Abstract
Base excision repair (BER) is an evolutionarily conserved DNA repair pathway that is critical for repair of many of the most common types of DNA damage generated both by endogenous metabolic pathways and exposure to exogenous stressors such as pollutants. C. elegans is an increasingly important model organism for the study of DNA damage-related processes including DNA repair, genotoxicity, and apoptosis, but BER is not well understood in this organism, and has not previously been measured in vivo. We report robust BER in the nuclear genome and slightly slower damage removal from the mitochondrial genome; in both cases the removal rates are comparable to those observed in mammals. However we could detect no deficiency in BER in the nth-1 strain, which carries a deletion in the only glycosylase yet described in C. elegans that repairs oxidative DNA damage. We also failed to detect increased lethality or growth inhibition in nth-1 nematodes after exposure to oxidative or alkylating damage, suggesting the existence of at least one additional as-yet undetected glycosylase.
Keywords: Caenorhabditis elegans, base excision repair, nth-1, hydrogen peroxide, methylmethane sulfonate
1. INTRODUCTION
Nuclear and mitochondrial DNA are constantly exposed to endogenous and exogenous agents that induce DNA damage, which if not repaired can block transcription and replication or result in mutations including point changes, deletions, and chromosomal rearrangements. Base excision repair (BER) is the primary mechanism for the removal of small and common types of DNA damage including oxidative damage, bases with small alkyl adducts, single strand breaks, deaminated bases, and abasic sites. This is a critical pathway because a large amount of such damage is generated endogenously. The amount of damage can be increased by exposure to xenobiotics, and faulty repair of such damage has been linked to cancer and aging [1–4].
BER repairs damage to both the nuclear and mitochondrial genomes. The initial step of BER involves the recognition and removal of a damaged or misincorporated base by DNA glycosylases. Base removal from the DNA strand generates an abasic site which serves as a substrate for an AP endonuclease. In the second step of BER, the AP endonuclease cleaves the phosphate backbone next to the abasic site, creating a single strand break. The remaining abasic sugar is excised by a deoxyribo-phosphodiesterase (dRPase). This cleavage and excision leaves a single nucleotide gap that is filled in by a polymerase and the remaining nick is sealed by a DNA ligase. BER operates similarly in the mitochondria, largely via alternatively-targeted BER proteins and in part via mitochondria-specific proteins [5–7]. Mitochondria and nuclear DNA also have a distinct long patch BER pathway that involves the insertion of a short nucleotide sequence rather than a single nucleotide (short-patch BER) extending from the incision site [8].
The multicellular nematode species Caenorhabditis elegans (C. elegans) is emerging as a powerful model system for the investigation of the DNA damage response, including mutagenesis [9–11], cell cycle arrest and apoptosis [12, 13], and DNA repair [14–17]. However, there are relatively few publications on BER in this organism. Two glycosylases [18–20] and two endonucleases [21, 22] have been identified, and in vitro biochemical analyses have revealed BER activities [20, 23] but a direct analysis of the in vivo rate of removal of DNA damage by BER has not been reported in either the nuclear or mitochondrial genomes. Interestingly, a mutant strain (RB877) carrying a deletion in the only identified C. elegans glycosylase that would handle oxidative DNA damage, nth-1, has had a detectable phenotype in two studies [24, 25] but no phenotype in two other studies [20, 26]. Human NTH-1 is responsible for carrying out the initial recognition step in BER and subsequently excises oxidized pyrimidine bases. Repair per se has never been measured in the nth-1-deficient C. elegans mutant.
To further characterize the critical BER pathway in C. elegans, we quantified removal of damage generated by oxidative and alkylating agents in both genomes, in wild-type as well as the nth-1 mutant strain of C. elegans. We also measured the relative sensitivity of the two strains to oxidative and alkylating agents. Using a sensitive and genome-specific QPCR assay, we report robust removal of both types of damage in the nuclear genome and slightly slower removal from the mitochondrial genome, but no detectable difference in removal in the nth-1 strain compared to wild-type. We also failed to detect a difference in lethality or growth between the strains after exposure to oxidative or alkylating damage.
2. MATERIALS AND METHODS
2.1 C. elegans strains and culture
C. elegans were cultured essentially as described [26]. Briefly, nematodes were subjected to age-synchronization via NaOH-bleach egg isolation and overnight hatch in K+ medium. C. elegans were otherwise cultured in petri dishes on K-agar seeded with OP50 strain Escherichia coli or in 96-well plates containing K+ medium with OP50 food (details for specific experiments provided below). All experiments were carried out at 20° C. In addition to wild-type (N2 Bristol), we used the mutant strain RB877 that carries a 793 bp deletion in the nth-1 gene, affecting three exons. We outcrossed this strain 3 times with N2 nematodes. Genotyping information, verifying the presence of the nth-1 deletion, is presented in Supplemental Data File 1. N2 and nth-1 nematodes were obtained from the Caenorhabditis Genetics Center (CGC; Minneapolis, MN, USA).
