Abstract
The passive stiffness of cardiac muscle plays a critical role in ventricular filling during diastole and is determined by the extracellular matrix and the sarcomeric protein titin. Titin spans from the Z-disk to the M-band of the sarcomere and also contains a large extensible region that acts as a molecular spring and develops passive force during sarcomere stretch. This extensible segment is titin's I-band region, and its force-generating mechanical properties determine titin-based passive tension. The properties of titin's I-band region can be modulated by isoform splicing and post-translational modification and are intimately linked to diastolic function. This review discusses the physical origin of titin-based passive tension, the mechanisms that alter titin stiffness, and titin's role in stress-sensing signaling pathways.
Keywords: passive tension, connectin, entropic force, mechanical signaling
1. Introduction
The cardiac sarcomere contains, in addition to actin-based thin filaments and myosin-based thick filaments, the giant protein titin (also called connectin). While the proteins that compose the thin and thick filaments are responsible for generating active force in the sarcomere, titin provides passive elasticity to muscle. This elastic property of titin, in addition to the extracellular matrix, defines the passive stiffness of muscle that resists sarcomere stretch. Most of the extracellular matrix's contribution to passive tension is due to collagen, with collagen playing a larger role towards the upper limit of the physiological sarcomere length range (Wu et al. 2000; Wu et al. 2002). While the extracellular matrix is also important to passive stiffness, this review will focus on titin. In addition to providing elasticity to muscle, titin is also essential for maintaining the structural integrity of the sarcomere. Diastolic filling of the beating heart is largely determined by titin-based passive tension, with changes in titin stiffness associated with diastolic dysfunction and cardiac disease.
A single titin molecule spans the half sarcomere, binding to the Z-disk and to the thin filament at its N-terminus, and binding to the thick filament and M-band towards its C-terminus (Fig. 1A). Between its thin and thick filament binding domains titin contains a large segment that behaves as a molecular spring that extends during sarcomere stretch. This spring-like segment is titin's I-band region; it centers the thick filament in the middle of the sarcomere and generates a passive restoring force that resists increases in sarcomere length. However, this portion of titin is not only a molecular spring but can also interact with thin filament proteins or possibly adjacent titin molecules. Nonetheless, it primarily defines titin-based passive tension. Titin's I-band region contains three extensible elements: 1) serially-linked immunoglobulin(Ig)-like domains, 2) the PEVK element (rich in proline (P), glutamate (E), valine (V), and lysine (K) residues), and 3) the N2B element. Changes in titin's I-band region, namely isoform splicing and post-translational modifications, directly influence titin-based passive tension. Although the I-band region of titin generates most titin-based passive tension, other regions of titin (Z-disk, A-band, and M-band) are involved in numerous cellular processes including maintaining sarcomere structure and force-dependent signaling. This review focuses on the physical concepts at the core of titin's elasticity, the physiological mechanisms that tune titin-based tension, and titin's role in mechanical stress sensing.
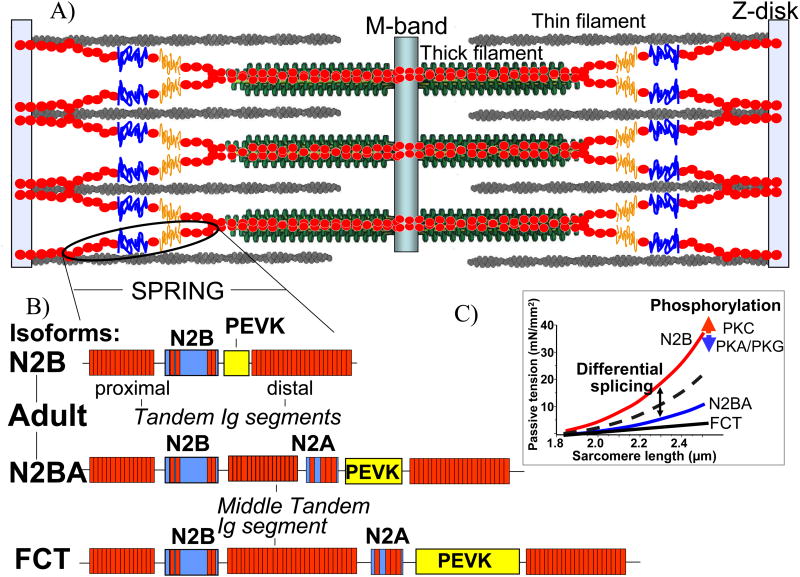
Figure 1.
A) A schematic of the sarcomere. A single titin molecule spans from the Z-disk to the M-band and contains a spring-like region that develops force upon sarcomere stretch. B) The I-band region composition of the three cardiac titin isoforms. The adult N2BA isoform and fetal cardiac titin isoform contain a middle tandem Ig segment, the N2A element, and a longer PEVK sequence than the short N2B isoform. C) Titin-based passive tension levels are determined by titin isoform composition and can also be modulated by phosphorylation of I-band elements.
2. Titin-based Passive Tension
The importance of titin-based passive tension cannot be overstated. Drastic changes in titin isoform expression occur during neonatal cardiac development as large, compliant titin isoforms are replaced by smaller, stiffer isoforms to meet the increased needs of the newborn heart (Lahmers et al. 2004; Opitz et al. 2004; Greaser et al. 2005). Certain cardiac diseases exhibit changes in titin isoform expression that can be compensatory or contribute to the disease phenotype (Neagoe et al. 2002; Warren et al. 2003; Nagueh et al. 2004). Stimulation of adrenergic pathways activates protein kinases that phosphorylate titin and change its elastic properties, which leads to changes in stiffness at the myocardial level (Kruger et al. 2006; Hidalgo et al. 2009; Kruger et al. 2009). Titin-based passive stiffness affects the pressure-volume relationship of the beating heart and can be a direct cause of diastolic dysfunction (Chung et al. 2011a). Changes in titin isoform expression and phosphorylation modulate titin-based stiffness in a sarcomere length dependent manner (Fig. 1C), and the presence of multiple mechanisms that alter titin elasticity suggests that fine-tuning titin stiffness is physiologically relevant.
Differential expression of titin isoforms significantly impacts titin-based passive tension because of the intimate relationship between the size of titin's I-band region and the force needed to extend it. Three classes of cardiac titin isoforms exist: N2B, N2BA, and fetal cardiac titin (FCT) (Fig. 1B). The primary difference between these three groups is the size of their extensible segments; the regions of titin that are necessary for binding to the Z-disk, thin filament, thick filament, and M-band are largely identical (Labeit et al. 1995). Since titin elasticity is primarily defined by its I-band region, the isoform-specific identification of I-band composition has been heavily researched, and numerous splicing pathways have been identified (Lahmers et al. 2004; Greaser et al. 2005). It is now clear that the N2B isoform has the shortest I-band region (and is therefore most “stiff”), FCT isoforms have the longest I-band region (most compliant), and N2BA titin is intermediate (Fig. 1C). The passive tension-sarcomere length relationship of the N2B, N2BA, and FCT isoforms is well-described by treating the respective I-band regions as molecular (entropic) springs of varying extensibility, and an in-depth understanding of the origin of titin-based passive tension is warranted.
3. Origin of Titin-Based Passive Tension
3.1 Entropic force
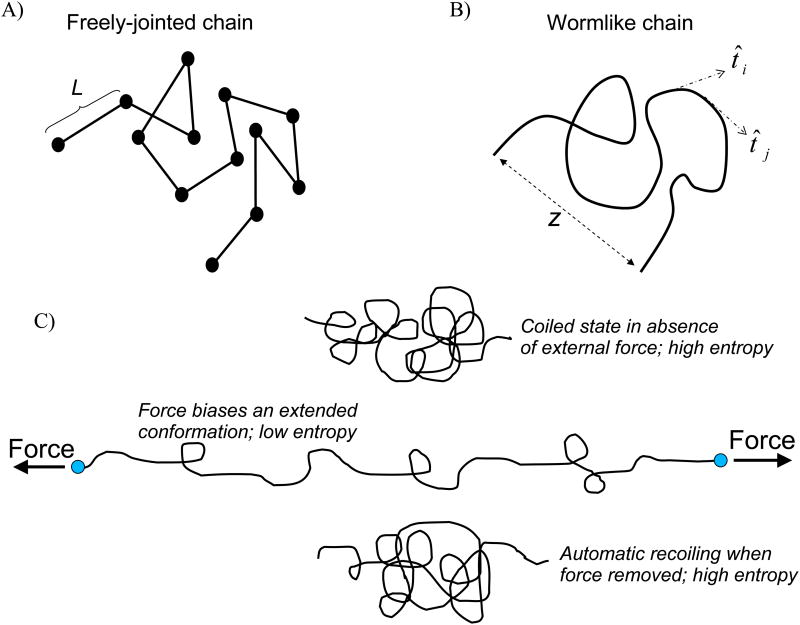
The passive tension generated when striated muscle is stretched comes from titin and the extracellular matrix (ECM) (Wu et al. 2000). The two types of resistance are different, however, as the ECM contribution is an elastic force derived primarily from collagen's high tensile strength and intertwined matrix orientation (Weber 1989), while titin-based passive tension is entropic in nature. This entropic force is due to titin's extensible I-band region that contains three distinct spring-like elements. The I-band region of titin is unconstrained in the sarcomere (i.e. not attached to actin or myosin) and freely interacts with the thermal energy of the surrounding aqueous medium. The thermal energy at room (and body) temperature is high enough that protein structure is constantly changing as a protein explores all the conformations accessible at that given temperature. The sum of all configurations a protein can occupy determines the phase-space volume of that protein, with each possible configuration of atomic coordinates and momenta corresponding to a unique point, or microstate, in the phase-space. Important to note is that each microstate has an associated probability of occupancy pi and energy Ei, and that in solution an ensemble of proteins occupies various microstates according to the probability of occupancy (note, microstates with equal Ei have equal pi). Consider the freely-jointed chain, a 3D random walk of monomers of fixed length L (Fig. 2A), which is a simple discrete model to describe an unstructured protein. Note that as L→ 0 the freely-jointed chain becomes continuous (wormlike chain; Fig. 2B). There is no interaction between the monomers, which means all configurations have the same energy. Therefore, all configurations are equally likely and the chain freely diffuses through different microstates as time progresses (for proteins, this diffusion is due to thermal energy). However, only one microstate (configuration) describes the chain being held fully-taut from end to end while numerous microstates are associated with the chain occupying a compact configuration. Because all microstates are occupied with equal probability, it is equally like that the fully-taut configuration and a single compact configuration will be occupied. But since there are vastly more compact configurations, probability alone dictates that the chain will almost always exist in a compact state. Now, lets consider the PEVK element and the unique sequence of the N2B element that are thought to occupy a largely random coil structure, as evidenced by single molecule force spectroscopy experiments that measured smooth (absence of unfolding events) force-extension profiles for both (Li et al. 2001; Watanabe et al. 2002b; Leake et al. 2006; Anderson et al. 2010). In an aqueous environment, these unstructured proteins tend to occupy a compact configuration (small radius of gyration)(Fig. 2C) because there are many more compact microstates compared to extended microstates. To increase the end-to-end length of the protein a force needs to be applied on the ends of the molecule to “bias” the extended conformations and reduce the conformational entropy (randomness) of the chain. The molecular force that resists the external force applied to extend the molecule is the entropic force generated by the random coil protein, which physically results from diffusion that tends to keep the protein compact. If the external force is removed, the protein will quickly re-assume a compact configuration because of this apparent entropic force, which, physically, is powered by diffusion. This entropic restoring force is precisely what is measured in single molecule experiments that stretch random coil proteins (although bond stretching becomes relevant at high fractional extensions) and is the basis for titin-based passive tension in the cardiac cell (Leake et al. 2006; Anderson et al. 2010).
