Abstract
This study reevaluates the putative advantages of microwave-assisted tryptic digests compared to conventionally heated protocols performed at the same temperature. An initial investigation of enzyme stability in a temperature range of 37–80 °C demonstrated that trypsin activity declines sharply at temperatures above 60 °C, regardless if microwave dielectric heating or conventional heating is employed. Tryptic digests of three proteins of different size (bovine serum albumin, cytochrome c and β-casein) were thus performed at 37 °C and 50 °C using both microwave and conventional heating applying accurate internal fiber-optic probe reaction temperature measurements. The impact of the heating method on protein degradation and peptide fragment generation was analyzed by SDS-PAGE and MALDI-TOF-MS. Time-dependent tryptic digestion of the three proteins and subsequent analysis of the corresponding cleavage products by MALDI-TOF provided virtually identical results for both microwave and conventional heating. In addition, the impact of electromagnetic field strength on the tertiary structure of trypsin and BSA was evaluated by molecular mechanics calculations. These simulations revealed that the applied field in a typical laboratory microwave reactor is 3–4 orders of magnitude too low to induce conformational changes in proteins or enzymes.
Keywords: Enzyme activity, Molecular mechanics calculations, Molecular modeling, Nonthermal microwave effects, Tryptic digest of proteins
Graphical abstract
Highlights
► First critical evaluation of microwave-assisted proteomic protocols. ► Often claimed “nonthermal effects” were not found. ► Electric field simulations indicate that proteins do not change structure in an applied field.
1. Introduction
The implementation of microwave technology into proteomics research has recently emerged as a popular tool to perform enzymatic protein digestions [1–22]. The frequently reported main benefits of this non-classical proteomics technique are significantly higher digest efficiencies, accelerated degradation rates, and higher sequence coverage [1–22]. In many of these cases, the observed differences between microwave-assisted and conventional digestion were ascribed to so-called non-thermal microwave effects [23–26], resulting from a direct interaction of the 2.45 GHz electromagnetic field with the protein or enzyme structure, not related to a macroscopic change in temperature [1–17]. Improvements in digest efficiency and/or sequence coverage have not only been observed in solution [1–17], but also for in-gel tryptic digests [18,19]. Similar enhancements were also achieved when digest solutions were loaded with magnetic components to increase the microwave absorptivity of the irradiated solutions [20–22].
In addition to the field of microwave-assisted proteomics, there are a number of other scientific disciplines where non-thermal interactions of the microwave field with proteins, enzymes, or other biological samples have been proposed. For example, non-thermal microwave effects have often been claimed in the field of biocatalysis [27–31], DNA hybridization [32], and in studies on enzyme stability under the influence of microwave irradiation [33–37]. Irradiation with microwave frequencies has also been proposed to cause unfolding or conformational changes in proteins [38–42]. Importantly, if these direct (non-thermal) effects of the electromagnetic field on protein structure and enzyme activity would genuinely exist, health risks resulting from the use of cellular phones or domestic microwave ovens, which both operate in the microwave frequency range (~ 800–2450 MHz), cannot be excluded [38–48]. Therefore, an examination of the influence of microwave irradiation on protein structure and enzyme activity is of considerable importance [49–54].
In order to address the question if protein or enzyme structure/activity can be directly influenced by 2.45 GHz electromagnetic fields we herein present a thorough reinvestigation of microwave-assisted proteomics protocols. In the present study we evaluate the influence of the electromagnetic field on the tryptic digest of three model proteins, paying meticulous attention to the accurate control of all relevant reaction parameters, most importantly, the reaction temperature during the irradiation process. In essence, the analysis of a tryptic protein digest under microwave conditions allows to simultaneously unravel the effect of the electromagnetic field on both enzyme activity and protein structure. Any differences in enzyme stability, digestion speed, or the observed peptide fragments comparing microwave and conventional conditions at the same temperature would indicate the presence of non-thermal microwave effects. We therefore have performed a comparative study on trypsin activity applying 2.45 GHz microwave irradiation (dielectric heating) and conventional heating at identical reaction temperatures. In addition, the overall rate and time dependent generation of tryptic peptides from three model proteins using microwave and conventional heating at 37 and 50 °C was investigated by MALDI-TOF analysis. Furthermore, the impact of increasing electromagnetic field strength on the tertiary structure of trypsin and bovine serum albumin (BSA) was evaluated by molecular mechanics simulations. The results of our combined experimental and computational studies reveal that neither the enzyme stability itself, nor the tryptic digest can be directly affected by the electromagnetic field. No evidence for the previously claimed non-thermal microwave effects was obtained.
2. Materials and methods
2.1. General section and materials
BSA (A8806), cytochrome c (C3131), and β-casein (C6905), Nα-benzoyl-l-arginine-4-nitroanilide hydrochloride, α-cyano-4-hydroxycinnamic acid, and sinapic acid were obtained from Sigma Aldrich. Bradykinin fragment 1–7, angiotensin II, P14R, and ACTH fragment 18–39 were provided in the ProteoMass™ Peptide MALDI-MS Calibration Kit which was obtained from Sigma Aldrich. Sequencing grade modified trypsin was obtained from Promega (V5111). Coomassie brilliant blue R 250 was from Serva (Heidelberg, Germany). Calcium chloride-2-hydrat (CaCl2), ammonium bicarbonate (NH4HCO3), all other chemicals and solvents were purchased from standard commercial sources and used without further purification.
2.2. Microwave instrumentation and conventional heating setup
Microwave-assisted heating experiments were performed in a Monowave 300 single-mode microwave reactor (Anton Paar GmbH, Graz, Austria) equipped with a fiber-optic (ruby) thermometer for internal online temperature monitoring (2.45 GHz, 850 W). All microwave reactions were performed utilizing G4 reaction vessels (0.5–2 mL reaction volume) and the corresponding stir bars (5 × 3 mm in diameter). Simultaneous cooling during the microwave heating experiments was conducted with compressed air (6 bar, 25 °C). Importantly, all microwave experiments in this work were performed under temperature control mode, not with constant power in order to be able to correlate the data with experiments performed by conventional heating. Conventional heating was performed in an oil bath combined with a standard hotplate/stirrer (RCT basic, IKA® Werke GmbH & Co. KG, Staufen, Germany). The oil bath temperature, controlled by an adjustable contact thermometer, was varied to obtain the desired internal reaction temperature monitored by an external fiber optic probe (OTG-F fiber optic probe) which was connected to a multi-channel conditioner (TempSens signal conditioner, Opsens, Quebec, Canada). G4 reaction vessels were also used for all oil bath experiments.
2.3. Photometric analysis for testing enzyme activity
Spectrophotometric analysis was performed using a Spectronic Genesys 5 spectrophotometer (Milton Roy, Ivyland, PA, USA) utilizing polystyrene semi-micro (1.5 mL) cuvettes. Measurements were performed at 405 nm.
2.4. Trypsin activity
A G4 Pyrex vessel, equipped with a stir bar, was filled with 838 μL NH4HCO3 buffer (50 mM), 42 μL CaCl2 solution (120 mM), and 20 μL of a trypsin aliquot containing 1.5 μg trypsin. For both, the conventional and the microwave-assisted experiments, enzyme mixtures were heated to internal reaction temperatures of 37 °C, 50 °C, 60 °C, 70 °C, and 80 °C, respectively. To reach the desired inside target temperatures the oil bath temperature was set to 39 °C, 52 °C, 62 °C, 73 °C, and 83 °C, respectively. Heating times for the five selected temperatures were 1 min, 5 min, and 10 min, ensuring identical ramp times (time required to reach the target temperatures) for all corresponding heating experiments. After heating, the reaction mixtures were cooled to ~ 35 °C, then 100 μL of a 1 mg/mL Nα-benzoyl-l-arginine-4-nitroanilide hydrochloride solution were added and subsequently the extinction change of each sample was measured over a period of 10 min (405 nm).
