SUMMARY
Chemical shift frequencies represent a time-average of all the conformational states populated by a protein. Thus, chemical shift prediction programs based on sequence and database analysis yield higher accuracy for rigid rather than flexible protein segments. Here we show that the prediction accuracy can be significantly improved by averaging over an ensemble of structures, predicted solely from amino acid sequence with the Rosetta program. This approach to chemical shift and structure prediction has the potential to be useful for guiding resonance assignments, especially in solid-state NMR structural studies of membrane proteins in proteoliposomes.
Keywords: chemical shift, prediction, Rosetta, conformational ensemble, ensemble averaging, structure, membrane protein
NMR methods for protein structure determination increasingly rely on measurements of dipolar coupling, chemical shift anisotropy and isotropic chemical shift to provide molecular orientation and backbone dihedral angle ( , ) restraints (Delaglio et al. 2000; Tian et al. 2001; Marassi and Opella 2003; Asbury et al. 2006; Cavalli et al. 2007; Shen et al. 2008; Wishart et al. 2008). This approach relieves the time-consuming burden of measuring multiple, long-range distances from side chains, and facilitates solution NMR and solid-state NMR structure determination by molecular fragment replacement for soluble proteins in water (Shen et al. 2008; Raman et al. 2010), membrane proteins in micelles (Berardi et al. 2011) and membrane proteins in lipid bilayers (Das et al. 2012; Marassi and Opella 2003; Sharma et al. 2010; Park et al. 2012).
While dipolar couplings and chemical shift anisotropies have the distinct advantage of directly reflecting molecular orientation relative to the direction of the magnetic field or the lipid bilayer membrane, isotropic chemical shifts are also important sources of structural information (Clayden and Williams 1982; Dalgarno et al. 1983; Saito 1986; Gross and Kalbitzer 1988; Szilagyi and Jardetzky 1989; Pastore and Saudek 1990; Osapay and Case 1991; Spera and Bax 1991; Wishart et al. 1991; Iwadate et al. 1999). Several approaches have been developed both for deriving backbone dihedral angles from chemical shifts (Luginbuhl et al. 1995; Cornilescu et al. 1999) and for generating protein structures using chemical shifts as the sole source of experimental restraints, including CHESHIRE (Cavalli et al. 2007), CS23D (Wishart et al. 2008) and CS-Rosetta (Shen et al. 2008; Shen et al. 2009).
The reverse process, where isotropic chemical shifts are predicted from a structural model is also possible. Chemical shifts can be calculated ab initio from molecular structures (de Dios et al. 1993; Xu and Case 2001; Vila et al. 2008; Vila et al. 2009) and methods based on neural networks also make good chemical shift predictions (Meiler 2003). However, methods based on sequence homology and database analysis, such as ShiftX (Neal et al. 2003), ShiftX2 (Han et al. 2011), SPARTA (Shen and Bax 2007), SPARTA+ (Shen and Bax 2010), CamShift (Kohlhoff et al. 2009), and shAIC (Nielsen et al. 2012), have been shown to yield better agreement between experimentally observed and predicted values.
As proteins are intrinsically dynamic, the experimentally measured isotropic chemical shifts represent both an ensemble and time average over all explored conformational states (Mittag and Forman-Kay 2007). Thus, significant improvements in chemical shift prediction have been obtained either by averaging the predicted chemical shifts over extended (~1 s) molecular dynamics trajectories (Li and Bruschweiler 2009), or by averaging over molecular dynamics trajectories obtained by enhanced conformational space sampling (Markwick et al. 2010).
Here we show that averaging over an ensemble of protein structural models, predicted de novo from their amino acid sequences, can also significantly improve the root-mean-square difference (RMSD) between predicted and experimental chemical shift values. We demonstrate this for two proteins from the bacterial mercury detoxification Mer operon: the soluble periplasmic protein MerP, whose structure has been determined by solution NMR in aqueous buffer (Steele and Opella 1997), and the integral membrane protein MerF, whose structure has been determined by solid-state NMR in proteoliposomes (Das et al. 2012), by solid-state NMR in magnetically aligned phospholipid bilayers (De Angelis et al. 2006), and by solution NMR in detergent micelles (Howell et al. 2005).
The Rosetta program is very successful for predicting atomic level three-dimensional structures of proteins from their amino acid sequences (Das and Baker 2008). Starting with the sequences of MerP (all 72 residues) and MerFt (residues 13–72 in the helix-loop-helix integral membrane core of 81-residue MerF), we performed blind structure predictions using Rosetta, by excluding the PDB structural coordinates of the two proteins as well as their homologues from the structure prediction database. For each protein, 10,000 coarse-grained structural models were generated and then refined by all-atom relaxation, performed with either the implicit aqueous environment protocol (for MerP) or the implicit membrane protocol (for MerFt) available in Rosetta 3.2 (Das and Baker 2008; Yarov-Yarovoy et al. 2006). The refined all-atom structures were clustered according to their overall energy and their backbone CA atom RMSD to the lowest energy structure with a cutoff of 5 Å. For each protein, the most populated cluster of molecular ensembles encompassed more than 40% of the entire sampling space and contained 5,928 structures for MerP and 4,273 structures for MerFt.
