Abstract
When plasminogen binds to cells its activation to plasmin is markedly enhanced compared to the reaction in solution. Thus, cells become armed with the broad spectrum proteolytic activity of plasmin. Cell-surface plasmin plays a key role in macrophage recruitment during the inflammatory response. Proteins exposing basic residues on the cell surface promote plasminogen activation on eukaryotic cells. We have used a proteomics approach combining targeted proteolysis with carboxypeptidase B and multidimensional protein identification technology, MudPIT, and a monocyte progenitor cell line to identify a novel transmembrane protein, the plasminogen receptor, Plg-RKT. Plg-RKT exposes a C-terminal lysine on the cell surface in an orientation to bind plasminogen and promote plasminogen activation. Here we review the characteristics of this new protein, with regard to membrane topology, conservation of sequence across species, the role of its C-terminus in plasminogen binding, its function in plasminogen activation, cell migration, and its role in macrophage recruitment in the inflammatory response.
1. Introduction
When plasminogen binds to cells its activation is markedly enhanced, compared to the reaction in the solution phase [1–7]. Active plasmin remains associated with the cell surface where its activity is protected from inhibitors [8, 9]. Localization of plasminogen on cell surfaces is a crucial control point for positive regulation of cell surface plasmin proteolytic activity that facilitates both physiological and pathological processes [10, 11], notably, macrophage recruitment during the inflammatory response [12–15]. Studies in plasminogen deficient mice have demonstrated that plasminogen plays a key role in macrophage recruitment in response to inflammatory stimuli. Plasmin-dependent cell migration is accomplished by direct degradation of extracellular matrix components by plasmin and also by activation of matrix metalloproteinases for further degradation of extracellular matrices [12–14, 16].
Among plasminogen-binding proteins, those exposing C-terminal basic residues on cell surfaces are predominantly responsible for the ability of eukaryotic cells to enhance plasminogen activation because carboxypeptidase B (CpB) treatment abrogates cell-surface-dependent plasminogen activation [7]. Furthermore, plasminogen-dependent macrophage recruitment is mediated by CpB-sensitive plasminogen-binding sites [21].
Several plasminogen-binding proteins with established intracellular functions that are synthesized with C-terminal lysines are known to associate with the monocytoid cell surface (e.g., α-enolase [22, 23], TIP49a [24], histone H2B, and p11 [25]). Other functional plasminogen binding proteins that are not synthesized with C-terminal basic residues are present on monocytoid cells, including annexin II [26], amphoterin [27], tissue factor [28] and αMβ2 [29]. However, no integral membrane plasminogen binding proteins synthesized with C-terminal basic residues had previously been identified. The identification of a receptor with such a structure would constitute a novel mechanism for stimulating plasminogen activation because its induction would endow cells with the ability to bind plasminogen and promote plasminogen activation, without requiring release and re-binding of intracellular proteins or proteolytic cleavage of a membrane protein to reveal C-terminal basic residues.
2. Rationale for the Use of a Proteomics Approach to Identify Integral Membrane Plasminogen Receptor(s) with C-Terminal Basic Residues
Previous characteristics of plasminogen binding proteins and the methods used for their identification may have precluded identification of an integral membrane plasminogen binding protein with a C-terminal basic residue. Previously, the identification of plasminogen receptors has relied on cell surface labeling followed by affinity chromatography on plasminogen-Sepharose columns and N-terminal sequencing of fractions eluted from SDS gels. Thus, many intracellular proteins that are also present on the cell surface were readily identified because protein fractions that bound to plasminogen Sepharose included the labeled, surface-associated protein, as well as nonlabeled and relatively abundant intracellular protein. Thus, a lower abundance integral membrane plasminogen-binding protein might not have been detectable.
Previously, we used a proteomics approach to examine monocytoid cell membranes for the presence of proteins exposing carboxyl terminal lysines on the extracellular side of the cell membrane [30]. We compared plasminogen ligand blots of 2D gels of membrane fractions of intact cells treated with CpB with untreated membranes. We eluted a prominent CpB-sensitive protein from the gels and obtained two peptide sequences using tandem mass spectrometry. Both peptide sequences were contained within TATA-binding protein-interacting protein (TIP49a) [24]. However, TIP49a is a member of the class of cell surface plasminogen-binding proteins synthesized with a C-terminal lysine and also having intracellular functions and is not an integral membrane protein.
The method used to identify TIP49a and other plasminogen receptors required elution of candidate proteins from 2D SDS polyacrylamide gels. However, many membrane proteins do not resolve well on SDS polyacrylamide gels. Therefore, recently we used an isolation method that used column chromatography instead of SDS polyacrylamide gels. We took advantage of the exquisite sensitivity of multidimensional protein identification technology (MudPIT) [31] to search for integral membrane plasminogen receptor(s) exposing a C-terminal basic residue on the cell surface.
3. Discovery of a Regulated Integral Membrane Plasminogen Receptor Exposing a C-Terminal Basic Amino Acid on the Cell Surface
First, to establish a system in which plasminogen receptors could be actively induced, we tested the effect of a differentiation-inducing agonist, macrophage colony stimulating factor (M-CSF) on plasminogen binding to a mouse monocyte progenitor cell line, Hoxa9-ER4. The Hoxa9-ER4 cell line is derived from primary murine bone marrow myeloid precursors immortalized with an estrogen-regulated conditional oncoprotein, HoxA94-ER [32]. The Hoxa9-ER4 line is factor dependent (GM-CSF) and differentiates to monocytes when estrogen is removed from the medium, thereby inactivating the Hoxa9-ER protein. The mature monocytes respond to M-CSF [33]. We could not detect specific plasminogen binding to undifferentiated Hoxa9-ER4 progenitor cells. However, as the cells differentiated along the monocytic pathway in response to M-CSF, specific plasminogen binding to the cells was observed [18].
We used specific proteolysis followed by MudPIT to probe the membrane proteome of differentiated, M-CSF-treated Hoxa9-ER4 cells for the presence of integral membrane plasminogen receptor(s) exposing a C-terminal basic residue on the cell surface, as outlined in Figure 1. First, the Hoxa9-ER4 monocyte progenitor cells were differentiated with M-CSF to induce plasminogen receptor expression [18]. Then intact cells were biotinylated using a biotinylation reagent that reacts with carboxyl groups, rather than basic groups (that would interfere with the plasminogen-binding function of C-terminal basic residues). Because early apoptotic and nonviable/necrotic cells exhibit markedly enhanced plasminogen-binding ability [34–36] we wished to focus on plasminogen receptors on viable cells and, therefore, passed the biotinylated cells over a dead cell removal column to enrich for live cells. The cells were then lysed and membrane fractions prepared and passed over a plasminogen-Sepharose affinity column and specifically eluted with ε-aminocaproic acid (EACA), a lysine analog that blocks the binding of plasminogen to cells [1]. Biotinylated cell surface proteins bound to the avidin column and were digested with trypsin while still on the column. The peptide digest was then subjected to MudPIT.
Figure 1.
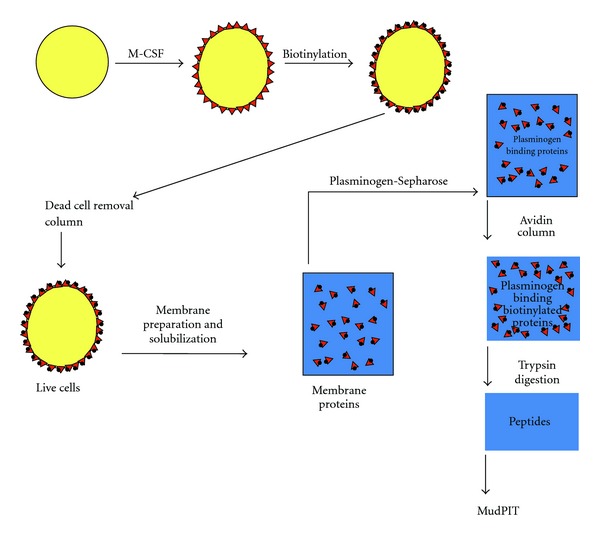
Isolation of plasminogen receptors. Monocyte (Hoxa9-ER4) progenitor cells were differentiated with macrophage colony stimulating factor (M-CSF), which induces plasminogen receptors (▲) on these cells. Then intact cells were biotinylated (●) and passed over a dead cell removal column. Live cells were then lysed and membrane fractions prepared and passed over a plasminogen-Sepharose affinity column and specifically eluted. Biotinylated plasminogen receptors (▲●) were then bound to an avidin column and digested with trypsin. This figure was originally published in [17].
In MudPIT, the peptide mixtures were first resolved by strong cation exchange liquid chromatography upstream of reversed phase liquid chromatography. The eluting peptides were electrosprayed onto an LTQ ion trap mass spectrometer and full MS spectra were recorded over a 400–1600 m/z range, followed by three tandem mass events. The resulting spectra were searched against a mouse protein database. Only one protein with a predicted transmembrane sequence and a C-terminal basic residue was identified: the hypothetical protein, C9orf46 homolog (IPI00136293), homologous to the protein predicted to be encoded by human chromosome 9, open reading frame 46. We have designated the protein, Plg-RKT, to indicate a plasminogen receptor with a C-terminal lysine and having a transmembrane domain (see below).
