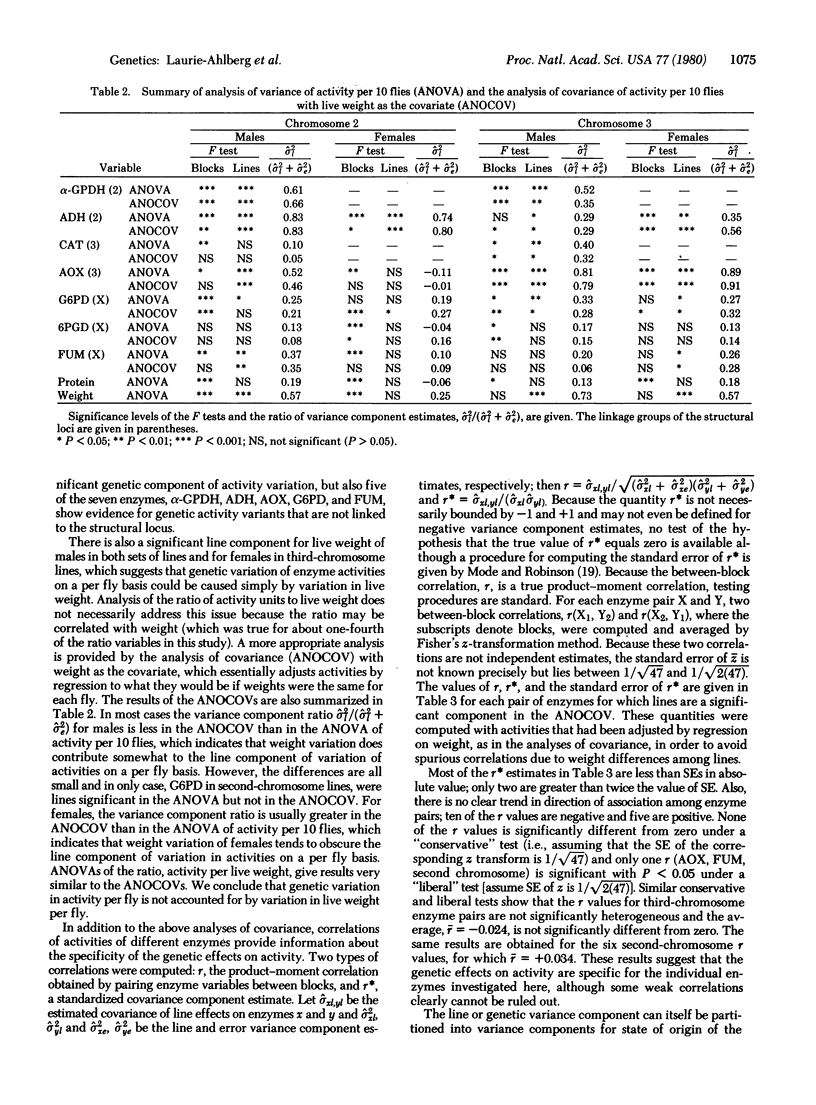
Abstract
The genetic component of variation of enzyme activity in natural populations of Drosophila melanogaster was investigated by using two sets of chromosome substitution lines. The constitution of a line of each type is: i1/i1;+2/ +2;i3/i3 and i1/i1;i2/ i2;+3/+3, where i refers to a chromosome from a highly inbred line and + refers to a chromosome from a natural population. The + but not the i chromosomes vary within a set of lines. By use of a randomized block design to test and estimate components of variance, 50 of the second- and 50 of the third- chromosome substitution lines have been screened for variation in the activity levels of seven enzymes. Six of the seven enzymes show a significant genetic component in at least one set of lines, and five of the seven enzymes show activity variations attributable to factors that are not linked to the structural gene. These unlinked activity modifiers identify possible regulatory elements. Analyses of covariance show that most of the genetic variation of enzyme activities cannot be accounted for by genetic variation of live weight or protein content. These results and the lack of strong correlations between the genetic effects on the activities of different enzymes indicate that the effects are mainly specific for individual enzymes.
