Abstract
The expression of the alternative oxidase (AOX) was investigated during cotyledon development in soybean (Glycine max [L.] Merr.) seedlings. The total amount of AOX protein increased throughout development, not just in earlier stages as previously thought, and was correlated with the increase in capacity of the alternative pathway. Each AOX isoform (AOX1, AOX2, and AOX3) showed a different developmental trend in mRNA abundance, such that the increase in AOX protein and capacity appears to involve a shift in gene expression from AOX2 to AOX3. As the cotyledons aged, the size of the mitochondrial ubiquinone pool decreased. We discuss how this and other factors may affect the alternative pathway activity that results from the developmental regulation of AOX expression.
AOX is a quinol oxidase located in the inner mitochondrial membrane of plants, some fungi, and protists (McIntosh, 1994; Day et al., 1995; Siedow and Umbach, 1995; Wagner and Krab, 1995). The presence of AOX provides an alternative to the Cyt pathway (complex III, Cyt c, and complex IV) for the transport of electrons from ubiquinol to molecular oxygen to form water. Unlike the Cyt pathway, operation of the alternative pathway is not linked to proton translocation, and the free energy released by electron transfer is lost as heat. Alternative pathway activity can be catalyzed by a single gene product (Kumar and Söll, 1992; Albury et al., 1996), with the functional AOX enzyme thought to be a dimer of two similar or identical subunits (Umbach and Siedow, 1993).
High levels of AOX activity accompany maturation of the spadices of thermogenic flowers such as voodoo lily (Rhoads and McIntosh, 1992), but AOX activity has also been detected in every other higher plant examined (Siedow and Umbach, 1995). Increases in AOX activity have been observed in plant cells after exposure to a range of stresses, including low temperature (Vanlerberghe and McIntosh, 1992b), wounding (Hiser and McIntosh, 1990), and the addition of compounds that inhibit protein synthesis (Morohashi et al., 1991; Zhang et al., 1996), amino acid synthesis (Aubert et al., 1997), and Cyt pathway activity (Vanlerberghe and McIntosh, 1992a, 1994). AOX activity also changes during the ripening of mango fruit (Cruz-Hernández and Gómez-Lim, 1995) and during the development of pea leaves (Lennon et al., 1995). Increases in AOX activity are usually paralleled by increases in the amount of AOX protein (McIntosh, 1994; Siedow and Umbach, 1995), which are manifested as an increase in immunoreactive signal produced by probing mitochondrial protein extracts with an anti-AOX antibody raised against voodoo lily AOX proteins (Elthon et al., 1989). The increase in protein is typically accompanied by an increase in the abundance of AOX transcripts (Rhoads and McIntosh, 1992; Vanlerberghe and McIntosh, 1994, 1996; Cruz-Hernández and Gómez-Lim, 1995; Aubert et al., 1997).
AOX activity in mitochondria from cotyledons of soybean (Glycine max) seedlings increases with seedling age (Tuquet and Dizengremel, 1984; Azcón-Bieto et al., 1989; Obenland et al., 1990). In one study this increase was found to correlate with an increase in AOX protein, but the magnitude of the protein increase was greater than the increase in activity (Obenland et al., 1990). This discrepancy may be explained by the recent findings that the AOX enzyme is activated by reduction of an intermolecular disulfide bond between the two subunits of the dimer (Umbach and Siedow, 1993, 1996; Umbach et al., 1994) and is stimulated by certain α-keto acids (Millar et al., 1993).
Two distinct AOX proteins can be detected in mitochondria from soybean cotyledons (Obenland et al., 1990; Kearns et al., 1992). The higher-mobility species (approximately 34 kD) was observed at all developmental stages, whereas the lower-mobility species (approximately 36 kD) did not appear until several days after germination (Obenland et al., 1990). Although the presence of multiple AOX proteins is common in plants (Elthon et al., 1989; Obenland et al., 1990; Kearns et al., 1992; Hiser and McIntosh, 1994; Cruz-Hernández and Gómez-Lim, 1995; Zhang et al., 1996), the molecular basis for this has only been resolved for soybean, in which three distinct AOX genes (GmAOX1, GmAOX2, and GmAOX3) have been identified (Whelan et al., 1996). Direct N-terminal sequencing of partially purified soybean AOX proteins has demonstrated that the 34- and 36-kD proteins found in the mitochondria of soybean cotyledons are the products of the AOX2 and AOX3 genes, respectively (Finnegan et al., 1997). Multiple AOX proteins in other species may also be due to multiple AOX genes, because AOX multigene families have been identified in tobacco (Whelan et al., 1996), Arabidopsis (Saisho et al., 1997), rice (Ito et al., 1997), mango (M. Considine and J. Whelan, unpublished data), and tomato (R. Holtzapffel, P.M. Finnegan, and D.A. Day, unpublished data). The mechanisms by which these different genes are regulated have not yet been elucidated.
We examined the expression of the AOX multigene family in cotyledons during soybean seedling development and reinvestigated AOX expression in this tissue in terms of protein abundance and alternative pathway capacity. In contrast to previous work on soybean cotyledons, but in line with other systems, increases in alternative pathway capacity during cotyledon development strongly correlated with increases in total AOX protein abundance. However, the increase in AOX activity and protein abundance was not due simply to the transcriptional up-regulation of an already active AOX gene, but, rather, to a developmentally regulated shift from the expression of one AOX subunit gene to the expression of another.