A key advantage of MudPIT is that proteins in a given proteome can be identified simultaneously. As proof of principle of our isolation method, peptides corresponding to other proteins previously identified as plasminogen-binding proteins on monocytes were also detected in the membrane preparations: α-enolase, gamma actin, S100A10, annexin 2, histone H2B, and β2 integrin. (A limitation of shotgun proteomics, such as MudPIT, is that they typically undersample a proteome because they use data-dependent data acquisition (a computer-driven data acquisition approach). This can lead to variations in the proteins identified, particularly among the lower abundance proteins. Thus, we cannot exclude the possibility that other membrane proteins exposing C-terminal basic residues were present in the membrane proteome.)
4. Conservation of the Plg-RKT Sequence
The C9orf46 homolog/Plg-RKT murine DNA sequence encodes a protein of 147 amino acids with a molecular mass of 17,261 Da and a C-terminal lysine (Table 1, first line). We blasted the C9orf46 homolog/Plg-RKT sequence against all species using NCBI Blast and obtained unique human, rat, dog, cow, giant panda, gibbon, horse, pig, rabbit, and rhesus monkey predicted orthologs, with high identity (e.g., human versus chimpanzee = 99% identity) and no gaps in the sequence (Table 1). Of key importance, a C-terminal lysine was predicted for all of the mammalian orthologs obtained in the blast search. In a query of the Ensembl Gene Report, DNA sequences of all 10 other sequenced mammalian orthologs encoded C-terminal lysines (Table 1).
Table 1.
Alignment of Mammalian Orthologs of Plg-RKT.
| 1 0 | 20 | 30 | 40 | 50 | 60 | 70 | 8 0 | |
|---|---|---|---|---|---|---|---|---|
| Mouse | MGFIFSKSMN | ENMKNQQEFM | VTHARLQLER | HLTMQNEMRE | RQMAMQIAWS | REFLKYFGTF | FGIATISLAT | GALKRKKPAF |
| Human | MGFIFSKSMN | ESMKNQKEFM | LMNARLQLER | QLIMQSEMRE | RQMAMQIAWS | REFLKYFGTF | FGLAAISLTA | GAIKKKKPAF |
| Rat | MGFIFSKSMN | ENMKNQQEFM | VMHARLQLER | QLIMQNEMRE | RQMAMQIAWS | REFLKYFGTF | FGIATISLAA | GAIKRKKPAF |
| Dog | MGFIFSKSMN | ENMKNQQEFM | LMNARLQMER | QLMMQNEMRE | RQMAMQIAWS | REFLKYFGTF | FGIAAISLTA | GAIRKKKPAF |
| Cow | MGFIFSKSMN | ENLKSQQEFM | LMNSRLQLER | QLIMQNEMRE | RQMAMQIAWS | REFLKYFGTF | FGITAVSLTA | GAIKGKKPVL |
| Alpaca | MGFIFSKSMN | ENMKSQQEFM | LMNARLQLER | QLMMQNEMRE | RQMAMQIAWS | REFLKYFGTF | FGIAAISLTA | GAIKRKKPAF |
| Chimpanzee | MGFIFSKSMN | ESMKNQKEFM | LMNARLQLER | QLIMQSEMRE | RQMAMQIAWS | REFLKYFGTF | FGLAAISLTA | GAIKKKKPAF |
| Dolphin | MGFIFSKSMN | ENMKSQQEFM | LMNARLQLER | QLMMQNETRE | RQMAMQIAWS | REFLKYFGTF | FGIAAISLTA | GAIKKKKPAF |
| Gibbon | MGFIFSKSMN | ESMKNQKEFM | LMNARLQLER | QLIMQSEMRE | RQMAMQIAWS | REFLKYFGTF | FGLAAISLTA | GAIKKKKPAF |
| Guinea Pig | MGFMLSKSMN | ENMKNQQEFM | LMNARLQLER | QLLLQNEMRE | RQMAMQIAWS | REFLKYFGTF | FGISAISLTA | RAIKQKKPAF |
| Horse | MGFIFSKSMN | ENMKNQQEFM | LMNARLQLER | QLTMQNEMRE | RQMAMQIAWS | REFLKYFGTF | FGIAAISLTA | GALKRKKPAF |
| Lemur | MGFIFSKSMK | ENAQNQQEFM | LMNARLQLER | QLTMQNEMRE | RQMAMQIAWS | REFMKYFGTF | FGITAISLTA | GAIKSKKPGF |
| Opossum | MGFLFSKHMN | ENMKQQQEFM | LMNARLQMER | QLTIQNEMRE | RQMAMQIAWT | REFLKYFGTF | FGIAAISLTA | GAIKKKQPGL |
| Orangutan | MGFIFSKSMN | ESMKNQKEFM | LMNARLQLER | QLIMQSEMRE | RQMAMQIAWS | REFLKYFGTF | FGLAAISLTA | GAIKKKKPAF |
| Panda (Giant) | MGFIFSKSMS | ENMKNQQEFM | LMNARLQLER | QLMMQNEMRE | RQMALQIAWS | REFLKYFGTF | FGITAISLTA | GAIRRKKPAF |
| Pig | MGFIFSKSMN | ENMKRQQEFM | LMNTRLQLER | QLIMQNEMRE | RQMAMQIAWS | REFLKYFGTF | FGIASVALTA | GAIKRKKPAF |
| Rabbit | MGFIFSKSMN | ENLKNQQEFM | LMNARLQLER | QLMLQNEMRE | RQMAMQIAWS | REFLKYFGTF | FGVATISLTA | GAMRRKKPAF |
| Rhesus Monkey | M GFIFSKSMN | ESMKNQKEFM | LMSARLQLER | QLIMQSEMRE | RQMAMQIAWS | REFLKYFGTF | FGFAAISLTA | GAIKKKKPAF |
| Tarsier | MGFIF- KSMN | ENMKHQQEFM | LMNAQLQLER | QLTMQNEMRE | RQMAMQIAWS | REFLKYFGTF | FGITAISLTA | GAIKRKKPAL |
|
| ||||||||
| Tree Shrew | MGFIFSKSMN | ENMKNQQEFM | LMNARLQLER | QLMMQNEMRE | RQMAMQIAWS | REFLKYFGTF | FGIAAISLTA | GAIKKKNPAF |
|
| ||||||||
| 90 | 100 | 110 | 120 | 130 | 140 | 147 | ||
|
| ||||||||
| Mouse | LVPIVPLSFI | FTYQYDLGYG | TLLQRMKSEA | EDILETEKTK | LELPKGLITF | ESLEKARREQ | SKLFSDK | |
| Human | LVPIVPLSFI | LTYQYDLGYG | TLLERMKGEA | EDILETEKSK | LQLPRGMITF | ESIEKARKEQ | SRFFIDK | |
| Rat | LIPIVPLSFI | FTYQYDLGYG | TLLQRMKSEA | EDILETEKTK | LELPKGLITF | ESLEKARREQ | SKFFSDK | |
| Dog | LFPIIPLSFI | FTYQYDLGYG | TLLQRMKGEA | ENILETEKSK | LQLPRGMITF | ESLEKARREQ | SKFFIDK | |
| Cow | IFPIVPLGFV | LAYQYDMGYG | TLIHRMKGEA | ENILETEKSK | LQLPKGMITF | ESLEKARKEQ | SKFFIDK | |
| Alpaca | IFPIVPLGFV | LTYQFDLGYG | TLLQRMKGEA | ENILETEKSK | LQLPKGIITF | ESLEKARKEQ | SKFFIDK | |
| Chimpanzee | LVPIVPLSFI | LTYQYDLGYG | TLLERMKGEA | EDILETEKSK | LQLPRGMITF | ESIEKARKEQ | SRFFIDK | |
| Dolphin | VFPIVPLGFV | LAYQYDMGYG | TLIQRMKGEA | DNILETEKSK | LQLPKGMITF | ENLEKARREQ | SKFFIDK | |
| Gibbon | LVPIVPLSFI | LTYQYDLGYG | TLLERMKGEA | EDILETEKSK | LQLPRGMITF | ESIEKARKEQ | SKFFIDK | |
| Guinea Pig | FIPIVPLSFV | LAYQYDLGYG | TLLQRMKGEA | EDILETEKNK | LELPKGVITF | ESLEKARREQ | SKFFLGK | |
| Horse | LFPIVPLGFV | LTYQYDLGYG | TLLQRMKGEA | ENILETEKSK | LQLPKGMITF | ESLEKARREQ | SKFFIDK | |
| Lemur | LFPIVPLSFV | LAYQYDLGYG | TLLQRMKGEA | EDILETEKSK | LQLPKGMITF | ESLEKARREQ | SKFFIEK | |
| Opossum | FFPIVPLSFI | LAYQYDMGYG | TLLQRMKGEA | ENILETENSK | LQLPRGSITF | ETLEKARKAQ | SKFFIEK | |
| Orangutan | LVPIVPLSFI | LTYQYDLGYG | TLLERMKGEA | EDILETEKSK | LQLPRGMITF | ESIEKARKEQ | SRFFIDK | |
| Panda (Giant) | LFPIIPLSFI | FTYQYDLGYG | TLLQRMKGEA | ENILETEKSK | LQLPRGMITF | ENLEKARREQ | SKFFIDK | |
| Pig | FLPIIPLGFV | FTYQYDLGYG | TLLQRMKGEA | ENILETETSK | LQLPKGMITF | EGLEKARREQ | SKFFIDK | |
| Rabbit | LLPIVPLSFI | FIYQCDLGYG | TLLQRMKGEA | EDILETEKSK | LQLPGGMITF | ESLEKARREQ | SKFFIDK | |
| Rhesus Monkey | LVPIVPLSFI | LTYQYDLGYG | TLLERMKGEA | EDILETEKSK | LQLPRGMITF | ESIEKARKEQ | SKFFIDK | |
| Tarsier | LLPIVPLSFI | FTYQYDLGYG | TLLERMKGEA | EEILEAEKNM | LQLPKGMITF | ESLEKTRREQ | SKFFTDK | |
| Tree Shrew | FFPIVPLSFI | LTYQYDLGYG | TLLPRMKSEA | EDILETEKSK | LELPRGMITF | ESLEKARREQ | SKFFVDK | |
In addition to mammals, the DNA sequences of xenopus and the green lizard also encoded C-terminal lysines (Table 2). Furthermore, Plg-RKT orthologs with 149 amino acids with a C-terminal lysine were encoded in bony fish (e.g., zebrafish) and the high similarity with a mammalian ortholog is illustrated in the alignment with the mouse protein in Table 2.
