Abstract
HOTAIR is a long intervening non-coding RNA (lincRNA) that associates with the Polycomb Repressive Complex 2 (PRC2) and overexpression is correlated with poor survival for breast, colon and liver cancer patients. In this study, we show that HOTAIR expression is increased in pancreatic tumors compared to non-tumor tissue and is associated with more aggressive tumors. Knockdown of HOTAIR (siHOTAIR) by RNA interference shows that HOTAIR plays an important role in pancreatic cancer cell invasion and as reported in other cancer cell lines. In contrast, HOTAIR knockdown in Panc1 and L3.6pL pancreatic cancer cells that overexpress this lincRNA decreased cell proliferation, altered cell cycle progression, and induced apoptosis, demonstrating an expanded function for HOTAIR in pancreatic cancer cells compared to other cancer cell lines. Results of gene array studies showed that there was minimal overlap between HOTAIR-regulated genes in pancreatic vs. breast cancer cells and HOTAIR uniquely suppressed several interferon-related genes and gene sets related to cell cycle progression in pancreatic cancer cells and tumors. Analysis of selected genes suppressed by HOTAIR in Panc1 and L3.6 pL cells showed by knockdown of EZH2 and chromatin immunoprecipitation assays that HOTAIR-mediated gene repression was both PRC2-dependent and -independent. HOTAIR knockdown in L3.6pL cells inhibited tumor growth in mouse xenograft model, further demonstrating the pro-oncogenic function of HOTAIR in pancreatic cancer.
Keywords: HOTAIR, invasion, cell cycle progression, pro-oncogenic, prognostic
INTRODUCTION
Studies on the human, mouse and other genomes have demonstrated that a large number of genes and their corresponding RNAs are non-coding RNAs (ncRNAs) that are differentially expressed in various organs and tissues.1–4 The functions of some ncRNAs have been characterized and it is evident that they are key factors in gene regulation and influence normal and cancer cell phenotypes.4–8 Among the different classes of ncRNAs, microRNAs (miRs) have been the most extensively investigated, and it is estimated that >1000 miRs regulate up to 30% of all protein encoding genes.7–10 Several miRs are overexpressed in different tumors and their functional pro-oncogenic activity is usually associated with induction of oncogenes or inhibition of multiple genes with tumor suppressor-like activity.11–13
Rinn and coworkers have identified up to 3000 human long intervening non-coding RNAs (lincRNAs),14, 15 and biological characterization studies suggest that lincRNAs have important functions in both normal and cancer tissues.14–19 There is evidence that many lincRNAs act as scaffolds that regulate molecular (protein, RNA, DNA) interactions required for various signaling networks15–17 and this is accomplished, in part, by association with chromatin-modifying complexes.
HOTAIR is a 2,158 bp lincRNA localized to a boundary in the HOXC gene cluster. HOTAIR is a negative prognostic factor for breast, colon and liver cancer patient survival, and increased HOTAIR expression in patients has been correlated with enhanced breast and colon cancer metastasis.14, 20–23 HOTAIR expression has been linked to increased breast, liver and colon cancer cell invasiveness, whereas RNA interference (RNAi) studies in liver cancer cells showed that HOTAIR alone had minimal effects alone on cell viability or apoptosis but enhanced the activities of other agents.22 The activity of HOTAIR is due, in part, to interaction of HOTAIR with the Polycomb Repressive Complex 2 (PRC2) (EZH2, SUZ12 and EED) which enhances H3K27 trimethylation to decrease expression of multiple genes. Other lincRNAs also associate with PRC2 and other chromatin complexes, suggesting potential gene repressive activity similar to that described for HOTAIR.15
Initial data mining studies of publically available data bases24–26 showed that HOTAIR was overexpressed in pancreatic tumors compared to the normal pancreas and more highly expressed in advanced tumors.24 HOTAIR exhibited variable expression in pancreatic cancer cells, and knockdown of HOTAIR in Panc1 and L3.6pL pancreatic cancer cells by RNAi showed that HOTAIR was associated with enhanced cell invasion, cell proliferation, modulation of cell cycle progression, and induction of apoptosis. HOTAIR knockdown in L3.6pL cells also inhibited tumor growth in a mouse xenograft model. Knockdown of HOTAIR in Panc1 cells resulted in significant (>1.5) changes in expression of 1006 genes and analysis of the data suggested that HOTAIR-mediated gene regulation plays a critical role in pancreatic cancer progression and will be a novel epigenetic molecular target for treating pancreatic cancer patients.
RESULTS
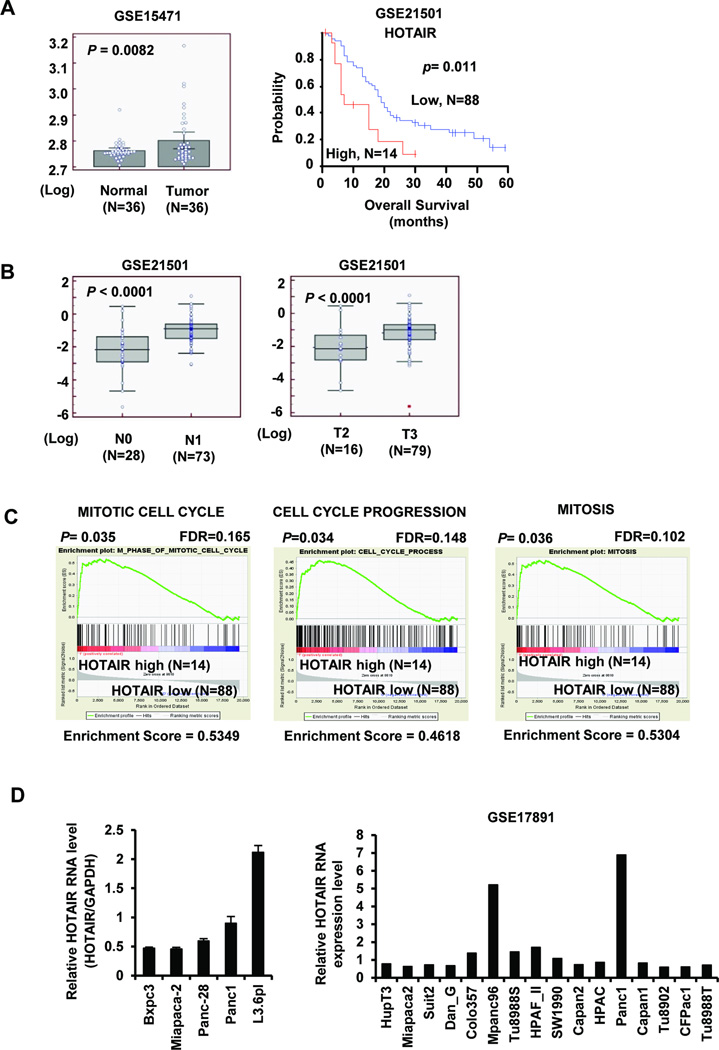
Data mining of publically available gene profiling results 24–26 showed that among 36 pancreatic cancer patients,25 HOTAIR was more highly expressed in pancreatic tumors compared to non-tumor tissue (Fig. 1A) and HOTAIR was more highly expressed in tumors extending beyond the pancreas (T3) compared to tumors only detected in the pancreas (T2) and more highly expressed in tumors spread to regional lymph nodes (N1) compared to tumors localized only in the pancreas (NO) (Fig. 1B).24 In this same study,24 Kaplan-Meier survival analysis showed that patients with low HOTAIR expression (bottom 85%) had significantly increased overall survival compared to patients with high HOTAIR expression (top 15%), and additional summaries of patient data are provided in Supplemental Figure S1. Furthermore, the result of Cox proportional hazard regression analysis consistently indicates that HOTAIR levels and N stage are strongly correlated with overall patient survival (p < 0.05) (Table S2). Figure 1C illustrates plots from gene set enrichment analysis (GSEA) using pancreatic patient gene profiling data (GSE21501) showing that gene set differences in HOTAIR high vs. low patients indicated that HOTAIR regulates gene sets mainly associated with cell proliferation and cell cycle progression (Supplemental Table S3).14 Figure 1D illustrates that HOTAIR was highly expressed in Panc1 and L3.6pL cells, whereas lower expression was observed in Panc28, MiaPac-2 and BxPC3 cells. These results overlapped with data mining from array data from several pancreatic cancer cell lines26 which also showed that HOTAIR was overexpressed in Panc1 cells (Fig. 1D).