For each of these structures, isotropic chemical shifts were predicted using ShiftX2 (v1.03) (Han et al. 2011) and SPARTA+ (Shen and Bax 2010), and then averaged over an increasing number of conformations, ranked from lowest to highest all-atom energy, in selected Rosetta ensembles. The results were compared to the experimental chemical shift values measured for MerP in aqueous buffer (Steele and Opella 1997) or the values measured with MAS solid-state NMR experiments on MerFt in proteoliposomes (Das et al. 2012).
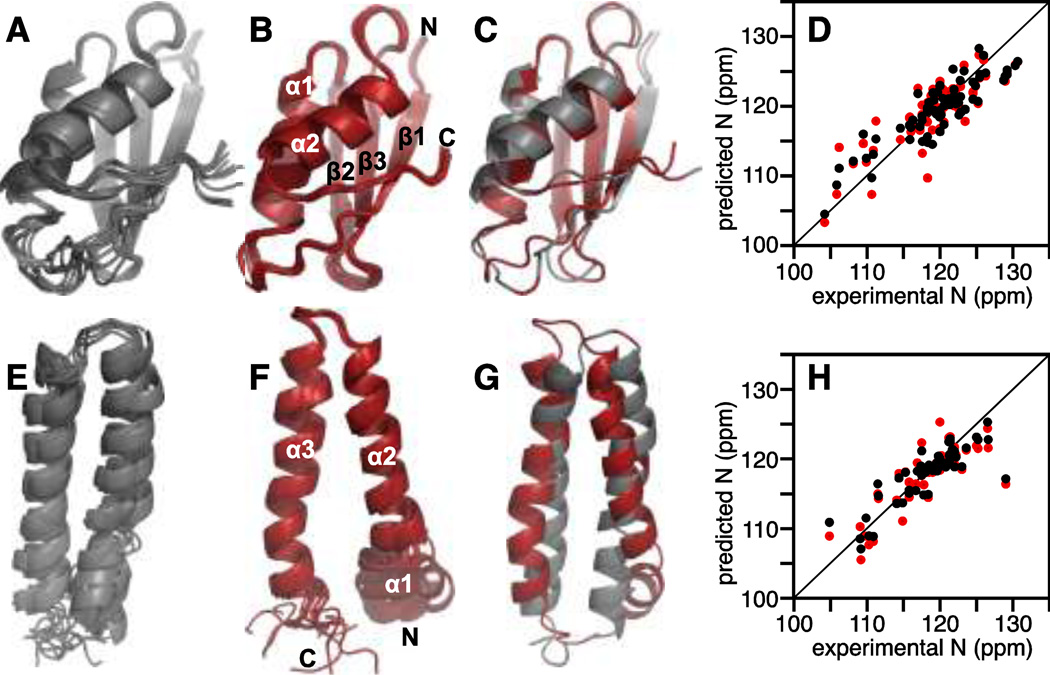
The protein structures predicted with Rosetta agree very well with those determined by NMR, with backbone RMSDs of 1.4 Å for MerP and 6.0 Å for MerFt. (Fig. 1). For MerFt, the higher RMSD reflects primarily conformational differences in the termini of the two structures. However, this does not seem to adversely affect the accuracy of chemical shift prediction. More substantial motions in the terminal regions could help mask any conformational differences through averaging, resulting in both more accurate chemical shift prediction and its improvement upon ensemble averaging (see below).
Fig. 1. Conformational ensemble averaging improves chemical shift prediction for the soluble periplasmic protein MerP (A–D) and the integral membrane protein MerFt (E–H).
(A, E) Overlay of 10 lowest energy Rosetta structures of MerP and MerFt.
(B, F) Overlay of 10 lowest energy structures determined by solution NMR for MerP (PDB: 1AFI) and by solid-state NMR for MerFt (PDB: 2LJ2).
(C, G) Overlay of lowest energy structures from Rosetta (gray) and NMR (red).
(D, H) Correlation between experimental and predicted 15N chemical shifts obtained for the single lowest energy Rosetta structure (red), or by averaging over an ensemble of 20 lowest energy Rosetta structures (black). Experimental chemical shifts and NMR coordinates were taken from the published data for MerP in aqueous solution (Steele and Opella 1997) and MerFt in proteoliposomes (Das et al. 2012).