Table 2.
Alignment of Mouse, Lizard (arborial), Frog (xenopus), and Zebrafish Plg-RKT Sequences.
| Mouse | 1 | MGFIFSKSMNENMKNQQEFMVTHARLQLERHLTMQNEMRERQMAMQIAWSREFLKYFGTF | 60 |
| Lizard | 1 | MGFIFSKSMNENLKNQQEFMIMNSRLQLERQLLMQNQMRERQMAMQIAWTREFLKYFGAF | 60 |
| Frog | 1 | MGSLISKATETQMKKQQELMQMNAQIQLERQIIMQNMMRERQMAMQIAWSREFLKYYGSF | 60 |
| Zebrafish | 1 | MGFVLSKGMEQNFQKQQEFMLLNARLQLERQLAMQNQMRERQMAMQLAWSREFLKYFGSF | 60 |
|
| |||
| Mouse | 61 | FGIATISLATGALKRKKPAFLVPIVPLSFIFTYQYDLGYGTLLQRMKSEAEDILETEKTK | 120 |
| Lizard | 61 | SGLAAVGLTVGAIKRKKPAFFLPMVPLSFILAYQYDMGYGSLLKRMKSEAESILDTESTT | 120 |
| Frog | 61 | FSLAVIGLTVGAVKNKKPALFTPVIPLTFVFAYQFDMGYGTLVTRMKGEAENILEKEHIL | 120 |
| Zebrafish | 61 | FGLATLGLTVGAVKKRKPALLAPVIPLSFILVYQMDAAYGTMLQRMRAEAESIMVSECER | 120 |
|
| |||
| Mouse | 121 | LELPKGLITFESLEKARREQSKL –– FSDK | 147 |
| Lizard | 121 | LEMPKGPLTFESIEKARRAQSKF –– FIEK | 147 |
| Frog | 121 | LEMPQGLPTFEGIEKTRKAHRSLLL ––– K | 147 |
| Zebrafish | 121 | LDVPHGMPTFESIEKSRRAKAHLTTLTEK | 149 |
The Plg-RKT sequence also encodes a putative conserved DUF2368 domain (encompassing amino acids 1–135), an uncharacterized protein with unknown function conserved from nematodes to humans. Notably, the DNA sequences of Plg-RKT orthologs of lower organisms predicted proteins of different lengths and did not consistently predict C-terminal lysines. It is interesting to note that the evolutionary origin of plasminogen is currently believed to originate with protochordates [37], so that lower organisms without plasminogen would not need the C-terminal lysine of Plg-RKT to bind plasminogen.
Within species, it is noteworthy that the primary sequence of C9orf46/Plg-RKT is apparently tightly conserved in humans, with no validated polymorphisms (cSNPs) within the 6 exons encoded by the gene (on chromosome 9p24.1) in the NCBI human genome sequence variation database (dbSNP, http://www.ncbi.nlm.nih.gov/SNP/).
5. Membrane Topology of Plg-RKT
We analyzed the C9orf46 homolog/Plg-RKT sequence in the TMpred site (http://sourceforge.net/projects/tmpred/files/). The strongly preferred model included two transmembrane helices extending from F53-L73 (secondary helix, oriented from outside the cell to inside the cell) and P78-Y99 (primary helix, oriented from inside the cell to outside the cell) (Figure 2). Thus, a 52 amino acid N-terminal region and a 48 amino acid C-terminal tail with a C-terminal lysine were predicted to be exposed on the cell surface.
Figure 2.
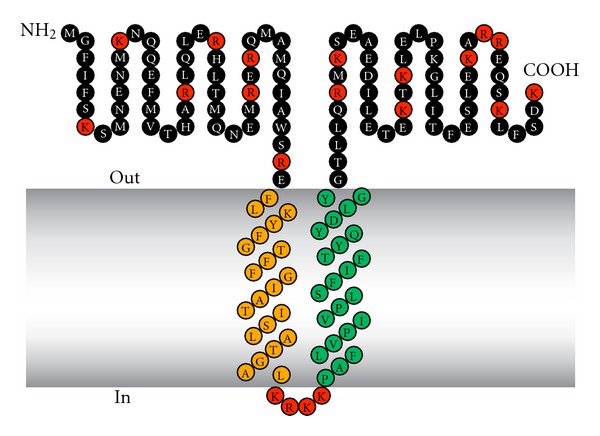
Structural model of Plg-RKT. Green indicates amino acids within the predicted primary transmembrane helix. Orange indicates amino acids within the predicted secondary transmembrane helix. Red indicates basic amino acids. This research was originally published in [18].
We experimentally tested predictions of the topology model. First, we raised a monoclonal antibody against a synthetic peptide, CEQSKLFSDK (corresponding to the nine C-terminal amino acids of murine Plg-RKT with an amino terminal cysteine added for coupling). The mAb reacted with the C-terminal peptide of murine Plg-RKT and blocked plasminogen binding to CEQSKLFSDK. To examine subcellular localization, membrane and cytoplasmic fractions from progenitor and differentiated Hoxa9-ER4 monocyte progenitor cells were electrophoresed and western blotted with anti-Plg-RKT mAb7H1 or isotype control mAb. A specific immunoreactive band migrating with an Mrapp of ~17,000 was detected in membrane fractions of differentiated monocyte progenitor cells, clearly demonstrating the existence of this new protein (Figure 3(a)). Plg-RKT was not detected in undifferentiated cells or in the cytoplasmic fraction of the differentiated cells, also demonstrating that Plg-RKT was an M-CSF-inducible protein.
Figure 3.

Plg-RKT behaves as a regulated integral membrane protein. (a) Membrane fractions or cytoplasmic fractions from either undifferentiated or M-CSF-treated Hoxa9-ER4 cells were electrophoresed on 12% sodium dodecyl sulfate polyacrylamide gels under reducing conditions and western blotted with either anti-Plg-RKT mAb, anti-α-enolase mAb as a loading control, or isotype control mAb. (b) M-CSF-treated Hoxa9-ER4 cell membranes were solubilized in 3% Triton X-114. After heating at 37°C and separation of the phases by centrifugation, an aliquot of both phases was electrophoresed and western blotted with anti-Plg-RKT mAb. This research was originally published in [18].
Because we had initially found Plg-RKT in murine monocyte progenitor cells, we also examined the expression and subcellular localization of Plg-RKT in human peripheral blood monocytes and human monocytoid cells using an anti-Plg-RKT mAb7H1. Plg-RKT (17,261 Da) was also observed in the membrane fractions of human peripheral blood monocytes, U937 cells and THP-1 cells, but was not detected in the cytoplasmic fractions of these cells [20]. Thus, Plg-RKT is markedly expressed in membranes of both normal human peripheral blood monocytes and human monocytoid cell lines. Plg-RKT was detected in other human peripheral blood cells, also. Plg-RKT was highly expressed in lymphocytes and less strongly expressed by granulocytes and was not detectable in rbc [20].
To further test the prediction that Plg-RKT is an integral membrane protein, membranes from differentiated monocyte progenitor cells were subjected to phase separation in Triton X-114 as described [38, 39]. In this method, integral membrane proteins form mixed micelles with the nonionic detergent and are subsequently recovered in the Triton X-114 detergent phase, whereas hydrophilic proteins remain in the aqueous phase. Plg-RKT was detected in the detergent phase in western blotting with anti-Plg-RKT mAb7H1, but was not detected in the aqueous phase (Figure 3(b)). These data support the prediction that Plg-RKT is an integral membrane protein.