Figure 1.
HOTAIR expression and prognostic significance in pancreatic cancer. (A) HOTAIR expression in normal pancreas and pancreatic tumors from patients (24) and Kaplan-Meier plot for survival of pancreatic cancer patients expressing high and low HOTAIR (25). (B) Relative expression of HOTAIR in pancreatic cancer patients with tumor localized in the pancreas (NO) or those with tumors already spread to regional lymph nodes (NO) or patients with tumor detected only in the pancreas (T2) and tumors extending beyond the pancreas (T3) (24). (C) GSEA analysis of GO terms showed that there was enriched expression of gene sets involved in cell cycle progression in pancreatic cancer patients. (D) Expression of HOTAIR in pancreatic cancer cells as determined by real time PCR or from array data (26).
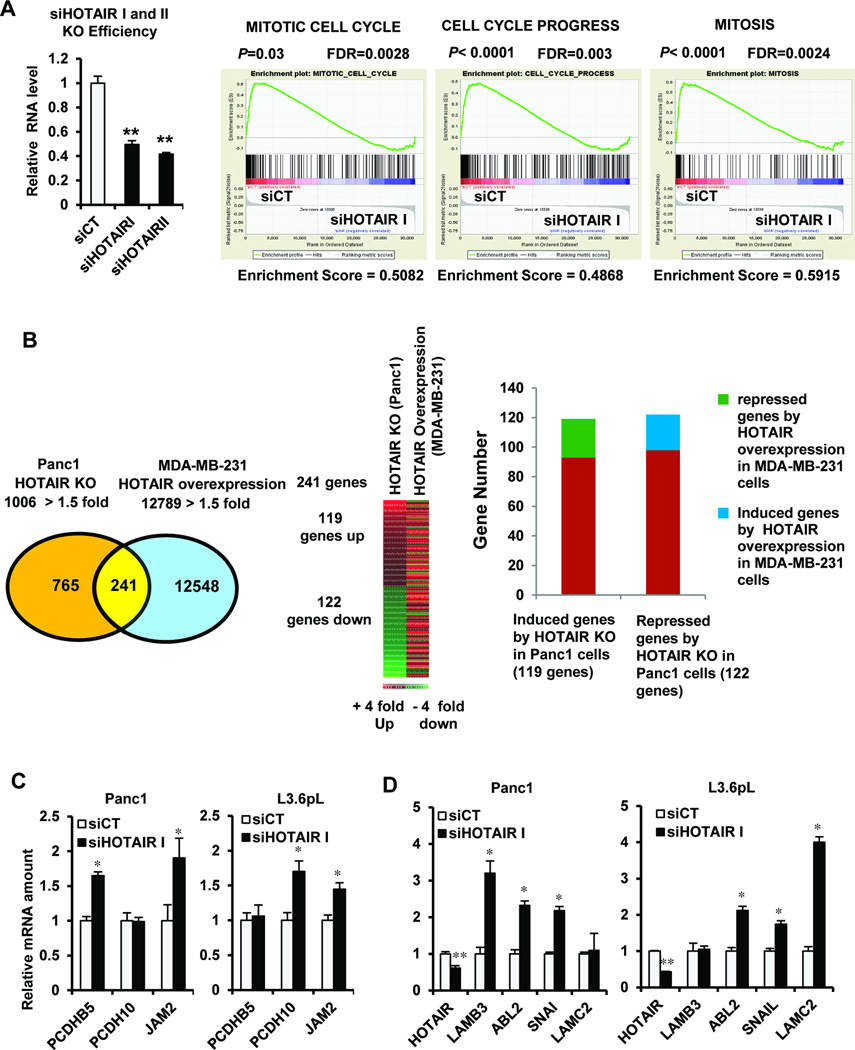
Panc1 cells are a highly aggressive “basal-like” pancreatic cancer cell line, and knockdown of HOTAIR by RNA interference (siHOTAIR) resulted in changes in expression (>1.5 fold) of 1006 genes in which expression of 454 genes was enhanced and expression of 552 genes was decreased (Supplemental Fig. S2A). HOTAIR knockdown was performed using two different siRNAs (siHOTAIR I and siHOTAIR II) to avoid off-target effects. Results from GSEA using gene ontology (GO) terms showed that 20 significant gene sets were affected by HOTAIR knockdown in Panc1 cells compared to control (siCT) cells (Supplemental Table S4). Since 10 out of the 20 GO terms were related to the cell cycle (Fig. 2A and Supplemental Table S3), we concluded that HOTAIR regulation of cell viability and cell cycle progression was important in both pancreatic cancer cells and patients (Fig. 1C).20–23
Figure 2.
