Abstract
The list of functions of long noncoding RNAs (lncRNAs) in human tissues is rapidly growing. To further underscore their critical role in human health, two reports in this issue of JCI associate altered expression of novel lncRNAs with the heritable syndromes HELLP and brachydactyly type E.
As technological advances such as next-generation sequencing and targeted RNA capture allow for a more refined view of the human transcriptome, the idea that the sole function of RNA is to transmit genetic information from DNA to ribosomes, facilitating protein expression, has faded. Studies like the Encyclopedia of DNA Elements (ENCODE) Project (1) are providing a more complete understanding of the transcribed genome, showing that noncoding intergenic and “desert” regions of the genome are in fact teeming with transcriptional life. Moreover, interesting and novel functions are being ascribed to a bewildering array of noncoding RNAs (ncRNAs) identified by such projects, and it is now clear that these comprise a large portion of the total RNA pool and perform critical cellular functions (2). One particular class, the long ncRNA (lncRNA), is of emerging interest as a major contributor to gene regulation in many biological processes and diseases. In this issue of the JCI, two papers identify lncRNAs as causal agents linked to important human inherited conditions — the hemolysis, elevated liver enzymes, and low platelets (HELLP) syndrome (3) and brachydactyly type E (BDE; ref. 4).
lncRNAs have many functions
Although many classes of functional ncRNAs have been recently described (including microRNAs [miRNAs], snRNAs, snoRNAs, piwiRNAs, and circular RNAs), lncRNAs are distinguished by their length (greater than 200 nucleotides). Often, lncRNAs are encoded in many exons spread over large, intergenic DNA regions (>50 kb) and undergo canonical RNA splicing to yield ncRNAs that are kilobases in length. Various diverse functions have been proposed for lncRNAs, including roles in regulating DNA metabolism, chromatin structure, and gene expression (reviewed in refs. 2, 5).
A useful framework has been suggested (2, 5) to generally classify lncRNAs functionally — as either scaffolds assembling interacting proteins, tethers linking proteins to areas of transcription (in cis), or guides bringing protein complexes to a regulated region of the genome (in trans). The best-known scaffold lncRNA is probably TERC, the RNA subunit of the telomerase holoenzyme complex. TERC both provides the RNA template for telomeric DNA extension and also binds TERT, the enzymatic protein subunit of telomerase that harbors reverse transcriptase activity. An example of a presumed tether ncRNA is Xist, which is expressed from the inactive X-chromosome (Xi) and is required for polycomb group–mediated (PcG-mediated) silencing of the Xi in cis. HOTAIR, on the other hand, is an example of a guide lncRNA, binding PcG proteins to lead this repressive machinery to repress loci on other chromosomes (acting in trans). Although helpful, this conceptual framework is not rigid, as these various activities are not mutually exclusive: for example, HOTAIR and TERC play scaffolding roles in the assembly of multiprotein enzymatic complexes, but also likely serve as guides to targeted regions of the genome.
It is important to mention that although several lncRNAs have been associated with epigenetic transcriptional repression, the mechanisms whereby lncRNAs cause gene silencing are distinct from those of the better-understood miRNAs, which target specific mRNAs posttranscriptionally to block translation and induce transcript degradation. In contrast, the binding of lncRNAs to DNA/chromatin can promote profound silencing of transcription, which is epigenetically enforced and stable through cellular replication, allowing for heritable gene regulation from mother to daughter cells. For example, Xist-mediated inactivation of an X-chromosome in a female somatic stem cell is reliably transmitted so that the same X-chromosome is silenced in that cell’s progeny. Similarly, other lncRNAs such as Airn, H19, and KCNQ1OT1 promote the regional and heritable silencing of neighboring genes as part of the process of genomic imprinting (6).