To experimentally test whether the C-terminal lysine of Plg-RKT was exposed on the cell surface, we treated intact biotinylated cells with carboxypeptidase B prior to performing our isolation procedure. Under this condition, C-terminal lysines exposed on the cell surface are removed but intracellular C-terminal lysines are protected. Under this condition, no peptides corresponding to Plg-RKT were obtained in the MudPIT analysis, consistent with cell surface exposure of the C-terminal lysine of Plg-RKT.
Other key aspects of our experimental paradigm support the topological model shown in Figure 2. First, recovery of peptides corresponding to C9orf46 homolog relied on accessibility to the biotinylation reagent in the context of intact cells, supporting the exposure of Plg-RKT domains on the cell surface. The biotinylation reagent we used reacts with the carboxyl moieties of the R-groups of either Asp or Glu. Thus, biotinylation of Plg-RKT on intact cells would not occur if the highly basic loop between the transmembrane helices (K74–K77) was exposed on the cell surface. Hence, a model in which the amino and C-termini are both on the cytoplasmic face of the membrane can be excluded.
In order to experimentally evaluate whether the N-terminus as well as the C-terminus of Plg-RKT was exposed on the cell surface, PC12 (rat pheochromocytoma) cells were stably transfected with V5-pCIneo-Plg-RKT that expressed a V5 tag at the N-terminus of Plg-RKT. A specific band migrating with a Mrapp of 17,000 was detected in cell membranes of the stably transfected cells with both anti-V5 mAb, which reacts with the N-terminus and anti-Plg-RKT mAb7H1, which reacts with the C-terminus of Plg-RKT (Figure 4, lane 1). The band was not detected by either mAb after trypsin digestion of isolated membrane fractions (lane 2). When intact cells were incubated with trypsin and the trypsin neutralized with SBTI prior to preparation of the membrane fraction, the majority of the band detectable with either anti-V5 or anti-Plg-RKT was lost (lane 4). (In controls, treatment with soybean trypsin inhibitor (SBTI) fully neutralized the ability of trypsin to degrade the V5-tagged Plg-RKT in purified membrane fractions (lane 3), demonstrating that the trypsin had been neutralized prior to membrane fractionation of the treated cells.) These results suggest that both the N-terminus and C-terminus of Plg-RKT are accessible to trypsin proteolysis of intact cells and are, therefore, exposed on the extracellular face.
Figure 4.
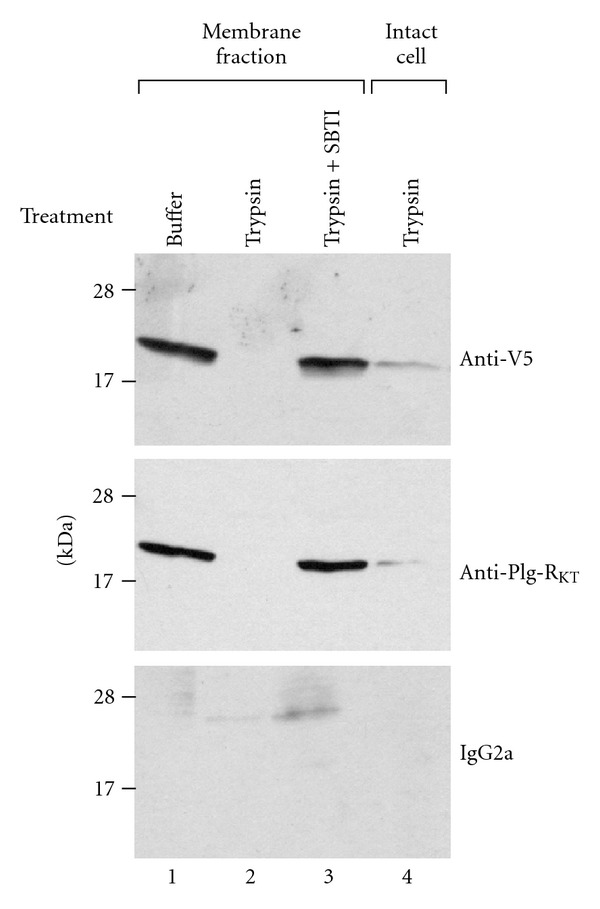
The N-termini and C-termini of Plg-RKT are exposed on the cell surface. Membrane fractions of PC12 cells stably transfected with V5-pCIneo-Plg-RKT were incubated with either buffer (lane 1), trypsin (1 mg/mL) (lane 2), or trypsin 1 mg/mL + soybean trypsin inhibitor (SBTI) (2 mg/mL) (lane 3) for 30 minutes at 37°C or intact PC12 cells were incubated with 1 mg/mL trypsin for 2 hr at 37°C, followed by 2 mg/mL SBTI for 15 min. Following neutralization of trypsin with SBTI, the membrane fraction was prepared from the treated, intact cells (lane 4). 30 μg/lane of membrane fractions was electrophoresed on 18% SDS page under reducing conditions and western blotted with either anti-V5, anti-Plg-RKT mAb, or isotype control. This research was originally published in [17]
6. Role of the C-Terminal Lysine of Plg-RKT in Plasminogen Binding
We further addressed the exposure of the C-terminus of Plg-RKT on the cell surface using confocal microscopy with anti-Plg-RKT mAb7H1 raised against the Plg-RKT C-terminal peptide. When cells were incubated with anti-Plg-RKT mAb and a polyclonal antiplasminogen antibody, Plg-RKT and plasminogen were both immunodetected in small aggregates dispersed over the cell surface [18], in a similar distribution to that published for confocal analyses of monocyte-associated plasminogen [15]. Most importantly, after preincubation of monocytes with plasminogen, immunodetection of Plg-RKT was reduced by half [18]. These results demonstrate that the C-terminus of Plg-RKT is exposed on the cell surface. Furthermore, these results show that plasminogen binds to the C-terminal domain of Plg-RKT on the cell surface.
To further explore the functional importance of the C-terminal lysine, we tested whether a synthetic peptide, corresponding to the C-terminus of Plg-RKT could interact with plasminogen. The peptide, CEQSKLFSDK, was coupled to BSA and coated onto wells of microtiter plates. Glu-plasminogen was incubated with the wells, followed by antiplasminogen mAb [19, 40] and detection with HRP-conjugated goat anti-mouse IgG. Glu-plasminogen bound to the peptide in a concentration-dependent manner, reaching half saturation at a concentration of 7.6 nM (Figure 5(a)). The binding was specific because it was blocked in the presence of EACA, consistent with the ability of EACA to inhibit plasminogen binding to cells. Lys-plasminogen also bound specifically to the peptide (Figure 5(b)) and the concentration for half saturation was ≤2.7 nM, consistent with the higher affinity of Lys-plasminogen compared to Glu-plasminogen for cell surfaces [41, 42]. We also investigated the interaction of t-PA with the C-terminal peptide because t-PA and plasminogen share binding sites on monocytoid cells and t-PA binding to monocytoid cells is sensitive to CpB [43]. Concentration-dependent binding of t-PA to the peptide was observed (Figure 5(c)) and the concentration for 50% saturation was 3.2 nM, consistent with the relative affinities of Glu-plasminogen and t-PA for the cell surface [44]. We noticed that the concentration for 50% saturation of plasminogen binding to immobilized CEQSKLFSDK-coupled to BSA was much greater than the Kd value we had determined for plasminogen binding to cells. The differences in apparent affinity when plasminogen bound to the immobilized peptide compared to the cell surface may have been due to our use of BSA-conjugated peptide to coat the plates. If multiple peptides were conjugated to the BSA that could have provided a higher affinity surface than the cell surface receptor, since plasminogen has multiple kringle domains that may interact in a cooperative lysine-dependent manner with several Plg-RKT peptides on a single BSA molecule. To resolve this issue, we tested the ability of the soluble C-terminal peptide to inhibit Glu-plasminogen binding under solution phase equilibrium conditions. The soluble peptide competed for Glu-plasminogen binding in a dose-dependent manner with an IC50 of 2 μM (Figure 5(d)), similar to the Kd values we had determined for Glu-plasminogen binding to M-CSF-treated Hoxa9-ER4 cells [18]. Furthermore, a mutated peptide with the C-terminal lysine substituted with alanine did not compete for plasminogen binding at concentrations up to 1 mM (Figure 7(d)), further supporting the role of the C-terminal lysine in the interaction of Plg-RKT with plasminogen.
Figure 5.