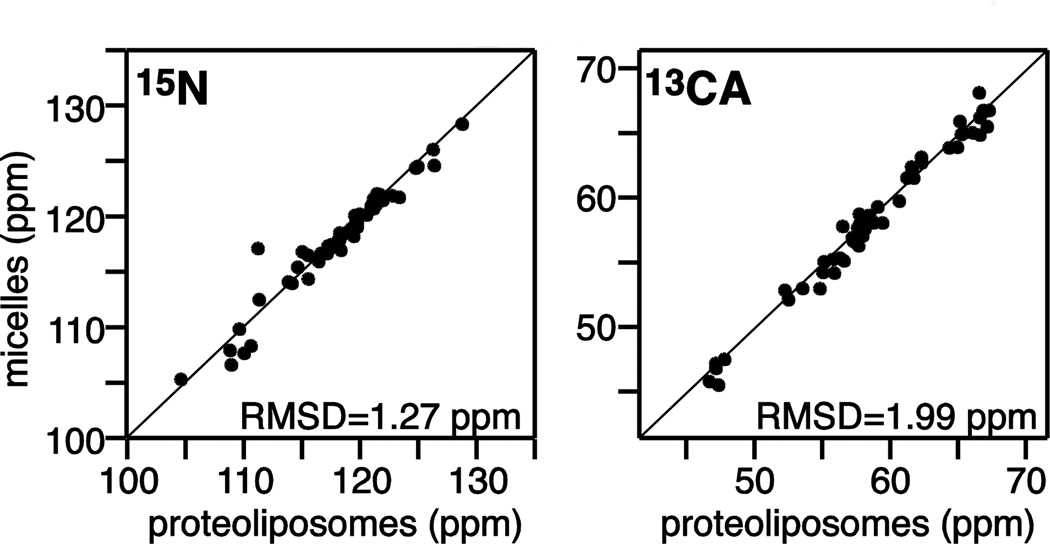
The chemical shifts measured for backbone 15N and 13CA sites of MerFt in proteoliposomes by solid-state NMR agree remarkably well with those measured from the same protein in micelles by solution NMR (Fig. 2), indicating that the overall transmembrane helical structure of the protein is preserved in the two different environments. It will be interesting to examine whether good correlations can also observed for lipid-water interfacial helices of membrane proteins where the environment is highly heterogeneous.
Fig. 2. Correlation between solution and solid-state NMR chemical shifts of MerFt.
The 15N and 13CA chemical shifts were measured for MerFt in micelles by solution NMR (Howell et al. 2005) and in proteoliposomes by solid-state NMR (Das et al. 2012). The RMSDs are 1.27 ppm for 15N and 1.99 ppm for 13CA.
Significant correlation has been found to exist between the accuracy of a Rosetta structural model and its agreement with experimental chemical shifts (Shen et al. 2008). Furthermore, the model quality required for accurate chemical shift prediction will likely depend on the extent of local dynamics and on the specific nature of the protein's structure. For example, mixed - structures, such as MerP, are more challenging to model than -helical structures, such as MerFt (Bonneau et al. 2001). Nevertheless, for both MerP and MerFt, a significant improvement in chemical shift prediction accuracy was observed when the predicted frequencies were averaged over an ensemble of conformations, rather than taken from a single lowest energy structural model (Figs. 1D, 1H; Table 1).
Table 1.
RMSDs between experimental and predicted shifts obtained for a single structure (n=1) or by averaging over an ensemble of n structures generated by Rosetta prediction or determined by NMR spectroscopy.
| RMSD (ppm) | |||||||
|---|---|---|---|---|---|---|---|
| Structure | n | HN | N | CA | CB | CO | cumulative |
| MerP (Rosetta) | 1 | 0.63 | 3.10 | 1.39 | 1.11 | 6.2 | |
| 20 | 0.61 | 2.81 | 1.23 | 1.04 | 5.7 | ||
| 100 | 0.61 | 2.84 | 1.24 | 1.05 | 5.7 | ||
| MerP (1AFI) | 1 | 0.71 | 4.93 | 1.61 | 1.65 | 8.9 | |
| 20 | 0.69 | 4.43 | 1.49 | 1.53 | 8.1 | ||
| MerFt (Rosetta) | 1 | 2.77 | 2.31 | 2.57 | 7.7 | ||
| 20 | 2.53 | 1.74 | 2.50 | 6.8 | |||
| 100 | 2.49 | 1.80 | 2.49 | 6.8 | |||
| MerFt (2LJ2) | 1 | 2.90 | 1.66 | 2.54 | 7.1 | ||
| 20 | 2.42 | 1.59 | 2.38 | 6.4 | |||
| 100 | 2.40 | 1.61 | 2.38 | 6.4 | |||
The chemical shifts predicted from the single lowest energy Rosetta structures correlate well with the experimental data, with cumulative RMSDs of 6.2 for MerP (N, HN, CA, CB) and 7.7 for MerFt (N, CA, CO). However, ensemble-averaging of the chemical shifts yielded lower cumulative RMSDs for both proteins, 5.7 for MerP and 6.8 for MerFt, representative of a ~12% improvement over single structure prediction. ShiftX2 (Table 1; Figs. 1, 3, 4) and SPARTA+ (data not shown) yielded similar results, although somewhat better prediction accuracy was obtained with ShiftX2.