2.2 Lethality assays
In order to establish a dose response for each chemical, 50–100 age-synchronized L1 worms of both strains were exposed for one hour to varying concentrations of hydrogen peroxide (H2O2) or methyl methanesulfonate (MMS) in 400 μL K medium in 48 well plates, rinsed 3 times in 2 mL K medium, then placed on 60 mm K agar plates seeded with OP50 and observed immediately and every 24 h thereafter for three days. The MMS was dissolved in DMSO. The DMSO concentration in the dosing solutions was <1% in all cases; we previously found that 1% DMSO does not inhibit growth or reproduction in C. elegans [27]. Experiments with ~300 nematodes per well resulted in indistinguishable results, indicating that the number of nematodes used was not so high as to deplete the dosing agent (data not shown). At each observation, worms on each plate were scored as unhealthy if they were non-motile unless prodded with a probe, and were considered dead if tapping failed to induce movement. Nematodes showing normal spontaneous movement were scored as healthy.
2.3 Growth assays
In order to test for a possible strain effect on growth in the presence of each chemical, age-synchronized, L1-staged N2 and nth-1 nematode strains were added to 1.7 ml microcentrifuge tubes and exposed for 1 hour at the following concentrations of chemical in K medium: 1.5 mM H2O2 or 2.5 mM MMS. K medium alone was used as a control. After 1 h, the nematodes were centrifuged and the dosing solution was removed and replaced with K medium; this rinse was carried out three times before transferring the nematodes to 96-well plates containing K+ medium and OP50 bacteria. 100 nematodes were added to each well in 100 μL K+. uvrA (DNA repair-deficient: [28]) bacteria were added and the extinction coefficient and time of flight were measured at 24, 48, and 72 h using the COPAS BioSort (Union Biometrica), as previously described [29]. The measured extinction coefficient and time of flight values are proxies for nematode size.
2.4 Measurement of DNA damage and repair
Worms were exposed to chemicals as in the growth assay, using 5mM MMS or 5mM H2O2, except that exposures were in 200 μL in 96-well plates. After rinsing, the worms were placed on 60 mm K agar plates seeded with OP50. As soon as the plates were dry, worms were picked into PCR tubes containing lysis buffer at a ratio of 1 nematode to 15 μl of buffer, with triplicate tubes for the controls and duplicate tubes for the treatments for each strain. The tubes were then stored at −80°C. Picking was repeated at 3, 6, and 24 h post-treatment. DNA damage and repair were measured using a quantitative PCR (QPCR)-based assay in which we employed previously-described primers to amplify long and short portions of the mitochondrial and nuclear genomes [30]. The unc-22 target was used as a long nuclear amplicon. This assay works on the principle that DNA damage inhibits the progression of the polymerase used in the QPCR reaction [31, 32]. Measurements of mtDNA amplification were normalized to mtDNA copy number before damage levels were calculated to ensure that amplification was not confounded by biogenesis or degradation of mitochondrial genomes.
2.5 Measurement of mitochondrial DNA copy number
Absolute mitochondrial copy number was measured using aliquots from samples collected for the measurement of DNA damage and repair (Section 2.4). Quantitative, real-time PCR measuring short nuclear and mitochondrial DNA amplicons was carried out as described in Bratic et al [33]. A single real-time PCR run produced three technical replicates for each nematode sample. The resulting values were averaged and the absolute mitochondrial copy number data obtained was normalized to the corresponding nuclear DNA copy number. The relative mitochondrial copy number of each sample was determined compared to the 0h time point.
2.6 Statistical analysis
All data were analyzed with Statview for Windows (Version 5.0.1, SAS Institute Inc., Cary, NC). DNA damage data were analyzed via multifactorial ANOVA. Initial four-factor ANOVA (chemical, strain, genome, and time point) of all DNA damage data showed main effects for time, treatment, and genome but not strain (p < 0.0001, p < 0.0001, p = 0.01, and p = 0.73, respectively). Significant interaction terms were time * treatment (p < 0.0001), treatment * genome (p < 0.0001), and time * treatment * genome (p = 0.008). Subsequent analyses described in Results were carried out using lower-order ANOVA or Fisher’s Protected Least Significant Differences (FPLSD) post hoc test. Growth data were not normally distributed (as assessed by the Kolmogorov-Smirnov Normality test) and so were analyzed using Mann-Whitney U or Kruskal-Wallis tests with Bonferroni corrections for multiple tests (chemical and time: H2O2 and MMS results were analyzed separately, as were the different timepoints). p values > 0.05 were considered insignificant. Box plots indicate 10th, 25th, 50th, 75th, and 90th percentiles, plus outliers.
3. RESULTS
3.1 Toxicity and Lethality of MMS and H2O2
To identify appropriate doses of MMS and H2O2 for our DNA damage and repair studies, we first treated the N2 (wild-type) C. elegans with increasing concentrations of each toxicant and monitored the nematodes for viability and lethality (Fig. S1). This experiment was replicated five times, with between 36 and 144 nematodes counted per dose and per chemical at 0, 24, 48, and 72 h post-exposure. As molecular damage is expected to precede and produce observable phenotypic changes, and because we did not want DNA repair to be inhibited by frank organismal toxicity, we chose concentrations for each toxicant that resulted in a mild phenotypic response in order to study DNA repair.