MATERIALS AND METHODS
Plant Growth, Tissue Collection, and Mitochondrial Isolation
Soybean (Glycine max [L.] Merr. cv Stevens) seeds were germinated in trays of vermiculite and grown with a photocycle of 12 h of light (120 or 380 μmol photons m−2 s−1) at 28°C and 12 h of dark at 19°C in controlled-environment cabinets with daily watering. Cotyledons were harvested 5, 7, 10, 14, or 20 d after planting. For each stage examined, cotyledons for RNA extraction and for isolation of mitochondria were taken from the same tray of plants. Cotyledons to be used for RNA isolation were frozen immediately in liquid nitrogen and stored at −80°C. Cotyledons from approximately 200 plants were harvested for isolation of mitochondria, which was done according to the method of Day et al. (1985).
mRNA Analysis
Frozen cotyledons from 5 to 30 plants were ground to a powder under liquid nitrogen using a mortar and pestle. Total RNA was isolated by the guanidine thiocyanate/organic extraction method of Chomczynski and Sacchi (1987) and extracted with LiCl (Puissant and Houdebine, 1990). Poly(A+) RNA was enriched from 0.35 to 1.0 mg of total RNA using a kit (Oligotex, Qiagen, Clifton Hill, Australia) and the protocol described by the supplier.
Poly(A+)-enriched RNA samples were mixed with sample buffer containing ethidium bromide, separated by electrophoresis through agarose gels containing formaldehyde (Sambrook et al., 1989), and transferred by capillary action to nylon membranes (Hybond-N+, Amersham) using 20× SSC (3 m NaCl and 0.3 m trisodium citrate) as the transfer buffer. RNA was fixed to the membrane by baking at 80°C for 2 h. Linearized clones of full-length soybean AOX cDNAs (Finnegan et al., 1997) were used as templates for the synthesis of digoxigenin-labeled antisense RNA probes using a commercial kit (Boehringer Mannheim). Prehybridization (2 h) and hybridization (approximately 16 h) were performed in hybridization buffer (DIG Easy Hyb, Boehringer Mannheim) at 65°C or 68°C. After hybridization, membranes were washed for 15 min twice with 1× SSC, 1% (w/v) SDS at ambient temperature, twice with 0.1× SSC, 1% (w/v) SDS at 65°C or 68°C, and twice with 0.1× SSC, 0.1% (w/v) SDS at ambient temperature. Hybrids were tagged with an alkaline phosphatase-conjugated anti-digoxigenin antibody and visualized by an alkaline-phosphatase-catalyzed reaction using a chemiluminescent substrate (CDP-Star, Boehringer Mannheim) following the supplier's instructions. A competitive reverse transcriptase-PCR assay specific for the soybean AOX multigene family (Finnegan et al., 1997) was used to estimate the relative abundance of AOX gene transcripts in the total RNA.
Protein Analysis
Protein concentrations were estimated by the method of Lowry et al. (1951). Mitochondrial proteins were solubilized in sample buffer, separated by SDS-PAGE (Laemmli, 1970), and transferred to a supported nitrocellulose membrane (Hybond-C Extra, Amersham) by the method of Towbin et al. (1979) using a semidry blotting apparatus (Millipore). AOX proteins were tagged with the AOA monoclonal antibody raised against AOX proteins from voodoo lily (Elthon et al., 1989), followed by a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Bio-Rad), and detected by chemiluminescence according to the instructions supplied with a commercial kit (Boehringer Mannheim). Signal intensities were estimated using ImageQuant software (version 1.11, Molecular Dynamics, Sunnyvale, CA) after digitization using an image scanner (UMAX, Hsinchu, Taiwan).
Assays
Oxygen uptake by isolated mitochondria was measured using an oxygen electrode (Rank Bros., Cambridge, UK) in 2 mL of reaction medium (0.3 m Suc, 5 mm KH2PO4, 10 mm NaCl, 2 mm MgSO4, 0.1% [w/v] BSA, and 10 mm Tes [pH 7.0]). Alternative pathway capacity was measured as the rate of oxygen uptake in the presence of 2 mm NADH, 10 mm succinate, 1 mm ADP, 2 mm pyruvate, and an inhibitor of the Cyt pathway (16 μm myxothiazol or 0.5 mm KCN). Subsequent addition of DTT to 10 mm was used to check that the capacity had not been limited by the presence of oxidized AOX. Alternative pathway activity was measured in the presence of 2 mm NADH, 1 mm ADP, and a Cyt pathway inhibitor (16 μm myxothiazol or 0.5 mm KCN), before and after pyruvate was added to a concentration of 2 mm. The capacity of the Cyt pathway was measured as oxygen uptake in the presence of 2 mm NADH, 10 mm succinate, 1 mm ADP, and 0.25 mm n-propyl gallate. The residual oxygen uptake (after addition of both n-propyl gallate and myxothiazol/KCN) was subtracted from all rates.
Ubiquinone was extracted from isolated cotyledon mitochondria and homologs were quantified by reverse-phase HPLC according to the method of Ribas-Carbo et al. (1995) as modified by Millar et al. (1997). Malic enzyme activity was assayed according to the method of Day et al. (1984).
