was paired with dT, which leads to the most frequent G→A base substitutions observed in E. coli, the NER activity was 40–50% more efficient than its normal partner, dC (Figure 3 a & b). More efficient dual incisions by the human NER proteins suggests that the departure from the Watson-Crick base pairing allows better recognition and/or dual incisions of S-cdG•dT by human NER. By contract, the repair of S-cdG•dA pair was 40–50% less efficient than the S-cdG•dC pair (Figure 3 a & b). The S-cdG•dA mismatch pair differs in conformation from the dG•dA mismatch pair. For the S-cdG•dA mismatch pair, both S-cdG and dA intercalate, but no hydrogen bonding is observed between S-cdG and dA. The duplex containing the S-cdG•dA pair exhibited a Tm of 31 °C, while the Tm of the unmodified dG•dA mismatch was 39 °C. Thus, the incorporation of S-cdG reduced the Tm by 8 °C. In spite of the lack of hydrogen bonding between S-cdG and dA and a significant drop in melting temperature, the S-cdG•dA mismatch pair was excised less efficiently than the S-cdG•dC pair.
Chart 1.
Schematic representation of the base pairing modes of S-cdG opposite dC, dT, and dA as determined from the solution NMR studies [13,14]. For the S-cdG•dA pair, both S-cdG and dA were intercalated without any hydrogen bonds. Please note that the base pairs shown here are not energy minimized. For the refined structures, please see ref [13,14].
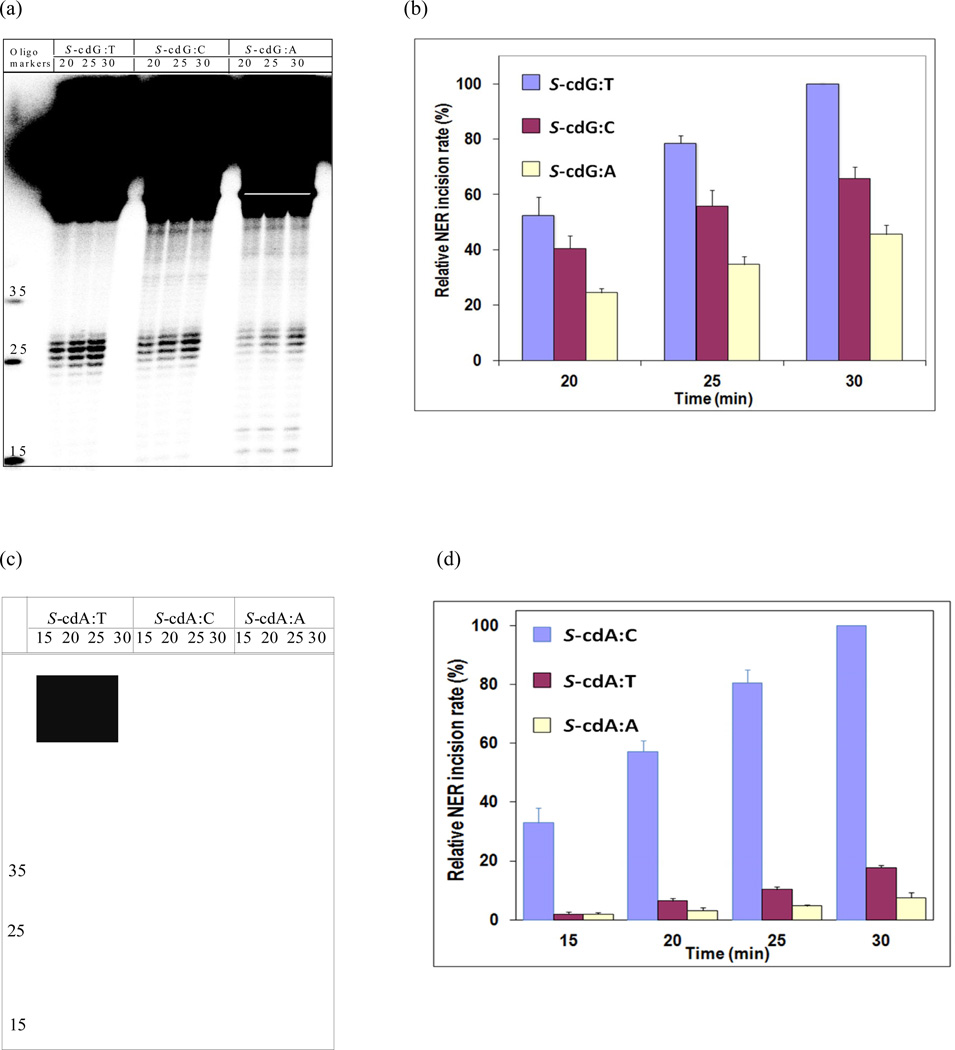
Figure 3.