Figure 2.
Simple models of random coil proteins. A) The freely-jointed chain is a 3D random walk of rigid monomers of length L, and B) the wormlike chain model represents a continuous, homogenous polymer. Both chains can occupy numerous conformations at low fractional extensions (end-to-end distance z ≪ molecular contour length) but only one conformation when z = Lc. C) External force is required to extend a flexible polymer. The molecular restoring force than opposes the external force is the entropic force generated by the polymer. This entropic force is powered by diffusion and is the apparent force that recoils the protein when external force is removed.
3.2 Wormlike Chain Equation
The force needed to extend a polymer (or random coil protein) is well-described by the wormlike chain (WLC) equation (Marko et al. 1995). The WLC equation is an approximate interpolation formula that describes the non-linear force needed to fractionally extend a polymer of given stiffness and length:
F is the force, kBT is thermal energy, Lp is the persistence length of the molecule, Lc is the contour length of the molecule (the distanced traced along the molecular backbone), and z is the end-to-end length of the molecule (the Euclidean distance between the molecule termini). The persistence length quantifies the “stiffness” of the molecule, or how easily the molecule bends; it defines the distance along the backbone of the molecule that vectors tangent to the backbone are correlated (Fig. 2B), and it is routinely used to quantify an unstructured protein's resistance to stretch. Qualitatively, a lower Lp describes a more flexible molecule. Note, however, that according to the WLC equation a low Lp is associated with a high force needed to stretch the polymer, which is counterintuitive considering that stiffness and force are proportional (|F|=kx) in mechanical springs. This microscopic phenomenon is readily understood when considering entropy. A more flexible molecule can occupy a compact structure in more ways (more microstates) than a more rigid molecule; therefore, there is a stronger entropic force that biases the molecule to exist in a compact state compared to an extended state, and hence a larger external force is needed to overcome this entropic force to increase the end-to-end length (stretch) of the molecule. Persistence length is defined for an ideal, homogenous polymer and therefore is only an approximation of the behavior of a random coil protein. Regardless, the Lp of a molecule is often determined by fitting experimental force-extension data to the WLC equation (Fig. 4C) and is very useful for summarizing the force-extension behavior of extensible proteins. The WLC equation also shows why N2B titin is the stiffest isoform: at a given sarcomere length the I-band regions of all titin isoforms have the same end-to-end length (z), but because the I-band region of the N2B isoform has the shortest contour length its fractional extension (z/Lc) is greater than the N2BA and FCT isoforms and, according to WLC equation, requires more force to stretch.
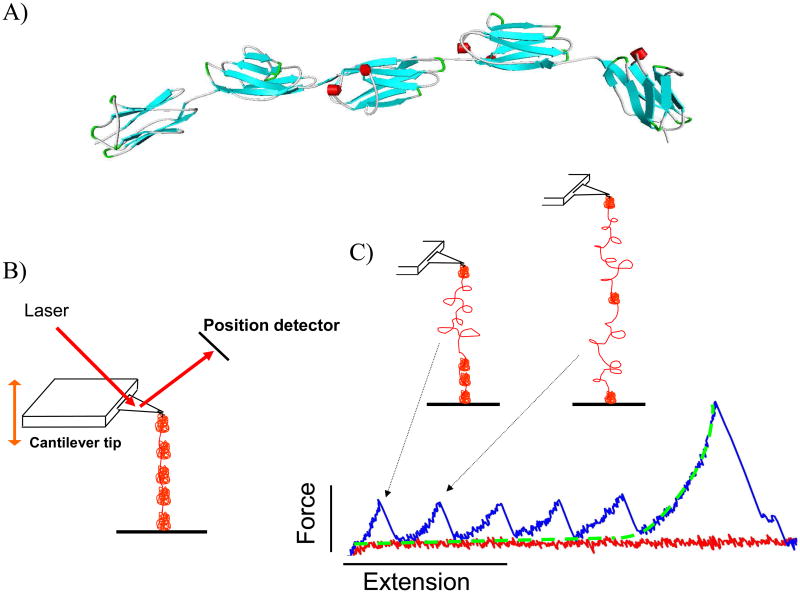
Figure 4.
A) A ribbon schematic of five serially-linked Ig domains. β-strands are colored blue and alpha helices are red. The atomic coordinates were downloaded from the Worldwide Protein Data Bank, PDB ID: 3B43 (Von Castelmur et al. 2008). Each folded Ig domain is 4-5 nm in diameter and connected by short linker sequences. B) Simplified AFM schematic. The cantilever tip probes a protein-coated surface until a molecule is tethered. The molecule is then stretched and the force that develops is determined by measuring the deflection of the laser off the cantilever. C) A tethered polyprotein is stretched to induce domain unfolding. Large structural transitions, such as complete unfolding of an Ig domain, result in a large contour length increase, a release of tension in the molecule, and a sharp unfolding force peak. In the example shown, the molecule is still attached after all five domains unfold; the last force peak is due to stretching the completely unfolded peptide that behaves as an entropic spring and is well-described by the WLC equation (fitted dashed line).
4. Modulating Titin Stiffness
The two most recognized mechanisms for tuning titin-based passive tension are changes in isoform expression and post-translational modifications of titin. Isoform changes affect titin stiffness because the cardiac titin isoforms (N2B, N2BA, FCT) contain I-band regions of different length that are generated by titin splicing pathways.
4.1 Isoform Splicing
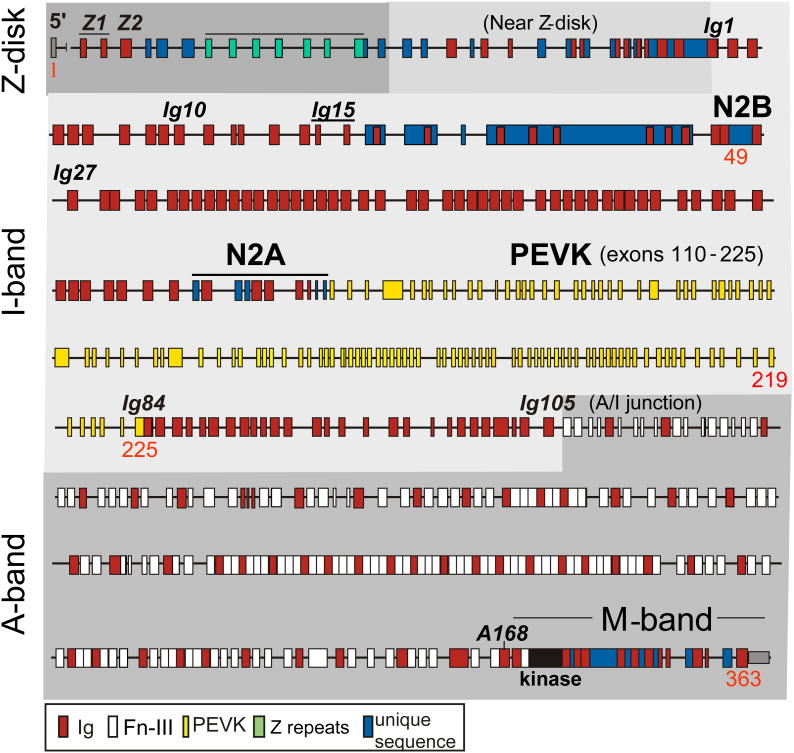
The specific I-band compositions of titin isoforms is processed by complex and poorly-understood pre-mRNA splice pathways that result in varying degrees of titin stiffness. In humans, the titin gene is located on chromosome 2, contains 363 exons, and has a maximal coding capacity of 38,138 residues (4.2 MDa) (Bang et al. 2001). Splicing events result in titin isoform expression being muscle specific (Wang et al. 1991; Labeit et al. 1995), and three classes of isoforms are found in the heart: adult N2B and N2BA isoforms and fetal cardiac titin (Lahmers et al. 2004; Greaser et al. 2005). The I-band region of the titin gene comprises ∼220 exons (Bang et al. 2001), with a subset of these exon gene products constitutively expressed in all cardiac isoforms (Fig. 3): the proximal tandem Ig segment (Ig1-15; some Ig domains coded by two exons), the N2B element (containing Ig24-26 and a 572 residue unique sequence), Ig27, the PEVK sequence encoded by exons 219-225, and the distal tandem Ig segment (Ig84-105). These domains are found in the I-band regions of all cardiac isoforms and are the entire I-band of the 2970 kDa N2B isoform (Fig. 1B). The N2BA isoform also contains a variable number of Ig domains in the so-called middle tandem Ig segment (Freiburg et al. 2000), the N2A element (which contains unique sequences and Ig80-83), and additional PEVK sequence (Freiburg et al. 2000). The additional I-band elements in N2BA titin make it larger (∼3.3 MDa) and more compliant than the N2B titin isoform. In adult left ventricle (LV), N2B titin is more abundant than N2BA (∼80% of mouse titin and ∼60% of human titin (Cazorla et al. 2000)), although in certain disease states these ratios are different. The third class of cardiac titin isoforms is the fetal cardiac titins (FCT; 3.5-3.6 MDa (Lahmers et al. 2004)) that express additional Ig domains in the middle tandem Ig segment and the longest PEVK sequence (Opitz et al. 2004; Greaser et al. 2005). FCT isoforms dominate at birth but are replaced by the shorter, stiffer adult isoforms during neonatal development (Lahmers et al. 2004). Titin isoform expression occurs at the level of the half-sarcomere (Trombitas et al. 2001), and the mechanics of each isoform depend on the size of its I-band region, with shorter I-band regions corresponding to higher titin-based passive tension. Titin isoform expression differences are responsible for myocardial passive tension variability across species, between muscle types, during development, and throughout disease phenotypes. For example: smaller mammals typically have faster heart rates, which correlates with stiffer myocardium and a lower N2BA:N2B expression ratio (Cazorla et al. 2000), skeletal muscle titin isoforms vary widely between muscle types and are much longer than cardiac isoforms (Neagoe et al. 2003), the large FCT cardiac isoforms are present in neonates but are replaced by N2B and N2BA during development (Lahmers et al. 2004), and cardiac titin isoform expressions are different in certain disease states of dog, rat, and humans (Neagoe et al. 2002; Wu et al. 2002; Warren et al. 2003; van Heerebeek et al. 2006). One hypothesis regarding the functional significance of changes in titin isoform expression is that the ratio of N2BA:N2B titin helps set an animal's optimal heart rate. For example, high levels of N2B, which results in stiffer myocardium, allows for rapid early diastolic filling and rapid setting of the end diastolic volume, both of which are necessary for the short diastolic filling times associated with high heart rates. This suggests that cardiac titin expression may help define the optimal heart beat frequency and that post-translational modifications, which can rapidly alter titin stiffness, may couple myocardial stiffness with acute cardiac demand.
Figure 3.
The exon structure of the human titin gene (based on (Bang et al. 2001)). Titin contains 363 exons, ∼220 of which are found in the I-band region. Titin's I-band region comprises immunoglobulin(Ig)-like domains, the PEVK element, and unique sequences. The thick filament bound A-band region of titin contains Ig and fibronectin-III (Fn-III) domains exclusively. Domains and exons alluded to in the text are labeled.