RESULTS
Changes in AOX mRNA and Protein during Cotyledon Expansion
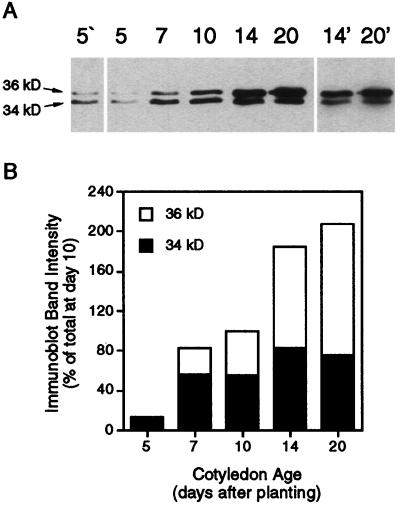
The expression of AOX was investigated at five stages of postgerminative cotyledon development. The amounts of AOX proteins in cotyledon mitochondria were determined by immunodetection (Fig. 1A). The two bands previously observed in cotyledon mitochondria (Obenland et al., 1990; Kearns et al., 1992) were detected in all samples. The 36-kD band contained AOX3 protein and the 34-kD band contained AOX2 protein (Finnegan et al., 1997). The linear range of signal intensity for AOX immunodetection was narrow; therefore, in a single exposure either the signals of highest intensity or lowest intensity were outside of this range. However, varying the exposure time enabled the changes to be seen clearly (Fig. 1A). By determining signal intensities from several exposures, we obtained estimates of the changes in the relative amounts of AOX2 and AOX3 and the total amount of AOX protein. Figure 1B shows signal-intensity data from one exposure (Fig. 1A, middle), which are representative of the qualitative changes observed.
Figure 1.
Analysis of the expression of AOX proteins during the development of soybean cotyledons. A, Immunoblot of AOX proteins in cotyledon mitochondria. Each lane contains 20 μg of mitochondrial protein. Numbers across the top refer to seedling age in days postimbibition. 5‘, Longer exposure of 5 min; 14′ and 20′, shorter exposures of 14 and 20 min, respectively. Results from one of two independent experiments are shown. B, Intensity of the bands on the immunoblot exposure shown in the central panel of A. Values are expressed as the percentages of the sum of the d-10 band intensities.
It was obvious that the total amount of AOX protein increased with cotyledon age, mostly as a result of increasing amounts of AOX3 throughout seedling growth (Fig. 1B). AOX2 protein abundance increased between d 5 and 7 but thereafter was relatively constant. AOX3 was less abundant than AOX2 at d 5 but by d 14 had increased to become the predominant form. An underlying assumption in these measurements was that the AOA antibody recognizes all forms of soybean AOX equally. The ability of the AOA antibody to cross-react with divergent AOX proteins from a range of plant and nonplant species following electrophoresis under denaturing conditions (Chaudhuri et al., 1995; Siedow and Umbach, 1995) suggests that the antibody recognizes a linear amino acid sequence epitope that is conserved across these proteins. Considering the high degree of similarity between soybean AOX proteins in the regions conserved across all AOX proteins (Finnegan et al., 1997), it is likely that their cross-reactivity with AOA is similar.
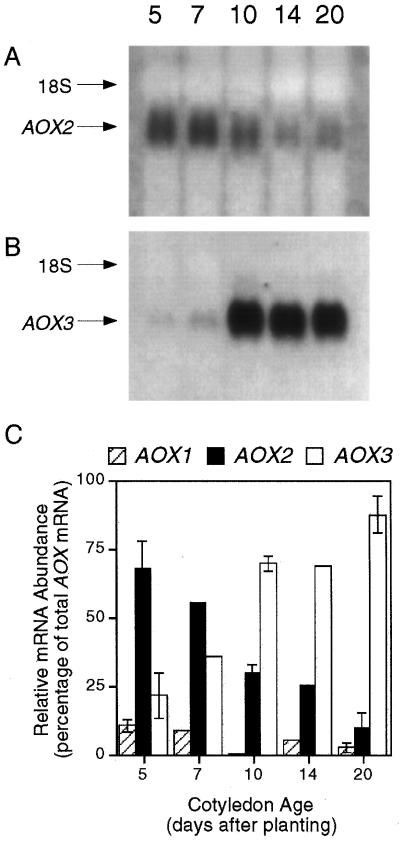
Northern analysis of cotyledon poly(A+)-enriched RNA using antisense RNA probes derived from the soybean AOX2 and AOX3 cDNAs produced the results shown in Figure 2. With each probe, only the hybridizing band shown in Figure 2 was observed. The intensity of the signal obtained using the AOX2 probe decreased during the growth of the seedlings (Fig. 2A); that obtained with the AOX3 probe was low in young cotyledons but became much greater in the later three stages (Fig. 2B). Each sample contained the same amount of nucleic acid, and we confirmed that the opposing trends were not due to differences between experiments by stripping the former membrane of the AOX2 probe and reprobing it with the AOX3 probe. Because of the low abundance of AOX1 mRNA in cotyledons (Finnegan et al., 1997; see below), no signal was detected by northern analysis of cotyledon RNA with an AOX1 probe. It should be noted that the absolute ratio of AOX2 and AOX3 mRNAs cannot be determined from these results because of different labeling efficiencies and exposure times. Nonetheless, a clear opposing trend in message abundance between the two genes was evident.
Figure 2.