Gene and gene sets regulated by HOTAIR in Panc1 cells and comparisons with MDAMB-231 breast cancer cells. (A) HOTAIR knockdown and GSEA analysis of GO terms showed that HOTAIR knockdown enriched expression of genes involved in cell cycle progression. (B) Venn diagram and heat map of genes regulated by HOTAIR in Panc 1 and MDA-MB-231 cells by HOTAIR knockdown and overexpression, respectively, and a summary of coordinate and independent regulation of genes by HOTAIR knockdown in Panc1 and HOTAIR overexpression (20) in MDA-MB-231 cells. Effects of siHOTAIR in Panc1 cells on genes suppressed (C) or induced (D) by HOTAIR overexpression in MDAMB-231 cells (20). Gene expression was determined by real time PCR and results are expressed as mean ± SE for 3 replicate determinations and significant (p < 0.05) is indicated (*). siHOTAIR was used in the knockdown studies.
Previous studies characterized HOTAIR-regulated genes by overexpression of this ncRNA in MDA-MB-231 breast cancer cells,20 and Figure 2B shows that there were 241 common genes among 1006 and 9,260 genes (>1.5 fold change) modulated by HOTAIR knockdown or overexpression in Panc1 and MDA-MB-231 cancer cells, respectively (summarized in Supplemental Tables S5 – S6). Since the HOTAIR-regulated genes were determined by opposite procedures (knockdown vs. overexpression) in Panc1 and MDA-MB-231 cells, respectively, we compared heat maps of the genes induced or repressed by HOTAIR knockdown or overexpression in both cell lines (Fig. 2B and Supplemental Table S7). The heat map illustrates the limited overlap between the 241 genes regulated by HOTAIR in Panc1 and MDA-MB-231 cells and this is further evidenced in the bar graphs (Fig. 2B) showing that among the 119 genes induced in Panc1 cells by siHOTAIR II, only 27 of these genes (green color) were repressed in MDA-MB-231 cells by HOTAIR overexpression (Supplemental Table S7) whereas among the 122 genes repressed after HOTAIR KO in Panc1 cells only 24 genes were induced (blue color). Moreover, siHOTAIR resulted in the repression of 122 genes in Panc1 cells and only 18 of these genes were induced by HOTAIR overexpression in MDA-MB-231 cells.20 HOTAIR plays an important role in PRC2-mediated gene suppression. Supplemental Figure S2 and Table S7 compares HOTAIR-regulated genes in Panc1 cells with HOTAIR/PRC2-coregulated genes in MDA-MB-231 (in a ChIP assay) and there were only 9 genes in common (Supplemental Fig. S2B and Table S7) but only minimal overlap between PRC2-regulated genes induced by siHOTAIR in Panc1 cells and genes suppressed by HOTAIR overexpression in MDA-MB-231 cells. We further investigated differences in HOTAIR regulated genes in Panc1 and MDA-MB-231 cells by comparing specific genes repressed and induced by HOTAIR overexpression in the latter cell line.20 Among 3 genes repressed by HOTAIR overexpression in MDA-MB-231 cells (PCDHB5, PCDH10 and JAM2), two were enhanced after HOTAIR knockdown in Panc1 and L3.6pL cells, but only JAM2 expression was enhanced in both cell lines (Fig. 2C). LAMB3, ABL2, SNA1 and LAMC2 were induced by HOTAIR overexpression in MDA-MB-231 cells;20 however, none of these genes were repressed by siHOTAIR in either Panc1 or L3.6pL cells (Fig. 2D), and two of the four genes (ABL2 and SNA1) were induced in both cell lines. These results confirm that HOTAIR regulates significantly different sets of genes in pancreatic vs. breast cancer cells.
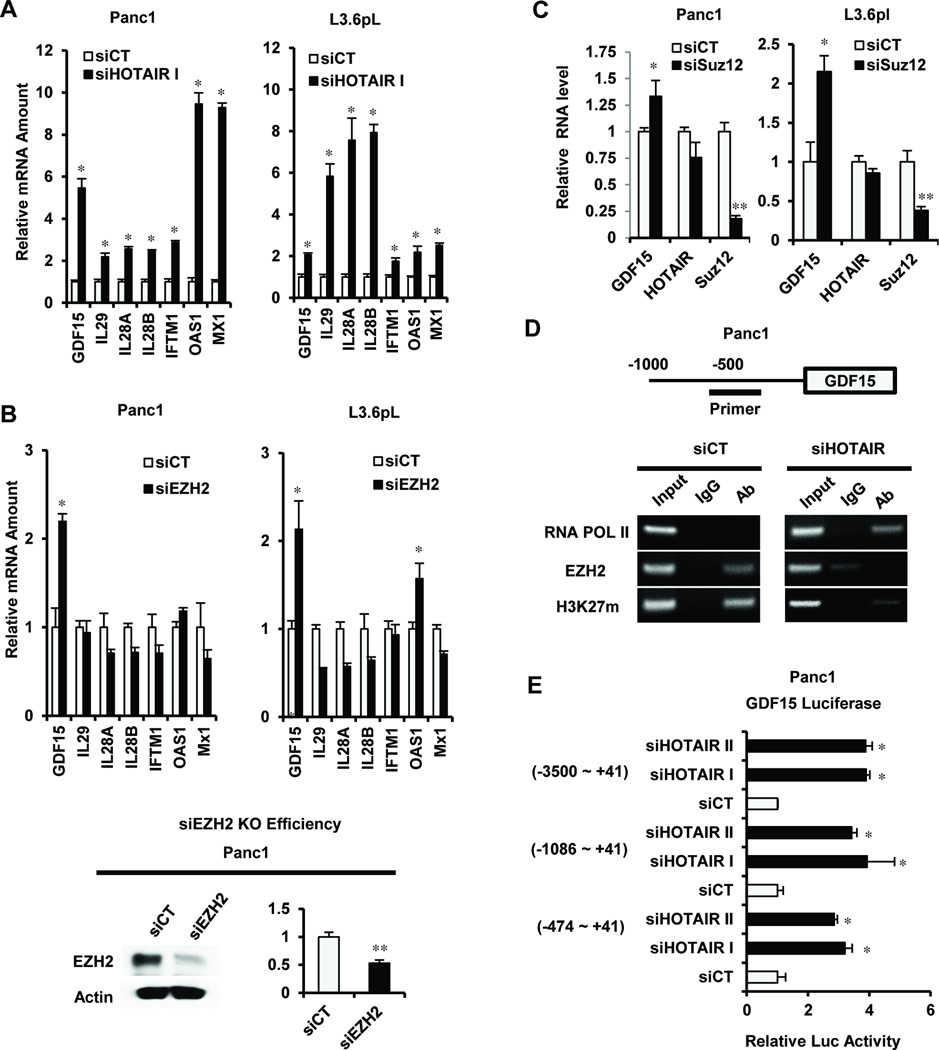