2.5. Conventional and microwave-assisted tryptic digest of different protein samples
For all protein digest experiments G4 Pyrex vessels were equipped with stir bars, 928 μL NH4HCO3 buffer (50 mM), 42 μL CaCl2 solution (120 mM), 20 μL of a trypsin aliquot containing 1.5 μg trypsin, and 10 μL of the protein stock solution (10 mg/mL solutions of BSA, cytochrome c, or β-casein). Heating experiments were performed at 37 °C and 50 °C, respectively, applying heating times of 0 min, 1 min, 3 min, 5 min, 10 min, 30 min, 2 h, and 16 h. The ramp times of oil bath and the corresponding microwave-assisted experiments were identical and the stirring speed was set to 150 rpm in all cases. The protease/protein ratio for all experiments was 1:67 (w/w, recommended protease/protein ratio by the vendor is 1:20 up to 1:100). After heating the mixtures were cooled to ~ 35 °C and the digest was stopped by adding 10 μL of formic acid. Subsequently 100 μL of the reaction mixture were removed and dried under a gentle stream of argon. The samples were subsequently analyzed utilizing SDS-PAGE or MALDI-TOF-MS.
2.6. SDS-PAGE
10 μg protein of tryptically digested BSA were resuspended in sample buffer and loaded onto self-cast 12% SDS gels. Proteins were separated by linear PAGE (150 V, reducing conditions) and were subsequently stained with Coomassie brilliant blue (0.1% (w/v) Coomassie brilliant blue R 250 in 40% ethanol, 10% (v/v) acetic acid) for 2 h. Destaining of SDS gels was performed by gently shaking the gels in 40% ethanol, 10% (v/v) acetic acid (20 min, twice). Then the gels were immersed in 500 mL distilled water containing 20 mL acetic acid for 24 h at 4 °C to further reduce background staining and to increase sensitivity followed by scanning of the destained SDS gels on a ScanMarker 8700 (Microtek).
2.7. MALDI-TOF analysis
Dried protein extracts containing 10 μg of trypsin digested peptides were dissolved and subsequently diluted in 0.1% (v/v) trifluoroacetic acid (TFA). 7.5 pmol protein were applied to a stainless steel target plate (Applied Biosystems, Foster City, CA, USA) as 1 μL aliquots followed by addition of 1 μL of a saturated matrix solution of 10 mg/mL α-cyano-4-hydroxycinnamic acid in 50% acetonitrile (ACN), 0.1% (v/v) TFA. Subsequently, samples were allowed to crystallize at room temperature. All MALDI-TOF mass spectra were acquired on a Voyager-DE STR BioSpectrometry workstation (Applied Biosystems). The system utilizes a pulsed nitrogen laser emitting at 337 nm, which was operated in positive ion mode. The extraction voltage was 20 kV and the “low mass gate” was set at 500 Da to prevent saturation of the detector by ions resulting from the matrix. All measurements were performed in duplicates and for each mass spectrum, 3 single spectra recorded with more than 100 single laser shots were averaged. To enhance the spectral resolution, all spectra were measured in the reflector mode. External mass calibration was carried out using bradykinin fragment 1–7 (human; 757.40 Da), angiotensin II (human; 1046.54 Da), P14R (synthetic peptide; 1533.85 Da), and ACTH fragment 18–39 (human; 2465.20 Da) as standards. Data Explorer version 4.0. (Applied Biosystems) was used for spectra analysis. Time-dependent tryptic cleavage of respective proteins was followed using the PeptideMass software (http://web.expasy.org/peptide_mass/; ExPASy) for comparison of generated tryptic fragments allowing a maximum of 1 missed cleavage. Sequence coverage of respective proteins was calculated using the Mascot program (http://www.matrixscience.com/search_form_select.html; Matrix Science).
2.8. BSA structure determination by molecular modeling
The structure of bovine serum albumin (BSA) was modeled based on the crystal structure of human serum albumin (PDB-entry: 2BXK) [55], using the program Yasara (http://yasara.org). The two proteins share an overall sequence identity of 76% and a similarity of 88%. The final homology model had an overall Yasara Z-score of about 0.4 indicating a high quality model as expected from the high sequence identity between target and template. During the preparation of this manuscript the structure of bovine serum albumin has been resolved by X-ray diffraction (PDB-entry: 3V03, DOI: http://dx.doi.org/10.2210/pdb3v03/pdb), although the results remain unpublished. Gratifyingly, the folding predicted by homology fits with the X-ray resolved structure.
2.9. Computational simulation of the effect of an increasing electric field strength on the protein structure
The starting geometry for trypsin was obtained from the crystal structure deposited in the Protein Data Bank (PDB ID: 1S83), corresponding to porcine trypsin complexed with 4-aminopropanol. The ligand and the ions present in the outer part of the protein were deleted using GaussView, while the buried calcium cation was kept in the structure. The initial structure for the BSA model was obtained as described above. Minor structural errors were checked and corrected via visual inspection also with the GaussView software. Protonation states of the titratable residues were estimated by PROPKA [56] and accordingly corrected by GaussView. In addition, the protonation state of histidine (δ or ε nitrogen) was decided from the local hydrogen-bonding network by visual inspection and the PROPKA output. The tuned structure was then solvated with an explicit water molecule environment constructed via the SOLVATE software (SOLVATE 1.0.1 © 1996–2010 Helmut Grubmüller). The minimum water shell thickness was set to 10 Å for trypsin. Therefore, nowhere will the protein be closer to the surface of the solvent than this value. This set up leads to a total number of 5479 molecules of water surrounding the protein (Fig. S1). Due to the large system derived from a solvation environment with a 10 Å thickness in the case of BSA, the minimum water shell thickness was reduced to 5 Å for this solute, giving rise to a solvent environment composed of 10,169 molecules of water. The explicit water solvation model showed improved accuracy for the optimized structure and allows assessing the possible effect of the water orientation within the electric field in the protein conformation (for further details see Fig. S2 in the Supporting information). The final entire system subjected to geometry optimizations in the presence and absence of electric fields consist of 19,678 atoms for the trypsin model and 39,689 atoms for BSA.
Geometry optimizations were performed with AMBER all-atom force field and the TIP3P water model, implemented in the Gaussian09 package. Since small changes in the system are expected for the smaller electric fields applied, the cutoffs on forces and step size that are used to determine convergence for geometry optimizations were tightened by using the Opt = Tight option, which provides an accuracy (convergence criteria) for the energy of 0.0016 (or 1.6 × 10− 3) kcal/mol. The initially optimized geometries were then reoptimized in the presence of static electric fields with strengths in the range of 3 × 104 to 1010 V/m, in the X, Y and Z directions, applying the same cutoffs for the geometry convergence for electric fields up to 10− 8 V/m. Due to the drastic changes in the structure provoked by higher electric fields, the convergence criteria was relaxed to a value of 0.01 kcal/mol.
3. Results and discussion
3.1. Preliminary microwave irradiation experiments
Similar to other areas of microwave chemistry, the use of dedicated microwave instruments with accurate internal reaction temperature control is essential for being able to perform reproducible experiments, and for studying the existence of microwave effects. Without knowledge of the exact internal reaction temperature, such investigations cannot be performed, and invariably will lead to erroneous results [57–59]. For the experiments described herein a single-mode high field-density microwave reactor (2.45 GHz, 850 W) with internal fiber-optic temperature control was employed, allowing accurate monitoring of the reaction temperature on sufficiently small scale (0.5–2 mL) (Fig. S3 in the Supporting information) [60,61]. Unfortunately, the majority of published work in the field of microwave-assisted proteomics has so far been carried out in domestic microwave ovens without reporting (reliable) reaction temperatures [13–17], or using microwave instrumentation which does not allow direct monitoring of the actual reaction temperature by internal probes. Such equipment generally relies on external IR sensors recording the surface temperature of the reaction vessel [62]. In particular for strongly absorbing or highly viscous reaction mixtures, the IR sensor technique has been shown to be unreliable [57–59].