The precise mechanisms whereby lncRNAs enforce transcriptional silencing are not well understood; therefore, even the question of whether lncRNAs act predominantly in cis or in trans has become controversial. Many lncRNAs with known target genes are transcribed near these targets, consistent with activity in cis. For example, a lncRNA expressed from the DHFR locus represses expression of the DHFR gene, presumably through cis action. Surprisingly, however, ectopic production of this lncRNA (i.e., in trans) is also able to cause repression of DHFR, suggestive of the capacity for trans activity, at least in the setting of experimental overexpression (7). Further complicating matters are lncRNAs that apparently act in cis to regulate distant targets on the same chromosome. Whereas HOTTIP is located over 40 kilobases from the HOXA genes it regulates, it appears to act in cis, being brought into proximity with its target genes through long-range chromosomal looping (8). In contrast, studies targeting lncRNAs with siRNAs have suggested that lncRNAs regulate gene expression at multiple loci, acting primarily in trans (9). This approach, however, may be biased toward the detection of trans activity, since siRNA-mediated transcript degradation is thought to predominantly occur in the cytosol, and would therefore not be expected to perturb the function of nuclear cis-acting transcripts.
lncRNAs are misregulated in heritable syndromes
Given the clear importance of lncRNAs in various aspects of mammalian cell biology, it is not surprising that mutations of lncRNAs have been associated with several heritable diseases. For example, mutations in TERC cause congenital telomerase insufficiency, leading to premature telomeric shortening in somatic stem cell compartments that manifests as dyskeratosis congenita (DKC), aplastic anemia, and pulmonary fibrosis (8, 10–15). Likewise, DBE-T is a lncRNA that is aberrantly expressed in the autosomal-dominant facio-scapulo-humeral dystrophy (FSHD) syndrome. Expression of DBE-T leads to epigenetic activation of several genes located in the FSHD locus, which generates the disease phenotype (16). Beyond rare disease like FSHD and DKC, single nucleotide polymorphisms and altered splicing/expression of ANRIL, an intriguing lncRNA located near the INK4/ARF (CDKN2a/b) tumor suppressor locus, have been linked to several common age-associated diseases, including atherosclerosis, type II diabetes, and several cancers (17), presumably through cis regulation of the transcripts encoded by the INK4/ARF or Interferon loci (18–20).
Against this background, two new studies have linked altered expression of lncRNAs to genetic syndromes. The HELLP syndrome is an uncommon but serious complication of late pregnancy comprised of the clinical triad of hemolysis, elevated liver enzymes, and low platelets. This condition is associated with placental dysfunction, and recent work has shown that it can be familial. Specifically, a 2001 genome-wide association study found that HELLP is genetically distinct from preeclampsia, a similar clinical entity, and linked HELLP to human chromosome 12q (21). Picking up on this prior work, van Dijk and colleagues made the clever assumption that placental (i.e., fetal) genotype might predispose the mother to HELLP (3). This assumption allowed the group to explain, for example, a pair of monozygotic sisters who were discordant for HELLP. In this instance, the fetus of the affected sister was homozygous for the risk allele, whereas the children of the unaffected sister were homozygous for non-disease alleles. In this way, the authors demonstrated that placental insufficiency attributable to fetal genetics dictated maternal phenotype. Importantly, this understanding explained why this condition does not have a clear recessive or dominant pattern of inheritance (at least when considered from the point of view of the affected mothers) and allowed for more precise mapping of the HELLP susceptibility locus, to a small intergenic region of chromosome 12q23.2 encoding a single lncRNA transcript.
The authors then pursued further experiments to associate this novel lncRNA with HELLP. They showed that this lncRNA was preferentially expressed in placental trophoblasts, consistent with their view that the 12q23 locus is associated with placental insufficiency. Knockdown of this transcript by siRNA and subsequent genome-wide RNA sequence analysis suggested that this lncRNA regulates multiple genes to control placental proliferation. Additionally, the authors used a morpholino approach to stabilize the lncRNA, which altered expression of the same transcripts affected by siRNA knockdown of this lncRNA. Based on these results, the authors proposed that the HELLP lncRNA regulates the cell cycle progression of placental trophoblasts. While van Dijk et al. did not identify a specific causal variant affecting expression or structure of this lncRNA, they believe that increased trophoblast proliferation is associated with reduced placental invasion of the maternal decidua, causing placental insufficiency and HELLP (Figure 1, A and B).