HOTAIR knockdown induces or represses multiple genes that could contribute to the functional pro-oncogenic activity of HOTAIR in Panc1 cells (Fig. 2A; Supplemental Tables S5 and S6). We further examined expression of 7 genes with tumor suppressor-like activity27–36 that are constitutively suppressed by HOTAIR and induced in Panc1 cells after transfection with siHOTAIR. Figure 3A demonstrates that siHOTAIR I significantly induced expression of GDF15, IL29, IL28A, IL28B, IFTM1, OAS1 and MX1 mRNA in Panc1 and L3.6pL cells. The role of the PRC2 complex in coregulating suppression of these HOTAIR-suppressed genes was investigated by EZH2 knockdown (siEZH2), and only GDF15 mRNA expression was induced in Panc1 and L3.6pL cells transfected with siEZH2 (Fig. 3B) and similar results were observed for knockdown of Suz12, another member of the PRC2 complex (Fig. 3C). GDF15 is a growth inhibitory/proapoptotic gene,27, 28 and ChIP analysis (Fig. 3D) shows that primers directed at the proximal region of the GDF15 promoter detected H3K27 trimethylation and EZH2 binding but not RNA pol II interactions in Panc1 cells transfected with siCT (control). In contrast, knockdown of HOTAIR resulted in the loss of H3K27 trimethylation and EZH2 binding but increased interaction of RNA pol II with the promoter region and this is consistent with cooperative PRC2/HOTAIR induction of GDF15 (Fig. 3A–3C). The effects of HOTAIR on GDF15 were further evaluated using two different siRNAs for HOTAIR in order to minimize possible off-target effects; both siHOTAIR I and II exhibited a similar level of knockdown efficiency. Both siRNAs for HOTAIR decreased luciferase activity in Panc1 cells transfected with constructs containing the proximal −3500 to +41, −1086 to +41, and −474 to +41 regions of the GDF15 promoter (Fig. 3E), whereas effects of siHOTAIR were minimal in Panc1 cells transfected with a construct containing only the −133 to +41 region of the promoter. These results suggest that HOTAIR/PRC2 coordinately regulate GDF15 in pancreatic cancer cells; in contrast, this gene was not affected by HOTAIR overexpression in MDA-MB-231 cells,20 further demonstrating cell context-dependent differences in HOTAIR/PRC2-regulated genes. Identification of other HOTAIR/PRC2-regulated genes in pancreatic cancer cells and tumors is currently being investigated and we expect that GDF15 is only one of several proapoptotic/growth inhibitory genes suppressed by this complex.
Figure 3.
Role of HOTAIR and PRC2 on gene/reporter gene expression and binding to the GDF15 promoter. (A) Panc1 and L3.6pL cells were transfected with siHOTAIR for 48 hr; gene expression was determined by real-time PCR and results are expressed as means ± SE for 3 replicate determinations. (B) The effects of siEZH on expression of the same sub-set of genes (A) was determined in Panc1 and L3.6pL cells as described above in (A) and the effects of siEZH2 and EZH2 mRNA protein are illustrated. Significant (p < 0.05) induction of gene expression (A and B) is indicated (*). (C) The effects of Suz12 knockdown (siSuz12) on GDF15 and Suz12 mRNA and HOTAIR were determined as described in the Materials and Methods. Significant (p < 0.05) induction (*) and inhibition (**) are indicated. (D) ChIP assay.Panc1 cells were transfected with siCT or siHOTAIR and, after 48 hr, the interaction of RNA pol II, EZH2 and H3K27m with the proximal region of the GDF15 promoter was determined in a ChIP assay. (E) Panc1 cells were transfected with siHOTAIR (I or II) and different GDF15-luc constructs containing GDF15 −3500 to +41, −1086 to +41, and −474 to +41 promoter inserts. Results are expressed as means ± SE (luciferase activity) for 3 replicate determinations. Significant (p < 0.05) induction of luciferase activity is indicated (*).
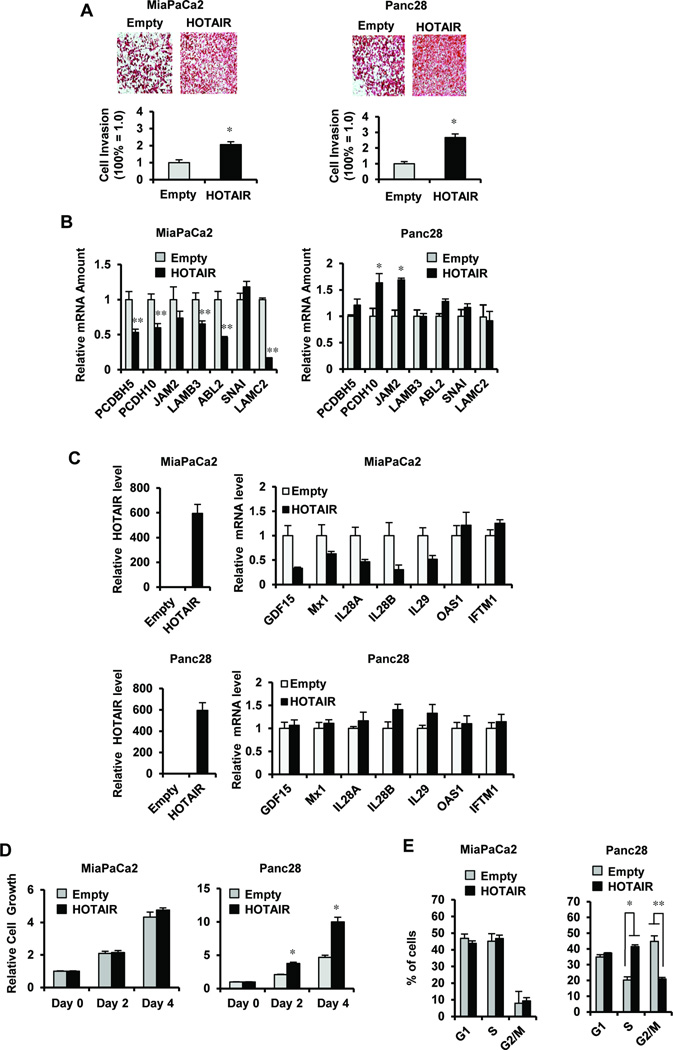