3.2 Characterization of BER in Wild-Type C. elegans
Nuclear (nucDNA) and mitochondrial DNA (mtDNA) damage was quantified using QPCR following exposure of N2 nematodes to 5mM MMS or 5mM H2O2. The QPCR assay allows the simultaneous measurement of DNA damage in the mitochondrial and nuclear genomes after in vivo exposures, without differential extraction of mitochondrial and nuclear DNA [32]. Repair can be measured by ending the chemical exposure and permitting time for removal of damage [31].
In agreement with previous studies, we observed more H2O2-induced damage in mtDNA than nucDNA (Fig. 1 and 2; p < 0.0001 for treatment by genome interaction). Immediately after treatment, we measured 1.4 lesions/10 kb in mtDNA, as compared to 0.7 lesions/10 kb in the nuclear genome. On the other hand, treating with MMS resulted in greater nucDNA damage (1.7 lesions/10 kb) than mtDNA damage (0.7 lesions/10 kb).
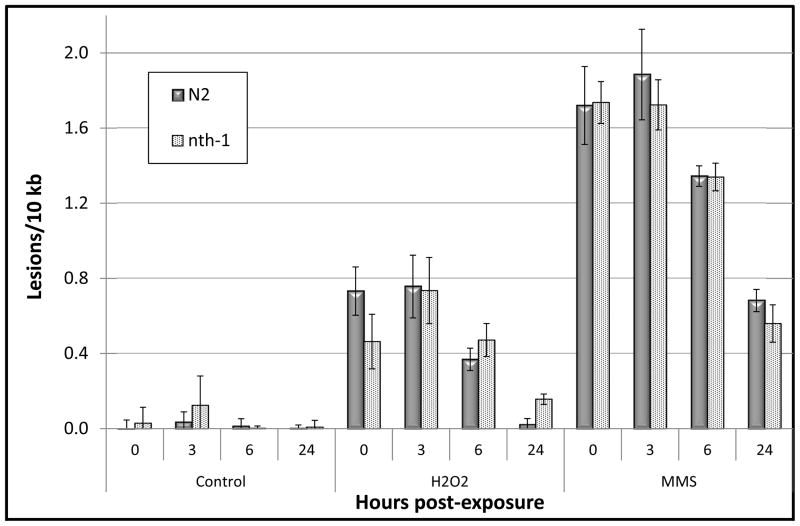
Figure 1.
N2 and nth-1 C. elegans show robust nuclear DNA repair following damage induced by 5mM H2O2 or 5 mM MMS. Nuclear DNA damage was analyzed following acute exposure of L1 larvae to H2O2 and MMS (0h). Subsequent repair was measured 3, 6, or 24h post-exposure. n = 6–11 nematodes per toxin per time point. Error bars represent the standard error of the mean.
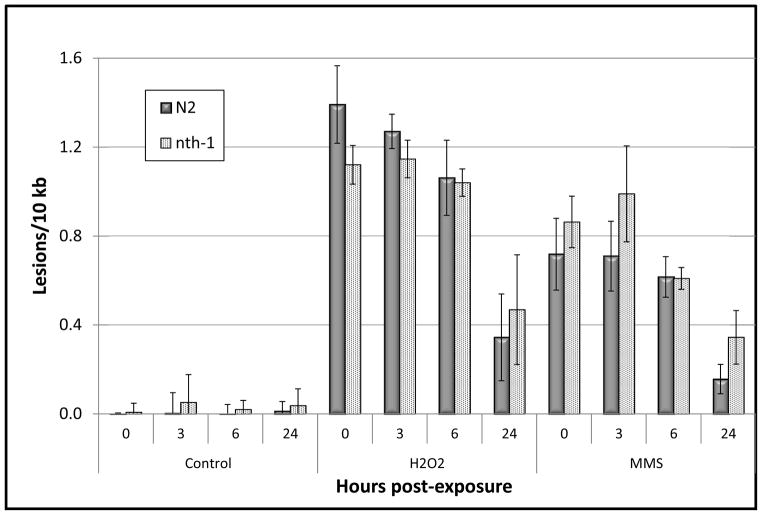
Figure 2.
N2 and nth-1 C. elegans show efficient repair of H2O2 and MMS-induced mitochondrial DNA damage. Mitochondrial DNA damage was measured in N2 (wild-type) and nth-1 C. elegans using QPCR following exposure of L1 larvae to 5mM H2O2 or 5mM MMS (time 0h). Repair of mitochondrial DNA was analyzed 3, 6, or 24h post-exposure. n = 6–11 nematodes per toxin per time point. Error bars represent the standard error of the mean.
We measured DNA repair in the nuclear and mitochondrial genomes following acute exposure to MMS and H2O2. We treated age-synchronized L1 nematodes with MMS and H2O2 in the absence of food. Following treatment, C. elegans were placed on plates with bacterial food and picked for analysis immediately after the exposure (0 h) and 3, 6, or 24h post-exposure to permit repair. Results reflect at least three experiments replicated in time for each strain and treatment group, with n = 2–3 tubes per experiment for a total n = 6–11 per strain per dose per time point.
Little to no repair of nuclear or mitochondrial DNA damage was observed 3h after exposure (p > 0.05 by FPLSD for all 0 to 3 h comparisons for both chemicals and both genomes). In the nuclear genome, repair of MMS-induced damage was detectable after 6 h, resulting in removal of 22% of the accumulated damage (Fig. 1; p = 0.008, FPLSD). H2O2-induced lesions were reduced by 50% after 6 h (p = 0.03, FPLSD). By 24h the levels of H2O2-induced nucDNA damage were comparable to baseline levels (97% reduction in lesions) and those of MMS were reduced by 60 percent (p < 0.0001 for 24 vs. 0h in both cases).