Comparison of excision of 24–32 base-pair fragments from 136-mer S-cdG opposite dC, dT, and dA in HeLa cell extracts. A typical experiment is shown in panel (a), which includes incubation times of 20, 25, and 30 min for each base pair. Panel (b) shows the time course of formation of the dual incision products from three separate experiments. The data points in panel (b) were normalized to the 30-min excision products from the S-cdG•dT duplex, which gave the maximum value. Similar comparison of excision from 136-mer S-cdA opposite dT, dC, and dA is shown in panel (c), and the time course of formation of the dual incision products is shown in panel (d).
In the case of S-cdA, the NER difference between the pair with its normal partner dT and the mispair with dC was found much more significant. The NER dual incisions of the S-cdA•dC mispair were 5–15-fold more efficient than the S-cdA•dT pair (Figure 3 c & d). As in the case of S-cdG, the NER of S-cdA•dA mispair was less efficient than the S-cdA•dT pair (Figure 3 c & d). Although the NMR structures of S-cdA with different partners have not yet been reported, the overall pattern by human NER was the same in that the dual incisions of the S-cdA•dC mismatch pair were much more efficient than the S-cdA•dT pair, whereas the S-cdA•dA mismatch pair was excised less efficiently than the latter.
The undamaged base opposite a lesion has been shown to exhibit a critical function in the incision complex in human NER [21,22]. It was suggested that the XPC protein initiates the NER by assessing oscillations across the lesion. The data in the current study shows that a departure from the Watson-Crick base pairing provides a better substrate for the NER proteins, but a complete lack of hydrogen bonding does not necessarily improve the repair. Increased thermodynamic stabilization due to more effective stacking in DNA, when the partner base has been deleted, results in NER resistance [23]. So, it is likely that both base pairing and base stacking are important, but the lack of correlation of NER with the thermal melting temperatures suggests that additional criteria such as bulge, twist, and stretch of the modified base pair play important roles in NER. The unusual effect of dA as a complementary base was also shown for the bulky cis benzo[a]pyrene adducts, where efficiency of excision dropped ten-fold when dC opposite the lesion was replaced with dA [24]. It was suggested that dA located across the cis-B[a]P-N2-dG adducts may involve less base displacement than dC and, in the cases of the helix-inserted cis adducts, this results in ten-fold-lower levels of excision repair activity. Future investigation on the interactions of S-cdG and S-cdA with the damage recognition proteins may shed light on these issues.
Supplementary Material
Acknowledgments
We are indebted to Professor Nicholas E. Geacintov and Dr. Konstantin Kropachev (New York University, New York, NY) for their kind help in training us the NER assay and their gift of the cis-anti- B[a]P-N2-dG adducted 11-mer. We thank Professor Sankar Mitra for his gift of human OGG1, NEIL1, and NEIL2. This work was supported by grants from the NIEHS and NCI of the NIH (grant ES013324 to AKB and CA 90860 to YWK).