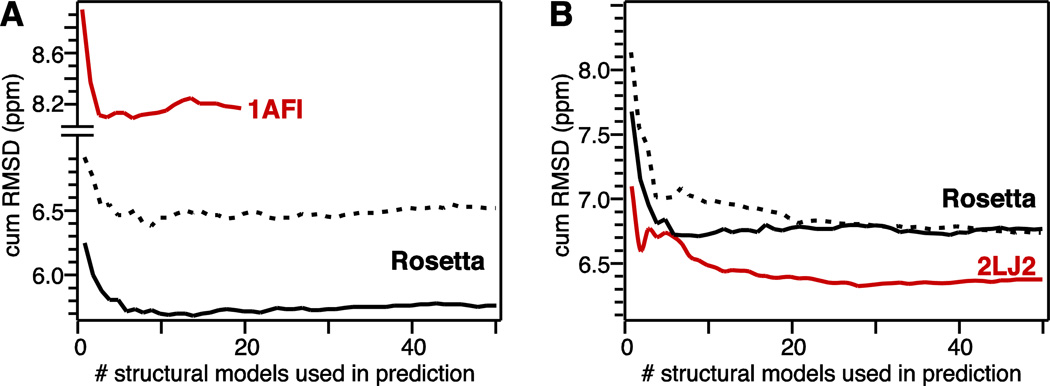
Fig. 3. Improvement in cumulative RMSD between experimental and predicted chemical shifts due to ensemble averaging for MerP (A) and MerFt (B).
Predicted chemical shifts were averaged over increasing numbers of ensemble structures ranked from lowest to highest energy. (Red) Ensembles of lowest energy NMR structures. (Black solid line) Ensembles of lowest energy Rosetta structures. (Black dashed line) Ensembles of 100 highest energy Rosetta structures.
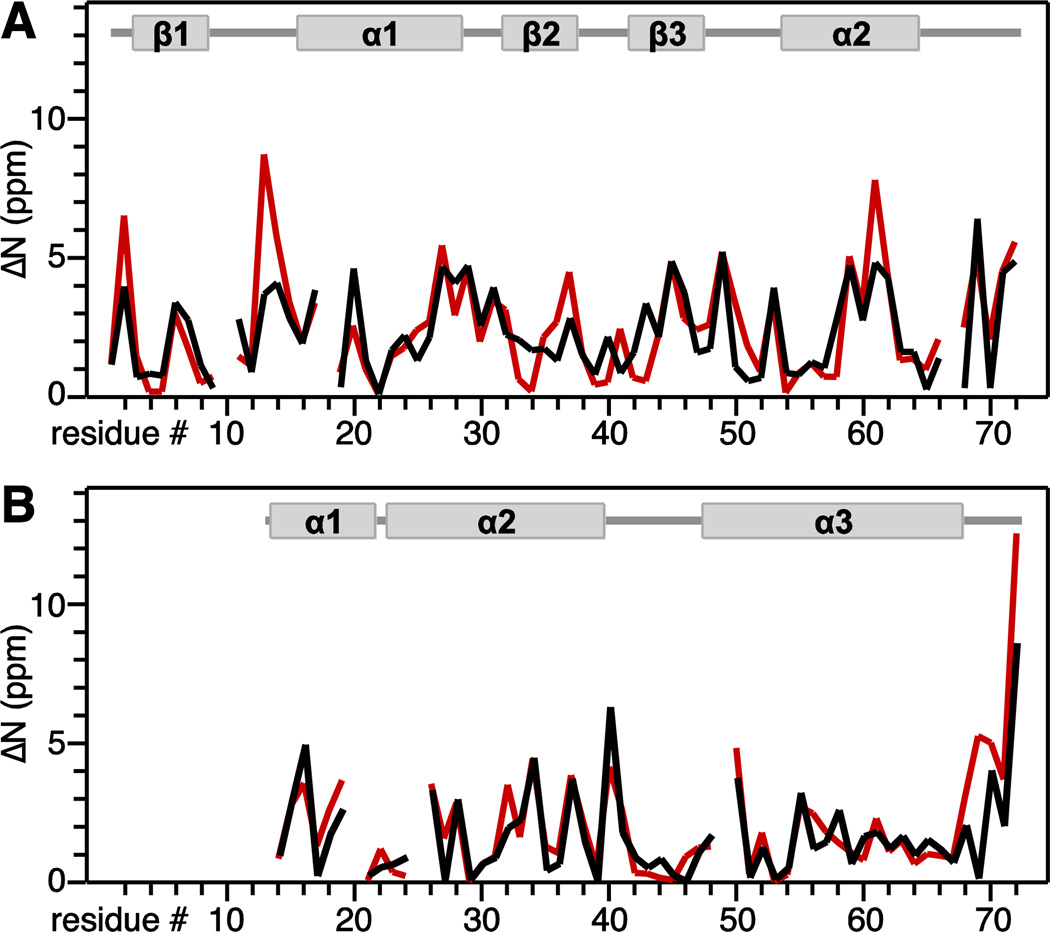
Fig. 4. Residue-specific improvement in 15N chemical shift prediction accuracy obtained by ensemble averaging for MerP (A) and MerFt (B).
The improvement value ( N) is the difference between experimentally observed chemical shifts and chemical shifts predicted from single lowest energy Rosetta structure (red), or chemical shifts predicted and averaged over 20 lowest energy Rosetta structures (black). Protein secondary structures are shown above the graphs.