As opposed to nucDNA, repair of the lesions induced by MMS and H2O2 in the mitochondrial genome was not detectable 6h after exposure (Fig. 2; p = 0.18 for both H2O2 and MMS, 0 vs. 6h). However, we measured significant repair of MMS and H2O2-induced mtDNA damage 24 h after treatment (78 and 75% repair, respectively, and p < 0.0001 for both). Thus, C. elegans has detectable BER in both genomes, but repair is more efficient in the nuclear genome, at least for removal of oxidative and alkylating damage.
3.3 Characterization of BER in nth-1 mutant C. elegans
To further characterize the genetic components of BER in C. elegans, we utilized the RB877 strain that carries a mutation in the nth-1 gene in which exons 2 through 4 have been deleted. The glycosylase NTH-1 is responsible for locating damaged bases and carrying out the initial step of BER [20], and the nth-1 strain accumulates mutations in the nuclear genome [25]. The nth-1 gene product has a 0.60 likelihood of mitochondrial localization based on the full-length protein sequence reported by Morinaga et al. [20]. Therefore, we expected to see an abrogation of nuclear and mitochondrial BER in nth-1 nematodes. We also hypothesized that we would observe an elevated level of lesions either before or immediately after treatment, due to accumulation of damage caused by endogenous processes. However, despite the elevated levels of mutations reported in the nth-1 genome, the quantity of lesions measured in unexposed nematodes or following acute exposure of the C. elegans nth-1 strain to the genotoxic stress was similar to that of wild-type N2 C. elegans (Figs. 1 and 2) (p > 0.05 for main effect of strain and all interaction terms including “strain” as an independent factor).
Surprisingly, DNA repair in the nth-1 nematodes was also statistically indistinguishable from that of the wild-type N2 strain (Figs. 1 and 2) (p > 0.05 for all N2 vs. nth-1 comparisons). Lesion frequencies decreased significantly in both nuclear and mitochondrial genomes. The apparent trend of slower repair of H2O2-induced nucDNA damage in nth-1 nematodes as compared to similarly treated N2 C. elegans (Fig. 1) was not statistically significant. Thus, the nth-1 deficiency did not detectably reduce repair in either genome.
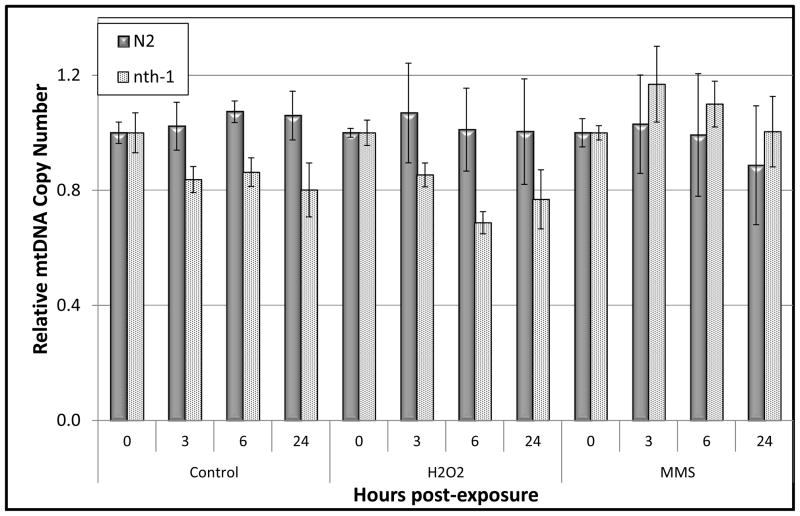
We measured the mitochondrial copy number in nematode samples obtained for the detection of DNA damage and repair. This measurement is important to the measurement of damage itself due to the need for normalization to mtDNA copy number. However, we also considered the possibility that mtDNA copy number would change due to either biogenesis or degradation of damaged genomes. However, as indicated in Figure 3 the mtDNA copy number values remain relatively stable throughout the duration of the repair time course (p > 0.05 for all main and interactive effects).
Figure 3.
mtDNA copy number is relatively stable over 24h in N2 and nth-1 C. elegans strains exposed to 5mM H2O2 or 5mM MMS. Mitochondrial DNA copy number was measured in N2 (wild-type) and nth-1 C. elegans using quantitative, real-time PCR following exposure of L1 larvae to H2O2 and MMS. Data represents the relative copy number in samples analyzed 3, 6, or 24h post-exposure as compared to the 0h time point. n = 6–11 nematodes per toxin per time point. Error bars represent the standard error of the mean.