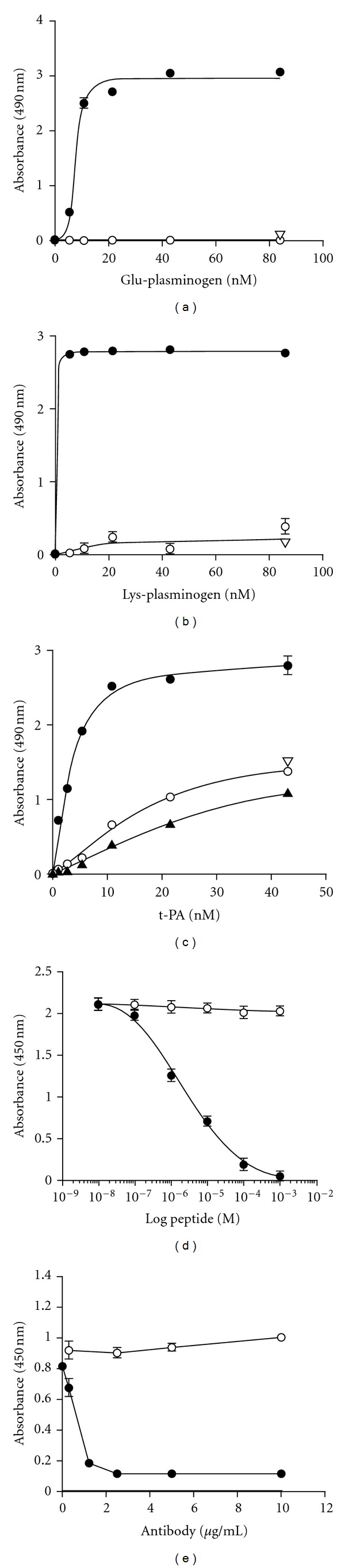
Plasminogen binds to the C-terminal peptide of Plg-RKT. The peptide, CEQSKLFSDK, corresponding to the amino terminus of Plg-RKT was coupled to BSA and coated onto wells of microtiter plates. Either Glu-plasminogen (a) or Lys-plasminogen (b) or t-PA (c) was then incubated with the wells, followed by antiplasminogen mAb [19] (a, b) or anti-t-PA polyclonal mAb (c) and detection with HRP-conjugated goat anti-mouse IgG (closed circles) as described in Materials and Methods. The binding was specific because it was blocked in the presence of 0.2 M EACA (open triangles), consistent with the ability of EACA to inhibit plasminogen binding to differentiated Hoxa9-ER4 cells. In additional controls, nonspecific binding to either BSA (closed triangles) or to the reverse peptide (open circles) was <10% of binding to CEQSKLFSDK. (At high input concentrations of t-PA nonspecific binding increased but was <10% of binding to CEQSKLFSDK at the concentration required for 50% saturation (3.2 nM). In controls for the detection method, O.D.490 values obtained using an isotype control antibody or in the absence of added plasminogen or t-PA were <5% of the values for plasminogen or t-PA binding to immobilized CEQSKLFSDK. Panels d and e: biotinylated-Glu-plasminogen (25 nM) was incubated with immobilized CEQSKLFSDK in the presence of increasing concentrations of (d) the C-terminal peptide, CEQSKLFSDK (closed circles), or a mutated C-terminal peptide with K147 substituted with alanine, CEQSKLFSDA (open circles) or (e) anti-Plg-RKT mAb35B10 (closed circles) or isotype control (open circles). Biotinylated Glu-plasminogen binding was detected with HRP-streptavidin and was 96% inhibited in the presence of 0.2 M EACA (not shown). Data are as mean ± SEM, n = 3, for each determination. This research was originally published in [18].
Figure 7.
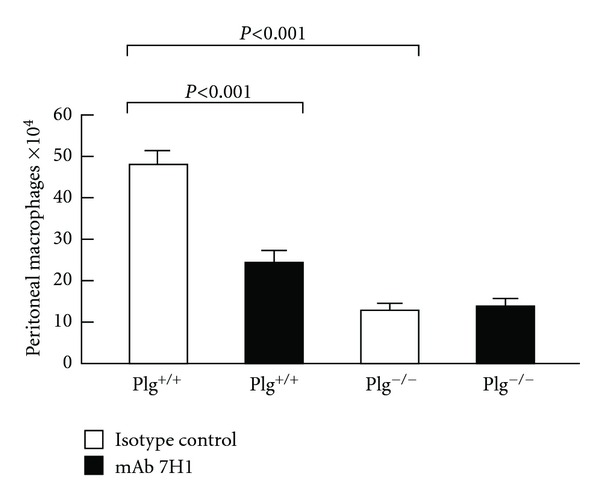
Effect of Plg-RKT on thioglycollate-induced monocyte recruitment. Both plasminogen-deficient (Plg−/−) and wild-type littermates (Plg+/+) mice were injected intravenously with either mAb7H1 (■) or isotype control (□) (500 μg). After 30 minutes thioglycollate was injected intraperitoneally. A second injection of antibody was given 24 hours later. After 72 hours, thioglycollate-recruited cells were collected by peritoneal lavage and macrophages were purified by adherence. The adherent cells were detached and counted using a hemocytometer. Data represent mean ± S.E.M. n = 5/group). This research was originally published in [20].
7. Plg-RKT Regulates Cell Surface Plasminogen Activation
Plasminogen activation was stimulated 12.7-fold in the presence of M-CSF-treated Hox9-ER4 cells, compared to the reaction in the absence of cells and cell-dependent plasminogen activation was stimulated 4.4-fold on differentiated cells, compared to undifferentiated cells (Figure 6(a)). In order to verify the role of Plg-RKT in plasminogen activation, we tested the effect of a monoclonal antibody (anti-Plg-RKT mAb35B10) raised in rats against the synthetic peptide, CEQSKLFSDK. The IgG fraction reacted with the C-terminal peptide of murine Plg-RKT and blocked plasminogen binding to CEQSKLFSDK (Figure 5(e)). Anti-Plg-RKT mAb35B10 substantially suppressed cell-dependent plasminogen activation by 46% and suppressed cell differentiation-dependent plasminogen activation by 58% (Figure 6(a)). In controls, plasminogen activation in the absence of cells or on undifferentiated cells was not affected by anti-Plg-RKT mAb.
Figure 6.
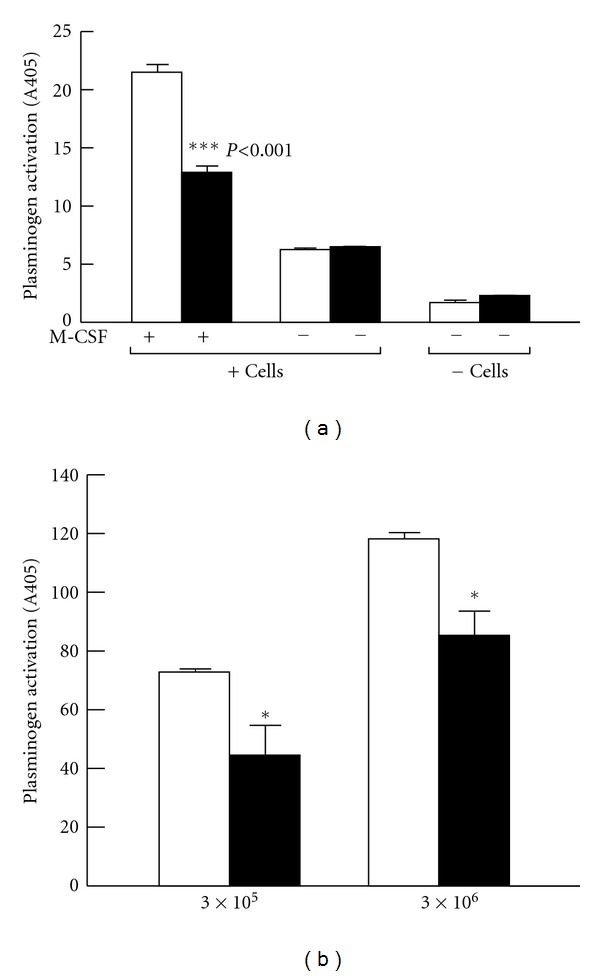
(a) Plg-RKT regulates cell surface plasminogen activation by t-PA. Plasminogen activation was determined after adding 2.7 μM Glu-plasminogen and 20 nM single chain recombinant t-PA in either the presence or absence of either undifferentiated Hoxa9-ER4 progenitor cells or M-CSF-differentiated Hoxa9-ER4 cells and in the presence of either rat anti-Plg-RKT mAb35B10 (filled bars) or isotype control rat IgG2a (open bars). ***P < 0.001, compared to the corresponding isotype control. This research was originally published in [18]. (b) Plg-RKT regulates cell surface plasminogen activation by uPA. Plasminogen activation was determined in the presence of different concentrations of U937 cells, as indicated, and in the presence of 2.7 μM Glu-Plasminogen and 20 nM uPA and in the presence of 170 nM of either anti-Plg-RKT mAb7H1 (filled bars) or mouse IgG2a isotype control (open bars). Cell-dependent plasminogen activation on the tripeptide substrate, S2251, is shown after subtracting plasminogen activation in the absence of cells. **P < .001 compared with the corresponding isotype control. This research was originally published in [20].
We examined whether Plg-RKT colocalized with uPAR, an additional key component of the cell-surface plasminogen activation system. Plg-RKT was markedly colocalized with uPAR on the surfaces of M-CSF-differentiated monocyte progenitor Hoxa9-ER4 cells, as revealed by merged confocal images. The extent of colocalization of Plg-RKT with uPAR was 73 ± 3% [18]. These results suggest that Plg-RKT and uPAR are present in very close proximity on the cell surface in an orientation to promote plasminogen activation.