Since MiaPaCa2 and Panc28 cells express relatively low levels of HOTAIR, we further investigated the functional and genomic effects of HOTAIR overexpression and compared these results with overexpression of HOTAIR as previously reported in MDA-MB-231 cells20 and knockdown of HOTAIR in Panc1 and L3.6pL cells (Figs. 3A and 3B). HOTAIR overexpression enhanced MiaPaCa2 and Panc28 cell invasion in a Boyden chamber assay (Fig. 4A) and this was consistent with previous functional studies in breast and colon cancer cells.20, 23 HOTAIR overexpression decreased PCDBH5 and PCDH10 [as observed in MDA-MB-31 cells20] but not JAM2 in MiaPaCa2 cells, whereas in Panc28 cells PCDH10 and JAM2 were induced and PCDBH5 was unchanged by HOTAIR overexpression (Fig. 4B). In contrast, overexpression of HOTAIR either decreased or did not affect LAMB3, ABL2, SNA1 or LAMC2 mRNA levels in MiaPaCa2 and Panc28 cells (Fig. 4B), whereas these genes were all induced by HOTAIR overexpression in MDAMB-231 cell.20 The HOTAIR repressed genes (e.g. Fig. 3A) were also investigated by overexpression of HOTAIR in MiaPaCa2 and Panc28 cells. HOTAIR downregulated GDF15, MX1, IL28, IL28A, IL28B and IL29 only in MiaPaCa2 cells (Fig. 4C) and the remaining genes were unchanged in both cell lines. Overexpression of HOTAIR in MiaPaCa2 and Panc28 cells increased cell proliferation (Fig. 4D) and the % of cells in S phase and decreased the % in G2/M (Fig. 4E) in Panc28 but not in MiaPaCa2 cells. Thus, results of both HOTAIR knockdown or overexpression in pancreatic cancer cells further demonstrate differences between HOTAIR regulated genes in pancreatic vs. breast cancer cells and also show differences in HOTAIR function in MiaPaCa2 and Panc28 cells that express low levels of this ncRNA.
Figure 4.
HOTAIR overexpression in pancreatic cancer cells. (A) MiaPaCa2 and Panc28 cells were transfected with HOTAIR expression plasmid, and the effects on cell invasion were determined in a Boyden chamber assay. Results are expressed as means ± SE for 3 separate determinations and significantly (p < 0.05) enhanced invasion is indicated. Cells were transfected with HOTAIR expression plasmid and expression of genes regulated by HOTAIR overexpression in MDA-MB-231 cells (20) (B) and the HOTAIR repressed genes investigated in Figure 3A (C) were determined by real time PCR. Cells were transfected with HOTAIR expression plasmid, and effects on MiaPaCa2 and Panc28 cell proliferation (D) and cell cycle progression (E) are indicated. Results are expressed as means ± SE for 3 replicate determinations and significant (p < 0.05) induction or inhibition is indicated (*).
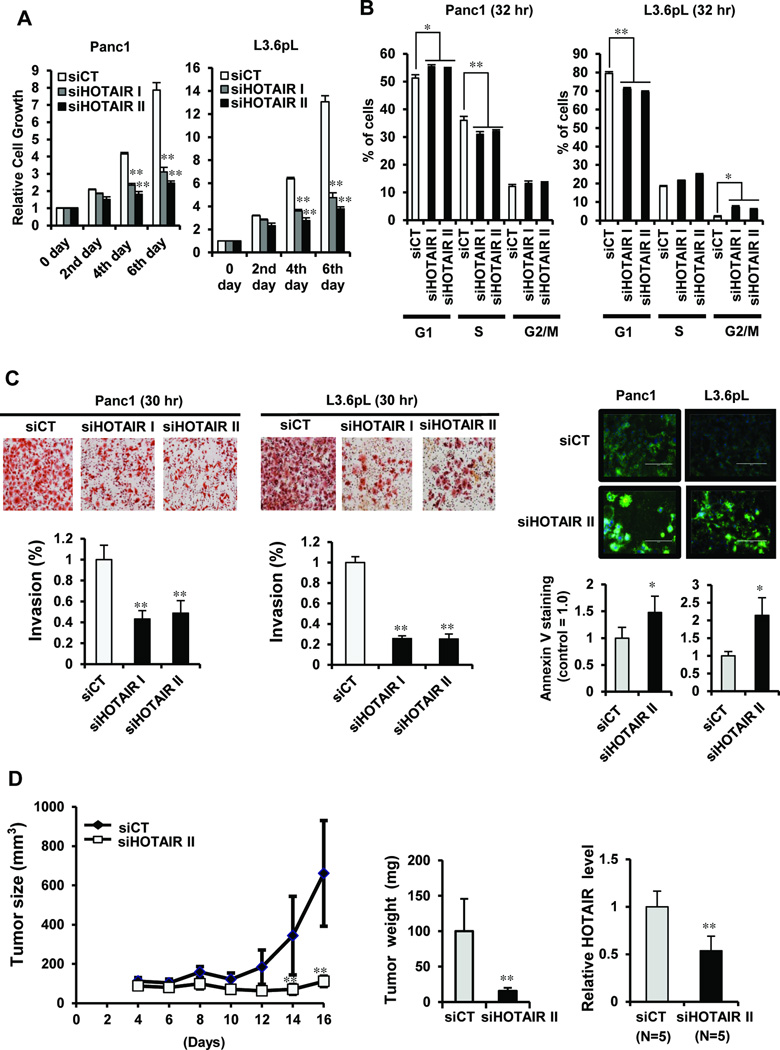
We further investigated the functional effects of siHOTAIR in Panc1 and L3.6pL cells that overexpressed this ncRNA. Transfection of siHOTAIR I and II inhibited Panc1 and L3.6pL growth which was significant after 4 days, and >50% growth inhibition was observed after 6 days (Fig. 5A). siHOTAIRs caused a G0/G1 to S phase arrest in Panc1 cells (32 hr), whereas in L3.6pL cells there was a decrease in the percent cells in G0/G1 and increase in G2/M, indicating pancreatic cancer cell context-dependent differences in the effects of HOTAIR on cell cycle progression (Fig. 5B). siHOTAIRs significantly decreased Panc1 and L3.6pL invasion using a Boyden chamber assay (Fig. 5C) and siHOTAIR II also enhanced Annexin V staining associated with induction of apoptosis in Panc1 cells (Fig. 5C) and siHOTAIR also induced PARP cleavage in both cell lines (data not shown). L3.6pL cells transfected with siHOTAIR II were used in a mouse xenograft model and, up to 16 days after knockdown of HOTAIR by RNA interference, there was a dramatic decrease in pancreatic tumor volume and weight and HOTAIR expression (Fig. 5D). Supplemental Figure 3 also shows that immunohistochemical staining of tumor tissue indicates an increase in TUNEL staining and a decrease in proliferation markers (Ki67 and PCNA) in siHOTAIR vs. siCT transfected tumors. These in vivo data complement the functional in vitro studies of HOTAIR and confirm the pro-oncogenic activity of this lincRNA in pancreatic cancer cells and tumors.