3.4 Effect of nth-1 deficiency on viability and growth after exposure to DNA damaging agents
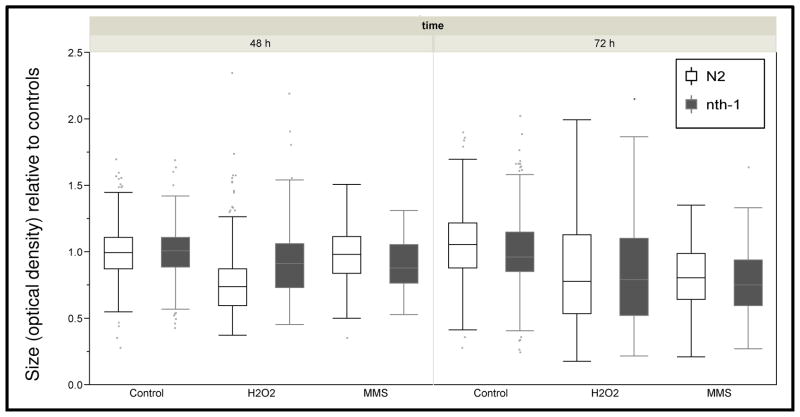
To test whether nth-1-deficient nematodes would show a phenotypic sensitivity to DNA damaging agents, we analyzed the viability of the nth-1 strain following exposure to MMS and H2O2, as we did for N2 nematodes (Fig. S1). The viability of the nth-1 strain following toxicant exposure was indistinguishable from that of N2s (Fig. S2). Reasoning that a mild repair deficiency might manifest as an alteration in a sublethal rather than lethal phenotype, we then compared growth in N2 and nth-1 nematodes following MMS and H2O2 exposure. We measured larval growth after two and three days (at which point the nematodes had reached adulthood), to allow any biological effects of unrepaired damage to integrate over time. Nematodes were treated with low amounts of toxicant, designed to produce mild growth inhibition such that potential exacerbation of that inhibition due to the nth-1 deletion would be detectable. After exposure, nematodes were placed on food plates so as to resume their development, and growth was quantified by measuring size daily. Results reflect three or four distinct experiments with 50 to 100 individuals measured per strain and treatment group each day (approximately 2700 nematodes measured total). The deficiency of nth-1 in C. elegans did not affect growth following treatment of MMS or H2O2 (Fig. 4; p > 0.05 for all comparisons). Thus, although exposure to MMS and H2O2 measurably stunted the growth of nematodes, nth-1 deficient C. elegans did not show an additional sensitivity to oxidative stress or alkylation damage.
Figure 4.
Growth inhibition by hydrogen peroxide (1.5 mM) and MMS (2.5 mM) is indistinguishable in the N2 and nth-1 strains. n = 74–334 nematodes per time point per strain per chemical; results include four separate (pooled) experiments.
4. DISCUSSION
4.1 C. elegans has robust BER capacity in vivo
We challenged C. elegans with exposure to the oxidative DNA damaging agent H2O2 and the DNA alkylating agent MMS. BER is the primary mechanism in both nuclear and mitochondrial DNA for the removal of both oxidative lesions and small alkyl DNA adducts. We observed robust removal of such damage in both the nuclear and mitochondrial genomes. Thus, BER, like NER [31, 34], appears to be functionally (as determined by rate of damage repair) conserved between C. elegans and higher eukaryotes. The rates of removal of H2O2- and MMS-induced nuclear and mitochondrial DNA damage are roughly comparable to those reported for oxidative-induced damage in post-mitotic mammalian cells in culture [35]. Similar to our observations of the C. elegans nuclear genome, previous studies have shown that human nuclear DNA displays rapid removal of oxidative lesions. Additionally, analogous to human mtDNA, the C. elegans mitochondrial genome shows less efficient repair of oxidative stress-induced damage compared to nucDNA. Mitochondria are constantly challenged by superoxide radicals released during oxidative phosphorylation and repair of mitochondrial DNA damage is increasingly recognized as being particularly important [36, 37].
4.2 Differential sensitivities of the nuclear and mitochondrial genomes
We observed more mtDNA than nucDNA damage after H2O2 exposure, as has been observed in other systems [38]. It is interesting to note that MMS caused more nucDNA than mtDNA damage; this same pattern was observed previously in cell culture [39]. Thus, while many genotoxins cause more mtDNA than nucDNA damage, this pattern is not universal.
4.3 The role of nth-1 in C. elegans BER
The relative importance of the nth-1 gene in BER in C. elegans is not clear. On the one hand, it has been shown biochemically to have glycosylase activity [20], and the RB877 strain with a deletion in the gene is somewhat mutation-prone [25] and shows transcriptional evidence of genotoxicity [24]. On the other hand, we [26] and others [20] detected no genotoxicant sensitivity phenotype, and we now report that we could not detect any difference in BER in nth-1 deficient nematodes compared to wild-type. If NTH-1 were the sole glycosylase responsible for BER then the loss-of-function mutation in nth-1 nematodes would be expected to inhibit repair. Thus, perhaps the most likely possibility is that there is another as-yet unidentified glycosylase present in C. elegans that complements the activity of NTH-1. This is commonly the case in bacteria [40, 41], yeast [42, 43] and mammals [44, 45] and Morinaga et al. presented evidence for such activity in C. elegans [20].