Of particular interest to researchers is how titin splicing is altered by hemodynamic demands on the heart. For example, a dilated cardiomyopathy (DCM) model was induced in dogs by imposing pacing tachycardia for two weeks that resulted in chamber dilation and increased chamber stiffness (Bell et al. 2000), and a study in which pacing was administered for four weeks showed down-regulation of compliant N2BA titin and up-regulation of stiff N2B titin in addition to, as expected, increased titin-based passive tension (Wu et al. 2002). A hypertensive rat model subject to pressure overload also showed reduced N2BA isoform expression (Warren et al. 2003), which is consistent with the elevated passive tension found in hypertensive rat cardiomyocytes ((Conrad et al. 1995; Cicogna et al. 1999).
In humans, patients with coronary artery disease (CAD) exhibited increased N2BA expression that accompanied decreased myofibrillar stiffness, although LV wall stiffness was higher in CAD patients and explained by increased collagen and desmin expression levels (Neagoe et al. 2002). Unlike the animal models mentioned above in which increases in titin-based stiffness contributed to the disease pathology, it seems that decreased titin stiffness in human CAD patients acts to compensate for the increased ECM-based stiffness that accompanies the disease (Porter et al. 2009). In patients with end-stage heart failure due to DCM, N2BA:N2B ratio was increased and corresponded with lowered passive stiffness (Nagueh et al. 2004). In addition, the physiological relevance of changes in titin isoform expression was shown by positive correlations between N2BA:N2B ratio and chamber compliance parameters, which suggests that DCM affects diastolic filling by lowering titin-based stiffness (Nagueh et al. 2004). Interestingly, the same study showed an increased N2BA:N2B ratio also correlated with improved exercise tolerance, which suggests that the titin isoform shift may be a beneficial adaptation in response to heart dilation. The work on animal models subject to short-term cardiac stress showed that changes in titin isoform expression contributed to the pathology, while chronic cardiac stress in human patients showed adaptation in titin expression to try to compensate for deleterious changes in the other determinant of myocardial stiffness (ECM). The relationship between cardiac insult and changes in titin is clearly complex, and a better understanding of titin is needed to more effectively confront cardiac disease.
4.2 Post-translational Modifications
Changes in titin isoform expression occur on the time scale of days to weeks depending on the stage of development and species (Wu et al. 2002; Lahmers et al. 2004), but adjustment of titin's contribution to passive tension can also occur rapidly via post-translational modifications (PTMs) of titin's I-band region. Cyclic AMP (cAMP)-dependent protein kinase A (PKA) is activated by β-adrenergic stimulation and has been shown to phosphorylate the unique sequence of the cardiac specific N2B element (N2B-Us). Phosphorylation by PKA reduced passive tension in skinned rat cardiomyocytes (Yamasaki et al. 2002) and human cardiac fibers (Kruger et al. 2006). The effect in human fibers was more pronounced when de-phosphorylation by protein phosphatase-1 (PP1) was performed before PKA treatment, which suggests that basal levels of phosphorylation play an important role in determining titin-based passive tension.
Protein kinase G (PKG), which is cGMP-dependent, also phosphorylates the N2B-Us and reduces passive tension, and the site of phosphorylation is the same residue that PKA targets (Kruger et al. 2009). Passive tension of skinned fiber bundles from human LV was significantly reduced only after PP1 treatment removed basal phosphorylation levels, which, similar to the PKA results, suggests that basal levels of titin PTMs play important roles in vivo. A single molecule study on N2B-Us treated with PKG suggested that phosphorylation increases the Lp of the N2B-Us (Kruger et al. 2009), which corresponds to a reduction of passive tension at the tissue level. It is worth noting that the decrease in passive tension seen at the tissue level is similar to the change in tension that would result from an increased N2B-Us Lp following PKG treatment. Because single molecule experiments are able to quantify the effect of a treatment on the mechanical properties of I-band elements, it can be surmised if the effect at the tissue level is fully explained by changes at the molecular level or if a more dynamic hypothesis is needed to explain how the treatment variable affects changes in cardiac stiffness. However, more single molecule measurements are needed to strengthen this result because of the limited data gathered in the initial study.
Most recently, it has been shown that protein kinase C (PKC) specifically phosphorylates titin at two highly-conserved serine residues in the constitutive PEVK element found in all cardiac titin isoforms (Hidalgo et al. 2009). PEVK phosphorylation by PKCα (the predominant isozyme in the heart and an important player in contractile dysfunction and heart failure (Molkentin et al. 2001; Belin et al. 2007)) increases titin-based passive tension, an effect that is also exacerbated by PP1 pre-treatment. Following PKCα phosphorylation, PP1 treatment reversed the passive tension increase (Hidalgo et al. 2009), which suggests that PKCα phosphorylation directly influences titin stiffness. In addition, single molecule experiments have shown that phosphorylation of PEVK lowers the Lp of PEVK in a site-specific manner (Anderson et al. 2010). This increased flexibility results in an entropically stiffer molecule that is predicted to increase titin-based passive tension to a level similar to what was found in skinned myocardium (Hidalgo et al. 2009). Further solidifying the link between PKCα phosphorylation and PEVK, it was shown that PKCα had no effect on the passive stiffness of mice missing the constitutive PEVK element (PEVK KO) (Hudson et al. 2010). These results, which span from the single molecule level to the tissue level, show that PKCα phosphorylation directly influences titin-based passive tension. This opens the possibility of using α1-adrenergic antagonists, which would minimize PKCα activity, as a potential therapy for cardiomyopathies characterized by increased myocardial stiffness.
Because titin can be phosphorylated by various protein kinases resulting in disparate effects, elucidating the phosphorylation status of titin is important when a comprehensive understanding of titin-based stiffness is desired. For example, in rodents PKA reduces passive tension by approximately 30% (depending on sarcomere length (Yamasaki et al. 2002)) while PKC increases passive tension by 30% (Hidalgo et al. 2009). These kinase effects can also be titin isoform dependent (Fukuda et al. 2005). A thorough analysis of titin isoform expression and PTMs must be performed when deconstructing passive tension determinants in healthy and diseased myocardium.
5. Studying Physical Properties of Titin I-band Elements
In order to gain a better fundamental understanding of how the spring-like elements of titin's I-band region contribute to titin-based passive tension and how phosphorylation of the spring-like elements leads to changes in titin stiffness, the physical properties of the PEVK, N2B, and tandem Ig domains have been studied in isolation using single molecule techniques. Although laser tweezers are an excellent tool for probing the biomechanics of single molecules, most single molecule titin studies have utilized the atomic force microscope because it can easily access high force regimes (to elicit Ig unfolding), stretch short molecular fragments without the need for long linkers (as required for laser tweezers), and generate multiple molecular force-extension traces quickly.
5.1 Atomic Force Microscopy
Direct measurement of the physical properties of single molecules requires force and length resolution on the order of piconewtons and nanometers, respectively. Because it is versatile and relatively simple to use, the atomic force microscope (AFM) (Binnig et al. 1986) has been the single molecule force spectroscopy choice for many. Although AFMs are also widely-used for surface imaging (xy plane scanning), its ability to stretch proteins in the z-direction (normal to a flat surface) is ideally-suited for high throughput generation of molecular force-extension curves (for a recent review on AFM and titin see (Linke et al. 2008)). The basic AFM protocol for stretching protein fragments is as follows (Fig. 4). Purified recombinant protein is spotted onto a microscope slide or coverslip and allowed to adsorb onto the surface. The AFM cantilever tip then probes the protein-coated surface vertically until an adsorbed molecule attaches to the tip (a non-specific interaction unless the cantilever has been functionally modified). This tethered protein is then stretched as the tip retracts from the surface. The force that develops in the molecule is measured by the deflection of a focused light source that reflects off the back of a cantilever tip of known stiffness. The force needed to stretch the molecule is equivalent to the force needed to bend the cantilever tip. Consequently, the force needed to stretch the molecule as a function of cantilever position (which can be converted to molecular end-to-end length, z) is measured to generate a force-extension trace. The force that acts on the molecule increases as the extension increases, and this process continues until a force-induced structural transition is realized. When stretching serially-linked Ig domains, for instance, this structural transition is the complete unfolding of a single Ig domain from its native, globular structure to an unfolded, random coil structure. This sudden increase in contour length removes the tension in the tethered molecule and is recorded as a clearly-identifiable unfolding force peak. Unstructured proteins, on the other hand, are mechanically unstable and do not exhibit large unfolding force peaks when stretched. In this case, AFM traces of flexible proteins are fit with the WLC equation to estimate an effective persistence length of the protein; this is a convenient way of quantifying the force needed to extend a flexible protein.
5.2 PEVK
Appropriately named due to its high density of proline (P), glutamate (E), valine (V), and lysine (K) residues, the PEVK element of titin's I-band region behaves as an entropic spring. Although some local secondary structure in the form of polyproline II helices has been suggested (Ma et al. 2001), long range cooperativity between the helices is apparently absent (Gutierrez-Cruz et al. 2001), and polyproline helix structure is uncommon in the PEVK element of the cardiac N2B isoform (Watanabe et al. 2002b). It is worth noting that the N2BA titin isoform contains two polyE (negatively-charged glutamate-dense regions) and 20 PEVK repeat motifs (26-28 residues), compared to zero and five, respectively, found in the N2B isoform (Greaser 2001). Therefore, it cannot be assumed that the properties of PEVK are identical in N2B and N2BA titin, although most research (such as that cited in this review) has studied the constitutive PEVK sequence that is the entire PEVK element of the N2B titin isoform because all mammalian titin isoforms contain these PEVK exons. The PEVK element found in the N2B isoform is 188 residues in length (assuming a random coil and maximal residue spacing of 0.38 nm/aa gives a Lc of 71 nm) while the N2BA isoform has a ∼800 residue PEVK element (∼300 nm). The absence of large stable structures in PEVK is suggested by the high concentration of proline residues and charge clusters (Labeit et al. 1995) and is evidenced by smooth force-extension traces that also lack hysteresis (Li et al. 2001; Watanabe et al. 2002b). The persistence length of PEVK is ∼1 nm (Li et al. 2001; Linke et al. 2002;; Watanabe et al. 2002b;; Anderson et al. 2010), but evidence suggests that it is not purely a random coil. Phosphorylation of PEVK by protein kinase C and mutation of the phosphorylated residues (S26 and S170) both reduce the apparent Lp of PEVK, which suggests that these residues are involved in a structural network with other amino acids (Anderson et al. 2010) because a simple mutation would not significantly change the Lp of a pure random coil. However, primary sequence does have some effect on elasticity (Cheng et al. 2010). In addition, the PEVK sequence around S26 and S170 is highly conserved, which also suggests some PEVK structure (Hidalgo et al. 2009), although as mentioned previously this structure must not be mechanically stable since AFM force-extension traces do not show unfolding force peaks. Although the PEVK exhibits a smooth force-extension trace that can be fit relatively well with the WLC equation to determine an “effective” Lp of the molecule, the PEVK element (like all biomolecules) is not homogenous and parameters used to describe the entire protein are approximations.