Northern analysis of changes in the level of expression of AOX2 (A) and AOX3 transcripts (B) during the development of soybean cotyledons. Each lane contained 2 μg of poly(A+)-enriched RNA. Numbers across the top refer to seedling age in days postimbibition. The position of 18S rRNA on each blot is indicated. Results from one of two independent experiments are shown. C, Relative abundance of AOX mRNAs as determined using a competitive reverse transcriptase-PCR assay. The results from one experiment are shown. For d 5, 10, and 20, n = 3, and for d 7 and 14, n = 1. Bars indicate se.
To quantify the relative amounts of AOX transcripts at each age, the mole-fraction contribution of each AOX gene to the AOX transcript pool was determined using a competitive reverse transcriptase-PCR assay (Finnegan et al., 1997). This assay allowed the detection of AOX1 transcripts in addition to those of AOX2 and AOX3 (Fig. 2C). At d 5, AOX2 transcripts were the most abundant and AOX3 transcripts were about twice as abundant as those from AOX1. During seedling development the relative abundance of AOX2 transcripts decreased, whereas the relative abundance of AOX3 transcripts increased, exceeding AOX2 by d 10 and increasing further through d 20. AOX1 transcripts remained only a small percentage of total AOX transcripts at every cotyledon age. These results correlate well with the protein analysis shown in Figure 1.
Alternative Pathway Capacity in Isolated Mitochondria
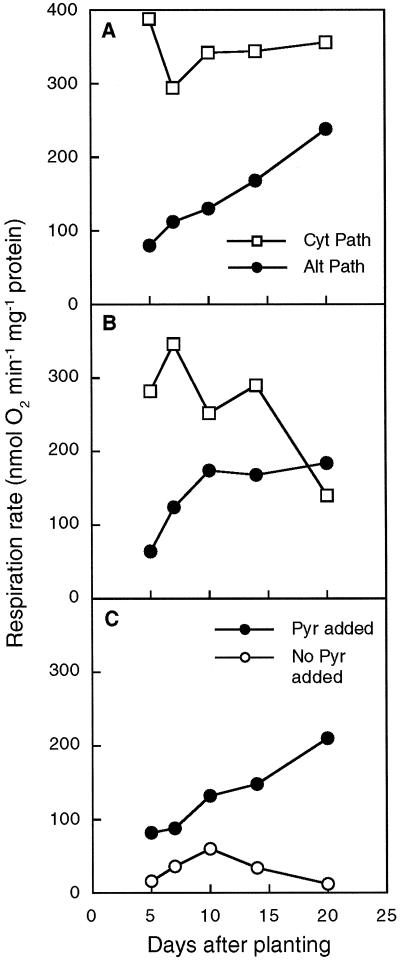
Measurements of oxygen uptake (Fig. 3) were made using aliquots of the mitochondrial samples used for AOX immunodetection. To estimate changes in the capacity of the alternative pathway, oxygen uptake via the alternative pathway was measured in reactions containing two respiratory substrates, NADH and succinate, with pyruvate added to activate AOX (Millar et al., 1993). A reducing agent, DTT, was added to check for the presence of oxidized AOX, which is inactive (Umbach and Siedow, 1993), but the change in the alternative pathway capacity was low (typically less than 5%). The capacity of the alternative pathway increased with cotyledon age (Fig. 3, A and B), which is consistent with previous reports (Gerard and Dizengremel, 1988; Azcón-Bieto et al., 1989; Obenland et al., 1990).
Figure 3.
Developmental changes in the capacity of respiratory pathways in cotyledon mitochondria. A, Changes in capacities of Cyt and alternative (Alt) pathways in mitochondria isolated from different-age cotyledons from plants grown at a light intensity of 120 μmol photons m−2 s−1. Results represent the means of two or three measurements on one mitochondrial sample. B, Changes in capacities of Cyt and alternative pathways in mitochondria isolated from different-age cotyledons from plants grown at a light intensity of 380 μmol photons m−2 s−1. Results represent the means of two to six measurements on one mitochondrial sample. C, Comparison of the activity of the alternative pathway in isolated mitochondria oxidizing NADH as a substrate in the absence or presence of exogenous pyruvate (Pyr). Growth conditions were as in A. Each data point represents one measurement on one mitochondrial sample.
However, our results differ from those of Obenland et al. (1990) in that all increases in capacity were accompanied by an increase in the total amount of AOX protein (Figs. 1 and 3A). This correspondence was observed in each of the experiments we performed, although the values for alternative pathway capacity were not exactly the same. In one experiment using a light intensity of 120 μmol photons m−2 s−1, the AOX capacity of mitochondria continued to increase until 20 d after planting (Fig. 3A); at higher light intensities (380 μmol photons m−2 s−1), AOX capacity appeared to have reached a plateau at about d 15 (Fig. 3B). This difference could have been due to the effect of the different growth conditions on the rate of seedling development.
To examine the dependency of the alternative pathway on the presence of AOX activators, AOX activity was measured before and after the addition of pyruvate, using NADH as the respiratory substrate (Fig. 3C). Succinate was omitted from these reactions, because it can lead to the production of pyruvate within the mitochondrion (Millar et al., 1996). In the presence of pyruvate, AOX activity with NADH was slightly less than that observed with both substrates, but the activity increased with cotyledon age (Fig. 3C), which is in agreement with increases in immunoreactive protein (Fig. 1). Pyruvate (and other metabolites) can be lost from the matrix during mitochondrial isolation; therefore, the matrix pyruvate concentration in isolated mitochondria will be less than or equal to the concentration in the original tissue. In the absence of exogenous pyruvate, AOX activity remained low throughout cotyledon aging (Fig. 3C). This indicates that the greater capacity of the alternative pathway at later stages of development will not be fully utilized unless the concentration of AOX activators in vivo is sufficient. This trend was the same in plants grown at light intensities of either 120 or 380 μmol photons m−2 s−1 (data not shown).