Our initial experiments focused on evaluating the microwave absorption characteristics of the components present in a typical enzymatic protein digestion cocktail (protein, enzyme, buffer components), and to identify those constituents which are mainly responsible for microwave absorption and thus for heat generation caused by 2.45 GHz microwave irradiation. Aqueous solutions (~ 1.0 mL) of the respective substrates were irradiated individually with a constant magnetron output power of 10 W and internal reaction temperature monitoring (Fig. S4). Identical heating profiles were recorded when distilled water, and samples of distilled water containing BSA (100 μg) or trypsin (1.5 μg), were irradiated, indicating that the comparatively low amounts of protein and enzyme present in the digestion cocktail do not change the overall heating behavior of the reaction mixture. This indicates that neither trypsin nor BSA is a significant absorber of microwave irradiation under the experimental conditions applied [63].
In contrast, CaCl2 (5 mM, 42 μL of a 120 mM solution in 958 μL water) and NH4HCO3 (1000 μL of a 50 mM solution) induced considerable changes to the heating profile (Fig. S4). It is well-known that dilute salt solutions significantly increase the microwave absorbance characteristics of aqueous media dramatically via ionic conduction heating mechanisms [62,64,65]. When the entire digest mixture was irradiated with 10 W constant microwave power a heating profile identical to the NH4HCO3 buffer solution was obtained. This points to the fact that the ionic components (NH4HCO3 and CaCl2) contribute the most to the microwave absorption characteristic of the digest solution; any direct interaction of the microwave field with the more or less microwave transparent enzyme or proteins in the digest solution therefore appears unlikely. In any event, owing to their ionic contents, the enzymatic digestion mixtures can be considered as rather strongly microwave absorbing, resulting in rapid heating when exposed to 2.45 GHz microwave irradiation.
As a general comment it must be stressed that the microwave power values stated herein cannot be directly compared with related data given in other publications, for example using domestic microwave ovens. This is due to the fact that all microwave cavities (multimode- or single-mode) are designed in a different way and consequently the electric field strength can be drastically different [62]. Importantly, the “microwave power” values given on the display of different instruments can at best be described as nominal magnetron output power levels and have very little physical meaning. For example, irradiating a sample with 100 W magnetron power does not mean that the sample has actually received 100 W. In fact most of the microwave power may have been reflected back to the magnetron and not been absorbed by the sample [78]. In order to accurately determine the amount of absorbed microwave power it would be necessary to measure so-called forward and reflected power data, a technology not implemented in laboratory scale microwave instruments. The high-field density single-mode microwave reactor used herein has the ability to very effectively couple microwave irradiation into samples placed inside the cavity (see Section 3.6) [60,61]. As can be seen in Fig. S4, irradiating the digest mixture with only 10 W of constant microwave power raises the temperature of the reaction mixture to 100 °C within less than 90 s. If the same experiment would be performed with 100 W (a value not untypical for other published microwave-assisted proteomics protocols using domestic microwave ovens) it would take less than 10 s to heat the sample to 100 °C and thus deactivate/destroy the enzyme (see Section 3.2). In the microwave proteomics literature, the following experimental conditions applying domestic microwave ovens are not untypical: 15 min of microwave irradiation with 30% power level (144 W) [8], 6 min with 20% power level (170 W) [13], 15 min with 30% power level (420 W) [15], or 4 min using 250 W of nominal magnetron power [16]. On the other hand, when dedicated microwave systems (with temperature control) are employed, significantly lower nominal magnetron power output levels have been reported, for example 2 W for 5–60 min [5]. While with the microwave instrumentation used herein the power values stated in other studies cannot be duplicated for the reasons explained above, there can be little doubt that the digestion samples are exposed intensely to microwave irradiation under these experimental conditions (as evidenced by effective dielectric heating, see Fig. S4).
3.2. Temperature-dependent trypsin activity
In organic transformations the general rule of thumb is that a temperature increase of 10 °C leads to a halving of the required heating time (Arrhenius law) [66,67]. This rough guideline is also applicable for enzymatic digests [68]. Therefore, the next set of experiments evaluated trypsin activity in a temperature range between 37 and 80 °C, utilizing both conventional as well as microwave heating. Trypsin specifically hydrolyses the amide bond of Nα-benzoyl-l-arginine-4-nitroanilide hydrochloride, generating 4-nitroaniline which can be measured photometrically at 405 nm [69,70]. The aim of these experiments was to compare the activity of the enzyme after exposure to microwave irradiation and conventional heating applying identical temperature-time profiles, since several previous reports have claimed that the activity/stability of an enzyme can be directly affected by microwave irradiation (i.e. by a non-thermal microwave effect) [33–37]. To ensure a scientifically valid and accurate comparison, the temperature profiles recorded by internal fiber-optic probes were matched as closely as possible between conventional (oil bath) and microwave heating experiments in order to guarantee that the enzyme samples were exposed to identical temperature–time profiles in both heating modes (ramp, hold, and cooling times, see Figs. S5a and S5b for representative examples). After hold times of 1, 5 or 10 min, the mixtures were cooled to ~ 35 °C applying similar cooling profiles. Both types of heating experiments were performed in identical reaction vessels, sample volumes and stirring speeds (Fig. S3).
As illustrated in Table S1 and Fig. S6 the highest activities for trypsin were obtained after 10 min at 50 °C and after 1 min at 60 °C. A slight increase of enzyme activity was observed when the enzyme was kept at 50 °C for longer time periods (10 min). A significant decrease of enzyme activity however was observed at 60 °C and incubation times > 1 min. The same trend was observed at 70 °C and at 80 °C where enzyme activity was generally very low. Since the enzyme retained its activity at 37 and 50 °C for the entire time course evaluated, all digestion experiments were performed at these temperatures.
Most importantly, trypsin activity was only a function of reaction temperature and time, but not dependent on the heating mode, since almost identical values were obtained regardless if microwave heating or conventional heating was employed. We therefore conclude that loss in enzyme activity is not caused by any specific, non-thermal microwave effect(s), but caused exclusively by elevated bulk reaction temperatures.
3.3. Kinetic studies of native BSA digestion utilizing SDS-PAGE analysis
Tryptic BSA digests have been investigated several times under microwave conditions, reporting mainly faster and more efficient digestion of proteins compared to standard protocols [8–17]. Additionally, a higher sequence coverage as well as peptide recovery, a higher number of missed cleavage sites, higher catalytic effectiveness, and even a reduced Michaelis–Menten constant have been proposed in the literature [8–17,35].
It cannot be excluded that microwave irradiation affects trypsin activity toward a more complex substrate. To clarify this issue, BSA was subjected to tryptic digestion in a time dependent manner at 37 and 50 °C using microwave and conventional heating (trypsin:BSA = 1:67; w/w). For microwave experiments at the comparatively low temperature of 37 °C the simultaneous cooling technique was applied [71,72]. Employing this protocol, compressed air is used to concurrently cool the reaction vial from the outside during the microwave heating process. This allows higher microwave field strengths to be applied which will enhance the occurrence of microwave effects which are dependent on the electromagnetic field (i.e. non-thermal microwave effects) [71,72]. With the present experimental set-up, a nominal magnetron output power of 1–2 W (for 37 °C), and ~ 10 W (50 °C) was achieved using the simultaneous cooling technique.