5.3 N2B-Us
The cardiac-specific N2B element is found in both the N2B and N2BA titin isoforms of the heart. It contains three Ig domains (Ig 24-26) and a 572 residue unique sequence (N2B-Us) that, like the PEVK, behaves as an entropic spring. If we assume a pure random coil, the Lc of the N2B-Us is 217 nm (572 aa × 0.38 nm/aa). AFM experiments have found the Lp of the N2B-Us to be ∼0.65 nm (Li et al. 2002; Watanabe et al. 2002b;; Zhu et al. 2009), which makes it more flexible (and harder to stretch) than the PEVK element. Also similar to the PEVK, the force-extension profile of the N2B-Us does not contain discernable force peaks or hysteresis (Watanabe et al. 2002b). Since the force resolution of most AFMs is 5-10 pN, low-force structural transitions cannot be excluded. Due to its presumably random coil conformation, the N2B-Us structure has not been solved, which makes structural transitions due to ligand binding (Zhu et al. 2009) and phosphorylation (Leake et al. 2006; Kruger et al. 2009) impossible to predict. This same difficulty is present when trying to predict the physical mechanism that results in changes in PEVK properties following phosphorylation (Anderson et al. 2010).
5.4 Tandem Ig segments
Titin's I-band region contains serially-linked immunoglobulin(Ig)-like domains that have a β-barrel fold characteristic of the intermediate I-set of the immunoglobulin superfamily (Pfuhl et al. 1995)(Fig. 4A). The proximal tandem Ig segment near the Z-disk contains 15 Ig domains and the distal tandem Ig segment near the A-band contains 22 domains. Ig domains also flank the N2B-Us and PEVK element (Fig. 1B). Unlike the N2B-Us and PEVK, Ig domains have well-defined structures that have been solved using NMR, x-ray crystallography, and small angle x-ray scattering techniques (Improta et al. 1996; Mayans et al. 2001; Von Castelmur et al. 2008; Stacklies et al. 2009). This has allowed for molecular dynamics experiments to study Ig domain dynamics at the atomic level (Lu et al. 1998; Lee et al. 2010), and, consequently, much more is known about the Ig domains than the PEVK and N2B-Us. AFM work on titin Ig domains is extensive and has shown that I-band Ig domains are mechanically very similar, although Ig unfolding force is domain-dependent (Marszalek et al. 1999; Li et al. 2000; Fowler et al. 2002; Watanabe et al. 2002a). Connected in series like beads on a string, globular Ig domains are 4-5 nm in diameter and are separated by short (2-4 residues) amino acid linkers (Marino et al. 2005). When stretched with an AFM, the domains first become taut while remaining in their folded state. When enough tension develops in the tandem protein, one of the Ig domains pops open, rapidly transitioning from its β-barrel structure to a fully unfolded (random coil) peptide chain. Ig domains comprise ∼80-90 amino acids (on average) which results in a ∼30 nm contour length increase following total unfolding. This sudden increase in contour length removes the tension in the polyprotein and results in a large force peak in the force-extension profile (Fig. 4C). Of particular interest to researchers is the force needed to unfold Ig domains and their kinetic rates of unfolding and refolding; knowing these parameters allows for prediction of how frequently Ig domains unfold, which is important for determining titin-based passive tension. Tandem Ig domains are physically not well described by the WLC equation because they are not random coils in their native state and they require much less force to extend when in their folded structure (for example, the effective Lp of folded Ig domains has been estimated at 10 nm (Granzier et al. 2005), which is reasonable considering that adjacent Ig domains must be correlated in orientation due to their short linker sequences). However, unfolded Ig domains are random coils that behave as entropic springs and would more significantly contribute to passive tension provided that they unfold under physiological forces. In AFM experiments, Ig domains typically unfold at forces between 150-250 pN (Li et al. 2000; Watanabe et al. 2002a; Zhu et al. 2009) (unfolding force is velocity dependent, see below), but in the sarcomere the force felt by each titin I-band at end diastole is estimated to be <5 pN when considering muscle mechanics (Nedrud et al. 2011), thick filament density (Irving et al. 2011), and working sarcomere length (Chung et al. 2011a) data.
Single molecule studies on Ig dynamics have also been used to elucidate the link between titin and cardiac disease. Recently, a mutation in the 10th Ig domain in the proximal tandem Ig segment has been linked with arrhythmogenic right ventricular cardiomyopathy (ARVC) (Taylor et al. 2011), which is characterized by fibrofatty replacement of the myocardium and an increased risk of cardiac arrhythmias and sudden death. It is surprising that a single point mutation in an Ig domain seemingly leads to cardiomyopathy, but the combination of a variety of experimental techniques has suggested a hypothesis that links altered Ig10 dynamics with degradation of healthy myocardium. Nuclear magnetic resonance (NMR) data and proteolysis assays have shown that Ig10 domains harboring the disease-linked mutation are structurally compromised and more prone to degradation (Taylor et al. 2011); AFM data also shows that mutant Ig10 unfolds at a lower force compared to native Ig10 (Anderson et al.), which is consistent with the idea that the mutation weakens the domain's β-barrel structure and results in a higher percentage of unfolded mutant Ig10 compared to native Ig10. This propensity to exist in an unfolded structure combined with the increased rate of degradation suggests that the mutation leads to cleavage in titin's I-band region, which would abolish titin's essential force-generating mechanism and likely lead to accelerated titin turnover and possibly even apoptosis. Clearly more work is needed to develop this hypothesis, but these interesting ideas are made possible because of the ability to study individual titin components on the single molecule level.
5.4.1 Analysis of Ig Unfolding
Substantial AFM work has investigated the properties of titin's I-band region, in particular the forced unfolding of Ig domains. Two phenomena related to Ig unfolding have been identified: the dependence of unfolding force on unfolding peak number and the dependence of unfolding force on pulling velocity. The conceptual ideas that explain these phenomena are worth discussing because they offer valuable insights regarding the dynamics that govern titin's I-band behavior. Furthermore, proper analysis of the dependence of unfolding force on peak number is needed to obtain accurate data, although only recently has theory been applied to accomplish this task.
In order to determine the force needed to unfold a globular domain, it is best to stretch a homopolyprotein (N identical domains connected in series). If a heteropolyprotein is stretched, it is impossible to assign unfolding force peaks to particular domains in the series because each domain unfolds stochastically and independently. However, if every domain in the series is the same, each unfolding force peak represents unfolding of the domain of interest. Previous studies cleverly used homopolyproteins to determine the properties and kinetic rates of Ig domains but usually combined all unfolding force data during analysis (i.e. the first unfolding force peak was not distinguished from the last unfolding force peak). This procedure assumes that the unfolding forces obtained from unfolding a tandem protein and a single protein are sampled from the same distribution, but this has been shown to be false (Zhmurov et al. 2010). Instead, the unfolding force depends on the number of domains being stretched, which leads to the first unfolding force of a force-extension trace being lower than the second peak, the second being lower than the third, and so on. This peak dependence of unfolding forces is a combinatorial phenomenon, and a method based on order statistics theory has been developed to remove this peak dependence and allow for unfolding force measurements to be combined when analyzing data (Zhmurov et al. 2010), which allows for more robust fitting procedures. This theory was published only recently, which explains why most unfolding force data of homopolymeric proteins in the literature is analyzed with all unfolding force peaks combined (Li et al. 2000; Watanabe et al. 2002a). Conceptually, it is simple to understand why unfolding force increases with peak number. Consider a string of five identical Ig domains held taut in solution. Due to thermal energy one of the domains will unfold first, say at time t1, and let us assume no refolding. A short while later another domain unfolds, say at t2, and this process continues for t3, t4, and t5. Now consider a string of 100 identical Ig domains and the subsequent t1′ through t100′. Because there are more Ig domains in this long string, the first Ig domain that stochastically unfolds will unfold, on average, quicker than the first domain that unfolded in the five Ig chain. In other words, t1′ < t1 because more domains had the potential to unfold in the longer Ig chain. Now consider an AFM pulling experiment with, for example, ten identical domains linked in series. All domains are folded prior to stretching and force develops in the molecule as the cantilever tip pulls away from the slide surface, with a force peak generated each time a domain unfolds. Note, however, that before the first force peak was generated ten domains had the potential to unfold, but before the final (10th) force peak only one domain had the potential to unfold. As mentioned above, the time that elapses before the first unfolding event takes place (t1) is shorter when ten domains are folded than when only one domain is folded. Therefore, the unfolding force of peak one is less than peak ten. From this same logic, it follows that F1<F2<…<FN on average for a serially-linked homopolymer of length N, which is what has been shown theoretically (Zhmurov et al. 2010) and experimentally (Anderson et al.).
Unfolding force also depends on the pulling velocity of the AFM tip (Li et al. 2000; Watanabe et al. 2002a), with higher pulling velocities resulting in larger unfolding forces, and intricate theories have been developed to extract kinetic information from data gathered at wide-ranging pulling speeds (Hummer et al. 2003; Dudko et al. 2008). Consider a serially-linked Ig homopolyprotein. Although the β-barrel structure imparts high stability to Ig domains, these domains still unfold under no external force with some finite probability (typically ∼10−6-10−4 s−1 (Li et al. 2000; Watanabe et al. 2002a)) due to thermal fluctuations. In an AFM pulling experiment, the domains are mechanically stretched which strains bond networks (hydrogen, disulfide, etc.) until the protein eventually denatures. For Ig domains, it has been shown that after the main rupture (force peak in AFM traces) the rest of the molecule unravels at a much lower force (Lu et al. 1998), which results in a single unfolding force peak being measured, although unfolding intermediates have also been seen (Marszalek et al. 1999). To understand why higher pulling speeds result in higher unfolding forces, consider the mechanical and thermal energy contributions. Both of these forms of energy can unfold the protein when the molecule is being stretched. When the stretch speed is fast, the force loading rate (N/s) acting on the molecule is increased relative to a slower stretch speed while the thermal energy bombarding the molecule is unchanged. This means that the energy that unfolds the molecule has a larger mechanical contribution than when the stretch speed is slow (i.e. when there is more time for diffusive thermal energy to unfold the protein). Of course, the mechanical force acting on the protein when a domain unfolds is measured by the bending of the AFM cantilever, which is output as the deflection signal (unfolding force peak). In summary, at faster pulling speeds there is less time for thermal energy to contribute towards unfolding the protein and more of a burden on mechanical energy.
5.4.2 Ig Kinetics
Although it is simple to determine the force needed to unfold titin Ig domains, this information is not particularly useful when determining how Ig domains contribute to titin-based passive tension because the forces experienced by the I-band are <5 pN while Ig unfolding force at physiological stretch speeds is much higher (150-250 pN depending on specific Ig domain (Watanabe et al. 2002a; Zhu et al. 2009)). More useful parameters for determining the fraction of Ig domains that are unfolded (behaving as force-generating entropic springs) are the kinetic rates of unfolding and refolding. The unfolding rate at zero force (α0) is the rate at which a protein transitions from a folded to an unfolded state under no external load, and the refolding rate (β0) is the opposite. Unfolding rate is often estimated by comparing the unfolding force distribution from AFM data with Monte Carlo simulations (Kellermayer et al. 1997; Rief et al. 1997; Li etal. 2000) where force-dependent unfolding is assumed to follow Bell's formula (Bell 1978) α(F) = α0eF⋅Δx/kBT and Δx is the distance along the reaction coordinate (i.e. molecular end-to-end length) from the free-energy minimum of the folded state to the transition barrier. For estimating unfolding rate at zero force (α0) Monte Carlo methods are used to simulate AFM pulling experiments and generate unfolding force distributions for various (α0, Δx) parameter pairs. The unfolding rate of the protein is then estimated to be the rate used in the simulation that best recreated the experimental data. The unfolding rate at zero force is difficult to measure directly using single molecule methods because of experimental limitations. For example, Ig unfolding is rare under no external force (∼10−5 s−1) and it is unlikely that a measurement of an unfolding event would occur prior to the molecule being stochastically displaced from the cantilever tip. Force-clamp techniques, in which a tethered molecule is held taut at a constant force (Oberhauser et al. 2001), have been used to directly measure force-dependent unfolding rates, although typically the hold forces are much higher than physiological force values (Garcia-Manyes et al. 2007). In this case, the unfolding rate at zero force is then extrapolated from the force-dependent rates by plotting ln α(F) vs. hold force. Determining α0 with more confidence would require α (F) values at forces closer to 0, although experimental force noise (typically ∼10 pN), thermal drift, and displacement of the molecule from the AFM tip prohibit direct measurement of unfolding rates at physiological force levels. Estimating low force unfolding rates can also be accomplished by assuming α(F) = α0eF⋅Δx/kBT with α0 and Δx determined from Monte Carlo simulations.