The kinetically favored substrate for uPAR-bound uPA is cell associated, rather than solution phase plasminogen [4]. Therefore, we tested whether Plg-RKT regulates uPA-dependent cell surface plasminogen activation using an antibody inhibition approach with anti-Plg-RKT mAb7H1. U937 human monocytoid cells were preincubated with either mAb7H1 or IgG2a isotype control, followed by incubation with plasminogen for 30 minutes. Plasmin generation was measured after addition of uPA and a chromogenic substrate for plasmin. Treatment with mAb7H1 markedly suppressed cell-dependent plasminogen activation by uPA and the extent of suppression was cell concentration-dependent. At a concentration of 3 × 105 cells/mL, cell surface dependent plasminogen activation by uPA was suppressed by 39% (Figure 6(b)). Therefore, Plg-RKT plays a major role in uPA-dependent plasminogen activation on monocytoid cells.
8. Regulation of Cell Migration by Plg-RKT
Because Plg-RKT promoted plasminogen activation and cell-associated plasmin plays a role in cell migration, we investigated the role of Plg-RKT in cell migration. Invasion of Matrigel by monocytoid cells in response to the chemotactic stimulus, MCP-1, is enhanced in the presence of plasminogen and also requires active plasmin [15, 45] and uPA [15] and is markedly suppressed in the presence of EACA, suggesting a key role of plasminogen receptors in this function [15, 45]. Plg-RKT played a major role in Matrigel invasion. Treatment of U937 cells or human peripheral blood monocytes with anti-Plg-RKT mAb markedly decreased migration of the cells through Matrigel (by 54% and 48%, respectively) compared to isotype control [20].
Plasmin promotes chemotactic cell migration across polycarbonate membranes in the absence of extracellular matrix [46]. We found that chemotactic migration in the absence of Matrigel was markedly regulated by Plg-RKT. Chemotactic migration was enhanced in the presence of plasminogen [20]. Migration was inhibited 65% by EACA consistent with plasminogen-receptor dependence of this function [20]. Cell migration was also inhibited by amiloride and aprotinin, consistent with a requirement for active plasmin and uPA in chemotactic migration [20]. Chemotactic migration of both U937 cells and human peripheral blood monocytes was maximally reduced by 64% and 39%, respectively, by treatment with anti-Plg-RKT mAb, compared to isotype control [20]. Regulation of chemotactic migration in the absence of extracellular matrix appears to be a unique property of Plg-RKT because other plasminogen receptors do not regulate chemotactic migration [15].
We tested whether, in addition to chemotaxis, Plg-RKT was involved in chemokinesis, that is, nondirectional cell motility in response to MCP-1. Checkerboard analysis showed that in the presence of plasminogen, a constant MCP-1 concentration and a positive concentration gradient induced U937 motility. Both processes were inhibited by anti- Plg-RKT antibody 7H1, with chemokinesis being suppressed almost to a background level [20].
9. Regulation of the Inflammatory Response by Plg-RKT
Having established the presence of Plg-RKT on human monocytoid cells and demonstrated its role in invasion and chemotactic migration in vitro, we examined the role of Plg-RKT in monocyte migration in vivo. Plg+/+ mice were injected intravenously with either mAb7H1 or IgG2a isotype control. After 30 minutes, the mice were injected intraperitoneally with thioglycollate to induce a sterile inflammatory response and a second dose of antibody was given intravenously 24 hours after the thioglycollate injection. After 72 hours (when most recruited leukocytes are macrophages [12, 47, 48]), cells were collected by peritoneal lavage and macrophages were purified by adherence. Macrophage recruitment was significantly (49%) impaired in the mice that were injected with mAb7H1 compared with the mice injected with the isotype control (2.46 × 105 ± 0.28 × 104 for mice injected with mAb7H1 versus 4.82 × 105 ± 0.33 × 104 for mice injected with isotype control, n = 5, P = .00048) (Figure 7) [20]. In 3 other experiments, C57Bl/6J mice injected with 7H1 had a 53 ± 4% reduction in peritoneal macrophages, compared to mice injected with the isotype control.
The decreased response in macrophage recruitment to the peritoneum could not be explained by a decreased level of monocytes in the circulation. Differences in blood levels of monocytes in animals treated with mAb7H1 compared with isotype control were not statistically significant [20].
We also examined the effect of mAb7H1 on recruitment of other leukocytes to the peritoneum. At 6 hr, when recruited granulocyte levels are maximal [12], neutrophil and eosinophil recruitment to the peritoneum was not statistically different in mice treated with mAb7H1 compared with isotype control. Total peritoneal neutrophil recruitment in mice treated with isotype control was 1.73 ± 0.58 × 106 and in mice injected with mAb7H1 was 1.8 ± 0.66 × 106 (n = 5). Total peritoneal eosinophil recruitment in mice injected with isotype control was 1.08 ± 0.4 × 106 and in mice treated with mAb7H1 was 1.45 ± 0.59 × 106 (n = 5). In contrast, there was an effect of mAb7H1 injection on lymphocyte recruitment. Total recruited peritoneal lymphocytes after 72 hr in mice treated with isotype control were 5.57 ± 0.63 × 105 and in mice treated with mAb7H1 were 2.17 ± 0.22 × 105 (n = 4, P = 0.002).
To assess whether the effect of mAb7H1 in the peritonitis model was consistent with the plasminogen binding function of Plg-RKT, we examined the effect of injection of anti-Plg-RKT mAb7H1 in Plg−/− mice. Thioglycollate recruitment in Plg−/− mice injected with isotype control was significantly decreased (by 73%) in Plg−/− compared to Plg+/+ littermates, as reported (56% [12]–65% [16]). When Plg−/− mice were treated with mAb7H1, there was no effect on the remaining macrophage recruitment in Plg−/− mice (Figure 7). Thus, the effect of the anti-Plg-RKT mAb7H1 was entirely dependent on plasminogen, consistent with Plg-RKT exhibiting plasminogen receptor function in vivo.
It is apparent that the sum of the effects of functional blockade of specific plasminogen receptors, that have been analyzed in the thioglycollate-induced peritonitis model, is greater than a 100% reduction in plasminogen-dependent macrophage recruitment. Treatment with specific antibodies to histone H2B results in 48% less macrophage recruitment [15], and injection of specific antibodies to α-enolase results in 24% less recruitment (compared to injection of nonimmune control) [15]. In S100A10−/− mice, macrophage recruitment in response to thioglycollate is 53% less than in wild-type mice [45]. Injection of mice with anti-Plg-RKT mAb7H1 resulted in 49% less macrophage recruitment compared to mice treated with isotype control [20]. Thus, it is likely that distinct plasminogen receptors may be required at different steps in the inflammatory response, for example chemotactic migration to the peritoneum, or, perhaps, crossing different layers of peritoneal tissue at which different contributions of direct plasmic cleavage of the extracellular matrix may predominate. The contribution of specific plasminogen receptors to macrophage recruitment may also be tissue and stimulus specific. For example in a model of LPS-mediated monocyte recruitment to the alveolar compartment, α-enolase appears to play a predominant role [49].
10. Tissue and Cellular Distribution of the Plg-RKT Transcript
We searched results of gene expression array analyses for possible expression of the C9orf46 homolog/anti-Plg-RKT transcript in other cells and tissues. The transcript is broadly expressed in both normal human and mouse tissues (as determined in high-throughput gene expression profiling in which RNA samples from human and murine tissues were hybridized to high-density gene expression arrays [50, 51]). The C9orf46 homolog/Plg-RKT transcript is present in spleen, lymph node, thymus, bone marrow, lung, intestine, adrenal, pituitary, and other endocrine tissues, vascular tissue, liver, kidney, stomach, bladder, and neuronal tissue (hippocampus, hypothalamus, cerebellum, cerebral cortex, olfactory bulb, and dorsal root ganglion).
In addition, we searched for C9orf46 homolog/Plg-RKT mRNA microarray expression data at http://www.ebi.ac.uk/microarray-as/aew/. C9orf46 homolog/Plg-RKT mRNA is present in monocytes, leukocytes, T cells, natural killer (NK) cells, myeloid, dendritic and plasmacytoid cells, breast cancer, acute lymphoblastic leukemia, and Molt-4 acute lymphoblastic leukemia cells.
These data are consistent with previous reports documenting expression of plasminogen-binding sites on peripheral blood leukocytes [52], breast cancer cells [53, 54], and other tissues (reviewed in [55]).
Other studies of transcript expression provide clues to additional potential functions of Plg-RKT. In a previously published genome-scale quantitative image analysis, overexpression of a cDNA that we now recognize to be the Plg-RKT cDNA resulted in dramatic increases in cell proliferation whereas knockdown of the corresponding mRNA resulted in apoptosis [56]. Consistent with an anti-apoptotic role of Plg-RKT, we have shown that cell-bound plasminogen inhibits TNFα-induced apoptosis [36]. In microarray studies, C9orf46 homolog mRNA expression has a high power to predict cervical lymph node metastasis in oral squamous cell carcinoma [57].
