Abstract
Intrahepatic cholangiocarcinoma (ICC) is the second most common primary malignancy in the liver. ICC has been classified as a malignant tumor arising from cholangiocytes; however, the co-occurrence of ICC and viral hepatitis suggests that ICC originates in hepatocytes. In order to determine the cellular origin of ICC, we used a mouse model of ICC in which hepatocytes and cholangiocytes were labeled with heritable, cell type–specific reporters. Our studies reveal that ICC is generated by biliary lineage cells derived from hepatocytes, rather than cholangiocytes. Additionally, we found that Notch activation is critical for hepatocyte conversion into biliary lineage cells during the onset of ICC and its subsequent malignancy and progression. These findings will help to elucidate the pathogenic mechanism of ICC and to develop therapeutic strategies for this refractory disease.
Introduction
ICC is the second most prevalent primary malignancy in the liver, and its incidence and mortality rates are increasing worldwide (1, 2). Intrahepatic cholangiocarcinoma (ICC) has been known as a tumor arising from malignant transformation of cholangiocytes (biliary epithelial cells) composing the intrahepatic bile ducts (3, 4). However, because ICC has been characterized by histological observations of dissected tumors, it is difficult to conclude whether ICC is truly derived from cholangiocytes. Indeed, it was reported that patients with viral hepatitis often develop ICC, suggesting that ICC is derived from transformed hepatocytes (5–7). For these reasons, it must be said that the cellular origin of ICC remains obscure. In this study, we sought to address this fundamental issue by following the lineage of two distinct types of hepatic epithelial cells, namely hepatocytes and cholangiocytes, during the development of ICC to determine which cell type is the source of ICC.
Results and Discussion
In our experimental system, hepatocytes or cholangiocytes expressing albumin (Alb) or cytokeratin 19 (CK19) in the liver, respectively, were heritably labeled in mutant mice with a tamoxifen-inducible (TM-inducible) Cre/loxP system. We used mouse lines expressing an inducible form of Cre recombinase (CreERT2) from the Alb or CK19 genomic locus (Alb-CreERT2 mice, ref. 8, or CK19-CreERT2 mice [Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI63065DS1], respectively). In mice resulting from crosses with R26RlacZ/lacZ or R26RYFP/YFP reporter mouse lines (9), administration of TM allowed permanent marking of Alb- or CK19-expressing cells and enabled us to follow the fate of the progeny of hepatocytes or cholangiocytes, respectively (Figure 1A and Supplemental Figures 2 and 3).
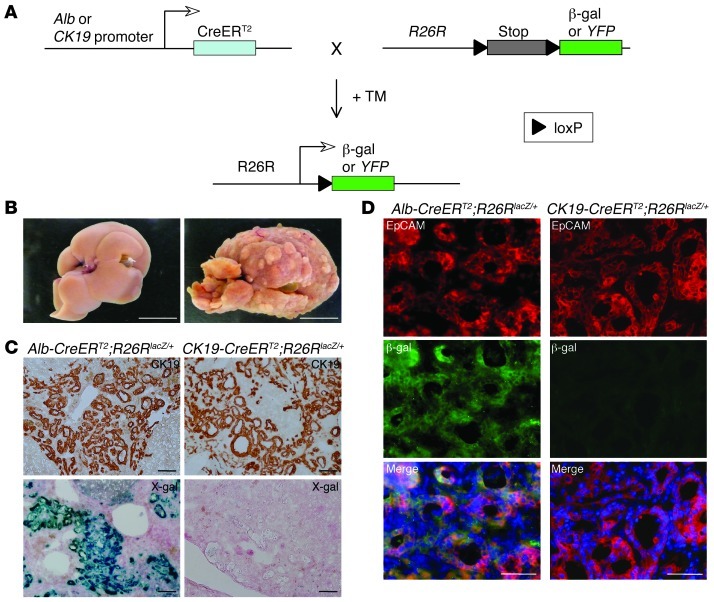
Figure 1. Primitive ductules formed in ICC are derived from hepatocytes, rather than cholangiocytes.