Abbreviations
- S-cdG and R-cdG
(5’S)- and (5’R)-8,5’-cyclo-2’-deoxyguanosine, respectively
- S-cdA and R-cdA
(5’S)- and (5’R)-8,5’-cyclo-2’-deoxyadenosine, respectively
- cis-anti-B[a]P-N2-dG
10R-(+)-cis-anti-B[a]P-N2-dG adduct formed by addition of dG to (+)-anti-B[a]PDE
- OHdU
5-hydroxy-2’-deoxyuridine
- BER
base excision repair
- NER
nucleotide excision repair
- PAGE
polyacrylamide gel electrophoresis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that there is no conflict of interest.
References
- 1.Chatgilialoglu C, Ferreri C, Terzidis MA. Purine 5',8-cyclonucleoside lesions: chemistry and biology. Chem Soc Rev. 2011;40:1368–1382. doi: 10.1039/c0cs00061b. [DOI] [PubMed] [Google Scholar]
- 2.Jaruga P, Dizdaroglu M. 8,5'-Cyclopurine-2'-deoxynucleosides in DNA: mechanisms of formation, measurement, repair and biological effects. DNA Repair (Amst) 2008;7:1413–1425. doi: 10.1016/j.dnarep.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Jaruga P, Xiao Y, Nelson BC, Dizdaroglu M. Measurement of (5'R)- and (5'S)-8,5'-cyclo-2'-deoxyadenosines in DNA in vivo by liquid chromatography/isotope-dilution tandem mass spectrometry. Biochem Biophys Res Commun. 2009;386:656–660. doi: 10.1016/j.bbrc.2009.06.107. [DOI] [PubMed] [Google Scholar]
- 4.Dizdaroglu M, Jaruga P, Rodriguez H. Identification and quantification of 8,5'-cyclo-2'-deoxy-adenosine in DNA by liquid chromatography/ mass spectrometry. Free Radic Biol Med. 2001;30:774–784. doi: 10.1016/s0891-5849(01)00464-6. [DOI] [PubMed] [Google Scholar]
- 5.Kuraoka I, Bender C, Romieu A, Cadet J, Wood RD, Lindahl T. Removal of oxygen free-radical-induced 5',8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc Natl Acad Sci U S A. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks PJ, Wise DS, Berry DA, Kosmoski JV, Smerdon MJ, Somers RL, Mackie H, Spoonde AY, Ackerman EJ, Coleman K, Tarone RE, Robbins JH. The oxidative DNA lesion 8,5'-(S)-cyclo-2'-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J Biol Chem. 2000;275:22355–22362. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 7.Brooks PJ. The 8,5'-cyclopurine-2'-deoxynucleosides: candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair. DNA Repair (Amst) 2008;7:1168–1179. doi: 10.1016/j.dnarep.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirkali G, de Souza-Pinto NC, Jaruga P, Bohr VA, Dizdaroglu M. Accumulation of (5'S)-8,5'- cyclo-2'-deoxyadenosine in organs of Cockayne syndrome complementation group B gene knockout mice. DNA Repair (Amst) 2009;8:274–278. doi: 10.1016/j.dnarep.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaruga P, Xiao Y, Vartanian V, Lloyd RS, Dizdaroglu M. Evidence for the involvement of DNA repair enzyme NEIL1 in nucleotide excision repair of (5'R)- and (5'S)-8,5'-cyclo-2'- deoxyadenosines. Biochemistry. 2010;49:1053–1055. doi: 10.1021/bi902161f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marietta C, Gulam H, Brooks PJ. A single 8,5'-cyclo-2'-deoxyadenosine lesion in a TATA box prevents binding of the TATA binding protein and strongly reduces transcription in vivo. DNA Repair (Amst) 2002;1:967–975. doi: 10.1016/s1568-7864(02)00148-9. [DOI] [PubMed] [Google Scholar]