Correlation with the experimental data improves as the predicted chemical shifts are averaged over an increasing number of ensemble structures (Fig. 3). The improvement levels off when >10 structural models are included in the averaging process, reflecting the onset of contributions from higher energy conformers and indicating that a Rosetta ensemble of 10 lowest energy structures is a better representation of the native structure than the single lowest energy model.
Notably, within a given conformational ensemble, the accuracy of chemical shift prediction is always better when the data are averaged over multiple conformations rather than taken individually, although averaging over the lowest energy structures (Fig. 3, black solid line) yields the most accurate prediction. For example, averaging the predicted chemical shifts over an ensemble of 100 highest Rosetta energy structures ranked from lowest to highest energy (Fig. 3, black dashed line) also yields an improvement in prediction accuracy. For MerFt, the improvement levels off at the same value as for prediction from the lowest energy ensemble, albeit after including at least twice as many conformations (Fig. 3B). For MerP, ensemble averaging over the higher energy structures improves the prediction but the accuracy never reaches the level obtained for the low energy ensembles (Fig. 3A). This difference between MerP and MerFt may reflect differences in protein dynamics, as well as the distinctly different conformations and environments of the two proteins (compact, soluble - structure for MerP; two transmembrane -helices for MerFt). Since MerP is water-soluble and MerF is an integral membrane protein, their respective models were predicted using either the all atom implicit solvent potential (MerP) or the all atom implicit membrane potential (MerFt) of Rosetta.
The most notable improvements in prediction accuracy from ensemble averaging were obtained for the 15N nuclei (Table 1). Furthermore, the degree of improvement correlates with protein dynamics (Fig. 4). Similar results were reported in a previous study where the predicted chemical shifts were averaged over molecular dynamics trajectories (Markwick et al. 2010). This is consistent with the observation that chemical shift prediction programs based on homology and database analysis yield higher prediction accuracy for rigid rather than flexible protein segments, and explains why they can be improved by ensemble averaging (Shen and Bax 2010). For MerP (Fig. 4A) the largest improvements in prediction accuracy coincide with residues in the 1- 1 metal-binding loop and the C-terminus, which are more flexible than the rest of the protein (Steele and Opella 1997). For MerFt (Fig. 4B), prediction improvements coincide primarily with the more flexible C-terminus of the protein.
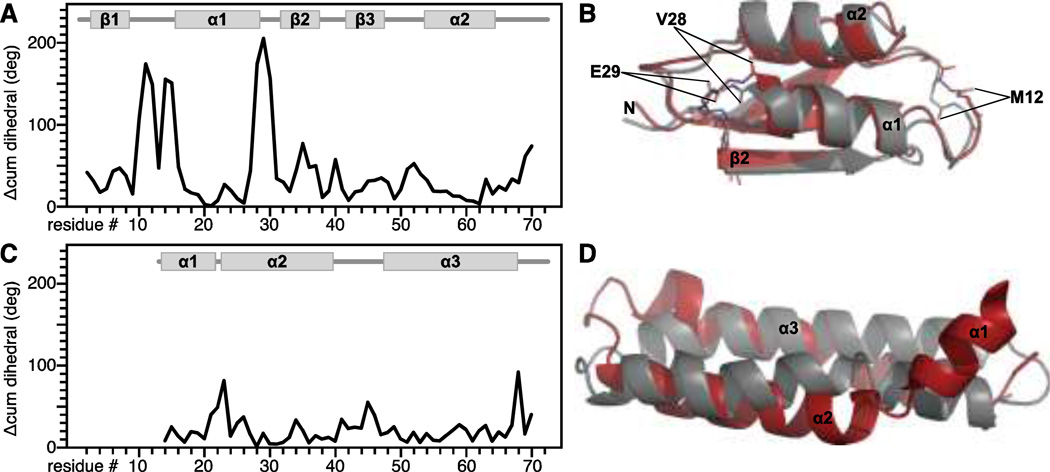
The effects of side-chain conformation on backbone chemical shifts are well recognized (de Dios et al. 1993; Wang and Jardetzky 2004; Vila et al. 2007; Villegas et al. 2007; London et al. 2008; Mulder 2009). For example, excluding 1 torsion angle input information from SPARTA+ chemical shift prediction has been shown to decrease the agreement with experimental data by 5% for 15N and by 1–2% for other nuclei (Shen and Bax 2010). In the case of MerFt, an additional 5% improvement in prediction accuracy is observed when the chemical shifts are predicted for and averaged over the experimentally determined solid-state NMR conformers (Fig. 3B, red). This likely coincides with refinement of side chain conformations by the NMR-restrained simulated annealing performed during structure refinement (Das et al. 2012). In the case of MerP, the accuracy of chemical shifts predicted from the NMR structure is poorer than that obtained from Rosetta (Fig. 3A, red). This reflects local differences in backbone dihedral angles between the NMR and Rosetta structures of MerP, found primarily in the 1- 1 and 1- 2 loops (Fig. 5A, B) and may be due to a localized distortion in the NMR structure. Conversely for MerFt, the local dihedral angles of the Rosetta and NMR structures are very similar (Fig. 5C, D) even though the overall backbone RMSD is poorer. The results in Fig. 3 and Fig. 5 indicate that local dihedral angles can have a substantial impact on the accuracy of chemical shift prediction even in cases, such as MerP, where the overall fold is accurate.