Figure 5.
Functional effects of HOTAIR knockdown in pancreatic cancer cells and tumors. Panc1 and L3.6pL cells were transfected with siHOTAIR, and effects on cell growth (A) and cell cycle progression (B) were determined at the indicated time points. (C) Panc 1 and L3.6pL cells were transfected with siHOTAIR and cell invasion was determined in a Boyden chamber assay and apoptosis was determined by measuring enhanced Annexin V staining. (D) siHOTAIR or siCT were transfected into L3.6pL cells which were then used in a xenograft model in athymic nude mice (6 per group), and tumor volumes and weights were determined. Quantitative results are means ± SE for at least 3 replicate determinations for each data point and significant (p < 0.05) induction (*) or inhibition (**) of a response by siHOTAIR (compared to siCT) are indicated.
DISCUSSION
HOTAIR was initially identified as one of 231 ncRNAs associated with human HOX loci, and HOTAIR resided in the HOXC locus but repressed transcription in the more distal HOXD locus in foreskin fibroblasts. HOTAIR interacted with the PRC2 complex and was required for PRC2-dependent histone H3 lysine 27 trimethylation and gene silencing. HOTAIRM1 and HOTTIP are lincRNAs associated with the HOXA locus, and both ncRNAs differentially modulate gene expression in various cell and tissue types, but genes/pathways modulated by these lincRNAs are PRC2-independent.37, 38
HOTAIR has also been characterized as a negative prognostic factor in breast, liver and colon cancer patients,20–23 and results of this study demonstrate that HOTAIR is also overexpressed in human pancreatic tumors compared to non-tumor tissue (Fig. 1). Moreover, there is also evidence that HOTAIR is more highly expressed in more aggressive and invasive pancreatic tumors (Figs. 1A and 1B). HOTAIR function was investigated in knockdown studies and indicates that this ncRNA enhances pancreatic cancer cell invasion, inhibits cell growth, modulates cell cycle progression, and induces apoptosis in vitro, and HOTAIR knockdown in L3.6pL cell inhibited tumor growth in athymic nude mice bearing these cells as xenografts (Fig. 5). Results of HOTAIR overexpression in MiaCaPa2 and Panc28 cells were also consistent with the pro-oncogenic activity of HOTAIR, although there were some cell context-dependent differences. Thus, HOTAIR not only plays a role in invasion of pancreatic, breast, colon and liver cancer cells 20, 22, 23 but also exerts distinct pro-oncogenic activities in pancreatic cancer associated with increased cell survival and proliferation and repression of interferon-related genes. This was further supported by a similar result from GSEA analysis of Panc1 cells (Fig. 2A) and human tumors (Fig. 1C), showing that at least 50% of the gene sets were associated with cell cycle progression and proliferation.
Modulation of HOTAIR expression in breast and colon cancer cells and tumors results in both enhanced and suppressed expression of genes, and a subset of genes repressed by HOTAIR in these cancer cells were also coregulated by PRC2.20, 23 A comparison of the gene expression data modulated by HOTAIR overexpression in MDA-MB-231 cells and HOTAIR knockdown in Panc1 cells showed some overlap in expression of individual genes; however, a heat map of induced/repressed genes illustrates significant differences between the cell lines (Fig. 2). Moreover, only 9 of the 854 genes coregulated by HOTAIR/PRC2 in MDA-MB-231 cells were also affected by HOTAIR knockdown in Panc1 cells (Supplemental Fig. S2B and Table S8). Among these genes, only OC1AD2 and RSAD2 were induced in Panc1 cells, whereas only minimal repression was observed after HOTAIR overexpression in MDA-MB-231 cells.20
HOTAIR-dependent gene regulation in pancreatic cancer cells was investigated by HOTAIR knockdown in Panc1 and L3.6pL cells and HOTAIR overexpression in Panc28 and MiaPaCa2 cells using a set of genes repressed (JAM2, PCDH10, PCDHB5) and induced (ABL2, SNAIL, LAMB3 and LAMC2) by HOTAIR overexpression in MDA-MB-231 cells.20 HOTAIR knockdown or overexpression gave variable results among the four different pancreatic cancer cell lines (Figs. 2D, 2E and 4B) which in turn exhibited minimal overlap with respect to HOTAIR-dependent regulation of this set of genes in MDA-MB-231 cells.20 These results were consistent with the differences observed in the gene arrays from Panc1 vs. MDA-MB-231 cells (Fig. 2B and Supplemental Fig. S2).
A second set of genes identified in the microarray that were significantly induced after HOTAIR knockdown were further investigated in Panc1 and L3.6pL cells transfected with siHOTAIR, and all were induced in both cell lines (Figs. 3A and 3B). Previous studies show that all seven genes exhibit tumor suppressor-like activities,27–36 and four genes, namely IL29, IL28A, IL28B and IFTM1, were among a subset of several interferon-regulated genes suppressed by HOTAIR in Panc1 cells (Supplemental Table S4). Transfection of Panc1 and L3.6pL cells with siEZH2, a key component of the PRC2 complex, or siSuz12, another PRC2 component, showed that only GDF15 was coregulated by HOTAIR and EZH2 (PRC2) (Figs. 3A, 3B and 3D), and ChIP assays in cells transfected with siCT vs. siHOTAIR confirmed that GDF15 was a HOTAIR/PRC2-regulated gene (Fig. 3C). In contrast, the interferon-related genes were not affected by EZH2 knockdown and the mechanisms of PRC2-independent but HOTAIR-mediated suppression of these genes are currently being investigated. These observations are consistent with recent reports showing that HOTAIR regulation of multiple genes is EZH2-indendent.21, 39 Consistent results were observed for the HOTAIR-regulated gene sets in pancreatic cancer patients (Fig. 1C) and after HOTAIR knockdown in pancreatic cancer cells (Fig. 2A), suggesting that knockdown of endogenous HOTAIR rather than overexpression may be preferable for investigating ncRNA-dependent gene regulation and function.