In contrast to the increase in capacity of AOX, Cyt pathway capacity changed little between d 5 and 20 in plants grown with a light intensity of 120 μmol photons m−2 s−1 (Fig. 3A). For these plants the capacity of the Cyt pathway was greater than the alternative pathway at each age. At 380 μmol photons m−2 s−1 light intensity, the capacity of the Cyt pathway was slightly less and decreased substantially after d 14, as reported previously (Azcón-Bieto et al., 1989), eventually becoming less than the alternative pathway capacity (Fig. 3B). The decrease in Cyt pathway capacity with age was accompanied by an appreciable yellowing of the cotyledons; this was not observed in the plants grown at low light, indicating that the decline in electron transport coincided with the onset of senescence.
Changes in Factors Affecting Alternative Pathway Activity
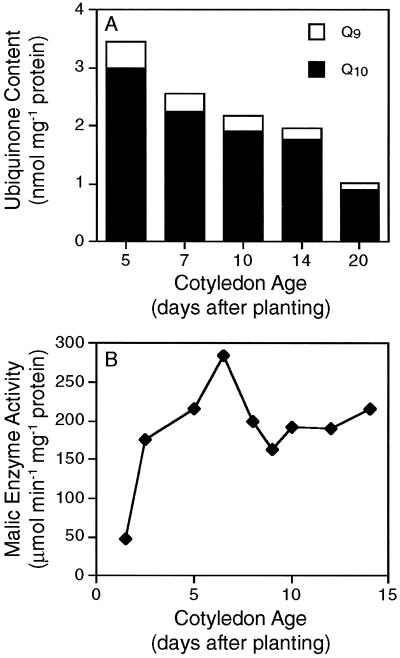
The amount of ubiquinone in the mitochondrial inner membrane may affect the rate at which electron transport occurs via the alternative pathway (Ribas-Carbo et al., 1995, 1997). Soybean mitochondria contain two ubiquinone homologs, ubiquinone 9 and 10 (Ribas-Carbo et al., 1995). Because AOX does not show a preference for either homolog, the sum of the concentrations is indicative of the size of the ubiquinone pool (Ribas-Carbo et al., 1995). Measurements were made of the amount of these ubiquinone homologs in the membranes of mitochondria from cotyledons at different ages (Fig. 4A). In all cases, ubiquinone 10 was the predominant homolog, as previously observed for soybean mitochondria (Ribas-Carbo et al., 1995). The amount of both ubiquinone homologs, and therefore total ubiquinone, decreased during seedling growth from d 5 to 20. Values for mitochondria from younger cotyledons were comparable to those previously reported for mitochondria from soybean cotyledons; however, the ubiquinone contents at d 20 were as low as values reported for mitochondria from soybean roots (Ribas-Carbo et al., 1995, 1997).
Figure 4.
Factors that may affect alternative pathway activity during development of soybean cotyledons. A, Changes in the amounts of ubiquinone 9 and 10 (Q9 and Q10) in the membranes of cotyledon mitochondria. Results from one of two independent experiments are shown. B, Mitochondrial malic enzyme activity during cotyledon development.
Alternative pathway activity is also affected by organic acid activators, as shown in Figure 3C. Pyruvate concentrations in whole cotyledons were below the level of detection by conventional techniques (data not shown); however, it is the presence of intramitochondrial pyruvate that is important for AOX activity (Day et al., 1994; Millar et al., 1996), and this can be either produced in the matrix by NAD-dependent malic enzyme or can enter from the cytosol via a transporter (Douce and Neuburger, 1989). In isolated mitochondria, malic enzyme activity is correlated with intramitochondrial pyruvate generation and consequent AOX activation (Day et al., 1994; Vanlerberghe et al., 1995). Figure 4B shows the developmental changes in the activity of malic enzyme in mitochondria from soybean cotyledons. Activity was detected at all ages and was low only in very young cotyledons (before d 5). The presence of malic enzyme activity throughout cotyledon development demonstrates the sustained potential for intramitochondrial production of pyruvate and the subsequent activation of AOX.
DISCUSSION
The results presented here agree with previous reports showing that mitochondrial respiration rates and AOX protein levels in soybean cotyledons change during seedling growth (Tuquet and Dizengremel, 1984; Azcón-Bieto et al., 1989; Obenland et al., 1990). We have now shown that the increase in AOX protein accompanies changes in AOX transcript levels, indicating a regulation of AOX transcription or mRNA turnover during seedling growth. In particular, our results indicate that there is a developmental regulation of the level of transcripts for different AOX genes, with AOX2 transcripts (and protein) being more abundant early in seedling growth and those for AOX3 appearing later. Since AOX2 protein levels remain more or less constant in the later stages, when transcript levels are barely detectable, it appears that AOX protein does not turn over rapidly in cotyledons.