In a previous study, mouse models lacking functional NTH-1 show neither phenotypic nor life span abnormalities; nor did they display enhanced sensitivity following exposure to oxidizing agents such as H2O2 due to redundancy in substrate specificity provided by thymine glycol glycosylases (TGG1 and TGG2) which putatively replace NTH-1 [44]. Additionally, bacterial NEI and their human homologs (NEIL 1 and NEIL 2) have been shown to excise oxidized pyrimidine bases [46]. In previous studies, nei and nth double mutant E. coli strains demonstrated extreme sensitivity to exogenous oxidative damage while cells lacking either single enzyme activity were similar to wild type [41]. Although NEI serves as a complementary glycosylase to NTH-1 in E. coli and mammalian cells, a homolog of NEI in C. elegans was not detected by BLAST search [20]. To date, only one nth homologue has been identified in C. elegans. Further studies need to be carried out to identify other potential C. elegans glycosylases that may be functioning in BER in NTH-1 deficient nematodes.
It has also been suggested that NER may serve as an important back-up pathway for BER in this species [24, 47]. In support of this theory, BER-deficient yeast cells only show partial sensitivity when exposed to the alkylating agent MMS while cells deficient in both the BER and NER pathways are severely compromised [48]. However, compensation by NER would not explain the robust removal of oxidative and alkylating damage in mtDNA, as NER is not operative in mitochondria [14, 22, 31]. Autophagy-mediated removal of mtDNA damage could play a role, but the kinetics of that process appear to be slower than what we observed here [49]. Alternatively, C. elegans appears to lack any homologue to DNA polymerase β, the polymerase required for the gap-filling step during BER in humans [23]. Additionally, C. elegans has been shown to express DNA polymerase θ, a polymerase with lesion bypass activity [50, 51]. Pol θ is also active in the repair of oxidative damage in vertebrate cells, having redundant function with pol β in BER [52]. It is possible that pol θ is similarly involved in BER in C. elegans. The absence of polymerase β along with the lack of the requirement for NTH-1 suggests that C. elegans may have an alternative molecular pathway involving pol θ that mediates the cellular response to oxidative and alkylating DNA damage.
5. CONCLUSIONS
Ours is the first study to directly measure BER in C. elegans. While we have provided evidence for general similarity for BER processes in C. elegans and higher eukaryotes, much remains to be discovered. For example, is BER developmentally regulated, as transcriptional profiling [30]would suggest? What is the DNA polymerase involved in BER in C. elegans, since it appears to lack DNA polymerase β [23]? What is the full complement of glycosylases in this species? Ultimately, the complete BER system needs to be elucidated for C. elegans. The ability to confidently extrapolate DNA damage-related results obtained in this model organism to humans awaits a fuller understanding of this organism’s response to endogenous and exogenous genotoxins.
Supplementary Material
Highlights.
We measured repair of oxidative and alkylating DNA damage in vivo in the mitochondrial and nuclear genomes of C. elegans.
Base excision repair of both types of damage in both genomes was comparable to what has been observed in mammals.
H2O2 caused more mitochondrial than nuclear DNA damage, but the reverse occurred with methylmethanesulfonate (MMS).
A deletion in the nth-1 glycosylase gene did not reduce DNA repair or sensitize C. elegans to exposure to H2O2 or MMS.
Acknowledgments
We thank Zhirui Zhu for outcrossing the strain nth-1, and Elena Turner for assistance with operation of the COPAS Biosort. This work was supported by the RJR-Duke Leon Golberg Toxicology Training Program, the National Institute of Environmental Health Sciences (1R01-ES017540-01A2), and the National Science Foundation (NSF) and the Environmental Protection Agency (EPA) under NSF Cooperative Agreement EF-0830093, Center for the Environmental Implications of Nano Technology (CEINT). Any opinions, findings, conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the NSF or the EPA. This work has not been subjected to EPA review and no official endorsement should be inferred. Strain RB877 was provided by the C. elegans Reverse Genetics Core Facility at UBC, which is part of the International C. elegans Gene Knockout Consortium.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutation research. 2003;531:37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annual review of genetics. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 4.Friedberg EC, Walker GC, Siede W, Shultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington, DC: 2006. [Google Scholar]
- 5.Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, Szczesny B. Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5′-EXO/endonuclease) in their repair. The Journal of biological chemistry. 2011;286:31975–31983. doi: 10.1074/jbc.M110.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu P, Demple B. DNA repair in mammalian mitochondria: Much more than we thought? Environmental and molecular mutagenesis. 2010;51:417–426. doi: 10.1002/em.20576. [DOI] [PubMed] [Google Scholar]
- 7.Gredilla R, Bohr VA, Stevnsner T. Mitochondrial DNA repair and association with aging--an update. Experimental gerontology. 2010;45:478–488. doi: 10.1016/j.exger.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sung JS, Demple B. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. The FEBS journal. 2006;273:1620–1629. doi: 10.1111/j.1742-4658.2006.05192.x. [DOI] [PubMed] [Google Scholar]