In summary, we show that HOTAIR is a negative prognostic factor for pancreatic cancer patients and exhibits pro-oncogenic activity in both in vitro and in vivo bioassays. HOTAIR-dependent gene regulation in pancreatic cancer cells is complex and differs significantly from a previous report in breast cancer cells.20 Nevertheless, HOTAIR knockdown in cells overexpressing this ncRNA gave consistent results using a subset of highly regulated genes, suggesting that HOTAIR-mediated suppression of genes in pancreatic cancer is both PRC2-dependent and PRC2-independent. Current studies are focused on mechanisms associated with suppression and activation of genes by HOTAIR in pancreatic cancer and development of therapeutic strategies that target HOTAIR.
MATERIALS AND METHODS
Cell lines
Human pancreatic cancer cell lines Panc1, MiaPaCa2 and Panc28 were obtained from American Type Culture Collection (Manassas, VA). L3.6pl pancreatic cancer cell line was kindly provided from Dr. I. J. Fidler in M.D. Anderson Cancer Center (Houston, TX). The cancer cell lines were grown and maintained in Dulbecco’s modified Eagle’s medium (DMEM) nutrient mixture (Hyclone, Logan, UT) supplemented with 0.22% sodium bicarbonate, 0.011% sodium pyruvate, 10% fetal bovine serum (FBS), and 10 ml/l 100× antibiotic antimycotic solution (Sigma Aldrich, St. Louis, MO).
Gene set enrichment analysis (GSEA)
Pancreatic cancer patient gene profiling data (GSE20501) was obtained from Gene Expression Omnibus (GEO) site. The patients are classified into two groups according to their HOTAIR expression level (top 15%: high vs. bottom 85%: low) and GSEA was carried out to assess the effects of HOTAIR expression level on various biological pathways using these two classified data sets. Similarly, GSEA was also performed using gene profiling data sets obtained from control siRNA control vs. HOTAIR siRNA (siHOTAIR I) transfected Panc1 cells. Significantly enriched biological pathways were identified, which produced nominal p-value < 0.05 and false discovery rates (FDR) < 0.25.
RNA isolation and quantative PCR
Total RNA was extracted either from pancreatic cancer cell lines or tissue samples. Five control and siRNA HOTAIR transfected tissue samples were analyzed, respectively, from xenograft study using mirVana™ RNA isolation kit from Ambion (Austin, TX), and quantative PCR was carried out using iCycler IQTM real time PCR detection system (Bio-Rad, Hercules, CA) after reverse transcription to cDNAs. The RT primer sets for HOTAIR, PCDHB5, PCDH10, JAM2, LAMB3, ABL2, SNAI and LAMC2 were used as previously described.20 All other primer sets and sequences are shown in Supplemental Table S1.
siRNA transfection and luciferase assay
Pancreatic cancer cells were transfected with 100 nM of control siRNA or HOTAIR siRNA I (SASI_Hs02_00380445) or HOTAIR siRNA II (SASI_Hs02_00380446) purchased from Sigma Aldrich (St. Louis, MO) using Lipofectamine 2000 (Invitrogen). Panc1 cells were cotransfected with control siRNA or HOTAIR siRNA I and HOTAIR II, and various length of GDF15 luciferase constructs. Luciferase activities were measured after 24 hr as previously described.40, 41 MISSION® siRNAs for EZH2 and Suz12 were also purchased from Sigma-Aldrich.
Chromatin immunoprecipitation assay (ChIP)
Antibodies for RNA polymerase, EZH2 and Histone H3 trimethyl Lys 27 antibodies were obtained from Active Motif (Carlsbad, CA), and ChIP assay was performed as previously described.41 The ChIP primer sequences: (forward) 5'-GGAGCACCCTGCTTAGACTG-3' and (reverse) 5'-GGGCCTCAGTATCCTCTTCC-3', which amplified 5' promoter region (−680 ~ −190) of GDF15 gene.
Boyden chamber cell invasion and apoptosis assays
Pancreatic cancer cells either transfected with siRNAs (HOTAIR vs. control) or expression vectors (empty vs. HOTAIR) and cell invasion assay was performed as described previously.41 Cells were transfected with siCT or siHOTAIR and after 72 hr, cells were stained for Annexin V using the Vibrant apoptosis assay kit as described.42
Cell proliferation and fluorescence-activated cell sorting (FACS) analysis and apoptosis
Cells were seeded in 12-well plates and transfected with either appropriate siRNA or expression vectors and cell numbers were counted at the indicated times using a Coulter Z1 cell counter (Beckman Coulter, Fullerton, CA). For FACS analysis, after pancreatic cancer cells transfected with siRNA either for control or HOTAIRs, cells were stained with propidium iodide solution and were analyzed on a FACS Calibur Flow Cytometer (Becton Dickinsin Systems, Franklin Lakes, NJ).
Xenograft study
L3.6pL cells (6×104 mL) were transfected with 100 nM siHOTAIR or siCT using Lipofectamine and after 48 hr, cells were collected and 1×106 cells injected into either side of the flank area of female nude mice (Harlan), and tumor volumes and weights were determined in mice from the siHOTAIR (6 mice) or siCT (6 mice) groups as described,43 and siHOTAIR levels were determined by real time PCR. Tumor volumes were measured (0.5 × length × width2) and after 16 days, the mice were sacrificed and tumor weights were measured and also used for further analysis as described.41–43
Immunohistochemistry and TUNEL assay
Tissue sections were deparaffinized in xylene and treated with graded series of alcohol and rehydrated in PBS. Antigen retrieval was done using 10 mM sodium citrate (pH 6.0–6.2) and endogenous peroxidase was blocked by 3% hydrogen peroxide in methanol for 6 min. Slides were then incubated with blocking serum (Vecstatin ABC Elite kit, Vector Laboratories, Burlingame, CA) for 45 min. Samples were then incubated overnight with Ki-67 and PCNA antibodies at 4°C. Sections were then washed in PBST and then incubated with biotinylated secondary antibody followed by streptavidin. The brown staining specific for antibody binding was developed by exposing the avidin and biotinylated peroxidase complex to diaminobenzidine reagent (Vector Laboratories) and sections were then counterstained with hematoxylin (Vector Laboratories). The in situ cell death detection POD kit was used for the terminal deoxyribonucleotide transferase-mediated nick-end labeling (TUNEL) assay according to the instructions in the protocol for tissue sections.