The increase in AOX transcripts and protein was correlated with an increase in AOX capacity in isolated mitochondria if pyruvate was present to activate the enzyme. It therefore appears that some coarse control of respiration at the protein/RNA level can occur during soybean seedling growth. Conversely, Cyt oxidase activity and capacity declined appreciably during cotyledon senescence. Despite an overall decline during seedling growth, Cyt chain capacity was still approximately equal to or greater than that of AOX in mitochondria from 20-d-old cotyledons (Fig. 3). However, preliminary estimates of oxidase activity in intact cotyledons (using oxygen-discrimination techniques; Robinson et al., 1995) indicate that at this stage of growth more oxygen uptake occurred via AOX than via Cyt oxidase (A.H. Millar, G. Farquhar, and D.A. Day, unpublished data). This implies that metabolic control of Cyt oxidase (e.g. by adenylate levels) also occurs in vivo in soybean cotyledons.
Whether metabolic control of AOX activity also occurred is difficult to judge. It is probable that intramitochondrial pyruvate levels were sufficient to activate AOX at all stages of growth; otherwise, AOX would not have been able to compete for electrons with the Cyt path (Hoefnagel et al., 1995), especially in view of the decline in ubiquinone content with time. The high levels of malic enzyme activity, which can generate pyruvate within the mitochondrial matrix at all stages supports this idea. We were unable to assess the redox status of AOX in cotyledons because the high general protein content of the tissue made accurate detection of AOX protein in whole-tissue extracts impossible. However, it should be noted that AOX protein in isolated mitochondria existed as the reduced monomer in addition to the oxidized dimer at all ages (not shown). Although changes in redox status can occur during isolation of mitochondria, to date only oxidation has been demonstrated (Umbach and Siedow, 1997).
The fact that AOX content and activity in older cotyledons increase at the same time that the activity of the Cyt pathway declines suggests that the cotyledon needs to maintain the flow of carbon through its mitochondria (or to oxidize excess cytosolic reductant) without the need for high levels of ATP synthesis. In young cotyledons there is a rapid turnover of storage compounds and a great deal of metabolite export to the growing seedling (Bewley and Black, 1994); therefore, it is likely that this phase requires large quantities of ATP to support these activities. As the reserves are depleted and the cotyledon greens and develops photosynthetic capability, the need for mitochondrial ATP synthesis presumably declines. Under these conditions, the synthesis and activation of AOX would allow for continued oxidation of carbon compounds in the mitochondria in the face of high cytosolic ATP/ADP. This would avoid the oxidative damage that might otherwise occur (Purvis and Shewfelt, 1993; Wagner and Krab, 1995; Millar and Day, 1997). Similar changes in AOX and Cyt oxidase activity have been observed in soybean roots during seedling growth (Millar et al., 1998).
Our results indicate that regulation of AOX gene expression contributes to the developmental increase in AOX respiration in cotyledons. A relationship between increased expression of AOX transcripts and proteins and increased AOX activity has also been observed in other systems (Rhoads and McIntosh, 1992; Vanlerberghe and McIntosh, 1992a, 1994, 1996; Cruz-Hernández and Gómez-Lim, 1995; Aubert et al., 1997). In cotyledons, AOX gene expression may be up-regulated in response to the decline in Cyt pathway function, as seen in other systems upon addition of Cyt pathway inhibitors (Vanlerberghe and McIntosh, 1992a, 1994; Saisho et al., 1997). However, when seedlings were grown under low-light conditions that retarded senescence, Cyt capacity did not decline but AOX capacity nonetheless increased, suggesting that AOX synthesis can occur in vivo independently of changes in Cyt pathway components.
The up-regulation of the alternative pathway seen in this study is a normal developmental process due to the up-regulation of a single AOX gene, not the entire gene family. Similarly, when the alternative pathway in Arabidopsis was up-regulated upon the addition of antimycin A, only a single gene was induced (Saisho et al., 1997). Whatever the signals responsible for the up-regulation, they appear to act on only a single gene, perhaps indicating that multiple pathways lead to the induction of AOX in plant cells. Unlike previous reports, our results using cotyledons demonstrate not only that an increase in expression of an AOX transcript (AOX3) occurs during the up-regulation of AOX activity but that expression of another AOX transcript (AOX2) concomitantly decreases. This shift from the expression of AOX2 to the expression of AOX3 may be the result of enhancing expression of one AOX gene during a period when a general down-regulation of gene expression is under way, and may suggest that the gene products have different properties. Examining the promoter regions of the various genes will give greater insight into these signals and into the roles of AOX in plant cells.
ACKNOWLEDGMENTS
Supply of the AOA antibody by Drs. Tom Elthon and Lee McIntosh is gratefully acknowledged.
Abbreviation:
- AOX
alternative oxidase
Footnotes
This work was supported by Australian Research Council grants to J.W. and D.A.D. and by an Australian postgraduate award from the Australian Commonwealth Government to T.C.M.