- 9.Denver DR, Morris K, Lynch M, Thomas WK. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature. 2004;430:679–682. doi: 10.1038/nature02697. [DOI] [PubMed] [Google Scholar]
- 10.Keightley PD, Charlesworth B. Genetic instability of C. elegans comes naturally. Trends in genetics: TIG. 2005;21:67–70. doi: 10.1016/j.tig.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 11.van Haaften G, Vastenhouw NL, Nollen EA, Plasterk RH, Tijsterman M. Gene interactions in the DNA damage-response pathway identified by genome-wide RNA-interference analysis of synthetic lethality. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12992–12996. doi: 10.1073/pnas.0403131101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stergiou L, Hengartner MO. Death and more: DNA damage response pathways in the nematode C. elegans. Cell death and differentiation. 2004;11:21–28. doi: 10.1038/sj.cdd.4401340. [DOI] [PubMed] [Google Scholar]
- 13.Gartner A, Boag PR, Blackwell TK. WormBook: the online review of C. elegans biology. 2008. Germline survival and apoptosis; pp. 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung MC, Williams PL, Benedetto A, Au C, Helmcke KJ, Aschner M, Meyer JN. Caenorhabditis elegans: an emerging model in biomedical and environmental toxicology. Toxicological sciences: an official journal of the Society of Toxicology. 2008;106:5–28. doi: 10.1093/toxsci/kfn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pothof J, van Haaften G, Thijssen K, Kamath RS, Fraser AG, Ahringer J, Plasterk RH, Tijsterman M. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes & development. 2003;17:443–448. doi: 10.1101/gad.1060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boulton SJ, Gartner A, Reboul J, Vaglio P, Dyson N, Hill DE, Vidal M. Combined functional genomic maps of the C. elegans DNA damage response. Science. 2002;295:127–131. doi: 10.1126/science.1065986. [DOI] [PubMed] [Google Scholar]
- 17.O’Neil N, Rose A. WormBook: the online review of C. elegans biology. 2006. DNA repair; pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shatilla A, Ramotar D. Embryonic extracts derived from the nematode Caenorhabditis elegans remove uracil from DNA by the sequential action of uracil-DNA glycosylase and AP (apurinic/apyrimidinic) endonuclease. The Biochemical journal. 2002;365:547–553. doi: 10.1042/BJ20020375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura N, Morinaga H, Kikuchi M, Yonekura S, Ishii N, Yamamoto K, Yonei S, Zhang QM. Cloning and characterization of uracil-DNA glycosylase and the biological consequences of the loss of its function in the nematode Caenorhabditis elegans. Mutagenesis. 2008;23:407–413. doi: 10.1093/mutage/gen030. [DOI] [PubMed] [Google Scholar]
- 20.Morinaga H, Yonekura S, Nakamura N, Sugiyama H, Yonei S, Zhang-Akiyama QM. Purification and characterization of Caenorhabditis elegans NTH, a homolog of human endonuclease III: essential role of N-terminal region. DNA repair. 2009;8:844–851. doi: 10.1016/j.dnarep.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 21.Shatilla A, Ishchenko AA, Saparbaev M, Ramotar D. Characterization of Caenorhabditis elegans exonuclease-3 and evidence that a Mg2+-dependent variant exhibits a distinct mode of action on damaged DNA. Biochemistry. 2005;44:12835–12848. doi: 10.1021/bi050195t. [DOI] [PubMed] [Google Scholar]
- 22.Shatilla A, Leduc A, Yang X, Ramotar D. Identification of two apurinic/apyrimidinic endonucleases from Caenorhabditis elegans by cross-species complementation. DNA repair. 2005;4:655–670. doi: 10.1016/j.dnarep.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Asagoshi K, Lehmann W, Braithwaite EK, Santana-Santos L, Prasad R, Freedman JH, Van Houten B, Wilson SH. Single-nucleotide base excision repair DNA polymerase activity in C. elegans in the absence of DNA polymerase {beta} Nucleic acids research. 2011 doi: 10.1093/nar/gkr727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fensgard O, Kassahun H, Bombik I, Rognes T, Lindvall JM, Nilsen H. A two-tiered compensatory response to loss of DNA repair modulates aging and stress response pathways. Aging. 2010;2:133–159. doi: 10.18632/aging.100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denver DR, Feinberg S, Steding C, Durbin M, Lynch M. The relative roles of three DNA repair pathways in preventing Caenorhabditis elegans mutation accumulation. Genetics. 2006;174:57–65. doi: 10.1534/genetics.106.059840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer JN, Lord CA, Yang XY, Turner EA, Badireddy AR, Marinakos SM, Chilkoti A, Wiesner MR, Auffan M. Intracellular uptake and associated toxicity of silver nanoparticles in Caenorhabditis elegans. Aquatic toxicology. 2010;100:140–150. doi: 10.1016/j.aquatox.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 27.Leung MC, Goldstone JV, Boyd WA, Freedman JH, Meyer JN. Caenorhabditis elegans generates biologically relevant levels of genotoxic metabolites from aflatoxin B1 but not benzo[a]pyrene in vivo. Toxicological sciences: an official journal of the Society of Toxicology. 2010;118:444–453. doi: 10.1093/toxsci/kfq295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Croteau DL, DellaVecchia MJ, Perera L, Van Houten B. Cooperative damage recognition by UvrA and UvrB: identification of UvrA residues that mediate DNA binding. DNA repair. 2008;7:392–404. doi: 10.1016/j.dnarep.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Gondikas AP, Marinakos SM, Auffan M, Liu J, Hsu-Kim H, Meyer JN. Mechanism of Silver Nanoparticle Toxicity Is Dependent on Dissolved Silver and Surface Coating in Caenorhabditis elegans. Environmental science & technology. 2012;46:1119–1127. doi: 10.1021/es202417t. [DOI] [PubMed] [Google Scholar]