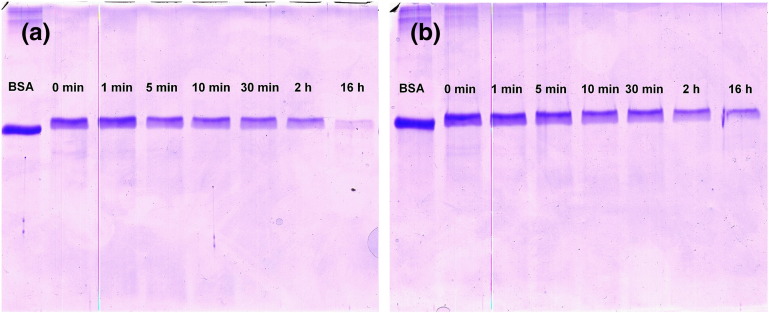
The time-dependent loss of native BSA subjected to tryptic digestion under conventional or microwave-assisted conditions is shown in Fig. 1.
Fig. 1.
Identification of remaining full-length/intact BSA after a conventional (a) and microwave-assisted tryptic digest (b) performed at 50 °C for the indicated time periods by SDS-PAGE (12% gels, reducing conditions). One representative experiment (out of three) is shown.
The results presented in Fig. 1 indicate that the loss of native BSA during microwave-assisted tryptic digestion proceeds at virtually identical rates as observed during conventional heating at the same temperature. Therefore, in contrast to several previous reports on microwave-assisted tryptic BSA digests (mostly involving domestic microwave ovens without accurate temperature control) [13–17], the presence of non-thermal microwave effects in these digests cannot be confirmed. The results for the identical experiments performed at 37 °C are shown in Fig. S7, also indicating the absence of significant differences between conventional and microwave-assisted heating.
3.4. Time-dependent degradation of BSA by trypsin
After analyzing the degradation of native BSA using SDS-PAGE, we monitored the generation of tryptic peptides over time by MALDI-TOF-MS [73–76]. For these experiments the same temperature/time regimes as for SDS-PAGE were selected. As expected, the longer the incubation time at 37 °C or 50 °C the more tryptic peptides were identified by MALDI-TOF-MS analysis (representative spectra from the conventional digest at 37 °C are shown in Fig. S8).
Peptide fragments were identified by comparison with an in silico digest (allowing 1 missed cleavage) of respective proteins using the PeptideMass software (http://web.expasy.org/peptide_mass/; ExPASy). The peptides generated during microwave-assisted and conventional digestion are compared in Table S2. In addition, Table 1 shows the comparison of identified BSA-derived tryptic peptides after a conventional or microwave-assisted digest for the time periods indicated (the corresponding sequence coverage is shown in Table 2). In the conventional digest a total number of 30 characteristic tryptic peptide fragments could be detected, out of which 29 were also obtained after microwave-assisted digestion appearing roughly at the same time point (Table 1 and Table S2).
Table 1.
Detected peptide fragments (including peptides containing one missed cleavage) obtained after conventional (black) and microwave-assisted (blue) tryptic digest of BSA performed at 37 °C for the indicated time periods.
aThe fragment including amino acids 161–168 was obtained after conventional heating and contains a missed cleavage position. Nevertheless, the corresponding fragment without a missed cleavage position was obtained utilizing both heating sources (161–167).
Table 2.
Sequence coverage comparison of conventional and microwave-assisted tryptic digest performed at 37 °C.
| Time | 0 min | 1 min | 3 min | 5 min | 10 min | 30 min | 120 min | 960 min |
|---|---|---|---|---|---|---|---|---|
| CONV | 10% | 14% | 16% | 22% | 26% | 27% | 32% | 34% |
| MW | 4% | 4% | 10% | 19% | 21% | 24% | 27% | 28% |
To get semi-quantitative information on the kinetics of tryptic peptide generation using conventional or microwave heating the intensities of seven randomly chosen peptides (covering the mass range of identified peptides) were summed up and set 100%. The relative contribution of individual peptides is shown as % of total (Fig. S9). These analyses revealed that the intensity contribution of individual peptides at different time points is very similar in tryptic digests generated by conventional and microwave heating.
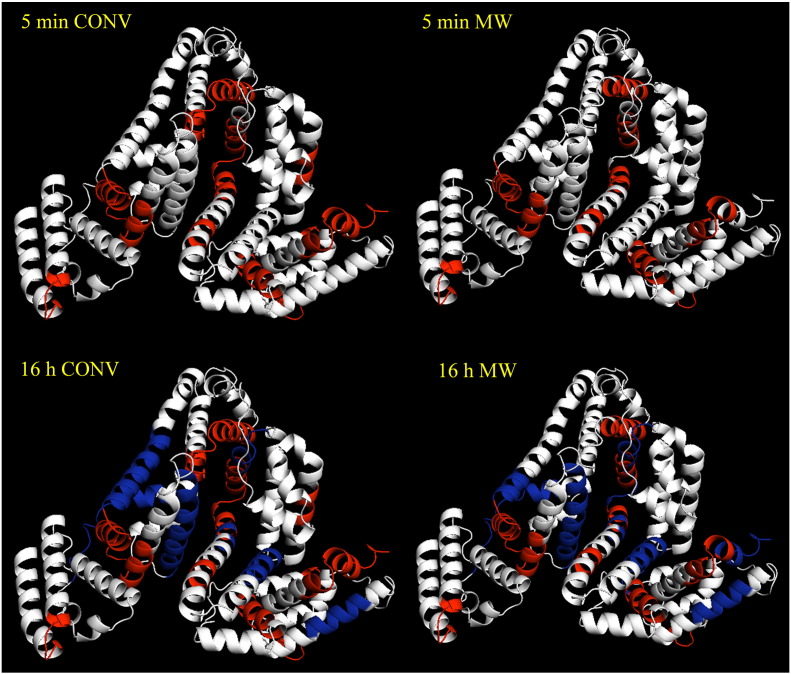
Since it is reasonable to assume that fragments appearing in the MS spectra after short heating periods are generated early in the digest, it was possible to establish a time-resolved overview of the tryptic digest of BSA for both the conventional (Table S3) and microwave-heated experiments (Table S4). A homology model of BSA was generated and visually analyzed using the program PyMOL (http://www.pymol.org/). This software allowed visualization of the obtained data by mapping the identified peptide fragments onto the 3D structure, thereby illustrating the time-dependent tryptic digest of BSA at 37 °C (Fig. S10, conventional heating; Fig. S11, microwave heating). The localization of tryptic peptides generated after a 5 min and 16 h incubation at 37 °C (microwave-assisted vs. conventional digest) within the 3D structure of BSA is shown in Fig. 2. For simplification, two time-points were selected for computational analysis representing the obtained peptide fragments after a short (5 min) and a long heating period (16 h). A comparison of conventional and microwave results is presented in Fig. 2.
Fig. 2.
Comparison of the time-dependent BSA digestion using the PyMOL software. Tryptic peptides obtained after a 5 min heating period at 37 °C are shown in red while additional fragments generated after 16 h at 37 °C are highlighted in blue.
A careful inspection of the data presented in Table 1, Table S2, Fig. 2 and Fig. S9, reveals that the 37 °C digests obtained in the microwave run closely match those of the conventionally heated experiments. These data suggest that the electromagnetic field does not inflict structural changes/damages to the BSA protein. Any significant conformational changes in the protein structure induced by an electromagnetic field would likely cause a change in the peptide sequences obtained in the enzymatic digest, as different regions of the protein would get exposed to the digesting enzyme. Similar results were obtained when the tryptic digest was performed at 50 °C. In general, the temperature increase from 37 °C to 50 °C causes a slightly faster reaction kinetics of trypsin towards BSA as evidenced by the earlier generation of BSA-specific fragments [68]. MALDI-TOF spectra comparing results from conventional BSA digestion experiments at 37 °C and 50 °C, respectively, are shown in Fig. S12 in the Supporting information. In contrast to literature reports describing an increase in digestion speed specifically caused by microwave irradiation [8–17], no significant differences between a microwave-assisted and a conventional digest were observed. The increased digest efficiency is ascribed exclusively to the elevated bulk temperature of the reaction mixture applying microwave dielectric heating, and can be readily reproduced by conventional heating to the same temperature regime.