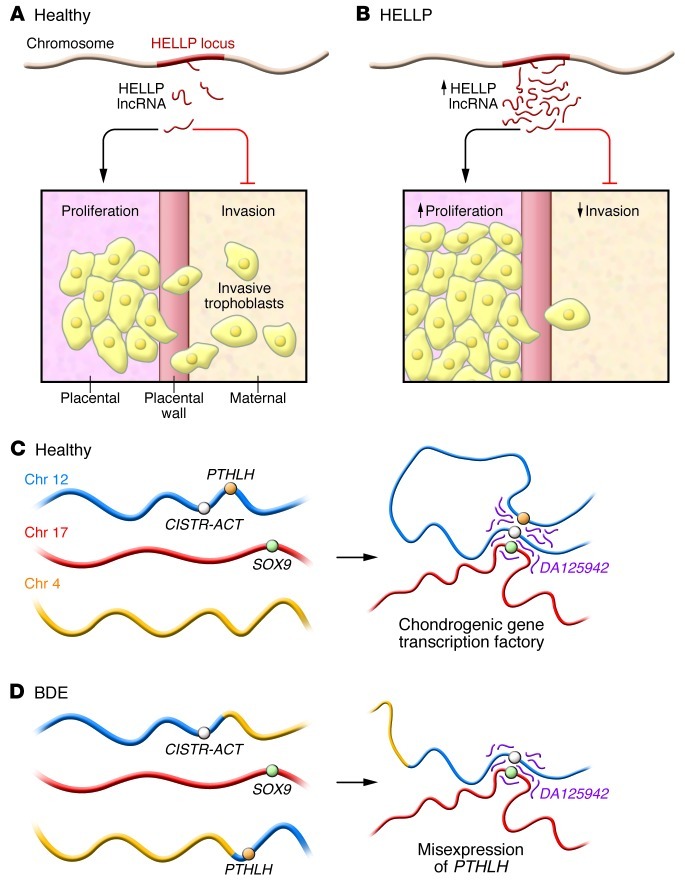
Figure 1. Dysregulation of two novel lncRNAs in heritable conditions.
(A) Placental trophoblasts normally invade the uterine wall as the placenta remodels the maternal vasculature. van Dijk et al. propose that a lncRNA transcribed from the HELLP locus regulates the transition from proliferation to invasion of trophoblasts (3). (B) Mutations predisposing to HELLP that are present in fetal tissue appear to promote increased expression of the HELLP lncRNA (pink) in placental trophoblasts, promoting trophoblast proliferation at the expense of differentiation and invasion. (C) The CISTR-ACT locus (purple) and DA125942, a lncRNA transcribed from the CISTR-ACT locus, interact in cis with PTHLH and in trans with SOX9. Maass et al. conclude that DA125942 organizes chromatin to coordinate transcription of multiple loci involved in cartilage differentiation (4). (D) A chromosomal translocation found in BDE patients disrupts the ability of DA125942 to bind the PTHLH locus in cis, potentially causing premature chondrocyte differentiation at the expense of proliferation and bone elongation.
In a separate study, Maass and colleagues studied families with BDE, a congenital skeletal malformation comprised of shortening of the bones of the hands and feet, sometimes in the setting of short stature. Several genes have been linked to the various subtypes of brachydactyly, but Maass et al. focused on two BDE families harboring balanced germline translocations involving chromosome 12p11.2. Prior work had shown that a chromosome 12 transcript, PTHLH, plays a role in the normal development of the digits, in part by regulating another gene, SOX9, on chromosome 17. Germline mutations of PTHLH or SOX9 have been associated with congenital skeletal malformations; in particular, Maass et al. had previously shown that a family with a balanced translocation on chromosome 12 harbored dysregulated expression of PTHLH (22).
In their present work, Maass et. al. characterize a second family with a balanced chromosome 12 translocation, identifying the breakpoint 145 kb downstream of PTHLH (4). Hypothesizing that the translocation disrupts the interaction between PTHLH and a cis-regulatory element (CRE), the authors used chromatin conformation capture to identify CREs of PTHLH and SOX9. In particular, the authors identified a CRE 24 Mb from PTHLH that associated in cis with PTHLH in cultured skeletal precursor cells. They also demonstrated that this same CRE interacted in trans with SOX9; given these cis and trans enhancer activities, the authors named this CRE CISTR-ACT. Upon further analysis, the authors discovered a conserved lncRNA, DA125942, originating from the CISTR-ACT region and showed that it interacted with both the PTHLH and SOX9 loci. Moreover, altered DA125942 expression resulted in misexpression of PTHLH and SOX9 mRNA. The authors suggest a model whereby DA125942 organizes chromatin so that chondrogenic genes are located in a common “transcription factory,” in which expression of SOX9 and PTHLH is appropriately coordinated (Figure 1C). Loss of the physiologic cis interaction of DA125942 with PTHLH in patients harboring balanced chromosome 12p translocations appeared to result in the BDE phenotype (Figure 1D).