11. Conclusions
In conclusion, MudPIT has allowed us to identify a new protein, Plg-RKT, a novel plasminogen receptor with unique characteristics: integral to the cell membrane and exposing a C-terminal lysine on the cell surface in an orientation to bind plasminogen. Furthermore, the ability of Plg-RKT to bind t-PA, as well as the colocalization of Plg-RKT with uPAR, brings the substrate, plasminogen, and its activators in close proximity on the cell surface in an orientation to promote plasminogen activation as shown in the model in Figure 8. Here we have reviewed emerging data establishing a role for Plg-RKT in plasminogen activation, macrophage invasion and migration, and macrophage recruitment in the inflammatory response.
Figure 8.
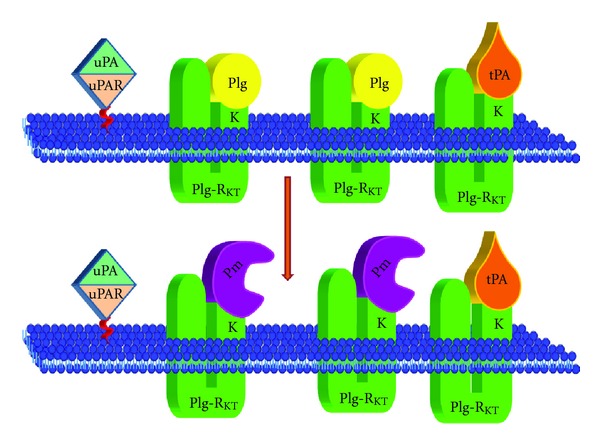
Model for Plg-RKT-dependent cell surface plasminogen activation. Plg-RKT is located on the monocyte surface in close physical proximity to the uPAR. The uPAR brings uPA in close proximity to plasminogen bound to Plg-RKT, thus promoting activation of the bound plasminogen to plasmin. In addition, t-PA also interacts specifically with Plg-RKT, thus mimicking the interaction of t-PA with cellular binding sites. Despite sharing a binding site on Plg-RKT, the relative concentrations of tPA and plasminogen in the circulation should permit simultaneous binding of both ligands to the cell surface, and each tPA molecule should be bound proximally to several plasminogen molecules, thus promoting plasminogen activation to plasmin on the cell surface.
The broad distribution of the Plg-RKT transcript and its regulation in tissues that have been demonstrated to express plasminogen binding sites suggests that Plg-RKT provides plasminogen receptor function that may serve to modulate plasmin proteolytic functions (both physiologic and pathologic) specific to a large number of tissues. Thus, Plg-RKT is likely to play a key role in plasminogen-dependent functions of cells including inflammation, wound healing, development, metastasis, neurite outgrowth fibrinolysis, myogenesis, and prohormone processing. Furthermore, the potential function of Plg-RKT in the regulation of apoptosis and proliferation may play a key role in cancer and metastasis. Future studies with knockout mice should build on our initial results using MudPIT to elucidate the role of Plg-RKT.
Acknowledgments
This paper is supported by National Institutes of Health Grants (HL38272, HL45934, and HL081046 to L. A. Miles, HL50398 to R. J. Parmer, NIH P41 RR011823, and GM103533 to J. R. Yates III, NIAID sub-contract Grant UCSD/MCB0237059 to E. I. Chen) and Department of Veterans Affairs to R. J. Parmer. The authors thank Dr. Ray Stevens at The Scripps Research Institute and Dr. Nuala Booth and Dr. Ian Booth, University of Aberdeen, for helpful discussions. The authors thank Ms. Linda Bonafede for paper preparation. This is publication no. 21717 from The Scripps Research Institute.
References
- 1.Miles LA, Plow EF. Binding and activation of plasminogen on the platelet surface. The Journal of Biological Chemistry. 1985;260(7):4303–4311. [PubMed] [Google Scholar]
- 2.Stricker RB, Wong D, Shiu DT. Activation of plasminogen by tissue plasminogen activator on normal and thrombasthenic platelets: effects on surface proteins and platelet aggregation. Blood. 1986;68(1):275–280. [PubMed] [Google Scholar]
- 3.Hajjar KA, Harpel PC, Jaffe EA, Nachman RL. Binding of plasminogen to cultured human endothelial cells. The Journal of Biological Chemistry. 1986;261(25):11656–11662. [PubMed] [Google Scholar]
- 4.Ellis V, Behrendt N, Dano K. Plasminogen activation by receptor-bound urokinase: a kinetic study with both cell-associated and isolated receptor. The Journal of Biological Chemistry. 1991;266(19):12752–12758. [PubMed] [Google Scholar]
- 5.Gonias SL, Braud LL, Geary WA, VandenBerg SR. Plasminogen binding to rat hepatocytes in primary culture and to thin slices of rat liver. Blood. 1989;74(2):729–736. [PubMed] [Google Scholar]
- 6.Duval-Jobe C, Parmely MJ. Regulation of plasminogen activation by human U937 promonocytic cells. The Journal of Biological Chemistry. 1994;269(33):21353–21357. [PubMed] [Google Scholar]
- 7.Félez J, Miles LA, Fábregas P, Jardí M, Plow EF, Lijnen RH. Characterization of cellular binding sites and interactive regions within reactants required for enhancement of plasminogen activation by tPA on the surface of leukocytic cells. Thrombosis and Haemostasis. 1996;76(4):577–584. [PubMed] [Google Scholar]
- 8.Plow EF, Freaney DE, Plescia J, Miles LA. The plasminogen system and cell surfaces: evidence for plasminogen and urokinase receptors on the same cell type. Journal of Cell Biology. 1986;103(6 I):2411–2420. doi: 10.1083/jcb.103.6.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall SW, Humphries JE, Gonias SL. Inhibition of cell surface receptor-bound plasmin by α2-antiplasmin and α2-macroglobulin. The Journal of Biological Chemistry. 1991;266(19):12329–12336. [PubMed] [Google Scholar]
- 10.Saksela O. Plasminogen activation and regulation of pericellular proteolysis. Biochimica et Biophysica Acta. 1985;823(1):35–65. doi: 10.1016/0304-419x(85)90014-9. [DOI] [PubMed] [Google Scholar]
- 11.Testa JE, Quigley JP. Protease receptors on cell surfaces: new mechanistic formulas applied to an old problem. Journal of the National Cancer Institute. 1988;80(10):712–713. doi: 10.1093/jnci/80.10.712. [DOI] [PubMed] [Google Scholar]
- 12.Ploplis VA, French EL, Carmeliet P, Collen D, Plow EF. Plasminogen deficiency differentially affects recruitment of inflammatory cell populations in mice. Blood. 1998;91(6):2005–2009. [PubMed] [Google Scholar]
- 13.Plow EF, Ploplis VA, Busuttil S, Carmeliet P, Collen D. A role of plasminogen in atherosclerosis and restenosis models in mice. Thrombosis and Haemostasis. 1999;82(1, supplement):4–7. [PubMed] [Google Scholar]
- 14.Busuttil SJ, Ploplis VA, Castellino FJ, Tang L, Eaton JW, Plow EF. A central role for plasminogen in the inflammatory response to biomaterials. Journal of Thrombosis and Haemostasis. 2004;2(10):1798–1805. doi: 10.1111/j.1538-7836.2004.00916.x. [DOI] [PubMed] [Google Scholar]
- 15.Das R, Burke T, Plow EF. Histone H2B as a functionally important plasminogen receptor on macrophages. Blood. 2007;110(10):3763–3772. doi: 10.1182/blood-2007-03-079392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong Y, Hart E, Shchurin A, Hoover-Plow J. Inflammatory macrophage migration requires MMP-9 activation by plasminogen in mice. Journal of Clinical Investigation. 2008;118(9):3012–3024. doi: 10.1172/JCI32750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miles LA, Andronicos NM, Chen EI, et al. Identification of the novel plasminogen receptor, Plg-RKT. In: Man TK, Flores RJ, editors. Proteomics—Human Diseases and Protein Functions. chapter 10. Intech; 2012. pp. 219–238. [Google Scholar]
- 18.Andronicos NM, Chen EI, Baik N, et al. Proteomics-based discovery of a novel, structurally unique, and developmentally regulated plasminogen receptor, Plg-RKT, a major regulator of cell surface plasminogen activation. Blood. 2010;115(7):1319–1330. doi: 10.1182/blood-2008-11-188938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pozzi A, Moberg PE, Miles LA, Wagner S, Soloway P, Gardner HA. Elevated matrix metalloprotease and angiostatin levels in integrin α1 knockout mice cause reduced tumor vascularization. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(5):2202–2207. doi: 10.1073/pnas.040378497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lighvani S, Baik N, Diggs JE, Khaldoyanidi S, Parmer RJ, Miles LA. Regulation of macrophage migration by a novel plasminogen receptor Plg-RKT. Blood. 2011;118(20):5622–5630. doi: 10.1182/blood-2011-03-344242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swaisgood CM, Schmitt D, Eaton D, Plow EF. In vivo regulation of plasminogen function by plasma carboxypeptidase B. Journal of Clinical Investigation. 2002;110(9):1275–1282. doi: 10.1172/JCI15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miles LA, Dahlberg CM, Plescia J, Felez J, Kato K, Plow EF. Role of cell-surface lysines in plasminogen binding to cells: identification of α-enolase as a candidate plasminogen receptor. Biochemistry. 1991;30(6):1682–1691. doi: 10.1021/bi00220a034. [DOI] [PubMed] [Google Scholar]