3.5. Tryptic digest of cytochrome c and β-casein
To verify results obtained with BSA (~ 66 kDa), microwave-assisted and conventional digests were also performed using cytochrome c (~ 12 kDa) and β-casein (~ 24 kDa) as substrates for trypsin. Using the same time regimes selected for the previous set of experiments (a short digestion period of 5 min and a longer digestion period of 2 h) allowed a comparison between microwave and conventional digest at 37 °C and 50 °C, respectively. Results including sequence coverage data are presented in Tables S5 and S6 in the Supporting information. Interestingly, an increase of reaction temperature from 37 °C to 50 °C seems to have more pronounced effects on the smaller protein (cytochrome c, ~ 12 kDa) compared to the larger BSA (~ 66 kDa). For details see Fig. S13 in the Supporting information. MALDI-TOF spectra of cytochrome c and β-casein digests are presented in Fig. S14 and are again in disagreement with previously published data [8].
In summary, this set of experiments conclusively demonstrated comparable efficacy for time- and temperature-dependent tryptic digestion of the three model proteins independent of the heating source used. Furthermore, unspecific cuts caused by microwave irradiation can be excluded since almost identical MALDI-TOF spectra were obtained in all cases. These results clearly indicate that nonthermal microwave effects during tryptic digestion are virtually absent.
3.6. Electric field strength simulations
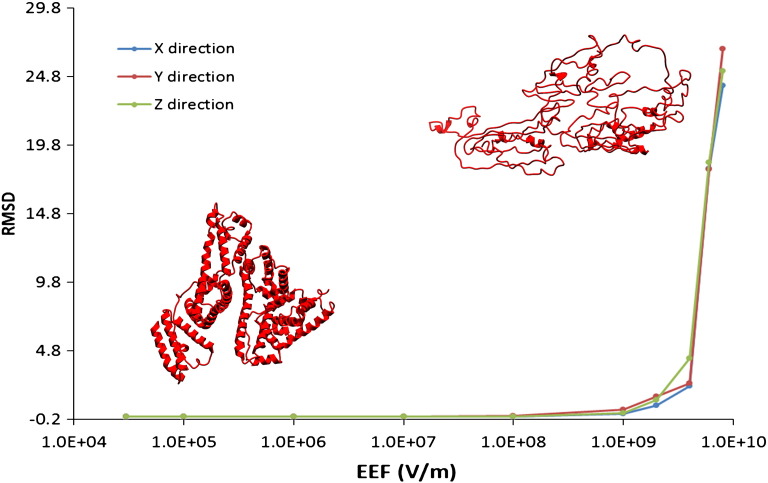
To further corroborate the findings presented above, molecular mechanics (MM) calculations were performed. In recent years, a number of publications investigated the mechanistic response of proteins towards an external electric field by means of molecular dynamics modeling [49–54]. These simulations aim to model and provide insights into altered protein conformation in response to electromagnetic fields resulting from e.g. cell phones or domestic microwave ovens [49–54]. Of note, the electric fields applied in these theoretical analyses (108–109 V/m) are many orders of magnitude stronger than the maximum electric field achievable in a commercial single-mode laboratory microwave instrument and/or multimode microwave ovens for domestic use (~ 3 × 104 V/m) [77–79]. A thorough computational study of the effect of external electric fields on protein conformations, ranging from the field strengths typically reachable in commercial laboratory microwave reactors/ovens to very high electric fields (~ 1010 V/m) has not been performed so far and can provide useful information. Any changes in protein structure due to the presence of the electric component of the microwave field, as suggested previously [49–54], would be revealed. In addition, a step-wise increase in field strength allows assessing the stability of a given protein structure toward increasing field intensity, thereby providing a measure for the stability of protein folding.
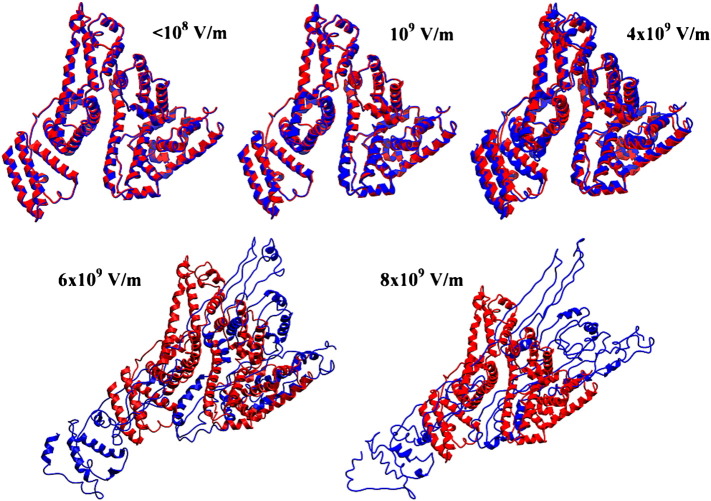
During these MM calculations, increasing static electric fields have been successively applied to the preoptimized structures of BSA and trypsin, respectively. The lowest field applied (3 × 104 V/m) during simulation can be considered as one of the highest fields that can be generated in a standard laboratory microwave cavity [77–79]. However, our calculations revealed no appreciable changes in atomic positions (RMSD = 0.000000, see Table S7 (BSA) and Table S8 (trypsin) in the Supporting information) under these conditions. The change in total energy due to the presence of the field is < 0.01 kcal/mol for the X, Y and Z directions. Notably, even under electric fields of 106 V/m, any change in the atom positions should be detectable, the RMSD value still being 0.000000. At this level of accuracy even small changes in the atom positions of the active site, which could lead to differences in the catalytic activity of the enzyme, can be discarded. This theoretical approach demonstrated that electromagnetic fields being 3–4 orders of magnitude higher than the microwave field are still without effect on BSA and trypsin structure (Figs. 3 and 4). The tertiary structure starts deteriorating at electric fields > 108 V/m, and trypsin as well as BSA is seriously damaged, inducing unfolding of the alpha helices, when electric fields of 8 × 109–1010 V/m are applied (Figs. 3 and 4). The corresponding results for trypsin are shown in the Supporting information (Fig. S15 and S16).
Fig. 3.
Effect of an external electric field on the structure of BSA. RMSD refers to the measurement of the average distance between the atoms of superimposed BSA structures in the presence and in the absence of the external electric field. Small changes can be seen above 107–108 V/m (see Table S7 in the Supporting information for further details).
Fig. 4.
Optimized structures for the BSA protein in the presence of various electric fields (blue color) in the X direction, overlapped with the structure of the native protein (red color).
4. Conclusion
In summary, the present study provides a comparison of tryptic digest efficacy performed by conventional and microwave heating under rigorously controlled thermal conditions. Due to the high microwave absorption characteristics of the ionic digestion buffers, temperatures up to 100 °C can be easily reached after a few seconds of microwave irradiation, even at comparatively low magnetron output powers. Therefore, digestion results obtained in microwave instruments without proper internal temperature control are not reliable and cannot be compared to results obtained by conventional heating where the digestion temperature can typically be determined more accurately. Acquiring the precise reaction temperature in a tryptic digest is, however, of critical importance, since our kinetic investigations have shown that at temperatures above 60 °C, trypsin activity in an aqueous buffer declines sharply. Importantly, the rapid decline of enzyme activity is identical, regardless if microwave irradiation or conventional heating is used.