- 11.Jasti VP, Das RS, Hilton BA, Weerasooriya S, Zou Y, Basu AK. (5'S)-8,5'-cyclo-2'- deoxyguanosine is a strong block to replication, a potent pol V-dependent mutagenic lesion, and is inefficiently repaired in Escherichia coli. Biochemistry. 2011;50:3862–3865. doi: 10.1021/bi2004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan B, Wang J, Cao H, Sun R, Wang Y. High-throughput analysis of the mutagenic and cytotoxic properties of DNA lesions by next-generation sequencing. Nucleic acids research. 2011;39:5945–5954. doi: 10.1093/nar/gkr159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang H, Das RS, Basu AK, Stone MP. Structure of (5'S)-8,5'-cyclo-2'-deoxyguanosine in DNA. J Am Chem Soc. 2011;133:20357–20368. doi: 10.1021/ja207407n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang H, Das RS, Basu AK, Stone MP. Structures of (5'S)-8,5'-Cyclo-2'-deoxyguanosine Mismatched with dA or dT. Chem Res Toxicol. 2012;25:478–490. doi: 10.1021/tx2005053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romieu A, Gasparutto D, Cadet J. Synthesis and characterization of oligonucleotides containing 5',8-cyclopurine 2'-deoxyribonucleosides: (5'R)-5',8-cyclo-2'-deoxyadenosine, (5'S)-5',8-cyclo-2'-deoxyguanosine, and (5'R)-5',8-cyclo-2'-deoxyguanosine. Chem Res Toxicol. 1999;12:412–421. doi: 10.1021/tx9802668. [DOI] [PubMed] [Google Scholar]
- 16.Kropachev K, Kolbanovskii M, Cai Y, Rodriguez F, Kolbanovskii A, Liu Y, Zhang L, Amin S, Patel D, Broyde S, Geacintov NE. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: insights into the structural factors that favor dual incisions. J Mol Biol. 2009;386:1193–1203. doi: 10.1016/j.jmb.2008.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smeaton MB, Miller PS, Ketner G, Hanakahi LA. Small-scale extracts for the study of nucleotide excision repair and non-homologous end joining. Nucleic Acids Res. 2007;35:e152. doi: 10.1093/nar/gkm974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odell ID, Newick K, Heintz NH, Wallace SS, Pederson DS. Non-specific DNA binding interferes with the efficient excision of oxidative lesions from chromatin by the human DNA glycosylase, NEIL1. DNA Repair (Amst) 2010;9:134–143. doi: 10.1016/j.dnarep.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, Geacintov NE. Probing for DNA damage with beta-hairpins: similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro. DNA Repair (Amst) 2011;10:684–696. doi: 10.1016/j.dnarep.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clement FC, Camenisch U, Fei J, Kaczmarek N, Mathieu N, Naegeli H. Dynamic two-stage mechanism of versatile DNA damage recognition by xeroderma pigmentosum group C protein. Mutat Res. 2010;685:21–28. doi: 10.1016/j.mrfmmm.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Buterin T, Meyer C, Giese B, Naegeli H. DNA quality control by conformational readout on the undamaged strand of the double helix. Chem Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Reeves DA, Mu H, Kropachev K, Cai Y, Ding S, Kolbanovskiy A, Kolbanovskiy M, Chen Y, Krzeminski J, Amin S, Patel DJ, Broyde S, Geacintov NE. Resistance of bulky DNA lesions to nucleotide excision repair can result from extensive aromatic lesion-base stacking interactions. Nucleic Acids Res. 2011;39:8752–8764. doi: 10.1093/nar/gkr537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hess MT, Gunz D, Luneva N, Geacintov NE, Naegeli H. Base pair conformation-dependent excision of benzo[a]pyrene diol epoxide-guanine adducts by human nucleotide excision repair enzymes. Mol Cell Biol. 1997;17:7069–7076. doi: 10.1128/mcb.17.12.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.