Fig. 5. Agreement between backbone dihedral angles predicted from Rosetta or determined experimentally for MerP (A, B) and MerFt (C, D).
(A, C) Cumulative difference between the backbone dihedral angles ( , ) determined by NMR and predicted from Rosetta ( cum = + ).
(B, D) Overlay of NMR (gray) and Rosetta (red) structures. Dihedral angles for MerP residues M12, V28 and E29 differ significantly between the NMR and Rosetta structures.
In summary, we show that the accuracy of chemical shift prediction can be significantly improved by averaging over an ensemble of conformations predicted de novo solely from the amino acid sequence. Ensemble averaging improves the prediction accuracy both for solution NMR data from a globular protein in water and for solid-state NMR data from an integral membrane protein in proteoliposomes. The resulting chemical shift prediction accuracy (cumulative RMSD ~6 ppm) is comparable to the accuracy obtained by ensemble averaging over MD trajectories of crystal structures (Markwick et al. 2010). It is not clear whether this accuracy is sufficient to assign the resonances of an entire protein, nonetheless, it definitely has the potential to resolve ambiguities and provide validation in the early stages of experimental structural determination by NMR
Estimates of the protein size and type compatible with de novo Rosetta structure prediction vary with protein secondary structure and the exact methods used in prediction (Das and Baker 2008). Small -helical proteins (up to ~120 residues) are typically well within the size and complexity limits of Rosetta, while mixed - structures are more challenging to model (Bonneau et al. 2001). The incorporation of chemical shifts in Rosetta structure prediction has been shown to dramatically improved correct structure convergence for proteins up to 100 residues (Shen et al. 2008) and the addition of RDCs further improves convergence for proteins up to 200 residues (Raman et al. 2010). Rosetta structure prediction works well for many proteins up to 100 amino acids using sequence information alone (Bradley et al. 2005) and for significantly larger proteins when assisted by a template (Das et al. 2007). The estimates may be even better for membrane proteins where the lipid bilayer membrane that is modeled implicitly in Rosetta (Yarov-Yarovoy et al. 2006) provides additional conformational and spatial restraints. Furthermore, recent methods for structure prediction based on evolutionary coupling can be used to provide very good models of both soluble and integral membrane proteins (Marks et al. 2011; Hopf et al. 2012)
In cases where good structural models can be predicted from amino acid sequence alone, or with assistance of a template derived from sequence homology, or by evolutionary coupling methods, the ensemble-averaging approach described here could provide useful information at a very early stage of a protein structure determination project. In particular, it could serve as a useful guide and validation tool for resonance assignment during de novo solid-state NMR structure determination of membrane proteins, where the process is complicated by broader or overlapped lines and, typically, the absence of 1H signals, and especially when combined with recently developed automated assignments methods (Moseley et al. 2010; Tycko and Hu 2010).
ACKNOWLEDGEMENTS
This research was supported by grants from the National Institutes of Health (R21GM094727; P01AI074805). It utilized the Biotechnology Research Center for NMR Molecular Imaging of Proteins at UCSD (P41EB002031).
REFERENCES
- Asbury T, Quine JR, Achuthan S, Hu J, Chapman MS, Cross TA, Bertram R. PIPATH: an optimized algorithm for generating alpha-helical structures from PISEMA data. J Magn Reson. 2006;183(1):87–95. doi: 10.1016/j.jmr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Berardi MJ, Shih WM, Harrison SC, Chou JJ. Mitochondrial uncoupling protein 2 structure determined by NMR molecular fragment searching. Nature. 2011;476(7358):109–113. doi: 10.1038/nature10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau R, Tsai J, Ruczinski I, Chivian D, Rohl C, Strauss CE, Baker D. Rosetta in CASP4: progress in ab initio protein structure prediction. Proteins Suppl. 2001;5:119–126. doi: 10.1002/prot.1170. [DOI] [PubMed] [Google Scholar]
- Bradley P, Misura KM, Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309(5742):1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. Protein structure determination from NMR chemical shifts. Proc Natl Acad Sci U S A. 2007;104(23):9615–9620. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayden NJ, Williams RJP. Peptide group shifts. J Magn Reson. 1982;49(3):383–396. [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13(3):289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Dalgarno DC, Levine BA, Williams RJ. Structural information from NMR secondary chemical shifts of peptide alpha C-H protons in proteins. Biosci Rep. 1983;3(5):443–452. doi: 10.1007/BF01121955. [DOI] [PubMed] [Google Scholar]