For the actual digestion results involving bovine serum albumin (BSA), cytochrome c and β-casein at 37 °C and 50 °C the results were similar. For all three cases, no differences between a microwave-assisted and a conventional tryptic digest were observed. SDS-PAGE analysis of the tryptic protein digests demonstrated that the proteins were digested at roughly the same rate regardless of the utilized heating source. MALDI-TOF monitoring of the protein-derived tryptic fragments at several time intervals during allowed the chronological investigation of the digest. Most importantly, for all three model proteins identical qualitative results with regard to peptide generation were obtained independent of the heating source. Apparently, only temperature determines the speed and progress of the digestion. Furthermore, molecular mechanics calculations of the electric field strength required for influencing the tertiary structure of trypsin and BSA have indicated that the electric field applied in commercial laboratory microwave instruments (~ 3 × 104 V/m) is 3–4 orders of magnitude too low to affect the structure of enzyme or protein molecules.
In summary, based on the results described herein, it appears evident that as in other fields of microwave chemistry [57–61], incorrect temperature monitoring has in the past led to erroneous conclusions about the involvement of non-thermal microwave effects in microwave-assisted proteomics experiments. The fact that no specific effects in the interaction of electromagnetic fields with proteins and enzymes could be experimentally verified in our investigations clearly has additional implications on the debate involving the use of cell phone technology and kitchen microwave ovens [38–45].
Acknowledgments
This work was supported by a grant from the Christian Doppler Research Society (CDG) and the Austrian Science Fund (FWF; grant no. F3007 to W. S.). Prof. Wolfgang Stadlbauer is thanked for the provision of the Photospectrometer. D. C. thanks the Ministry of Education and Science (CTQ2010-18938/BQU) and the Research, Technological Innovation and Supercomputing Center of Extremadura (CénitS) for supporting the use of LUSITANIA computer resources.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jprot.2012.07.043.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Lill J.R. RSC Publishing; Cambridge: 2009. Microwave assisted proteomics. [Google Scholar]
- 2.Zhao H. Microwave-assisted enzymatic reactions in aqueous media. In: Polshettiwar V., Varma R.S., editors. Aqueous microwave assisted chemistry. RSC Publishing; Cambridge: 2010. pp. 123–144. [Google Scholar]
- 3.Lill J.R., Ingle E.S., Liu P.S., Pham V., Sandoval W.N. Microwave-assisted proteomics. Mass Spectrom Rev. 2007;26:657–671. doi: 10.1002/mas.20140. [DOI] [PubMed] [Google Scholar]
- 4.Sandoval W.N., Pham V., Ingle E.S., Liu P.S., Lill J.R. Applications of microwave-assisted proteomics in biotechnology. Comb Chem High Throughput Screen. 2007;10:751–765. doi: 10.2174/138620707783018504. [DOI] [PubMed] [Google Scholar]
- 5.Sandoval W.N., Pham V.C., Lill J.R. Recent developments in microwave-assisted protein chemistries — can this be integrated into the drug discovery and validation process? Drug Discov Today. 2008;13:1075–1081. doi: 10.1016/j.drudis.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Collins J.M., Leadbeater N.E. Microwave energy: a versatile tool for biosciences. Org Biomol Chem. 2007;5:1141–1150. doi: 10.1039/b617084f. [DOI] [PubMed] [Google Scholar]
- 7.Capelo J.L., Carreira R., Diniz M., Fernandes L., Galesio M., Lodeiro C. Overview on modern approaches to speed up protein identification workflows relying on enzymatic cleavage and mass spectrometry-based techniques. Anal Chim Acta. 2009;650:151–159. doi: 10.1016/j.aca.2009.07.034. [DOI] [PubMed] [Google Scholar]
- 8.Pramanik B.N., Mirza U.A., Ing Y.H., Liu Y.H., Bartner P.L., Weber P.C. Microwave-enhanced enzyme reaction for protein mapping by mass spectrometry: a new approach to protein digestion in minutes. Protein Sci. 2002;11:2676–2687. doi: 10.1110/ps.0213702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin S.S., Wu C.H., Sun M.C., Sun C.M., Ho Y.P. Microwave-assisted enzyme-catalyzed reactions in various solvent systems. J Am Soc Mass Spectrom. 2005;16:581–588. doi: 10.1016/j.jasms.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Vesper H.W., Mi L., Enada A., Myers G.L. Assessment of microwave-assisted enzymatic digestion by measuring glycated haemoglobin A1c by mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:2865–2870. doi: 10.1002/rcm.2135. [DOI] [PubMed] [Google Scholar]
- 11.Reddy P.M., Hsu W.Y., Hu J.F., Ho Y.P. Digestion completeness of microwave-assisted and conventional trypsin-catalyzed reactions. J Am Soc Mass Spectrom. 2010;21:421–424. doi: 10.1016/j.jasms.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Izquierdo F.J., Penas E., Baeza M.L., Gómez R. Effects of combined microwave and enzymatic treatment on the hydrolysis and immunoreactivity of dairy whey proteins. Int Dairy J. 2008;18:918–922. [Google Scholar]
- 13.Vaezzadeh A.R., Deshusses J.M.P., Waridel P., Francois P., Zimmermann-Ivol C.G., Lescuyer P. Accelerated digestion for high-throughput proteomics analysis of whole bacterial proteomes. J Microbiol Methods. 2010;80:56–62. doi: 10.1016/j.mimet.2009.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Hahn H.W., Rainer M., Ringer T., Huck C.W., Bonn G.K. Ultrafast microwave-assisted in-tip digestion of proteins. J Proteome Res. 2009;8:4225–4230. doi: 10.1021/pr900188x. [DOI] [PubMed] [Google Scholar]
- 15.Lesur A., Varesio E., Hopfgartner G. Accelerated tryptic digestion for the analysis of biopharmaceutical monoclonal antibodies in plasma by liquid chromatography with tandem mass spectrometric detection. J Chromatogr A. 2010;1217:57–64. doi: 10.1016/j.chroma.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 16.Ha N.Y., Kim S.H., Lee T.G., Han S.Y. Rapid characterization of protein chips using microwave-assisted protein tryptic digestion and MALDI mass spectrometry. Langmuir. 2011;27:10098–10105. doi: 10.1021/la201812a. [DOI] [PubMed] [Google Scholar]
- 17.Liu X., Chan K., Chu I.K., Li J. Microwave-assisted nonspecific proteolytic digestion and controlled methylation for glycomics applications. Carbohydr Res. 2008;343:2870–2877. doi: 10.1016/j.carres.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Sun W., Gao S., Wang L., Chen Y., Wu S., Wang X. Microwave-assisted protein preparation and enzymatic digestion in proteomics. Mol Cell Proteomics. 2006;5:769–776. doi: 10.1074/mcp.T500022-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Juan H.F., Chang S.C., Huang H.C., Chen S.T. A new application of microwave technology to proteomics. Proteomics. 2005;5:840–842. doi: 10.1002/pmic.200401056. [DOI] [PubMed] [Google Scholar]