Keywords: population genetics, modifier loci, regulatory elements, protein polymorphism
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham I., Doane W. W. Genetic regulation of tissue-specific expression of amylase structural genes in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4446–4450. doi: 10.1073/pnas.75.9.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley G. C., Rawls J. M., Jr, Lucchesi J. C. Alpha-glycerophosphate dehydrogenase in Drosophila melanogaster: kinetic differences and developmental differentiation of the larval and adult isozymes. J Insect Physiol. 1974 Jan;20(1):153–165. doi: 10.1016/0022-1910(74)90130-9. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Davidson E. H. Gene regulation for higher cells: a theory. Science. 1969 Jul 25;165(3891):349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- Chovnick A., Gelbart W., McCarron M., Osmond B. Organization of the rosy locus in Drosophila melanogaster: evidence for a control element adjacent to the xanthine dehydrogenase structural element. Genetics. 1976 Oct;84(2):233–255. doi: 10.1093/genetics/84.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., Britten R. J. Regulation of gene expression: possible role of repetitive sequences. Science. 1979 Jun 8;204(4397):1052–1059. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- Day T. H., Hillier P. C., Clarke B. The relative quantities and catalytic activities of enzymes produced by alleles at the alcohol dehydrogenase locus in Drosophila melanogaster. Biochem Genet. 1974 Feb;11(2):155–165. doi: 10.1007/BF00485771. [DOI] [PubMed] [Google Scholar]
- Dickinson W. J. A genetic locus affecting the developmental expression of an enzyme in Drosophilia melanogaster. Dev Biol. 1975 Jan;42(1):131–140. doi: 10.1016/0012-1606(75)90319-x. [DOI] [PubMed] [Google Scholar]
- Dickinson W. J. Aldehyde oxidase in Drosophila melanogaster: a system for genetic studies on developmental regulation. Dev Biol. 1971 Sep;26(1):77–86. doi: 10.1016/0012-1606(71)90109-6. [DOI] [PubMed] [Google Scholar]
- Hughes M. B., Lucchesi J. C. Genetic rescue of a lethal "null" activity allele of 6-phosphogluconate dehydrogenase in Drosophila melanogaster. Science. 1977 Jun 3;196(4294):1114–1115. doi: 10.1126/science.404711. [DOI] [PubMed] [Google Scholar]
- Laurie-Ahlberg C. C., Weir B. S. Allozymic Variation and Linkage Disequilibrium in Some Laboratory Populations of DROSOPHILA MELANOGASTER. Genetics. 1979 Aug;92(4):1295–1314. doi: 10.1093/genetics/92.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinsky S., Bewley G. C. Genetics of Catalase in DROSOPHILA MELANOGASTER: Rates of Synthesis and Degradation of the Enzyme in Flies Aneuploid and Euploid for the Structural Gene. Genetics. 1979 Apr;91(4):723–742. doi: 10.1093/genetics/91.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi J. C., Rawls J. M., Jr Regulation of gene function: a comparison of enzyme activity levels in relation to gene dosage in diploids and triploids of Drosophila melanogaster. Biochem Genet. 1973 May;9(1):41–51. doi: 10.1007/BF00485589. [DOI] [PubMed] [Google Scholar]
- Maroni G. Genetic control of alcohol dehydrogenase levels in Drosophila. Biochem Genet. 1978 Jun;16(5-6):509–523. doi: 10.1007/BF00484215. [DOI] [PubMed] [Google Scholar]
- McCarron M., O'Donnell J., Chovnick A., Bhullar B. S., Hewitt J., Candido E. P. Organization of the rosy locus in Drosophila melanogaster: further evidence in support of a cis-acting control element adjacent to the xanthine dehydrogenase structural element. Genetics. 1979 Feb;91(2):275–293. doi: 10.1093/genetics/91.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. F., Ayala F. J. Genetic and biochemical basis of enzyme activity variation in natural populations. I. Alcohol dehydrogenase in Drosophila melanogaster. Genetics. 1978 Jun;89(2):371–388. doi: 10.1093/genetics/89.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]