- 23.Redlitz A, Fowler BJ, Plow EF, Miles LA. The role of an enolase-related molecule in plasminogen binding to cells. European Journal of Biochemistry. 1995;227(1-2):407–415. doi: 10.1111/j.1432-1033.1995.tb20403.x. [DOI] [PubMed] [Google Scholar]
- 24.Hawley SB, Tamura TA, Miles LA. Purification, cloning, and characterization of a profibrinolytic plasminogen-binding protein, TIP49a. The Journal of Biological Chemistry. 2001;276(1):179–186. doi: 10.1074/jbc.M004919200. [DOI] [PubMed] [Google Scholar]
- 25.Herren T, Burke TA, Das R, Plow EF. Identification of histone H2B as a regulated plasminogen receptor. Biochemistry. 2006;45(31):9463–9474. doi: 10.1021/bi060756w. [DOI] [PubMed] [Google Scholar]
- 26.Falcone DJ, Borth W, Khan KMF, Hajjar KA. Plasminogen-mediated matrix invasion and degradation by macrophages is dependent on surface expression of annexin II. Blood. 2001;97(3):777–784. doi: 10.1182/blood.v97.3.777. [DOI] [PubMed] [Google Scholar]
- 27.Rouhiainen A, Kuja-Panula J, Wilkman E, et al. Regulation of monocyte migration by amphoterin (HMGB1) Blood. 2004;104(4):1174–1182. doi: 10.1182/blood-2003-10-3536. [DOI] [PubMed] [Google Scholar]
- 28.Fan Z, Larson PJ, Bognacki J, et al. Tissue factor regulates plasminogen binding and activation. Blood. 1998;91(6):1987–1998. [PubMed] [Google Scholar]
- 29.Pluskota E, Soloviev DA, Bdeir K, Cines DB, Plow EF. Integrin αMβ2 orchestrates and accelerates plasminogen activation and fibrinolysis by neutrophils. The Journal of Biological Chemistry. 2004;279(17):18063–18072. doi: 10.1074/jbc.M310462200. [DOI] [PubMed] [Google Scholar]
- 30.Hawley SB, Green MA, Miles LA. Discriminating between cell surface and intracellular plasminogen-binding proteins: heterogeneity in profibrinolytic plasminogen-binding proteins on monocytoid cells. Thrombosis and Haemostasis. 2000;84(5):882–890. [PubMed] [Google Scholar]
- 31.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 32.Wang GG, Calvo KR, Pasillas MP, Sykes DB, Häcker H, Kamps MP. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nature Methods. 2006;3(4):287–293. doi: 10.1038/nmeth865. [DOI] [PubMed] [Google Scholar]
- 33.Odegaard JI, Vats D, Zhang L, et al. Quantitative expansion of ES cell-derived myeloid progenitors capable of differentiating into macrophages. Journal of Leukocyte Biology. 2007;81(3):711–719. doi: 10.1189/jlb.0906590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Mullane MJ, Baker MS. Loss of cell viability dramatically elevates cell surface plasminogen binding and activation. Experimental Cell Research. 1998;242(1):153–164. doi: 10.1006/excr.1998.4067. [DOI] [PubMed] [Google Scholar]
- 35.O’Mullane MJ, Baker MS. Elevated plasminogen receptor expression occurs as a degradative phase event in cellular apoptosis. Immunology and Cell Biology. 1999;77(3):249–255. doi: 10.1046/j.1440-1711.1999.00823.x. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell JW, Baik N, Castellino FJ, Miles LA. Plasminogen inhibits TNFα-induced apoptosis in monocytes. Blood. 2006;107(11):4383–4390. doi: 10.1182/blood-2005-07-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu M, Zhang S. A kringle-containing protease with plasminogen-like activity in the basal chordate Branchiostoma belcheri. Bioscience Reports. 2009;29(6):385–395. doi: 10.1042/BSR20080173. [DOI] [PubMed] [Google Scholar]
- 38.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. The Journal of Biological Chemistry. 1981;256(4):1604–1607. [PubMed] [Google Scholar]
- 39.Estreicher A, Wohlwend A, Belin D, Schleuning WD, Vassalli JD. Characterization of the cellular binding site for the urokinase-type plasminogen activator. The Journal of Biological Chemistry. 1989;264(2):1180–1189. [PubMed] [Google Scholar]
- 40.Han J, Baik N, Kim K-H, et al. Monoclonal antibodies detect receptor-induced binding sites in Glu-plasminogen. Blood. 2011;118(6):1653–1662. doi: 10.1182/blood-2010-11-316943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hajjar KA, Hamel NM, Harpel PC, Nachman RL. Binding of tissue plasminogen activator to cultured human endothelial cells. Journal of Clinical Investigation. 1987;80(6):1712–1719. doi: 10.1172/JCI113262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miles LA, Dahlberg CM, Plow EF. The cell-binding domains of plasminogen and their function in plasma. The Journal of Biological Chemistry. 1988;263(24):11928–11934. [PubMed] [Google Scholar]
- 43.Felez J, Chanquia CJ, Fabregas P, Plow EF, Miles LA. Competition between plasminogen and tissue plasminogen activator for cellular binding sites. Blood. 1993;82(8):2433–2441. [PubMed] [Google Scholar]
- 44.Felez J, Chanquia CJ, Levin EG, Miles LA, Plow EF. Binding of tissue plasminogen activator to human monocytes and monocytoid cells. Blood. 1991;78(9):2318–2327. [PubMed] [Google Scholar]
- 45.O’Connell PA, Surette AP, Liwski RS, Svenningsson P, Waisman DM. S100A10 regulates plasminogen-dependent macrophage invasion. Blood. 2010;116(7):1136–1146. doi: 10.1182/blood-2010-01-264754. [DOI] [PubMed] [Google Scholar]
- 46.Syrovets T, Tippler B, Rieks M, Simmet T. Plasmin is a potent and specific chemoattractant for human peripheral monocytes acting via a cyclic guanosine monophosphate-dependent pathway. Blood. 1997;89(12):4574–4583. [PubMed] [Google Scholar]
- 47.Hurley JV, Ryan GB, Friedman A. The mononuclear response to intrapleural injection in the rat. The Journal of pathology and bacteriology. 1966;91(2):575–587. doi: 10.1002/path.1700910234. [DOI] [PubMed] [Google Scholar]
- 48.Melnicoff MJ, Horan PK, Morahan PS. Kinetics of changes in peritoneal cell populations following acute inflammation. Cellular Immunology. 1989;118(1):178–191. doi: 10.1016/0008-8749(89)90367-5. [DOI] [PubMed] [Google Scholar]
- 49.Wygrecka M, Marsh LM, Morty RE, et al. Enolase-1 promotes plasminogen-mediated recruitment of monocytes to the acutely inflamed lung. Blood. 2009;113(22):5588–5598. doi: 10.1182/blood-2008-08-170837. [DOI] [PubMed] [Google Scholar]
- 50.Su AI, Cooke MP, Ching KA, et al. Large-scale analysis of the human and mouse transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(7):4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su AI, Wiltshire T, Batalov S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(16):6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miles LA, Plow EF. Receptor mediated binding of the fibrinolytic components, plasminogen and urokinase, to peripheral blood cells. Thrombosis and Haemostasis. 1987;58(3):936–942. [PubMed] [Google Scholar]
- 53.Correc P, Fondaneche MC, Bracke M, Burtin P. The presence of plasmin receptors on three mammary carcinoma MCF-7 sublines. International Journal of Cancer. 1990;46(4):745–750. doi: 10.1002/ijc.2910460432. [DOI] [PubMed] [Google Scholar]
- 54.Ranson M, Andronicos NM, O’Mullane MJ, Baker MS. Increased plasminogen binding is associated with metastatic breast cancer cells: differential expression of plasminogen binding proteins. British Journal of Cancer. 1998;77(10):1586–1597. doi: 10.1038/bjc.1998.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miles LA, Hawley SB, Baik N, Andronicos NM, Castellino FJ, Parmer RJ. Plasminogen receptors: the sine qua non of cell surface plasminogen activation. Frontiers in Bioscience. 2005;10(2):1754–1762. doi: 10.2741/1658. [DOI] [PubMed] [Google Scholar]
- 56.Harada JN, Bower KE, Orth AP, et al. Identification of novel mammalian growth regulatory factors by genome-scale quantitative image analysis. Genome Research. 2005;15(8):1136–1144. doi: 10.1101/gr.3889305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen ST, Hasegawa S, Tsuda H, et al. Identification of a predictive gene expression signature of cervical lymph node metastasis in oral squamous cell carcinoma. Cancer Science. 2007;98(5):740–746. doi: 10.1111/j.1349-7006.2007.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