- Das BB, Nothnagel HJ, Lu GJ, Son WS, Tian Y, Marassi FM, Opella SJ. Structure Determination of a Membrane Protein in Proteoliposomes. J Am Chem Soc. 2012;134(4):2047–2056. doi: 10.1021/ja209464f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Das R, Qian B, Raman S, Vernon R, Thompson J, Bradley P, Khare S, Tyka MD, Bhat D, Chivian D, Kim DE, Sheffler WH, Malmstrom L, Wollacott AM, Wang C, Andre I, Baker D. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins. 2007;69(Suppl 8):118–128. doi: 10.1002/prot.21636. [DOI] [PubMed] [Google Scholar]
- De Angelis AA, Howell SC, Nevzorov AA, Opella SJ. Structure determination of a membrane protein with two trans-membrane helices in aligned phospholipid bicelles by solid-state NMR spectroscopy. J Am Chem Soc. 2006;128(37):12256–12267. doi: 10.1021/ja063640w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Dios AC, Pearson JG, Oldfield E. Secondary and tertiary structural effects on protein NMR chemical shifts: an ab initio approach. Science. 1993;260(5113):1491–1496. doi: 10.1126/science.8502992. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Kontaxis G, Bax A. Protein Structure Determination Using Molecular Fragment Replacement and NMR Dipolar Couplings. J Am Chem Soc. 2000;122:2142–2143. [Google Scholar]
- Gross K-H, Kalbitzer HR. Distribution of chemical shifts in 1H nuclear magnetic resonance spectra of proteins. J Magn Reson. 1988;76(1):87–99. [Google Scholar]
- Han B, Liu Y, Ginzinger SW, Wishart DS. SHIFTX2: significantly improved protein chemical shift prediction. J Biomol NMR. 2011;50(1):43–57. doi: 10.1007/s10858-011-9478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf TA, Colwell LJ, Sheridan R, Rost B, Sander C, Marks DS. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012;149(7):1607–1621. doi: 10.1016/j.cell.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell SC, Mesleh MF, Opella SJ. NMR structure determination of a membrane protein with two transmembrane helices in micelles: MerF of the bacterial mercury detoxification system. Biochemistry. 2005;44(13):5196–5206. doi: 10.1021/bi048095v. [DOI] [PubMed] [Google Scholar]
- Iwadate M, Asakura T, Williamson MP. C alpha and C beta carbon-13 chemical shifts in proteins from an empirical database. J Biomol NMR. 1999;13(3):199–211. doi: 10.1023/a:1008376710086. [DOI] [PubMed] [Google Scholar]
- Kohlhoff KJ, Robustelli P, Cavalli A, Salvatella X, Vendruscolo M. Fast and accurate predictions of protein NMR chemical shifts from interatomic distances. J Am Chem Soc. 2009;131(39):13894–13895. doi: 10.1021/ja903772t. [DOI] [PubMed] [Google Scholar]
- Li D-W, Bruschweiler R. Certification of Molecular Dynamics Trajectories with NMR Chemical Shifts. The Journal of Physical Chemistry Letters. 2009;1(1):246–248. [Google Scholar]
- London RE, Wingad BD, Mueller GA. Dependence of amino acid side chain 13C shifts on dihedral angle: application to conformational analysis. J Am Chem Soc. 2008;130(33):11097–11105. doi: 10.1021/ja802729t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luginbuhl P, Szyperski T, Wuthrich K. Statistical Basis for the Use of 13Cα Chemical Shifts in Protein Structure Determination. J Magn Reson B. 1995;109(2):229–233. [Google Scholar]
- Marassi FM, Opella SJ. Simultaneous assignment and structure determination of a membrane protein from NMR orientational restraints. Protein Sci. 2003;12(3):403–411. doi: 10.1110/ps.0211503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, Sander C. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011;6(12):e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwick PR, Cervantes CF, Abel BL, Komives EA, Blackledge M, McCammon JA. Enhanced conformational space sampling improves the prediction of chemical shifts in proteins. J Am Chem Soc. 2010;132(4):1220–1221. doi: 10.1021/ja9093692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiler J. PROSHIFT: protein chemical shift prediction using artificial neural networks. J Biomol NMR. 2003;26(1):25–37. doi: 10.1023/a:1023060720156. [DOI] [PubMed] [Google Scholar]
- Mittag T, Forman-Kay JD. Atomic-level characterization of disordered protein ensembles. Curr Opin Struct Biol. 2007;17(1):3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Moseley HN, Sperling LJ, Rienstra CM. Automated protein resonance assignments of magic angle spinning solid-state NMR spectra of beta1 immunoglobulin binding domain of protein G (GB1) J Biomol NMR. 2010;48(3):123–128. doi: 10.1007/s10858-010-9448-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder FA. Leucine side-chain conformation and dynamics in proteins from 13C NMR chemical shifts. Chembiochem. 2009;10(9):1477–1479. doi: 10.1002/cbic.200900086. [DOI] [PubMed] [Google Scholar]
- Neal S, Nip AM, Zhang H, Wishart DS. Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J Biomol NMR. 2003;26(3):215–240. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- Nielsen JT, Eghbalnia HR, Nielsen NC. Chemical shift prediction for protein structure calculation and quality assessment using an optimally parameterized force field. Prog Nucl Magn Reson Spectrosc. 2012;60:1–28. doi: 10.1016/j.pnmrs.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osapay K, Case DA. A new analysis of proton chemical shifts in proteins. J Am Chem Soc. 1991;113(25):9436–9444. [Google Scholar]