LITERATURE CITED
- Albury MS, Dudley P, Watts FZ, Moore AL. Targeting the plant alternative oxidase protein to Schizosaccharomyces pombe mitochondria confers cyanide-insensitive respiration. J Biol Chem. 1996;271:17062–17066. doi: 10.1074/jbc.271.29.17062. [DOI] [PubMed] [Google Scholar]
- Aubert S, Bligny R, Day DA, Whelan J, Douce R. Induction of alternative oxidase synthesis by herbicides inhibiting branched-chain amino acid synthesis. Plant J. 1997;11:649–657. [Google Scholar]
- Azcón-Bieto J, Salom CL, Mackie ND, Day DA. The regulation of mitochondrial activity during greening and senescence of soybean cotyledons. Plant Physiol Biochem. 1989;27:827–836. [Google Scholar]
- Bewley JD, Black M (1994) Seeds: Physiology of Development and Germination, Ed 2. Plenum Press, New York
- Chaudhuri M, Ajayi W, Temple S, Hill GC. Identification and partial purification of a stage-specific 33 kDa mitochondrial protein as the alternative oxidase of Trypanosoma brucei brucei bloodstream trypomastigotes. J Euk Microbiol. 1995;42:467–472. doi: 10.1111/j.1550-7408.1995.tb05892.x. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cruz-Hernández A, Gómez-Lim MA. Alternative oxidase from mango (Mangifera indica, L.) is differentially regulated during fruit ripening. Planta. 1995;197:569–576. doi: 10.1007/BF00191562. [DOI] [PubMed] [Google Scholar]
- Day DA, Millar AH, Wiskich JT, Whelan J. Regulation of alternative oxidase activity by pyruvate in soybean mitochondria. Plant Physiol. 1994;106:1421–1427. doi: 10.1104/pp.106.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DA, Neuberger M, Douce R. Activation of NAD-linked malic enzyme in intact plant mitochondria by exogenous coenzyme A. Arch Biochem Biophys. 1984;231:233–242. doi: 10.1016/0003-9861(84)90383-7. [DOI] [PubMed] [Google Scholar]
- Day DA, Neuberger M, Douce R. Biochemical characterisation of chlorophyll-free mitochondria from pea leaves. Aust J Plant Physiol. 1985;12:219–228. [Google Scholar]
- Day DA, Whelan J, Millar H, Siedow JN, Wiskich JT. Regulation of the alternative oxidase in plants and fungi. Aust J Plant Physiol. 1995;22:497–509. [Google Scholar]
- Douce R, Neuburger M. The uniqueness of plant mitochondria. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:371–414. [Google Scholar]
- Elthon TE, Nickels RL, McIntosh L. Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol. 1989;89:1311–1317. doi: 10.1104/pp.89.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan PM, Whelan J, Millar AH, Zhang Q, Smith MK, Wiskich JT, Day DA. Differential expression of the multigene family encoding the soybean mitochondrial alternative oxidase. Plant Physiol. 1997;114:455–466. doi: 10.1104/pp.114.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard J, Dizengremel P. Properties of mitochondria isolated from greening soybean and lupin tissues. Plant Sci. 1988;56:1–7. [Google Scholar]
- Hiser C, McIntosh L. Alternative oxidase of potato is an integral membrane protein synthesized de novo during aging of tuber slices. Plant Physiol. 1990;93:312–318. doi: 10.1104/pp.93.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiser C, McIntosh L (1994) Potato alternative oxidase: detection of mRNA by PCR and tissue-specific differences in the protein levels. In WR Belknap, ME Vayda, WD Park, eds, The Molecular and Cellular Biology of the Potato, Ed 2. CAB International, Wallingford, CT, pp 143–150
- Hoefnagel MHN, Millar AH, Wiskich JT, Day DA. Cytochrome and alternative respiratory pathways compete for electrons in the presence of pyruvate in soybean mitochondria. Arch Biochem Biophys. 1995;318:394–400. doi: 10.1006/abbi.1995.1245. [DOI] [PubMed] [Google Scholar]
- Ito Y, Saisho D, Nakazono M, Tsutsumi N, Hirai A. Transcript levels of tandem-arranged alternative oxidase genes in rice are increased by low temperature. Gene. 1997;203:121–129. doi: 10.1016/s0378-1119(97)00502-7. [DOI] [PubMed] [Google Scholar]
- Kearns A, Whelan J, Young S, Elthon TE, Day DA. Tissue-specific expression of the alternative oxidase in soybean and siratro. Plant Physiol. 1992;99:712–717. doi: 10.1104/pp.99.2.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar AM, Söll D. Arabidopsis alternative oxidase sustains Escherichia coli respiration. Proc Natl Acad Sci USA. 1992;89:10842–10846. doi: 10.1073/pnas.89.22.10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lennon AM, Pratt J, Leach G, Moore AL. Developmental regulation of respiratory activity in pea leaves. Plant Physiol. 1995;107:925–932. doi: 10.1104/pp.107.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurements with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- McIntosh L. Molecular biology of the alternative oxidase. Plant Physiol. 1994;105:781–786. doi: 10.1104/pp.105.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Atkin OK, Menz RI, Henry B, Farquhar G, Day DA. Analysis of respiratory chain regulation in roots of soybean seedlings. Plant Physiol. 1998;117:1083–1093. doi: 10.1104/pp.117.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Day DA. Alternative solutions to radical problems. Trends Plant Sci. 1997;2:289–290. [Google Scholar]
- Millar AH, Finnegan PM, Whelan J, Drevron JJ, Day DA. Expression and kinetics of the mitochondrial alternative oxidase in nitrogen-fixing nodules of soybean roots. Plant Cell Environ. 1997;20:1273–1282. [Google Scholar]