- 20.Lo C.Y., Chen W.Y., Chen C.T., Chen Y.C. Rapid enrichment of phosphopeptides from tryptic digests of proteins using iron oxide nanocomposites of magnetic particles coated with zirconia as the concentrating probes. J Proteome Res. 2007;6:887–893. doi: 10.1021/pr060333g. [DOI] [PubMed] [Google Scholar]
- 21.Li Y.C., Lin Y.S., Tsai P.J., Chen C.T., Chen W.Y., Chen Y.C. Nitrilotriacetic acid-coated magnetic nanoparticles as affinity probes for enrichment of histidine-tagged proteins and phosphorylated peptides. Anal Chem. 2007;79:7519–7525. doi: 10.1021/ac0711440. [DOI] [PubMed] [Google Scholar]
- 22.Chen W.Y., Chen Y.C. Acceleration of microwave-assisted enzymatic digestion reactions by magnetite beads. Anal Chem. 2007;79:2394–2401. doi: 10.1021/ac0614893. [DOI] [PubMed] [Google Scholar]
- 23.Perreux L., Loupy A. A tentative rationalization of microwave effects in organic synthesis according to the reaction medium, and mechanistic considerations. Tetrahedron. 2001;57:9199–9223. [Google Scholar]
- 24.Perreux L., Loupy A. Nonthermal effects of microwaves in organic synthesis. In: Loupy A., editor. Microwaves in organic synthesis. 2nd edition. Wiley-VCH; Weinheim: 2006. pp. 134–218. [Google Scholar]
- 25.De La Hoz A., Diaz-Ortiz A., Moreno A. Microwaves in organic synthesis. Thermal and non-thermal microwave effects. Chem Soc Rev. 2005;34:164–178. doi: 10.1039/b411438h. [DOI] [PubMed] [Google Scholar]
- 26.De La Hoz A., Diaz-Ortiz A., Moreno A. Selectivity under the action of microwave irradiation. In: Loupy A., editor. Microwaves in organic synthesis. 2nd edition. Wiley-VCH; Weinheim: 2006. pp. 219–277. [Google Scholar]
- 27.Réjasse B., Lamare S., Legoy M.D., Besson T. Stability improvement of immobilized Candida antarctica lipase B in an organic medium under microwave radiation. Org Biomol Chem. 2004;2:1086–1089. doi: 10.1039/b401145g. [DOI] [PubMed] [Google Scholar]
- 28.Réjasse B., Besson T., Legoy M.D., Lamare S. Influence of microwave radiation on free Candida antarctica lipase B activity and stability. Org Biomol Chem. 2006;4:3703–3707. doi: 10.1039/b610265d. [DOI] [PubMed] [Google Scholar]
- 29.Yadav G.D., Borkar I.V. Kinetic and mechanistic investigation of microwave-assisted lipase catalyzed synthesis of citronellyl acetate. Ind Eng Chem Res. 2009;48:7915–7922. [Google Scholar]
- 30.de Souza R.O.M.A., Antunes O.A.C., Kroutil W., Kappe C.O. Kinetic resolution of rac-1-phenylethanol with immobilized lipases: a critical comparison of microwave and conventional heating protocols. J Org Chem. 2009;74:6157–6162. doi: 10.1021/jo9010443. [DOI] [PubMed] [Google Scholar]
- 31.Leadbeater N.E., Stencel L.M., Wood E.C. Probing the effects of microwave irradiation on enzyme-catalysed organic transformations: the case of lipase-catalysed transesterification reactions. Org Biomol Chem. 2007;5:1052–1055. doi: 10.1039/b617544a. [DOI] [PubMed] [Google Scholar]
- 32.Edwards W.F., Young D.D., Deiters A. The effect of microwave irradiation on DNA hybridization. Org Biomol Chem. 2009;7:2506–2508. doi: 10.1039/b903609a. [DOI] [PubMed] [Google Scholar]
- 33.Rejasse B., Lamare S., Legoy M.D., Besson T. Influence of microwave irradiation on enzymatic properties: applications in enzymatic chemistry. J Enzyme Inhib Med Chem. 2007;22:518–526. doi: 10.1080/14756360701424959. [DOI] [PubMed] [Google Scholar]
- 34.Young D.D., Nichols J., Kelly R.M., Deiters A. Microwave activation of enzymatic catalysis. J Am Chem Soc. 2008;130:10048–10049. doi: 10.1021/ja802404g. [DOI] [PubMed] [Google Scholar]
- 35.Izquierdo F.J., Alli I., Yaylayan V., Gómez R. Microwave-assisted digestion of β-lactoglobulin by pronase, α-chymotrypsin and pepsin. Int Dairy J. 2007;17:465–470. [Google Scholar]
- 36.Porcelli M., Cacciapuoti G., Fusco S., Massa R., d'Ambrosio G., Bertoldo C. Non-thermal effects of microwaves on proteins: thermophilic enzymes as model systems. FEBS Lett. 1997;402:102–106. doi: 10.1016/s0014-5793(96)01505-0. [DOI] [PubMed] [Google Scholar]
- 37.La Cara F., Scarfi M.R., D'Auria S., Massa R., d'Ambrosio G., Franceschetti G. Different effects of microwave energy and conventional heat on the activity of a thermophilic β-galactosidase from Bacillus acidocaldarius. Bioelectromagnetics. 1999;20:172–176. [PubMed] [Google Scholar]
- 38.Laurence J.A., French P.W., Lindner R.A., McKenzie D.R. Biological effects of electromagnetic fields — mechanism for the effects of pulsed microwave radiation on protein conformation. J Theor Biol. 2000;206:291–298. doi: 10.1006/jtbi.2000.2123. [DOI] [PubMed] [Google Scholar]
- 39.Bohr H., Bohr J. Microwave-enhanced folding and denaturation of globular proteins. Phys Rev E. 2000;61:4310–4314. doi: 10.1103/physreve.61.4310. [DOI] [PubMed] [Google Scholar]
- 40.Bohr H., Bohr J. Microwave enhanced kinetics observed in ORD studies of a protein. Bioelectromagnetics. 2000;21:68–72. [PubMed] [Google Scholar]
- 41.de Pomerai D.I., Smith B., Dawe A., North K., Smith T., Archer D.B. Microwave radiation can alter protein conformation without bulk heating. FEBS Lett. 2003;543:93–97. doi: 10.1016/s0014-5793(03)00413-7. [DOI] [PubMed] [Google Scholar]
- 42.de Pomerai D.I., Daniells C., David H., Allan J., Duce I., Mutwakil M. Non-thermal heat-shock response to microwaves. Nature. 2000;405:417–418. doi: 10.1038/35013144. [DOI] [PubMed] [Google Scholar]
- 43.World Health Organization, International Agency for Research on Cancer The potential carcinogenic hazards from exposure to radiofrequency electromagnetic fields. IARC Monogr. 2011:102. [Google Scholar]
- 44.Mancinelli F., Caraglia M., Abbruzzese A., d'Ambrosio G., Massa R., Bismuto E. Non-thermal effects of electromagnetic fields at mobile phone frequency on the refolding of an intracellular protein: myoglobin. J Cell Biochem. 2004;93:188–196. doi: 10.1002/jcb.20164. [DOI] [PubMed] [Google Scholar]
- 45.Inskip P.D., Tarone R.E., Hatch E.E., Wilcosky T.C., Shapiro W.R., Selker R.G. Cellular-telephone use and brain tumors. N Eng J Med. 2001;344:79–86. doi: 10.1056/NEJM200101113440201. [DOI] [PubMed] [Google Scholar]
- 46.Michaelson S.M., Lin J.C. Plenum Press; New York and London: 1987. Biological effects and health implications of radiofrequency radiation. [Google Scholar]
- 47.Takashima S. Studies on the effect of radio-frequency waves on biological macromolecules. IEEE Trans Biomed Eng. 1966;BME-13:28–31. [Google Scholar]
- 48.Fortune J.A., Wu B.I., Klibanov A.M. Radio frequency radiation causes no nonthermal damage in enzymes and living cells. Biotechnol Prog. 2010;26:1772–1776. doi: 10.1002/btpr.462. [DOI] [PubMed] [Google Scholar]