(A) Experimental procedure to follow the lineage of hepatocytes and cholangiocytes in mouse liver. In the presence of TM, CreERT2 expressed from the Alb or CK19 genomic locus translocates into the nucleus and removes the loxP-flanked stop cassette from the R26R allele, leading to permanent heritable expression of the β-gal or YFP gene. (B) Representative images of liver from mice that drank normal water (left) or TAA-containing water (right) for 30 weeks. Tumoral macronodules only formed after TAA administration. (C and D) Immunohistochemical staining of CK19 (C, top row), X-gal staining (C, bottom row), and co-immunofluorescence staining of β-gal and EpCAM (D) were conducted for neoplastic nodules formed in the liver of Alb-CreERT2;R26RlacZ/+ mice and CK19-CreERT2;R26RlacZ/+ mice after 30 weeks’ TAA administration. DNA was stained with DAPI. Scale bars: 10 mm (B), 100 μm (C), and 50 μm (D).
At 1 week after TM injection, we administered thioacetamide (TAA) to Alb-CreERT2;R26RlacZ/+ and CK19-CreERT2;R26RlacZ/+ mice to induce ICC (10, 11). After 30 weeks of TAA administration, all of the treated mice developed macronodular liver cirrhosis (Figure 1B). Immunohistochemical and histochemical analyses revealed that many primitive ductules composed of CK19-positive cells and containing cells with cytoplasmic mucin granules were formed into neoplastic nodules, showing the typical histology of ICC (Figure 1C and Supplemental Figure 4). In contrast, the absence of hepatocyte nuclear factor 4α (Hnf4α) expression in these primitive ductular cells eliminated the possibility of the development of hepatocellular carcinoma with pseudoglandular formation (Supplemental Figure 5). X-gal staining conducted to detect β-gal enzyme activity revealed that the primitive ductules found in Alb-CreERT2;R26RlacZ/+ mice, but not in CK19-CreERT2;R26RlacZ/+ mice, consisted of lacZ-positive cells (Figure 1C). Co-immunofluorescence staining of β-gal and epithelial cell adhesion molecule (EpCAM), an antigen for biliary lineage cells in the normal and neoplastic liver (12), revealed that EpCAM-expressing primitive ductules in Alb-CreERT2;R26RlacZ/+ mice, but not in CK19-CreERT2;R26RlacZ/+ mice, were positive for β-gal expression (Figure 1D). In control experiments, we kept Alb-CreERT2;R26RlacZ/+ and CK19-CreERT2;R26RlacZ/+ mice without TAA administration for 30 weeks after TM injection. In these mice, liver tissue morphology was normal, and β-gal was only detectable in hepatocytes and cholangiocytes, respectively (data not shown). These data indicate that the primitive ductules formed into neoplastic nodules of ICC are derived from hepatocytes, rather than cholangiocytes.
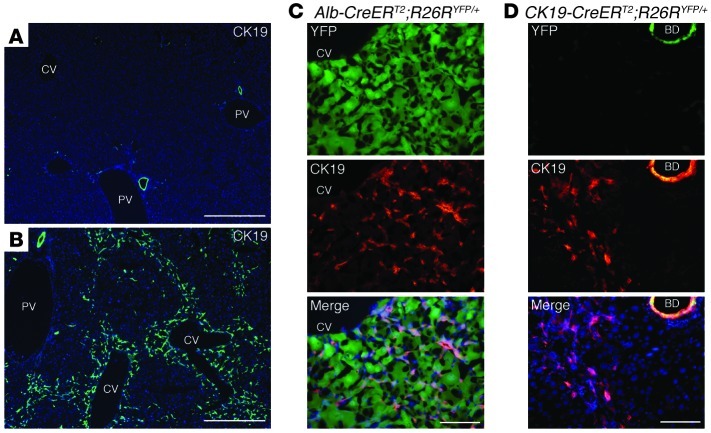
Given these unexpected findings, we next examined the mechanism controlling the generation of ICC from hepatocytes. After 14 weeks of TAA administration, CK19-expressing cells appeared in the hepatic lobules around the central veins in the liver, which were distinguished from cholangiocytes lining the intrahepatic biliary apparatus (Figure 2, A and B). Immunofluorescence analyses revealed that these newly generated CK19-positive cells expressed YFP in Alb-CreERT2;R26RYFP/+ mice, but not in CK19-CreERT2;R26RYFP/+ mice (Figure 2, C and D), indicating that hepatocytes residing around the central veins were initially converted into CK19-positive biliary lineage cells. These CK19-positive cells had an elongated morphology, but were clearly distinct from the surrounding desmin-positive stellate cells (Supplemental Figure 6) and were not derived from stellate cells that exhibited unexpected Cre activity (Supplemental Figure 7).