Statistical Analysis
Statistical significance of differences between different groups was determined using Student's t-test. Gene profiling data was either analyzed by BRB-Array Tools44 or Gene Pattern software.45 Cluster and Treeview programs were employed for generation of heat map and for gene clustering.46 Kaplan-Meier analysis and log-rank test were applied to evaluate a prognostic significance of HOTAIR expression level in terms of patient survival. Cox proportional hazard regression model was also used to evaluate independent prognostic factors correlated with tumor stage and lymph node metastasis.
Supplementary Material
Acknowledgements
This research was supported by National Institutes of Health (CA136571) and Texas AgriLife.
Footnotes
Disclosures: The authors declare no conflict of interest.
REFERENCES
- 1.Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- 2.Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 3.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: noncoding RNAs. Biochim Biophys Acta. 2005;1756:65–75. doi: 10.1016/j.bbcan.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the 'genome complexity' conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 5.Perez DS, Hoage TR, Pritchett JR, Ducharme-Smith AL, Halling ML, Ganapathiraju SC, et al. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet. 2008;17:642–655. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- 6.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 9.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Berezikov E, Plasterk RH. Camels and zebrafish, viruses and cancer: a microRNA update. Hum Mol Genet. 2005;14(Spec No. 2):R183–R190. doi: 10.1093/hmg/ddi271. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 12.Nelson KM, Weiss GJ. MicroRNAs and cancer: past, present, and potential future. Mol Cancer Ther. 2008;7:3655–3660. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 13.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6:539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa FF. Non-coding RNAs: Meet thy masters. Bioessays. 2010;32:599–608. doi: 10.1002/bies.200900112. [DOI] [PubMed] [Google Scholar]
- 19.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 23.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 24.Stratford JK, Bentrem DJ, Anderson JM, Fan C, Volmar KA, Marron JS, et al. A six-gene signature predicts survival of patients with localized pancreatic ductal adenocarcinoma. PLoS Med. 2010;7:e1000307. doi: 10.1371/journal.pmed.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 26.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baek SJ, Kim JS, Moore SM, Lee SH, Martinez J, Eling TE. Cyclooxygenase inhibitors induce the expression of the tumor suppressor gene EGR-1, which results in the up-regulation of NAG-1, an antitumorigenic protein. Mol Pharmacol. 2005;67:356–364. doi: 10.1124/mol.104.005108. [DOI] [PubMed] [Google Scholar]
- 28.Baek SJ, Horowitz JM, Eling TE. Molecular cloning and characterization of human nonsteroidal anti-inflammatory drug-activated gene promoter. Basal transcription is mediated by Sp1 and Sp3. J Biol Chem. 2001;276:33384–33392. doi: 10.1074/jbc.M101814200. [DOI] [PubMed] [Google Scholar]
- 29.Wongthida P, Diaz RM, Galivo F, Kottke T, Thompson J, Pulido J, et al. Type III IFN interleukin-28 mediates the antitumor efficacy of oncolytic virus VSV in immune-competent mouse models of cancer. Cancer Res. 2010;70:4539–4549. doi: 10.1158/0008-5472.CAN-09-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Numasaki M, Tagawa M, Iwata F, Suzuki T, Nakamura A, Okada M, et al. IL-28 elicits antitumor responses against murine fibrosarcoma. J Immunol. 2007;178:5086–5098. doi: 10.4049/jimmunol.178.8.5086. [DOI] [PubMed] [Google Scholar]
- 31.Steen HC, Gamero AM. Interferon-λ as a potential therapeutic agent in cancer treatment. J Interferon Cytokine Res. 2010;30:597–602. doi: 10.1089/jir.2010.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, Liu X, Zhou Y, Su SB. Interferon-lambdas: the modulators of antivirus, antitumor, and immune responses. J Leukoc Biol. 2009;86:23–32. doi: 10.1189/jlb.1208761. [DOI] [PubMed] [Google Scholar]
- 33.Maia CJ, Socorro S, Schmitt F, Santos CR. Characterization of oligoadenylate synthetase-1 expression in rat mammary gland and prostate: effects of 17β-estradiol on the regulation of OAS1g in both tissues. Mol Cell Biochem. 2008;314:113–121. doi: 10.1007/s11010-008-9771-z. [DOI] [PubMed] [Google Scholar]
- 34.Hatano H, Kudo Y, Ogawa I, Tsunematsu T, Kikuchi A, Abiko Y, et al. IFN-induced transmembrane protein 1 promotes invasion at early stage of head and neck cancer progression. Clin Cancer Res. 2008;14:6097–6105. doi: 10.1158/1078-0432.CCR-07-4761. [DOI] [PubMed] [Google Scholar]
- 35.Haller O, Kochs G, Weber F. Interferon, Mx, and viral countermeasures. Cytokine Growth Factor Rev. 2007;18:425–433. doi: 10.1016/j.cytogfr.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu F, Ng SS, Chow BK, Sze J, Lu G, Poon WS, et al. Knockdown of interferon-induced transmembrane protein 1 (IFITM1) inhibits proliferation, migration, and invasion of glioma cells. J Neurooncol. 2011;103:187–195. doi: 10.1007/s11060-010-0377-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chintharlapalli S, Papineni S, Baek SJ, Liu S, Safe S. 1,1-Bis(3'-indolyl)-1-(p-substitutedphenyl)methanes are peroxisome proliferator-activated receptor gamma agonists but decrease HCT-116 colon cancer cell survival through receptor-independent activation of early growth response-1 and nonsteroidal anti-inflammatory drug-activated gene-1. Mol Pharmacol. 2005;68:1782–1792. doi: 10.1124/mol.105.017046. [DOI] [PubMed] [Google Scholar]
- 41.Kim K, Chadalapaka G, Lee SO, Yamada D, Sastre-Garau X, Defossez PA, et al. Identification of oncogenic microRNA-17-92/ZBTB4/specificity protein axis in breast cancer. Oncogene. 2011 doi: 10.1038/onc.2011.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lei P, Abdelrahim M, Cho SD, Liu X, Safe S. Structure-dependent activation of endoplasmic reticulum stress-mediated apoptosis in pancreatic cancer by 1,1-bis(3'- indoly)-1-(p-substituted phenyl)methanes. Mol Cancer Ther. 2008;7:3363–3372. doi: 10.1158/1535-7163.MCT-08-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jutooru I, Chadalapaka G, Lei P, Safe S. Inhibition of NFκB and pancreatic cancer cell and tumor growth by curcumin is dependent on specificity protein down-regulation. J Biol Chem. 2010;285:25332–25344. doi: 10.1074/jbc.M109.095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 45.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 46.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.