Future questions regarding lncRNAs and human disease
As these studies illustrate, complex developmental events such as placentation and digit formation require tightly regulated transcriptional changes, often requiring coordinate expression of genes located in spatially distinct parts of the genome. Given that lncRNAs play critical roles in the epigenetic control of gene expression, it should not be a surprise that these studies and others have implicated lncRNA deregulation in heritable disease. While showing a novel mode of inheritance of HELLP susceptibility and explaining why chromosome 12p translocations cause BDE are satisfying to the geneticist, they also lead to several new questions. For example, the mechanism whereby the newly identified HELLP RNA affects cellular proliferation and invasion is unclear. It is possible that this lncRNA influences gene expression by guiding regulatory protein complexes in trans to regulated genes, as has been more thoroughly demonstrated for other lncRNAs; however, given the myriad emerging functions ascribed to lncRNAs, one should stay open-minded to the possibility of even more exotic activities. Likewise, for BDE, while the data seem fairly strong linking both cis and trans interactions of DA125942 with regulated target genes, the physical basis for these interactions is unknown, as is the mechanism whereby these interactions control the activity of the transcriptional complexes.
In summary, these reports add to the rapidly growing list of diseases and syndromes associated with misregulated expression or structure of lncRNAs. Clearly, lncRNAs are more common and more important than previously appreciated, and these species appear to play various roles in coordinating transcription and other crucial cellular functions. Several features have made the study of lncRNAs difficult, such as poor sequence conservation in model organisms and difficulties in predicting RNA structure, but given these important examples of human syndromes linked to aberrant lncRNA function, we can hope the increased interest in lncRNAs will accelerate our understanding of their role in human biology and disease.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(11):3837–3840. doi:10.1172/JCI66645.
See the related articles beginning on pages 3990 and 4003.
References
- 1.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Dijk M, et al. HELLP babies link a novel lincRNA to the trophoblast cell cycle. J Clin Invest. 2012;122(11):4003–4011. doi: 10.1172/JCI65171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maass PG, et al. A misplaced lncRNA causes brachydactyly in humans. J Clin Invest. 2012;122(11):3990–4002. doi: 10.1172/JCI65508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23(16):1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koerner MV, Pauler FM, Huang R, Barlow DP. The function of non-coding RNAs in genomic imprinting. Development. 2009;136(11):1771–1783. doi: 10.1242/dev.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445(7128):666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 8.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia CK, Wright WE, Shay JW. Human diseases of telomerase dysfunction: insights into tissue aging. Nucleic Acids Res. 2007;35(22):7406–7416. doi: 10.1093/nar/gkm644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsakiri KD, et al. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci U S A. 2007;104(18):7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armanios MY, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med. 2007;356(13):1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi H, et al. Mutations of the human telomerase RNA gene (TERC) in aplastic anemia and myelodysplastic syndrome. Blood. 2003;102(3):916–918. doi: 10.1182/blood-2003-01-0335. [DOI] [PubMed] [Google Scholar]
- 14.Vulliamy T, et al. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413(6854):432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 15.Vulliamy T, Marrone A, Dokal I, Mason PJ. Association between aplastic anaemia and mutations in telomerase RNA. Lancet. 2002;359(9324):2168–2170. doi: 10.1016/S0140-6736(02)09087-6. [DOI] [PubMed] [Google Scholar]
- 16.Cabianca DS, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149(4):819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeck WR, Siebold AP, Sharpless NE. Review: a meta-analysis of GWAS and age-associated diseases. Aging Cell. 2012;11(5):727–731. doi: 10.1111/j.1474-9726.2012.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harismendy O, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011;470(7333):264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4(4):e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lachmeijer AM, et al. A genome-wide scan for preeclampsia in the Netherlands. Eur J Hum Genet. 2001;9(10):758–764. doi: 10.1038/sj.ejhg.5200706. [DOI] [PubMed] [Google Scholar]
- 22.Maass PG, et al. A cis-regulatory site downregulates PTHLH in translocation t(8;12)(q13;p11.2) and leads to Brachydactyly Type E. Hum Mol Genet. 2010;19(5):848–860. doi: 10.1093/hmg/ddp553. [DOI] [PMC free article] [PubMed] [Google Scholar]