Figure 2. Hepatocytes on the periphery of central veins in the hepatic lobules are converted into biliary lineage cells at the onset of ICC.
(A–D) Immunofluorescence staining of CK19 was conducted in the liver of control wild-type mice (A) and the liver of wild-type mice (B), Alb-CreERT2;R26RYFP/+ mice (C), and CK19-CreERT2;R26RYFP/+ mice (D) after 14 weeks of TAA administration. CV, cetral vein; PV, portal vein; BD, bile duct. DNA was stained with DAPI. Scale bars: 500 μm (A and B) and 100 μm (C and D).
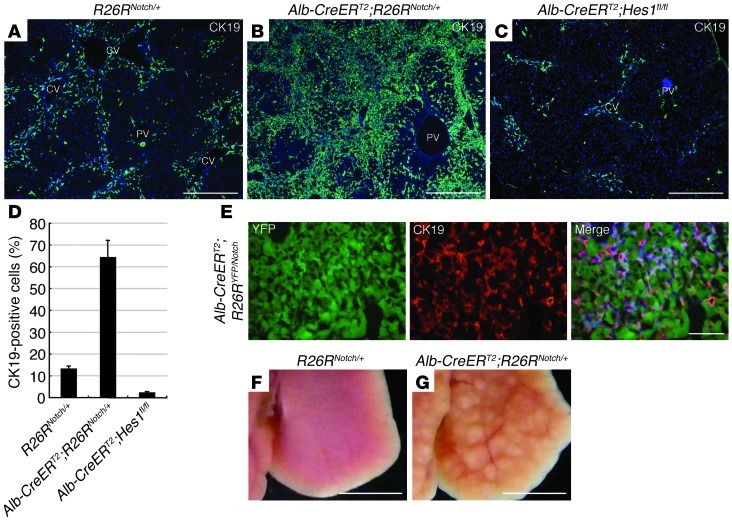
Previous studies have shown that activation of Notch signaling is important for induction of the biliary program not only in hepatic progenitor cells during development but also in postnatal hepatocytes (13, 14). Thus, our findings suggested that Notch signaling is also important for the conversion of hepatocytes into biliary lineage cells in the early stages of ICC formation. To address this issue, we investigated the process of ICC formation in the liver of TM-administered Alb-CreERT2;R26RNotch/+ mice, in which the hepatocytes express an intracellular fragment of mouse Notch1 (NotchIC) (15) that exhibits constitutive Notch signaling activity, and Alb-CreERT2;Hes1fl/fl mice, in which the hepatocytes lack the gene encoding the Notch effector Hes1 (16). After 14 weeks of TAA administration, immunofluorescence analyses revealed that the number of CK19-positive cells in the hepatic lobules was significantly increased or decreased in Alb-CreERT2;R26RNotch/+ or Alb-CreERT2;Hes1fl/fl mice, respectively (Figure 3, A–D). Lineage-tracing analyses using Alb-CreERT2;R26RYFP/Notch mice confirmed that the CK19-positive cells that were increased in response to Notch signal activation were derived from hepatocytes (Figure 3E). Moreover, neoplastic nodules rapidly formed in the liver of Alb-CreERT2;R26RNotch/+ mice even after 14 weeks of TAA administration, but were not observed in the liver of R26RNotch/+ and Alb-CreERT2;Hes1fl/fl mice (Figure 3, F and G, and Supplemental Table 1). Indeed, the number of developing primitive ductules formed by CK19-positive cells was considerably increased in the liver of Alb-CreERT2;R26RNotch/+ mice after 14 weeks of TAA administration (Supplemental Figure 8). Thus, the results of our gain-of-function and loss-of-function experiments demonstrate that ICC arises through Notch-mediated conversion of hepatocytes into biliary lineage cells and that its malignancy and progression depend on the intensity of Notch signaling in hepatocytes.