Low force refolding rate, however, can be directly measured with AFM refolding protocols. First, a tethered multidomain protein is stretched and unfolded although the protein is not detached from the slide surface or cantilever tip. Then the molecule is relaxed as the tip hovers above the slide surface. Because the unfolded domains are essentially random coils held at very small fractional extension, there is very low force acting on the molecule at this point. The unfolded domains are allowed to refold as the low force environment is maintained for a set amount of time, and then the molecule is completely stretched and unfolded to generate a full force-extension trace. By counting the number of “overlapping” force peaks one can infer how many domains refolded during the low force hold. From this information, refolding rate is directly determined (Li et al. 2000). In addition to determining how the large tandem Ig segments contribute to titin-based passive tension, measuring the kinetic rates of unfolding and refolding can also be important for quantifying how a mutation affects the structure of Ig domains in the context of identifying disease mechanisms, as was recently demonstrated with Ig10 and ARVC (Anderson et al.; Taylor et al. 2011).
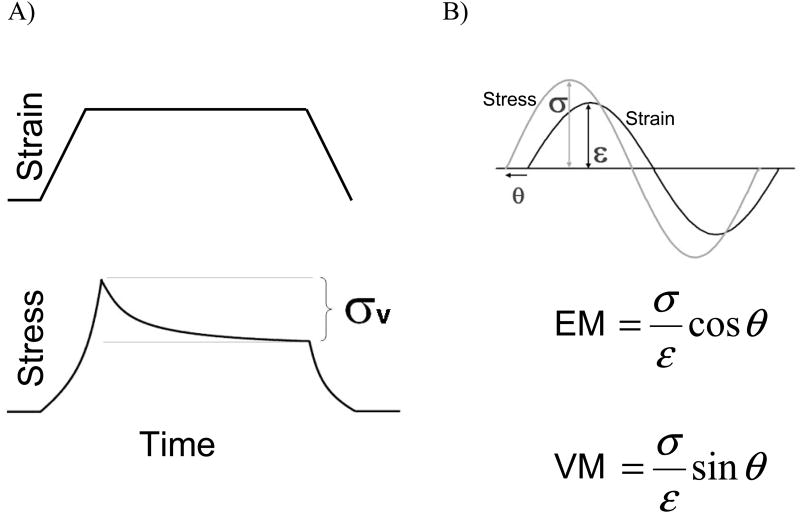
6. Viscoelasticity
Titin's I-band region has been treated as a non-linear spring so far, but sources of viscosity also exist within cardiac myocytes. The presence of viscosity creates a phase shift between stress (force) and strain (length) as the muscle is stretched and increases the force needed to stretch the myocardium because viscosity resists motion and must be overcome during sarcomere length changes (de Tombe et al. 1992). A schematic of how viscosity is measured is shown in Figure 5. The viscous modulus, which quantifies a material's resistance to stretch, is calculated as the ratio of the stress amplitude to strain amplitude times the sine of the phase angle difference between the two. The elastic modulus is calculated by replacing sin(θ) with cos(θ) in the viscous modulus equation. If the stress and strain traces superimpose there is no phase difference and the material is purely elastic. A 90° phase difference is present in purely viscous materials. In cardiac muscle preparations, the stress leads the strain as a result of viscosity. Recall Newton's equation of motion in one dimension: F = mα +γv + kx, where γ represents a viscous parameter that resists motion. When stretching muscle, this viscous resistance must be overcome, and the viscous force is largest when the rate of length change (velocity) is maximum. For sinusoidal strain, ΔL/Δt is largest when the strain switches from negative to positive values (i.e. when the sine wave that describes strain crosses the x-axis during stretch). The external force that imposes length changes on the muscle tissue must do work to oppose this viscous force. This is why a positive stress value is measured when there is no strain in the prep (Fig. 5B) and why stress leads strain in tissue and cell mechanics: the myocardium is viscoelastic.
Figure 5.
A) The relationship between stress and strain in cardiac muscle. The difference between peak stress and steady-state stress is due to viscosity. B) Stress leads strain when sinusoidal length changes are imposed on myocardial tissue and is due to the presence of viscosity. The elastic modulus (EM) and viscous modulus (VM) are determined from the amplitudes of stress and strain and the phase difference between them.
The primary source of titin-based viscosity in the cardiac myocyte comes from the interaction between the PEVK element and actin. In-vitro motility assays (Yamasaki et al. 2001; Linke et al. 2002), single molecule studies (Bianco et al. 2007), and myocyte mechanics (Yamasaki et al. 2001) have established that the interaction between PEVK and actin retards filament sliding during systole and diastole. It has been shown that the PEVK-actin interaction is present in all cardiac isoforms and is a source of hysteresis (Fukushima et al. 2010). Recently, studies using PEVK KO mice showed that the PEVK-actin interaction significantly contributes to viscosity in vivo (Chung et al. 2011b). The affinity between PEVK and actin is thought to be an electrostatic effect between negatively charged actin (Kabsch et al. 1990) and PEVK. The complete PEVK region found in N2B titin (which is also found in the N2BA isoform) comprises five basic (pI 9-10) ∼28 residue PEVK repeats (Greaser 2001). Therefore, at physiological pH cardiac PEVK has a net positive charge that may promote actin interaction. The hypothesis that PEVK-actin interaction is electrostatic is also supported by the fact that the interaction is dependent on ionic strength (Yamasaki et al. 2001; Nagy et al. 2004; Bianco et al. 2007) and lattice spacing (Chung et al. 2011b), with increased ionic strength (charge shielding) and increased lattice spacing (electrostatic force is inversely proportional to distance squared according to Coulomb's Law) reducing the binding of PEVK and actin. In addition, S100A1, a calcium-binding protein found in the heart, inhibits the interaction between PEVK and actin in a calcium-dependent manner (Yamasaki et al. 2001), which suggests that PEVK-actin forces may depend on the physiological state of the heart. PEVK-actin interaction has been well-studied for the constitutive PEVK element that is the entire PEVK sequence in the N2B isoform, but evidence for interaction between the additional N2BA isoform PEVK exons and actin is lacking. However, differential binding occurs between actin and PEVK segments of varying glutamate density (Nagy et al. 2004), which suggests that PEVK-actin interaction may differ between N2B and N2BA titin isoforms. Viscosity in the cardiac cell depends on the speed that intra-sarcomeric proteins slide past each other and is directly related to heart rate. Therefore, titin-based viscosity is predicted to be higher in small mammals (i.e. mice) with high heart rates compared to humans. However, the human heart rate can triple during exercise while rodent heart rates only slightly increase, which suggests that viscosity may be physiologically more relevant in larger mammals. It should be noted, however, that viscosity is greatest in cells and tissues following rest and that repeated stretches remove much of the viscous contribution (Helmes et al. 1999; Nedrud et al. 2011), although a significant steady-state level of viscosity remains. Repetitive stretches also reduced viscosity in isolated single titin molecules (Kellermayer et al. 2001).
7. Titin and Signaling Pathways
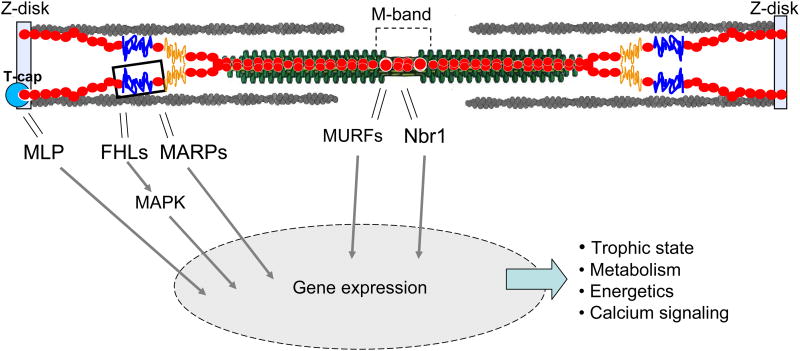
It is clear that titin isoform composition largely defines titin-based passive stiffness and that changes in hemodynamic load affect titin isoform expression, but what is less clear, and extremely interesting, is the pathways responsible for this adaptive dynamic. Significant contributions have recently been made towards a better understanding of titin's role in stretch-sensing and mechanical signaling. Although more work is needed to fully elucidate the details of titin's involvement, it has become clear that various regions along titin (Z-disk, I-band, and M-band) play a role in complex cell signaling pathways (Fig.6).
Figure 6.
Signaling hotspots in titin. At the Z-disk, the titin-binding protein T-cap interacts with muscle LIM protein (MLP), a nuclear regulator of myogenesis. In the I-band, the N2B and N2A elements interact with four-and-a-half-LIM protein (FHL) and muscle ankyrin-repeat proteins (MARPs), respectively. The N2B element and FHL are thought to form a force-dependent stretch-sensing complex that also involves components of MAPK signaling, which regulates hypertrophy. The interaction of the N2A element and MARPs is hypothesized to influence transcription as a function of mechanical strain. The M-band region of titin interacts with MURFs and Nbr1, and these interactions may play an important role in calcium signaling and hypertrophy.
7.1 Z-disk Structure and Signaling
The Z-disk is a protein-dense region in the sarcomere that is responsible for maintaining sarcomere structure and mechanically connecting adjacent half-sarcomeres. The N-terminus of titin contains two Ig domains (Z1/Z2) that embed titin in the Z-disk via strong contacts with proteins including T-cap (also called telethonin) and α-actinin, the latter which crosslinks titin and the thin filament (Ohtsuka et al. 1997; Sorimachi et al. 1997). Also, the near Z-disk portion of titin binds strongly to the thin filament and helps to separate titin's I-band region from interacting with the Z-disk (Granzier et al. 1997; Linke et al. 1997). Within the Z-disk, a single T-cap protein connects two titin molecules from the same half-sarcomere in an anti-parallel, sandwiched structure (Zou et al. 2006), which leads to strong β-strand crosslinking that likely functions to anchor titin at the Z-disk (Gregorio et al. 1998; Lee et al. 2006). A proposed mechanism for titin-based stretch sensing stems from T-cap's interaction with muscle LIM protein (MLP). MLP is an essential nuclear regulator of myogenic differentiation (Arber et al. 1994) and promotes myogenesis (Kong et al. 1997). Stretching cardiac myocytes in culture induces expression of brain natriuretic peptide (BNP) and atrial natriuretic factor (ANF), which are well-known stretch response markers, and this response was absent in MLP-null mice (Knoll et al. 2002); this suggests that MLP could be involved in stretch sensing through an interaction with T-cap and titin in the Z-disk. The coupling between sarcomere stress and Z-disk strain is shown by the fact that lattice spacing between thin filaments anchored in the Z-disk is altered by actomyosin-based active tension (Irving et al. 1998) and titin-based passive tension (Irving et al. 2011). In addition, T-cap mutations have been linked to hypertrophic cardiomyopathy (HCM) and dilated cardiomyopathy (DCM) with the disease phenotype hypothesized to arise from impaired binding between T-cap, titin, and other Z-disk proteins (Hayashi et al. 2004; Bos et al. 2006). MLP mutations that lead to mechanical stress signaling impairment have also been linked to HCM (Knoll et al. 2002; Geier et al. 2008), and MLP KO mice exhibit enlarged failing hearts (Arber et al. 1997).