- Millar AH, Hoefnagel MHN, Day D, Wiskich J. Specificity of the organic acid activation of alternative oxidase in plant mitochondria. Plant Physiol. 1996;111:613–618. doi: 10.1104/pp.111.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Wiskich JT, Whelan J, Day DA. Organic acid activation of the alternative oxidase of plant mitochondria. FEBS Lett. 1993;329:259–262. doi: 10.1016/0014-5793(93)80233-k. [DOI] [PubMed] [Google Scholar]
- Morohashi Y, Seto T, Matsushima H. Appearance of alternative respiration in cucumber cotyledon mitochondria after treatment with cycloheximide. Physiol Plant. 1991;83:640–646. [Google Scholar]
- Obenland D, Diethelm R, Shibles R, Stewart C. Relationship of alternative oxidase respiratory capacity and alternative oxidase amount during soybean seedling growth. Plant Cell Physiol. 1990;31:897–901. [Google Scholar]
- Puissant C, Houdebine L-M. An improvement of the single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Biotechniques. 1990;8:148–149. [PubMed] [Google Scholar]
- Purvis AC, Shewfelt RL. Does the alternative pathway ameliorate chilling injury in sensitive plant tissues? Physiol Plant. 1993;88:712–718. doi: 10.1111/j.1399-3054.1993.tb01393.x. [DOI] [PubMed] [Google Scholar]
- Rhoads DM, McIntosh L. Salicylic acid regulation of respiration in higher plants: alternative oxidase expression. Plant Cell. 1992;4:1131–1139. doi: 10.1105/tpc.4.9.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas-Carbo M, Lennon AM, Robinson SA, Giles L, Berry JA, Siedow JN. The regulation of electron partitioning between the cytochrome and alternative pathways in soybean cotyledon and root mitochondria. Plant Physiol. 1997;113:903–911. doi: 10.1104/pp.113.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas-Carbo M, Wiskich JT, Berry JA, Siedow JN. Ubiquinone redox behaviour in plant mitochondria during electron transport. Arch Biochem Biophys. 1995;317:156–160. doi: 10.1006/abbi.1995.1148. [DOI] [PubMed] [Google Scholar]
- Robinson SA, Ribas-Carbo M, Yakir D, Giles L, Reuveni Y, Berry JA. Beyond SHAM and cyanide: opportunities for studying the alternative oxidase in plant respiration using oxygen isotope discrimination. Aust J Plant Physiol. 1995;22:487–496. [Google Scholar]
- Saisho D, Nambara E, Naito S, Tsutsumi N, Hirai A, Nakazono M. Characterization of the gene family for alternative oxidase from Arabidopsis thaliana. Plant Mol Biol. 1997;35:585–596. doi: 10.1023/a:1005818507743. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Siedow JN, Umbach AL. Plant mitochondrial electron transfer and molecular biology. Plant Cell. 1995;7:821–831. doi: 10.1105/tpc.7.7.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuquet C, Dizengremel P. Changes in respiratory processes in soybean cotyledons during development and senescence. Z Pflanzenphysiol. 1984;114:355–359. [Google Scholar]
- Umbach AL, Siedow JN. Covalent and noncovalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiol. 1993;103:845–854. doi: 10.1104/pp.103.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN. The reaction of the soybean cotyledon mitochondrial cyanide-resistant oxidase with sulfhydryl reagents suggests that α-keto acid activation involves the formation of a thiohemiacetal. J Biol Chem. 1996;271:25019–25026. doi: 10.1074/jbc.271.40.25019. [DOI] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN. Changes in the redox state of the alternative oxidase regulatory sulfhydryl/disulfide system during mitochondrial isolation: implications for inferences of activity in vivo. Plant Sci. 1997;123:19–28. [Google Scholar]
- Umbach AL, Wiskich JT, Siedow JN. Regulation of alternative oxidase kinetics by pyruvate and intermolecular disulfide bond redox status in soybean seedling mitochondria. FEBS Lett. 1994;348:181–184. doi: 10.1016/0014-5793(94)00600-8. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, Day DA, Wiskich JT, Vanlerberghe AE, McIntosh L. Alternative oxidase activity in tobacco leaf mitochondria: dependence on tricarboxylic acid cycle-mediated redox regulation and pyruvate activation. Plant Physiol. 1995;109:353–361. doi: 10.1104/pp.109.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L. Coordinate regulation of cytochrome and alternative pathway respiration in tobacco. Plant Physiol. 1992a;100:1846–1851. doi: 10.1104/pp.100.4.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L. Lower growth temperature increases alternative pathway capacity and alternative oxidase protein in tobacco. Plant Physiol. 1992b;100:115–119. doi: 10.1104/pp.100.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L. Mitochondrial electron transport regulation of nuclear gene expression: studies with the alternative oxidase gene of tobacco. Plant Physiol. 1994;105:867–874. doi: 10.1104/pp.105.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L. Signals regulating the expression of the nuclear gene encoding alternative oxidase of plant mitochondria. Plant Physiol. 1996;111:589–595. doi: 10.1104/pp.111.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AM, Krab K. The alternative respiration pathway in plants: role and regulation. Physiol Plant. 1995;95:318–325. [Google Scholar]
- Whelan J, Millar AH, Day DA. The alternative oxidase is encoded in a multigene family in soybean. Planta. 1996;198:197–201. doi: 10.1007/BF00206244. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Mischis L, Wiskich JT. Respiratory responses of pea and wheat seedlings to chloramphenicol treatment. Aust J Plant Physiol. 1996;23:583–592. [Google Scholar]