Figure 3. Notch signal activation is significant not only for conversion of hepatocytes into biliary lineage cells at the onset of ICC, but also the malignancy and progression of ICC.
(A–C) Immunofluorescence staining of CK19 was conducted in the liver of R26RNotch/+ mice (A), Alb-CreERT2;R26RNotch/+ mice (B), and Alb-CreERT2;Hes1fl/fl mice (C) after 14 weeks of TAA administration. (D) The percentages of cells immunoreactive for CK19 in the liver of R26RNotch/+, Alb-CreERT2;R26RNotch/+, and Alb-CreERT2;Hes1fl/fl mice were calculated after counting approximately 3,000 cells per field of vision in 3 discontinuous liver tissue slides for 3 individual mice after 14 weeks of TAA administration. The data represent mean ± SD. (E) Immunofluorescence staining of CK19 was conducted in the liver of Alb-CreERT2;R26RYFP/Notch mice after 8 weeks of TAA administration. (F and G) Representative images of the liver from R26RNotch/+ mice (F) and Alb-CreERT2;R26RNotch/+ mice (G) after 14 weeks of TAA administration. Developing neoplastic nodules are only found in the liver of Alb-CreERT2;R26RNotch/+ mice. CV, central vein; PV, portal vein. DNA was stained with DAPI. Scale bars: 500 μm (A–C), 100 μm (E), and 5 mm (F and G).
Although it has been generally accepted that ICC is a tumor derived from cholangiocytes, our present data indicate that, at least in our mouse TAA model, ICC actually arises from hepatocytes, rather than cholangiocytes, through Notch-mediated conversion of hepatocytes into biliary lineage cells. However, it remains unclear why hepatocytes residing around the central veins are actively converted into biliary lineage cells in this model of ICC. In the portal areas of the hepatic lobule, it has been reported that hepatocytes can change into biliary lineage cells when intrahepatic bile duct regeneration is induced, but cholangiocytes cannot proliferate owing to toxic influences (17). Thus, hepatocytes may be capable of changing their fate to biliary lineage cells regardless of their position in the hepatic lobule, while the particular location of lineage-converting hepatocytes is likely decided by the nature of the toxins used. In the case of TAA, its toxicity depends on the activity of cytochrome P450 2B and 2E1, which are expressed in hepatocytes around the central veins in the liver (18–22). This may likely explain why CK19-positive biliary lineage cells appeared in the pericentral zone of the hepatic lobule after TAA administration.
Although it remains uncertain whether human ICC also arises from hepatocytes, our data may provide an answer to the question of why patients with viral hepatitis often develop ICC. In these patients, hepatocytes infected with hepatitis viruses may be converted into biliary lineage cells via Notch signal activation and become the source of ICC. Therefore, suppression of Notch signaling, which could inhibit the conversion of hepatocytes into biliary lineage cells at the onset of ICC, would be a novel strategy in the treatment of ICC. The identification of the cellular origin of ICC will be instrumental not only for elucidating the pathogenic mechanism of ICC, but also developing therapeutic approaches to this refractory disease.
Methods
Mice.
C57BL/6 mice (Clea), Alb-CreERT2 mice (a gift from Pierre Chambon and Daniel Metzger, Institute of Genetics and Molecular and Cellular Biology, Illkirch, R26RlacZ/lacZ mice (Jackson Laboratory), R26RYFP/YFP mice (a gift from Frank Costantini, Columbia University, New York, New York, USA), R26RNotch/+ mice (a gift from Douglas A. Melton, Harvard University, Cambridge, Massachusetts, USA), and Hes1fl/fl mice (a gift from Ryoichiro Kageyama, Kyoto University, Kyoto, Japan) were used in this study. To generate CK19-CreERT2 mice, we performed targeted insertion of the CreERT2 coding sequence (a gift from P. Chambon and D. Metzger) into the first exon of the CK19 gene by homologous recombination in embryonic stem cells. Genotyping by PCR was conducted using the following primers: CK19-wt-F, 5′-TCTCGCCTCCTACTTGGACAA-3′; CK19-mut-F, 5′-CTATCGCCTTCTTGACGAGTT-3′; CK19-R, 5′-ATATCCCTGACTATCCAAGCA-3′. For wild-type CK19, CK19-wt-F and CK19-R were used, and the product was 400 bp. For the mutant allele, CK19-mut-F and CK19-R were used, and the product was 720 bp.