Various other binding partners of titin's Z-disk region imply that titin's presence in the Z-disk is structurally very important. Obscurin, another giant sarcomeric protein, interacts with the N-terminus of titin via two Ig-like domains in obscurin (Young et al. 2001) and plays an important role in Z-disk formation (Bowman et al. 2008). Titin's Z-disk region also interacts with filamin (Labeit et al. 2006), which links titin to integrin and the focal adhesion complex (Samarel 2005; Romer et al. 2006) and suggests that titin behavior may be a stimulus that affects other cellular processes. To summarize, the protein-rich Z-disk of the sarcomere involves complex interplay between various proteins, including the N-terminus of titin. Disruption of this network is present in numerous cardiomyopathies and likely disrupts the role of titin in mechanotransduction.
7.2 I-band Signaling
The signaling hotspots within the I-band region of cardiac titin are the N2B and N2A elements. The cardiac-specific N2B element interacts with two members of the four-and-a-half-LIM domain protein family, FHL1 (Sheikh et al. 2008) and FHL2 (Lange et al. 2002), that can localize in the nucleus to act as transcription co-activators (Scholl et al. 2000). FHL1 is suggested to interact with the N2B element to form a stretch-sensing complex that is downstream of G-protein-coupled receptor (GPCR) signaling (Sheikh et al. 2008). This complex also involves Raf1, MEK2, and ERK2, which are components of the MAPK signaling pathway; the MAPK pathway is highly-involved in hypertrophy signaling (Muslin 2008). FHL1 KO mice displayed an attenuated hypertrophic response and a beneficial functional response to transverse aortic constriction (TAC)-induced pressure overload that leads to hypertrophy in control mice (Sheikh et al. 2008). This suggests that the N2B/FHL1 complex is a stretch sensor in cardiomyocytes. FHL2 has numerous binding partners that play a wide-ranging functional role in the cell (Johannessen et al. 2006) and may play a role in stretch-induced hypertrophy signaling through interaction with the N2B element (Granzier et al. 2009). In a mouse model where exon 49 was deleted (N2B KO; deletion of entire N2B element), FHL2 levels were significantly down-regulated and the hearts were atrophied (Radke et al. 2007). However, when the full PEVK element of the N2B titin isoform was removed (PEVK KO), which necessarily increases the strain placed on the N2B element, FHL2 (as well as FHL1) was up-regulated and cardiac hypertrophy was present (Granzier et al. 2009). It was hypothesized that strain on the N2B spring element induces structural changes that favor FHL binding and the assembly of a signaling complex that induces hypertrophy, and that elimination of this complex accounts for the atrophy response seen in the N2B KO model. The signaling network that connects this proposed mechanosensor to the trophic response requires further investigation.
The N2A element, which is found in the N2BA titin isoform but not the N2B isoform, is also involved in signaling pathways. The unique sequence between Ig80 and Ig81 within the N2A element binds to cardiac ankyrin-repeat protein (CARP), diabetes-related ankyrin-repeat protein (DARP), and ankyrin-repeat domain-protein-2 (Ankrd2) (Miller et al. 2003), all of which belong to the muscle ankyrin-repeat protein family (MARPs). MARPs are thought to relocate from the I-band to the nucleus following mechanical stress to regulate transcription (Kojic et al. 2004). It has been proposed that the N2A element forms a stretch sensing complex that includes MARPs, myopalladin, and the calpain protease p94. In cultured cardiomyocytes from rat, external stretch induced differential localization of CARP and DARP, including increased DARP staining at the intercalated disks (Miller et al. 2003), which are protein-dense structures that mechanically-link neighboring cells and interact with titin at terminal sarcomeres (Bennett et al. 2006).
7.3 M-band Signaling
The M-band region of titin, which binds to the thick filament near the middle of the sarcomere, is also involved in signaling pathways. Although the thick filament bears force in the sarcomere, it is only mildly compliant (Irving et al. 2011). Titin domains A168-A170 (comprised of two Ig-like domains and one fibronectin type 3 domain; Fig. 3), which are located immediately N-terminal to titin's kinase domain, bind to MURF-1 (McElhinny et al. 2002; Mrosek et al. 2007), an E3 ubiquitin ligase that targets muscle proteins for proteasomal degradation. MURF-1 also binds to glucocorticoid modulatory element binding protein-1 (GMEB-1) (McElhinny et al. 2002), a transcriptional regulator. This titin-MURF interaction is thought to regulate myofibril turnover and the trophic state of striated muscle, although ubiquitination levels of titin were unchanged in a MURF-1 KO mouse (Witt et al. 2005). However, MURF-1 and MURF-2 double KO mice developed extreme cardiac hypertrophy (Witt et al. 2008), which suggests that MURFs are important for regulating myogenesis in the heart.
The titin kinase (TK) domain is adjacent to the MURF binding site and has been thoroughly researched because, as titin's only catalytic domain, it has the potential to act as a direct biological force sensor. In-depth study has also been aroused by the determination of TK's crystal structure (Mayans et al. 1998). Although it is suggested that TK phosphorylates T-cap (Mayans et al. 1998), supporting data is lacking. This is likely due to TK's dual autoinhibitory mechanism: for the catalytic site of TK to be solvent-accessible, the C-terminal tail needs to be removed from its native state and tyrosine-170 needs to be phosphorylated (Mayans et al. 1998). AFM experiments (Puchner et al. 2008) and molecular dynamics simulations (Grater et al. 2005) have determined that external force acting on the termini of TK removes the regulatory C-terminal tail from blocking the ATP-binding site, which suggests that TK activity may be force activated; however this has not been shown directly. Two-hybrid assays and precipitation experiments with various truncated TK constructs showed that absence of the regulatory tail was necessary for interaction with Nbr1 (Lange et al. 2005), a zinc-finger protein. Such proteins have been found to act as scaffolds for signalosome assembly (Pawson et al. 1997), and it was also found that p62, another zinc-finger protein, bound to Nbr1; in vitro assays also found that Nbr1 and p62 (to a much lower level) are TK substrates (Lange et al. 2005). MURF-2, which is involved in myofibrillogenesis (Spencer et al. 2000), was also identified as a ligand of p62. It was suggested that Nbr1 acts as a scaffolding protein that assembles sarcomeric signalosomes that include p62, MURF-2, and titin kinase (Lange et al. 2005). In neonatal mice, deletion of TK results in cardiomyopathy and early death (Gotthardt et al. 2003), and a MerCreMer transgenic model, which allowed removal of TK in adult mice, exhibited an attenuated response to adrenergic stimulation and extracellular calcium, severe cardiac hypertrophy, and congestive heart failure (Peng et al. 2007). This collection of data suggests that titin kinase plays a force-dependent role in structural development and hypertrophy signaling.
8. Future Directions
Due to its immense size and multi-functional role, there is still much to be learned about titin's contribution in cardiac health and disease. The relationship between titin isoform composition and passive tension is well-understood, but the splicing pathways that respond to cardiac insult and bias expression of titin isoforms are largely unknown. Elucidation of these splice pathways will unlock the possibility of using pharmacological therapies to modulate titin isoform expression in chronic pathologies. In addition, the physics of titin-ligand interactions needs to be better understood, in particular the force-dependence of these interactions. It is likely that sarcomere stress influences titin-ligand binding differently along the titin molecule; for instance, the I-band region of titin may be more suited to sense strain than the Z-disk and M-band regions of titin because it is not strongly bound to other sarcomeric filaments, and integrative experiments will be required to determine the physical catalysts and physiological responses of titin-ligand interactions involved in signaling pathways. Although great progress has been made in the study of titin, more work is needed to shed light on titin biomechanics. Fortunately, various cutting-edge techniques, including single molecule force spectroscopy and generation of novel mouse models, are being successfully applied to titin research, and it seems to be only a matter of time before the mechanisms that link titin strain and the cellular response are critically understood.
Acknowledgments
This review was supported by NIH training grant GM084905 and an award from the American Heart Association 11PRE7370083 to B.A and by NIH HL062881 to H.G.