- 30.Boyd WA, Crocker TL, Rodriguez AM, Leung MC, Lehmann DW, Freedman JH, Van Houten B, Meyer JN. Nucleotide excision repair genes are expressed at low levels and are not detectably inducible in Caenorhabditis elegans somatic tissues, but their function is required for normal adult life after UVC exposure. Mutation research. 2010;683:57–67. doi: 10.1016/j.mrfmmm.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer JN, Boyd WA, Azzam GA, Haugen AC, Freedman JH, Van Houten B. Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome biology. 2007;8:R70. doi: 10.1186/gb-2007-8-5-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunter SE, Jung D, Di Giulio RT, Meyer JN. The QPCR assay for analysis of mitochondrial DNA damage, repair, and relative copy number. Methods. 2010;51:444–451. doi: 10.1016/j.ymeth.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bratic I, Hench J, Henriksson J, Antebi A, Burglin TR, Trifunovic A. Mitochondrial DNA level, but not active replicase, is essential for Caenorhabditis elegans development. Nucleic acids research. 2009;37:1817–1828. doi: 10.1093/nar/gkp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lans H, Vermeulen W. Nucleotide Excision Repair in Caenorhabditis elegans. Molecular biology international. 2011;2011:542795. doi: 10.4061/2011/542795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarrett SG, Albon J, Boulton M. The contribution of DNA repair and antioxidants in determining cell type-specific resistance to oxidative stress. Free radical research. 2006;40:1155–1165. doi: 10.1080/10715760600876613. [DOI] [PubMed] [Google Scholar]
- 36.LeDoux SP, Druzhyna NM, Hollensworth SB, Harrison JF, Wilson GL. Mitochondrial DNA repair: a critical player in the response of cells of the CNS to genotoxic insults. Neuroscience. 2007;145:1249–1259. doi: 10.1016/j.neuroscience.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- 38.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santos JH, Meyer JN, Van Houten B. Mitochondrial localization of telomerase as a determinant for hydrogen peroxide-induced mitochondrial DNA damage and apoptosis. Human molecular genetics. 2006;15:1757–1768. doi: 10.1093/hmg/ddl098. [DOI] [PubMed] [Google Scholar]
- 40.Melamede RJ, Hatahet Z, Kow YW, Ide H, Wallace SS. Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry. 1994;33:1255–1264. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- 41.Saito Y, Uraki F, Nakajima S, Asaeda A, Ono K, Kubo K, Yamamoto K. Characterization of endonuclease III (nth) and endonuclease VIII (nei) mutants of Escherichia coli K-12. Journal of bacteriology. 1997;179:3783–3785. doi: 10.1128/jb.179.11.3783-3785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alseth I, Eide L, Pirovano M, Rognes T, Seeberg E, Bjoras M. The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Molecular and cellular biology. 1999;19:3779–3787. doi: 10.1128/mcb.19.5.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.You HJ, Swanson RL, Harrington C, Corbett AH, Jinks-Robertson S, Senturker S, Wallace SS, Boiteux S, Dizdaroglu M, Doetsch PW. Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry. 1999;38:11298–11306. doi: 10.1021/bi991121i. [DOI] [PubMed] [Google Scholar]
- 44.Takao M, Kanno S, Shiromoto T, Hasegawa R, Ide H, Ikeda S, Sarker AH, Seki S, Xing JZ, Le XC, Weinfeld M, Kobayashi K, Miyazaki J, Muijtjens M, Hoeijmakers JH, van der Horst G, Yasui A. Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease III homolog for repair of thymine glycols. The EMBO journal. 2002;21:3486–3493. doi: 10.1093/emboj/cdf350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ocampo MT, Chaung W, Marenstein DR, Chan MK, Altamirano A, Basu AK, Boorstein RJ, Cunningham RP, Teebor GW. Targeted deletion of mNth1 reveals a novel DNA repair enzyme activity. Molecular and cellular biology. 2002;22:6111–6121. doi: 10.1128/MCB.22.17.6111-6121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang QM, Yonekura S, Takao M, Yasui A, Sugiyama H, Yonei S. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the hNEIL1 and hNTH1 enzymes in human cells. DNA repair. 2005;4:71–79. doi: 10.1016/j.dnarep.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Hyun M, Bohr VA, Ahn B. Biochemical characterization of the WRN-1 RecQ helicase of Caenorhabditis elegans. Biochemistry. 2008;47:7583–7593. doi: 10.1021/bi800197m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rowe LA, Degtyareva N, Doetsch PW. DNA damage-induced reactive oxygen species (ROS) stress response in Saccharomyces cerevisiae. Free radical biology & medicine. 2008;45:1167–1177. doi: 10.1016/j.freeradbiomed.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bess AS, Crocker TL, Ryde IT, Meyer JN. Mitochondrial dynamics and autophagy aid in removal of persistent mitochondrial DNA damage in Caenorhabditis elegans. Nucleic acids research. 2012 doi: 10.1093/nar/gks532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic acids research. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. The EMBO journal. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, Lan L, Seki M, Wood RD, Arakawa H, Buerstedde JM, Hochegger H, Okada T, Hiraoka M, Takeda S. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Molecular cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.