Chemical administration.
For induction of Cre activity, mice (8–10 weeks old) were given a single intraperitoneal injection of TM (6.5–8.0 mg/mouse; Sigma-Aldrich) dissolved in olive oil (Nacalai Tesque) at a concentration of 50 mg/ml. To induce ICC, we provided the mice with drinking water containing TAA (300 mg/l; Wako).
Immunostaining.
Liver tissues with YFP expression were fixed with 2% paraformaldehyde at 4°C overnight. After treatment with 30% sucrose at 4°C overnight, the tissues were embedded in O.C.T. compound (Tissue-Tek), and frozen sections were prepared. Liver tissues without YFP expression were directly embedded in O.C.T. compound, and frozen sections of these tissues were initially fixed with 4% paraformaldehyde for 5 minutes and then fixed with methanol for 5 minutes at room temperature. After washing in phosphate-buffered saline containing 0.1% Tween-20 and blocking, the sections were incubated with the following primary antibodies: rat anti-EpCAM (1:200; eBioscience); rabbit anti–β-gal (1:2,000; Abcam); goat anti-albumin (1:200; Bethyl); rabbit anti-keratin (1:2,000; Dako); mouse anti-Hnf4α (1:200; Perseus Proteomics Inc.); mouse anti-desmin (1:500; Sigma-Aldrich); and rabbit anti-GFP/YFP (1:1500; MBL). As a primary antibody, we also used a rabbit anti–mouse CK19 antibody generated against the peptide HYNNLPTPKAI (Genenet Co.). After washing, the sections were incubated with horseradish peroxidase–conjugated secondary antibodies (1:500; Dako) for immunohistochemistry or Alexa 488– and/or Alexa 555–conjugated secondary antibodies (1:200; Molecular Probes) with DAPI for immunofluorescence staining.
Study approval.
The experiments were approved by the Kyushu University Animal Experiment Committee, and the care of the animals was in accordance with institutional guidelines.
Supplementary Material
Acknowledgments
We thank Pierre Chambon, Daniel Metzger, Frank Costantini, Douglas A. Melton, and Ryoichiro Kageyama for sharing reagents and providing mice; and Maiko Terada, Eriko Gunshima, and Masato Tanaka for technical assistance. This work was supported in part by the Program for Improvement of the Research Environment for Young Researchers from the Special Coordination Funds for Promoting Science and Technology commissioned by the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT); Grants-in-Aid for Scientific Research from MEXT; a Health Labour Sciences Research Grant in Japan; the Precursory Research for Embryonic Science and Technology Program of the Japan Science and Technology Agency; the Core Research for Evolutional Science and Technology Program of the Japan Science and Technology Agency; and the Clinical Research Foundation.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(11):3914–3918. doi:10.1172/JCI63065.