Footnotes
Please see also related communications in this issue by AUTHOR-1 et al. (2012) and AUTHOR-2 et al. (2012)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson B, Bogomolovas J, et al. preparation [Google Scholar]
- Anderson BR, Bogomolovas J, et al. The effects of PKCalpha phosphorylation on the extensibility of titin's PEVK element. J Struct Biol. 2010;170(2):270–277. doi: 10.1016/j.jsb.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber S, Halder G, et al. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79(2):221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- Arber S, Hunter JJ, et al. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88(3):393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- Bang ML, Centner T, et al. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89(11):1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- Belin RJ, Sumandea MP, et al. Augmented protein kinase C-alpha-induced myofilament protein phosphorylation contributes to myofilament dysfunction in experimental congestive heart failure. Circ Res. 2007;101(2):195–204. doi: 10.1161/CIRCRESAHA.107.148288. [DOI] [PubMed] [Google Scholar]
- Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200(4342):618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Bell SP, Nyland L, et al. Alterations in the determinants of diastolic suction during pacing tachycardia. Circ Res. 2000;87(3):235–240. doi: 10.1161/01.res.87.3.235. [DOI] [PubMed] [Google Scholar]
- Bennett PM, Maggs AM, et al. The transitional junction: a new functional subcellular domain at the intercalated disc. Mol Biol Cell. 2006;17(4):2091–2100. doi: 10.1091/mbc.E05-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P, Nagy A, et al. Interaction forces between F-Actin and titin PEVK domain measured with optical tweezers. Biophysical J. 2007;93(6):2102–2109. doi: 10.1529/biophysj.107.106153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnig G, Quate CF, et al. Atomic Force Microscope. Physical Review Letters. 1986;56(9):930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- Bos JM, Poley RN, et al. Genotype-phenotype relationships involving hypertrophic cardiomyopathy-associated mutations in titin, muscle LIM protein, and telethonin. Mol Genet Metab. 2006;88(1):78–85. doi: 10.1016/j.ymgme.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman AL, Catino DH, et al. The Rho-guanine nucleotide exchange factor domain of obscurin regulates assembly of titin at the Z-disk through interactions with Ran binding protein 9. Molecular Biology of the Cell. 2008;19(9):3782–3792. doi: 10.1091/mbc.E08-03-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazorla O, Freiburg A, et al. Differential expression of cardiac titin isoforms and modulation of cellular stiffness. Circ Res. 2000;86(1):59–67. doi: 10.1161/01.res.86.1.59. [DOI] [PubMed] [Google Scholar]
- Cheng SM, Cetinkaya M, et al. How Sequence Determines Elasticity of Disordered Proteins. Biophysical J. 2010;99(12):3863–3869. doi: 10.1016/j.bpj.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CS, Granzier HL. Contribution of titin and extracellular matrix to passive pressure and measurement of sarcomere length in the mouse left ventricle. J Mol Cell Cardiol. 2011a;50(4):731–739. doi: 10.1016/j.yjmcc.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CS, Methawasin M, et al. Titin based viscosity in ventricular physiology: an integrative investigation of PEVK-actin interactions. J Mol Cell Cardiol. 2011b;51(3):428–434. doi: 10.1016/j.yjmcc.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicogna AC, Robinson KG, et al. Direct effects of colchicine on myocardial function: studies in hypertrophied and failing spontaneously hypertensive rats. Hypertension. 1999;33(1):60–65. doi: 10.1161/01.hyp.33.1.60. [DOI] [PubMed] [Google Scholar]
- Conrad CH, Brooks WW, et al. Myocardial fibrosis and stiffness with hypertrophy and heart failure in the spontaneously hypertensive rat. Circulation. 1995;91(1):161–170. doi: 10.1161/01.cir.91.1.161. [DOI] [PubMed] [Google Scholar]
- de Tombe PP, ter Keurs HE. An internal viscous element limits unloaded velocity of sarcomere shortening in rat myocardium. J Physiol. 1992;454:619–642. doi: 10.1113/jphysiol.1992.sp019283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudko OK, Hummer G, et al. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc Natl Acad Sci U S A. 2008;105(41):15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler SB, Best RB, et al. Mechanical unfolding of a titin Ig domain: structure of unfolding intermediate revealed by combining AFM, molecular dynamics simulations, NMR and protein engineering. J Mol Biol. 2002;322(4):841–849. doi: 10.1016/s0022-2836(02)00805-7. [DOI] [PubMed] [Google Scholar]
- Freiburg A, Trombitas K, et al. Series of exon-skipping events in the elastic spring region of titin as the structural basis for myofibrillar elastic diversity. Circ Res. 2000;86(11):1114–1121. doi: 10.1161/01.res.86.11.1114. [DOI] [PubMed] [Google Scholar]
- Fukuda N, Wu Y, et al. Phosphorylation of titin modulates passive stiffness of cardiac muscle in a titin isoform-dependent manner. J Gen Physiol. 2005;125(3):257–271. doi: 10.1085/jgp.200409177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima H, Chung CS, et al. Titin-isoform dependence of titin-actin interaction and its regulation by S100A1/Ca2+ in skinned myocardium. J Biomed Biotechnol. 2010;2010:727239. doi: 10.1155/2010/727239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Manyes S, Brujic J, et al. Force-clamp spectroscopy of single-protein monomers reveals the individual unfolding and folding pathways of I27 and ubiquitin. Biophys J. 2007;93(7):2436–2446. doi: 10.1529/biophysj.107.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier C, Gehmlich K, et al. Beyond the sarcomere: CSRP3 mutations cause hypertrophic cardiomyopathy. Hum Mol Genet. 2008;17(18):2753–2765. doi: 10.1093/hmg/ddn160. [DOI] [PubMed] [Google Scholar]
- Gotthardt M, Hammer RE, et al. Conditional expression of mutant M-line titins results in cardiomyopathy with altered sarcomere structure. J Biol Chem. 2003;278(8):6059–6065. doi: 10.1074/jbc.M211723200. [DOI] [PubMed] [Google Scholar]
- Granzier H, Kellermayer M, et al. Titin elasticity and mechanism of passive force development in rat cardiac myocytes probed by thin-filament extraction. Biophys J. 1997;73(4):2043–2053. doi: 10.1016/S0006-3495(97)78234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzier HL, Labeit S. Titin and its associated proteins: the third myofilament system of the sarcomere. Adv Protein Chem. 2005;71:89–119. doi: 10.1016/S0065-3233(04)71003-7. [DOI] [PubMed] [Google Scholar]
- Granzier HL, Radke MH, et al. Truncation of titin's elastic PEVK region leads to cardiomyopathy with diastolic dysfunction. Circ Res. 2009;105(6):557–564. doi: 10.1161/CIRCRESAHA.109.200964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grater F, Shen J, et al. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys J. 2005;88(2):790–804. doi: 10.1529/biophysj.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaser M. Identification of new repeating motifs in titin. Proteins. 2001;43(2):145–149. doi: 10.1002/1097-0134(20010501)43:2<145::aid-prot1026>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Greaser ML, Krzesinski PR, et al. Developmental changes in rat cardiac titin/connectin: transitions in normal animals and in mutants with a delayed pattern of isoform transition. J Muscle Res Cell Motil. 2005;26(6-8):325–332. doi: 10.1007/s10974-005-9039-0. [DOI] [PubMed] [Google Scholar]
- Gregorio CC, Trombitas K, et al. The NH2 terminus of titin spans the Z-disc: its interaction with a novel 19-kD ligand (T-cap) is required for sarcomeric integrity. J Cell Biol. 1998;143(4):1013–1027. doi: 10.1083/jcb.143.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Cruz G, Van Heerden AH, et al. Modular motif, structural folds and affinity profiles of the PEVK segment of human fetal skeletal muscle titin. J Biol Chem. 2001;276(10):7442–7449. doi: 10.1074/jbc.M008851200. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Arimura T, et al. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J Am Coll Cardiol. 2004;44(11):2192–2201. doi: 10.1016/j.jacc.2004.08.058. [DOI] [PubMed] [Google Scholar]
- Helmes M, Trombitas K, et al. Mechanically driven contour-length adjustment in rat cardiac titin's unique N2B sequence: titin is an adjustable spring. Circ Res. 1999;84(11):1339–1352. doi: 10.1161/01.res.84.11.1339. [DOI] [PubMed] [Google Scholar]
- Hidalgo C, Hudson B, et al. PKC phosphorylation of titin's PEVK element: a novel and conserved pathway for modulating myocardial stiffness. Circ Res. 2009;105(7):631–638. doi: 10.1161/CIRCRESAHA.109.198465. 617 p following 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BD, Hidalgo CG, et al. Excision of titin's cardiac PEVK spring element abolishes PKCalpha-induced increases in myocardial stiffness. J Mol Cell Cardiol. 2010;48(5):972–978. doi: 10.1016/j.yjmcc.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummer G, Szabo A. Kinetics from nonequilibrium single-molecule pulling experiments. Biophysical J. 2003;85(1):5–15. doi: 10.1016/S0006-3495(03)74449-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Improta S, Politou AS, et al. Immunoglobulin-like modules from titin I-band: extensible components of muscle elasticity. Structure. 1996;4(3):323–337. doi: 10.1016/s0969-2126(96)00036-6. [DOI] [PubMed] [Google Scholar]
- Irving T, Li Q, et al. Z/I and A-band lattice spacings in frog skeletal muscle: effects of contraction and osmolarity. J Muscle Res Cell Motil. 1998;19(7):811–823. doi: 10.1023/a:1005459605964. [DOI] [PubMed] [Google Scholar]
- Irving T, Wu Y. Thick-filament strain and interfilament spacing in passive muscle: effect of titin-based passive tension. Biophys J. 2011;100(6):1499–1508. doi: 10.1016/j.bpj.2011.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen M, Moller S, et al. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cellular and Molecular Life Sciences. 2006;63(3):268–284. doi: 10.1007/s00018-005-5438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Mannherz HG, et al. Atomic structure of the actin:DNase I complex. Nature. 1990;347(6288):37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Kellermayer MS, Smith SB, et al. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276(5315):1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- Kellermayer MS, Smith SB, et al. Mechanical fatigue in repetitively stretched single molecules of titin. Biophys J. 2001;80(2):852–863. doi: 10.1016/S0006-3495(01)76064-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll R, Hoshijima M, et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111(7):943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- Kojic S, Medeot E, et al. The Ankrd2 protein, a link between the sarcomere and the nucleus in skeletal muscle. Journal of Molecular Biology. 2004;339(2):313–325. doi: 10.1016/j.jmb.2004.03.071. [DOI] [PubMed] [Google Scholar]
- Kong Y, Flick MJ, et al. Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD. Mol Cell Biol. 1997;17(8):4750–4760. doi: 10.1128/mcb.17.8.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger M, Linke WA. Protein kinase-A phosphorylates titin in human heart muscle and reduces myofibrillar passive tension. J Muscle Res Cell Motil. 2006;27(5-7):435–444. doi: 10.1007/s10974-006-9090-5. [DOI] [PubMed] [Google Scholar]
- Kruger M, Kotter S, et al. Protein kinase G modulates human myocardial passive stiffness by phosphorylation of the titin springs. Circ Res. 2009;104(1):87–94. doi: 10.1161/CIRCRESAHA.108.184408. [DOI] [PubMed] [Google Scholar]
- Labeit S, Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270(5234):293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- Labeit S, Lahmers S, et al. Expression of distinct classes of titin isoforms in striated and smooth muscles by alternative splicing, and their conserved interaction with filamins. Journal of Molecular Biology. 2006;362(4):664–681. doi: 10.1016/j.jmb.2006.07.077. [DOI] [PubMed] [Google Scholar]
- Lahmers S, Wu Y, et al. Developmental control of titin isoform expression and passive stiffness in fetal and neonatal myocardium. Circ Res. 2004;94(4):505–513. doi: 10.1161/01.RES.0000115522.52554.86. [DOI] [PubMed] [Google Scholar]
- Lange S, Auerbach D, et al. Subcellular targeting of metabolic enzymes to titin in heart muscle may be mediated by DRAL/FHL-2. J Cell Sci. 2002;115(Pt 24):4925–4936. doi: 10.1242/jcs.00181. [DOI] [PubMed] [Google Scholar]