- Park SH, Das BB, Casagrande F, Tian Y, Nothnagel HJ, Chu M, Kiefer H, Maier K, De Angelis A, Marassi FM, Opella SJ. Structure of the Chemokine Receptor CXCR1 in Phospholipid Bilayers. Nature. 2012 doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore A, Saudek V. The relationship between chemical shift and secondary structure in proteins. J Magn Reson. 1990;90(1):165–176. [Google Scholar]
- Raman S, Lange OF, Rossi P, Tyka M, Wang X, Aramini J, Liu G, Ramelot TA, Eletsky A, Szyperski T, Kennedy MA, Prestegard J, Montelione GT, Baker D. NMR structure determination for larger proteins using backbone-only data. Science. 2010;327(5968):1014–1018. doi: 10.1126/science.1183649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H. Conformation-dependent 13C chemical shifts: A new means of conformational characterization as obtained by high-resolution solid-state 13C NMR. Magn Reson Chem. 1986;24(10):835–852. [Google Scholar]
- Sharma M, Yi M, Dong H, Qin H, Peterson E, Busath DD, Zhou HX, Cross TA. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330(6003):509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A. Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. J Biomol NMR. 2007;38(4):289–302. doi: 10.1007/s10858-007-9166-6. [DOI] [PubMed] [Google Scholar]
- Shen Y, Bax A. SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J Biomol NMR. 2010;48(1):13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci U S A. 2008;105(12):4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Vernon R, Baker D, Bax A. De novo protein structure generation from incomplete chemical shift assignments. J Biomol NMR. 2009;43(2):63–78. doi: 10.1007/s10858-008-9288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spera S, Bax A. Empirical correlation between protein backbone conformation and C.alpha. and C.beta. 13C nuclear magnetic resonance chemical shifts. J Am Chem Soc. 1991;113(14):5490–5492. [Google Scholar]
- Steele RA, Opella SJ. Structures of the reduced and mercury-bound forms of MerP, the periplasmic protein from the bacterial mercury detoxification system. Biochemistry. 1997;36(23):6885–6895. doi: 10.1021/bi9631632. [DOI] [PubMed] [Google Scholar]
- Szilagyi L, Jardetzky O. α-Proton chemical shifts and secondary structure in proteins. J Magn Reson. 1989;83(3):441–449. [Google Scholar]
- Tian F, Valafar H, Prestegard JH. A dipolar coupling based strategy for simultaneous resonance assignment and structure determination of protein backbones. J Am Chem Soc. 2001;123(47):11791–11796. doi: 10.1021/ja011806h. [DOI] [PubMed] [Google Scholar]
- Tycko R, Hu KN. A Monte Carlo/simulated annealing algorithm for sequential resonance assignment in solid state NMR of uniformly labeled proteins with magic-angle spinning. J Magn Reson. 2010;205(2):304–314. doi: 10.1016/j.jmr.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila JA, Aramini JM, Rossi P, Kuzin A, Su M, Seetharaman J, Xiao R, Tong L, Montelione GT, Scheraga HA. Quantum chemical 13C(alpha) chemical shift calculations for protein NMR structure determination, refinement, and validation. Proc Natl Acad Sci U S A. 2008;105(38):14389–14394. doi: 10.1073/pnas.0807105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila JA, Arnautova YA, Martin OA, Scheraga HA. Quantum-mechanics-derived 13Calpha chemical shift server (CheShift) for protein structure validation. Proc Natl Acad Sci U S A. 2009;106(40):16972–16977. doi: 10.1073/pnas.0908833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila JA, Villegas ME, Baldoni HA, Scheraga HA. Predicting 13Calpha chemical shifts for validation of protein structures. J Biomol NMR. 2007;38(3):221–235. doi: 10.1007/s10858-007-9162-x. [DOI] [PubMed] [Google Scholar]
- Villegas ME, Vila JA, Scheraga HA. Effects of side-chain orientation on the 13C chemical shifts of antiparallel beta-sheet model peptides. J Biomol NMR. 2007;37(2):137–146. doi: 10.1007/s10858-006-9118-6. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jardetzky O. Predicting 15N chemical shifts in proteins using the preceding residue-specific individual shielding surfaces from phi, psi i-1, and chi 1 torsion angles. J Biomol NMR. 2004;28(4):327–340. doi: 10.1023/B:JNMR.0000015397.82032.2a. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Arndt D, Berjanskii M, Tang P, Zhou J, Lin G. CS23D: a web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res. 2008;36(Web Server issue):W496–W502. doi: 10.1093/nar/gkn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol. 1991;222(2):311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- Xu XP, Case DA. Automated prediction of 15N, 13Calpha, 13Cbeta and 13C' chemical shifts in proteins using a density functional database. J Biomol NMR. 2001;21(4):321–333. doi: 10.1023/a:1013324104681. [DOI] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, Schonbrun J, Baker D. Multipass membrane protein structure prediction using Rosetta. Proteins. 2006;62(4):1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]