References
- 1.Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD. Cholangiocarcinoma. Lancet. 2005;366(9493):1303–1314. doi: 10.1016/S0140-6736(05)67530-7. [DOI] [PubMed] [Google Scholar]
- 2.Goodman ZD. Neoplasms of the liver. Mod Pathol. 2007;20(suppl 1):S49–S60. doi: 10.1038/modpathol.3800682. [DOI] [PubMed] [Google Scholar]
- 3.Blechacz B, Gores GJ. Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology. 2008;48(1):308–321. doi: 10.1002/hep.22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gatto M, et al. Cholangiocarcinoma: update and future perspectives. Dig Liver Dis. 2010;42(4):253–260. doi: 10.1016/j.dld.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Shaib YH, El-Serag HB, Davila JA, Morgan R, McGlynn KA. Risk factors of intrahepatic cholangiocarcinoma in the United States: a case-control study. Gastroenterology. 2005;128(3):620–626. doi: 10.1053/j.gastro.2004.12.048. [DOI] [PubMed] [Google Scholar]
- 6.Lee CH, Chang CJ, Lin YJ, Yeh CN, Chen MF, Hsieh SY. Viral hepatitis-associated intrahepatic cholangiocarcinoma shares common disease processes with hepatocellular carcinoma. Br J Cancer. 2009;100(11):1765–1770. doi: 10.1038/sj.bjc.6605063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sempoux C, et al. Intrahepatic cholangiocarcinoma: new insights in pathology. Semin Liver Dis. 2011;31(1):49–60. doi: 10.1055/s-0031-1272839. [DOI] [PubMed] [Google Scholar]
- 8.Schuler M, Dierich A, Chambon P, Metzger D. Efficient temporally controlled targeted somatic mutagenesis in hepatocytes of the mouse. Genesis. 2004;39(3):167–172. doi: 10.1002/gene.20039. [DOI] [PubMed] [Google Scholar]
- 9.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeh CN, Maitra A, Lee KF, Jan YY, Chen MF. Thioacetamide-induced intestinal-type cholangiocarcinoma in rat: an animal model recapitulating the multi-stage progression of human cholangiocarcinoma. Carcinogenesis. 2004;25(4):631–636. doi: 10.1093/carcin/bgh037. [DOI] [PubMed] [Google Scholar]
- 11.Mansuroglu T, et al. Expression of stem cell factor and its receptor c-Kit during the development of intrahepatic cholangiocarcinoma. Lab Invest. 2009;89(5):562–574. doi: 10.1038/labinvest.2009.15. [DOI] [PubMed] [Google Scholar]
- 12.de Boer CJ, van Krieken JH, Janssen-van Rhijn CM, Litvinov SV. Expression of Ep-CAM in normal, regenerating, metaplastic, and neoplastic liver. J Pathol. 1999;188(2):201–206. doi: 10.1002/(SICI)1096-9896(199906)188:2<201::AID-PATH339>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 13.McCright B, Lozier J, Gridley T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development. 2002;129(4):1075–1082. doi: 10.1242/dev.129.4.1075. [DOI] [PubMed] [Google Scholar]
- 14.Zong Y, et al. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136(10):1727–1739. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100(25):14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imayoshi I, Shimogori T, Ohtsuka T, Kageyama R. Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development. 2008;135(15):2531–2541. doi: 10.1242/dev.021535. [DOI] [PubMed] [Google Scholar]
- 17.Michalopoulos GK, Barua L, Bowen WC. Transdifferentiation of rat hepatocytes into biliary cells after bile duct ligation and toxic biliary injury. Hepatology. 2005;41(3):535–544. doi: 10.1002/hep.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter AL, Holscher MA, Neal RA. Thioacetamide-induced hepatic necrosis. I. Involvement of the mixed-function oxidase enzyme system. J Pharmacol Exp Ther. 1977;200(2):439–448. [PubMed] [Google Scholar]
- 19.Porter WR, Neal RA. Metabolism of thioacetamide and thioacetamide S-oxide by rat liver microsomes. Drug Metab Dispos. 1978;6(4):379–388. [PubMed] [Google Scholar]
- 20.Wang T, Shankar K, Ronis MJ, Mehendale HM. Potentiation of thioacetamide liver injury in diabetic rats is due to induced CYP2E1. J Pharmacol Exp Ther. 2000;294(2):473–479. [PubMed] [Google Scholar]
- 21.Oinonen T, Nikkola E, Lindros KO. Growth hormone mediates zone-specific gene expression in liver. FEBS Lett. 1993;327(2):237–240. doi: 10.1016/0014-5793(93)80176-U. [DOI] [PubMed] [Google Scholar]
- 22.Anundi I, Lähteenmäki T, Rundgren M, Moldeus P, Lindros KO. Zonation of acetaminophen metabolism and cytochrome P450 2E1-mediated toxicity studied in isolated periportal and perivenous hepatocytes. Biochem Pharmacol. 1993;45(6):1251–1259. doi: 10.1016/0006-2952(93)90277-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.