- Lange S, Xiang F, et al. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308(5728):1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- Leake MC, Grutzner A, et al. Mechanical properties of cardiac titin's N2B-region by single-molecule atomic force spectroscopy. J Struct Biol. 2006;155(2):263–272. doi: 10.1016/j.jsb.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Lee EH, Gao M. Mechanical strength of the titin Z1Z2-telethonin complex. Structure. 2006;14(3):497–509. doi: 10.1016/j.str.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Lee EH, Hsin J, et al. Tertiary and secondary structure elasticity of a six-Ig titin chain. Biophys J. 2010;98(6):1085–1095. doi: 10.1016/j.bpj.2009.12.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Carrion-Vazquez M, et al. Point mutations alter the mechanical stability of immunoglobulin modules. Nat Struct Biol. 2000;7(12):1117–1120. doi: 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- Li H, Oberhauser AF, et al. Multiple conformations of PEVK proteins detected by single-molecule techniques. Proc Natl Acad Sci U S A. 2001;98(19):10682–10686. doi: 10.1073/pnas.191189098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Linke WA, et al. Reverse engineering of the giant muscle protein titin. Nature. 2002;418(6901):998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- Linke WA, Ivemeyer M, et al. Actin-titin interaction in cardiac myofibrils: probing a physiological role. Biophys J. 1997;73(2):905–919. doi: 10.1016/S0006-3495(97)78123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke WA, Kulke M, et al. PEVK domain of titin: an entropic spring with actin-binding properties. J Struct Biol. 2002;137(1-2):194–205. doi: 10.1006/jsbi.2002.4468. [DOI] [PubMed] [Google Scholar]
- Linke WA, Grutzner A. Pulling single molecules of titin by AFM - recent advances and physiological implications. Pflugers Archiv-European Journal of Physiology. 2008;456(1):101–115. doi: 10.1007/s00424-007-0389-x. [DOI] [PubMed] [Google Scholar]
- Lu H, Isralewitz B, et al. Unfolding of titin immunoglobulin domains by steered molecular dynamics simulation. Biophys J. 1998;75(2):662–671. doi: 10.1016/S0006-3495(98)77556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Kan L, et al. Polyproline II helix is a key structural motif of the elastic PEVK segment of titin. Biochemistry. 2001;40(12):3427–3438. doi: 10.1021/bi0022792. [DOI] [PubMed] [Google Scholar]
- Marino M, Svergun DI, et al. Poly-Ig tandems from I-band titin share extended domain arrangements irrespective of the distinct features of their modular constituents. J Muscle Res Cell Motil. 2005;26(6-8):355–365. doi: 10.1007/s10974-005-9017-6. [DOI] [PubMed] [Google Scholar]
- Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- Marszalek PE, Lu H, et al. Mechanical unfolding intermediates in titin modules. Nature. 1999;402(6757):100–103. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- Mayans O, van der Ven PF, et al. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. 1998;395(6705):863–869. doi: 10.1038/27603. [DOI] [PubMed] [Google Scholar]
- Mayans O, Wuerges J, et al. Structural evidence for a possible role of reversible disulphide bridge formation in the elasticity of the muscle protein titin. Structure. 2001;9(4):331–340. doi: 10.1016/s0969-2126(01)00591-3. [DOI] [PubMed] [Google Scholar]
- McElhinny AS, Kakinuma K, et al. Muscle-specific RING finger-1 interacts with titin to regulate sarcomeric M-line and thick filament structure and may have nuclear functions via its interaction with glucocorticoid modulatory element binding protein-1. Journal of Cell Biology. 2002;157(1):125–136. doi: 10.1083/jcb.200108089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MK, Bang ML, et al. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. Journal of Molecular Biology. 2003;333(5):951–964. doi: 10.1016/j.jmb.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Dorn GW. Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annual Review of Physiology. 2001;63:391–426. doi: 10.1146/annurev.physiol.63.1.391. [DOI] [PubMed] [Google Scholar]
- Mrosek M, Labeit D, et al. Molecular determinants for the recruitment of the ubiquitin-ligase MuRF-1 onto M-line titin. Faseb Journal. 2007;21(7):1383–1392. doi: 10.1096/fj.06-7644com. [DOI] [PubMed] [Google Scholar]
- Muslin AJ. MAPK signalling in cardiovascular health and disease: molecular mechanisms and therapeutic targets. Clinical Science. 2008;115(7-8):203–218. doi: 10.1042/CS20070430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagueh SF, Shah G, et al. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation. 2004;110(2):155–162. doi: 10.1161/01.CIR.0000135591.37759.AF. [DOI] [PubMed] [Google Scholar]
- Nagy A, Cacciafesta P, et al. Differential actin binding along the PEVK domain of skeletal muscle titin. J Cell Sci. 2004;117(Pt 24):5781–5789. doi: 10.1242/jcs.01501. [DOI] [PubMed] [Google Scholar]
- Neagoe C, Kulke M, et al. Titin isoform switch in ischemic human heart disease. Circulation. 2002;106(11):1333–1341. doi: 10.1161/01.cir.0000029803.93022.93. [DOI] [PubMed] [Google Scholar]
- Neagoe C, Opitz CA, et al. Gigantic variety: expression patterns of titin isoforms in striated muscles and consequences for myofibrillar passive stiffness. J Muscle Res Cell Motil. 2003;24(2-3):175–189. doi: 10.1023/a:1026053530766. [DOI] [PubMed] [Google Scholar]
- Nedrud J, Labeit S, et al. Mechanics on Myocardium Deficient in the N2B Region of Titin: The Cardiac-Unique Spring Element Improves Efficiency of the Cardiac Cycle. Biophys J. 2011;101(6):1385–1392. doi: 10.1016/j.bpj.2011.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhauser AF, Hansma PK, et al. Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc Natl Acad Sci U S A. 2001;98(2):468–472. doi: 10.1073/pnas.021321798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka H, Yajima H, et al. Binding of the N-terminal 63 kDa portion of connectin/titin to alpha-actinin as revealed by the yeast two-hybrid system. Febs Letters. 1997;401(1):65–67. doi: 10.1016/s0014-5793(96)01432-9. [DOI] [PubMed] [Google Scholar]
- Opitz CA, Leake MC, et al. Developmentally regulated switching of titin size alters myofibrillar stiffness in the perinatal heart. Circ Res. 2004;94(7):967–975. doi: 10.1161/01.RES.0000124301.48193.E1. [DOI] [PubMed] [Google Scholar]
- Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278(5346):2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Peng J, Raddatz K, et al. Cardiac hypertrophy and reduced contractility in hearts deficient in the titin kinase region. Circulation. 2007;115(6):743–751. doi: 10.1161/CIRCULATIONAHA.106.645499. [DOI] [PubMed] [Google Scholar]
- Pfuhl M, Pastore A. Tertiary structure of an immunoglobulin-like domain from the giant muscle protein titin: a new member of the I set. Structure. 1995;3(4):391–401. doi: 10.1016/s0969-2126(01)00170-8. [DOI] [PubMed] [Google Scholar]
- Porter KE, Turner NA. Cardiac fibroblasts: At the heart of myocardial remodeling. Pharmacology & Therapeutics. 2009;123(2):255–278. doi: 10.1016/j.pharmthera.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Puchner EM, Alexandrovich A, et al. Mechanoenzymatics of titin kinase. Proc Natl Acad Sci U S A. 2008;105(36):13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke MH, Peng J, et al. Targeted deletion of titin N2B region leads to diastolic dysfunction and cardiac atrophy. Proc Natl Acad Sci U S A. 2007;104(9):3444–3449. doi: 10.1073/pnas.0608543104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief M, Gautel M, et al. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276(5315):1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- Romer LH, Birukov KG, et al. Focal adhesions: paradigm for a signaling nexus. Circ Res. 2006;98(5):606–616. doi: 10.1161/01.RES.0000207408.31270.db. [DOI] [PubMed] [Google Scholar]
- Samarel AM. Costameres, focal adhesions, and cardiomyocyte mechanotransduction. Am J Physiol Heart Circ Physiol. 2005;289(6):H2291–2301. doi: 10.1152/ajpheart.00749.2005. [DOI] [PubMed] [Google Scholar]
- Scholl FA, McLoughlin P, et al. DRAL is a p53-responsive gene whose four and a half LIM domain protein product induces apoptosis. Journal of Cell Biology. 2000;151(3):495–505. doi: 10.1083/jcb.151.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh F, Raskin A, et al. An FHL1-containing complex within the cardiomyocyte sarcomere mediates hypertrophic biomechanical stress responses in mice. J Clin Invest. 2008;118(12):3870–3880. doi: 10.1172/JCI34472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorimachi H, Freiburg A, et al. Tissue-specific expression and alpha-actinin binding properties of the Z-disc titin: implications for the nature of vertebrate Z-discs. J Mol Biol. 1997;270(5):688–695. doi: 10.1006/jmbi.1997.1145. [DOI] [PubMed] [Google Scholar]
- Spencer JA, Eliazer S, et al. Regulation of microtubule dynamics and myogenic differentiation by MURF, a striated muscle RING-finger protein. J Cell Biol. 2000;150(4):771–784. doi: 10.1083/jcb.150.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacklies W, Vega MC, et al. Mechanical network in titin immunoglobulin from force distribution analysis. PLoS Comput Biol. 2009;5(3):e1000306. doi: 10.1371/journal.pcbi.1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Graw S, et al. Genetic variation in titin in arrhythmogenic right ventricular cardiomyopathy-overlap syndromes. Circulation. 2011;124(8):876–885. doi: 10.1161/CIRCULATIONAHA.110.005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombitas K, Wu Y, et al. Cardiac titin isoforms are coexpressed in the half-sarcomere and extend independently. Am J Physiol Heart Circ Physiol. 2001;281(4):H1793–1799. doi: 10.1152/ajpheart.2001.281.4.H1793. [DOI] [PubMed] [Google Scholar]
- van Heerebeek L, Borbely A, et al. Myocardial structure and function differ in systolic and diastolic heart failure. Circulation. 2006;113(16):1966–1973. doi: 10.1161/CIRCULATIONAHA.105.587519. [DOI] [PubMed] [Google Scholar]
- von Castelmur E, Marino M, et al. A regular pattern of Ig super-motifs defines segmental flexibility as the elastic mechanism of the titin chain. Proc Natl Acad Sci U S A. 2008;105(4):1186–1191. doi: 10.1073/pnas.0707163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Mccarter R, et al. Regulation of Skeletal-Muscle Stiffness and Elasticity by Titin Isoforms - a Test of the Segmental Extension Model of Resting Tension. Proc Natl Acad Sci U S A. 1991;88(16):7101–7105. doi: 10.1073/pnas.88.16.7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CM, Jordan MC, et al. Titin isoform expression in normal and hypertensive myocardium. Cardiovasc Res. 2003;59(1):86–94. doi: 10.1016/s0008-6363(03)00328-6. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Muhle-Goll C, et al. Different molecular mechanics displayed by titin's constitutively and differentially expressed tandem Ig segments. J Struct Biol. 2002a;137(1-2):248–258. doi: 10.1006/jsbi.2002.4458. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Nair P, et al. Molecular mechanics of cardiac titin's PEVK and N2B spring elements. J Biol Chem. 2002b;277(13):11549–11558. doi: 10.1074/jbc.M200356200. [DOI] [PubMed] [Google Scholar]
- Weber KT. Cardiac interstitium in health and disease: the fibrillar collagen network. J Am Coll Cardiol. 1989;13(7):1637–1652. doi: 10.1016/0735-1097(89)90360-4. [DOI] [PubMed] [Google Scholar]
- Witt SH, Granzier H, et al. MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: Towards understanding MURF-dependent muscle ubiquitination. Journal of Molecular Biology. 2005;350(4):713–722. doi: 10.1016/j.jmb.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Witt CC, Witt SH, et al. Cooperative control of striated muscle mass and metabolism by MuRF1 and MuRF2. Embo Journal. 2008;27(2):350–360. doi: 10.1038/sj.emboj.7601952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Cazorla O, et al. Changes in titin and collagen underlie diastolic stiffness diversity of cardiac muscle. J Mol Cell Cardiol. 2000;32(12):2151–2162. doi: 10.1006/jmcc.2000.1281. [DOI] [PubMed] [Google Scholar]
- Wu Y, Bell SP, et al. Changes in titin isoform expression in pacing-induced cardiac failure give rise to increased passive muscle stiffness. Circulation. 2002;106(11):1384–1389. doi: 10.1161/01.cir.0000029804.61510.02. [DOI] [PubMed] [Google Scholar]
- Yamasaki R, Berri M, et al. Titin-actin interaction in mouse myocardium: passive tension modulation and its regulation by calcium/S100A1. Biophys J. 2001;81(4):2297–2313. doi: 10.1016/S0006-3495(01)75876-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki R, Wu Y, et al. Protein kinase A phosphorylates titin's cardiac-specific N2B domain and reduces passive tension in rat cardiac myocytes. Circ Res. 2002;90(11):1181–1188. doi: 10.1161/01.res.0000021115.24712.99. [DOI] [PubMed] [Google Scholar]
- Young P, Ehler E, et al. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J Cell Biol. 2001;154(1):123–136. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhmurov A, Dima RI, et al. Order statistics theory of unfolding of multimeric proteins. Biophys J. 2010;99(6):1959–1968. doi: 10.1016/j.bpj.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Bogomolovas J, et al. Single molecule force spectroscopy of the cardiac titin N2B element: effects of the molecular chaperone alphaB-crystallin with disease-causing mutations. J Biol Chem. 2009;284(20):13914–13923. doi: 10.1074/jbc.M809743200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P, Pinotsis N, et al. Palindromic assembly of the giant muscle protein titin in the sarcomeric Z-disk. Nature. 2006;439(7073):229–233. doi: 10.1038/nature04343. [DOI] [PubMed] [Google Scholar]