- 49.Budi A., Legge F.S., Treutlein H., Yarovsky I. Electric field effects on insulin chain-B conformation. J Phys Chem B. 2005;109:22641–22648. doi: 10.1021/jp052742q. [DOI] [PubMed] [Google Scholar]
- 50.Budi A., Legge F.S., Treutlein H., Yarovsky I. Effect of frequency on insulin response to electric field stress. J Phys Chem B. 2007;111:5748–5756. doi: 10.1021/jp067248g. [DOI] [PubMed] [Google Scholar]
- 51.Budi A., Legge F.S., Treutlein H., Yarovsky I. Comparative study of insulin chain-B in isolated and monomeric environments under external stress. J Phys Chem B. 2008;112:7916–7924. doi: 10.1021/jp800350v. [DOI] [PubMed] [Google Scholar]
- 52.Astrakas L., Gousias C., Tzaphlidou M. Electric field effects on chignolin conformation. J Appl Phys. 2011;109:094702. [Google Scholar]
- 53.Lugli F., Toschi F., Biscani F., Zerbetto F. Effects of electric field stress on a β-amyloid peptide. J Phys Chem B. 2009;113:369–376. doi: 10.1021/jp807896g. [DOI] [PubMed] [Google Scholar]
- 54.Lugli F., Toschi F., Biscani F., Zerbetto F. Electric field effects on short fibrils of β-amyloid peptides. J Chem Theory Comput. 2010;6:3516–3526. doi: 10.1021/ct1001335. [DOI] [PubMed] [Google Scholar]
- 55.Ghuman J., Zunszain P.A., Petitpas I., Bhattacharya A.A., Otagiri M., Curry S. Structural basis of the drug-binding specificity of human serum albumin. J Mol Biol. 2005;353:38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 56.Li H., Robertson A.D., Jensen J.H. Very fast empirical prediction and rationalization of protein pKa values. Proteins Struct Funct Bioinformatics. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 57.Herrero M.A., Kremsner J.M., Kappe C.O. Nonthermal microwave effects revisited: on the importance of internal reaction monitoring and agitation in microwave chemistry. J Org Chem. 2008;73:36–47. doi: 10.1021/jo7022697. [DOI] [PubMed] [Google Scholar]
- 58.Obermayer D., Kappe C.O. On the importance of simultaneous infrared/fiber-optic temperature monitoring in the microwave-assisted synthesis of ionic liquids. Org Biomol Chem. 2010;8:114–121. doi: 10.1039/b918407d. [DOI] [PubMed] [Google Scholar]
- 59.Bacsa B., Horvati K., Bosze S., Andreae F., Kappe C.O. Solid-phase synthesis of difficult peptide sequences at elevated temperatures — a critical comparison of microwave and conventional heating technologies. J Org Chem. 2008;73:7532–7542. doi: 10.1021/jo8013897. [DOI] [PubMed] [Google Scholar]
- 60.Obermayer D., Gutmann B., Kappe C.O. Microwave chemistry in silicon carbide reaction vials: separating thermal from nonthermal effects. Angew Chem Int Ed. 2009;48:8321–8324. doi: 10.1002/anie.200904185. [DOI] [PubMed] [Google Scholar]
- 61.Gutmann B., Obermayer D., Reichart B., Prekodravac B., Irfan M., Kremsner J.M. Sintered silicon carbide: a new ceramic vessel material for microwave chemistry in single-mode reactors. Chem Eur J. 2010;16:12182–12194. doi: 10.1002/chem.201001703. [DOI] [PubMed] [Google Scholar]
- 62.Kappe C.O., Stadler A., Dallinger D. 2nd edition. Wiley-VCH; Weinheim: 2012. Microwaves in organic and medicinal chemistry. [Google Scholar]
- 63.Grant E.H., Keefe S.E., Takashima S. The dielectric behaviour of aqueous solutions of bovine serum albumin from radiowave to microwave frequency. J Phys Chem. 1968;72:4373–4380. doi: 10.1021/j100859a004. [DOI] [PubMed] [Google Scholar]
- 64.Gabriel C., Gabriel S., Grant E.H., Halstead B.S.J., Mingos D.M.P. Dielectric parameters relevant to microwave dielectric heating. Chem Soc Rev. 1998;27:213–224. [Google Scholar]
- 65.Mingos D.P.M., Baghurst D.R. Application of microwave dielectric heating effects to synthetic problems in chemistry. Chem Soc Rev. 1991;20:1–47. [Google Scholar]
- 66.Kappe C.O. Controlled microwave heating in modern organic synthesis. Angew Chem Int Ed. 2004;43:6250–6284. doi: 10.1002/anie.200400655. [DOI] [PubMed] [Google Scholar]
- 67.Kappe C.O. Microwave dielectric heating in synthetic organic chemistry. Chem Soc Rev. 2008;37:1127–1139. doi: 10.1039/b803001b. [DOI] [PubMed] [Google Scholar]
- 68.Bisswanger H. 3rd edition. Wiley-VCH; Weinheim: 2000. Enzyme kinetics: principles and methods. [Google Scholar]
- 69.Havlis J., Thomas H., Sebela M., Shevchenko A. Fast-response proteomics by accelerated in-gel digestions of proteins. Anal Chem. 2003;75:1300–1306. doi: 10.1021/ac026136s. [DOI] [PubMed] [Google Scholar]
- 70.Finehout E.J., Cantor J.R., Lee K.H. Kinetic characterization of sequencing grade modified trypsin. Proteomics. 2005;5:2319–2321. doi: 10.1002/pmic.200401268. [DOI] [PubMed] [Google Scholar]
- 71.Hosseini M., Stiasni N., Barbieri V., Kappe C.O. Microwave-assisted asymmetric organocatalysis. A probe for non-thermal microwave effects and the concept of simultaneous cooling. J Org Chem. 2007;72:1417–1424. doi: 10.1021/jo0624187. [DOI] [PubMed] [Google Scholar]
- 72.Hayes B.L. Recent advances in microwave-assisted synthesis. Aldrichim Acta. 2004;37:66–77. [Google Scholar]
- 73.Agudelo R.A., Gauthier S.F., Pouliot Y., Marin J., Savoie L. Kinetics of peptide fraction release during in vitro digestion of casein. J Sci Food Agric. 2004;84:325–332. [Google Scholar]
- 74.Gauthier S.F., Vachon C., Savoie L. Enzymatic conditions of an in vitro method to study protein digestion. J Food Sci. 1986;51:960–964. [Google Scholar]
- 75.Urban P.L., Goodall D.M., Bruce N.C. Enzymatic microreactors in chemical analysis and kinetic studies. Biotechnol Adv. 2006;24:42–57. doi: 10.1016/j.biotechadv.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 76.Miranda G., Pelissier J.P. Kinetic studies of in vivo digestion of bovine unheated skim-milk proteins in the rat stomach. J Dairy Res. 1983;50:27–36. doi: 10.1017/s0022029900032490. [DOI] [PubMed] [Google Scholar]
- 77.Gutmann B., Schwan A.M., Reichart B., Gspan C., Hofer F., Kappe C.O. Activation and deactivation of a chemical transformation by an electromagnetic field — evidence for specific microwave effects in the formation of Grignard reagents. Angew Chem Int Ed. 2011;50:7636–7640. doi: 10.1002/anie.201100856. [DOI] [PubMed] [Google Scholar]
- 78.Robinson J., Kingman S., Irvine D., Licence P., Smith A., Dimitrakis G. Understanding microwave heating effects in single mode type cavities — theory and experiment. Phys Chem Chem Phys. 2010;12:4750–4758. doi: 10.1039/b922797k. [DOI] [PubMed] [Google Scholar]
- 79.Robinson J., Kingman S., Irvine D., Licence P., Smith A., Dimitrakis G. Electromagnetic simulations of microwave heating experiments using reaction vessels made out of silicon carbide. Phys Chem Chem Phys. 2010;12:10793–10800. doi: 10.1039/c0cp00080a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.