Abstract
Nuclear-encoded precursors of chloroplast proteins are synthesized with an amino-terminal cleavable transit sequence, which contains the information for chloroplastic targeting. To determine which regions of the transit sequence are most important for its function, the chloroplast uptake and processing of a full-length ferredoxin precursor and four mutants with deletions in adjacent regions of the transit sequence were analyzed. Arabidopsis was used as an experimental system for both in vitro and in vivo import. The full-length wild-type precursor translocated efficiently into isolated Arabidopsis chloroplasts, and upon expression in transgenic Arabidopsis plants only mature-sized protein was detected, which was localized inside the chloroplast. None of the deletion mutants was imported in vitro. By analyzing transgenic plants, more subtle effects on import were observed. The most N-terminal deletion resulted in a fully defective transit sequence. Two deletions in the middle region of the transit sequence allowed translocation into the chloroplast, although with reduced efficiencies. One deletion in this region strongly reduced mature protein accumulation in older plants. The most C-terminal deletion was translocated but resulted in defective processing. These results allow the dissection of the transit sequence into separate functional regions and give an in vivo basis for a domain-like structure of the ferredoxin transit sequence.
Among eukaryotes perhaps the most complex subcellular organization is found in higher plants. In addition to routing cytosolically synthesized proteins to peroxisomes, mitochondria, and the secretory machinery, plants also route proteins to their plastids. The targeting of proteins in the plant cell is therefore of particular interest. Because most chloroplast proteins are nuclear encoded and synthesized in the cytoplasm, chloroplast biogenesis depends upon a protein-import apparatus that recognizes these proteins and translocates them across the chloroplast's double-membrane envelope (for reviews, see de Boer and Weisbeek, 1991; Gray and Row, 1995; Schnell, 1995). The in vitro reconstruction of chloroplast import has indicated that the translocation process can be separated into at least three steps: an initial binding step (Cline et al., 1985), full envelope transfer, and proteolytic processing of the transit sequence (Robinson and Ellis, 1984). The initial stages of envelope translocation in pea are mediated by a complex of outer envelope membrane proteins (Tocs) that includes two GTP-binding proteins, a putative channel component (Waegemann and Soll, 1991; Hirsch et al., 1994; Kessler et al., 1994; Perry and Keegstra, 1994; Schnell et al., 1994; Tranel et al., 1995), and at least two membrane-associated molecular chaperones (Waegemann and Soll, 1991; Schnell et al., 1994; Wu et al., 1994). Several candidates for components of the inner membrane translocation machinery (Tics) and stromal components involved in processing have been identified (Oblong and Lamppa, 1992; Vandervere et al., 1995; Kessler and Blobel, 1996; Lubeck et al., 1996).
In unraveling the molecular mechanism of translocation it will be essential to understand the interactions between the import machinery and the transit sequence of the precursor. Such an understanding requires that the content and architecture of the topogenic information in the precursor are known. When analyzing this information only in vitro, possible shortcomings of the experimental system are difficult to discriminate from differences in topogenic information. In the intact plant cell mistargeting needs to be prevented or minimized, which may require the presence of additional information. In addition, developmental aspects of targeting and routing in different tissues need to be understood. Thus, the in vitro analysis of topogenic information may result in a simplified and incomplete picture. This prompted us to analyze the information content of a transit sequence both in vitro and in vivo, in a homologous system.
We chose as a marker for transport, pre-Fd from Silene pratensis, which has been extensively characterized. In vitro this precursor is imported without the need for cytosolic factors, indicating that the precursor directly recognizes the chloroplast envelope (Pilon et al., 1992a). The transit sequence and mature part of the precursor are structurally independent (Pilon et al., 1992b). This partly explains the functional dominance of the transit sequence in gene fusions, e.g. the Fd transit sequence directs many foreign proteins into chloroplasts (Smeekens et al., 1986, 1987; Hageman et al., 1990; de Boer et al., 1991). An extensive in vitro analysis of a collection of deletion mutants covering the full length of the 47-residue-long transit sequence in import, binding, and competition assays with pea chloroplasts revealed four functional domains in the transit sequence contributing to different steps of the translocation process (Pilon et al., 1995).
The use of transgenic plants in this study allowed only a limited set of deletion mutants to be analyzed. As the amino-terminal sequence of the protein might be very important for the efficiency of translation, deletions in the first five amino acids of the transit sequence were not used. Deletions that affect the sequence of the mature part of the protein were also excluded. The four deletion mutants that were chosen span the transit sequence from residues 6 to 42 and are comparable in length. The analysis of import with pea chloroplasts had indicated that each deleted region contains a specific function of the transit sequence. Deletion mutant Δ6–14 was found to be fully defective in the initial binding reaction and import. Deletion mutant Δ15–25 was not translocated but it recognized the envelope, and at this step could compete for the binding of the full-length precursor. Deletion mutant Δ26–34 was imported by pea chloroplasts, although with a decreased efficiency, suggesting that the deleted region is relatively unimportant. Deletion mutant Δ35–42 was imported with a low efficiency but it was not processed (Pilon et al., 1995).
Arabidopsis was used as a model plant. The endogenous Arabidopsis Fd has a higher electrophoretic mobility than the S. pratensis protein and therefore can be distinguished on western blots. Also, for this plant species efficient transformation protocols are available (Valvekens et al., 1988). An in vitro chloroplast import assay was developed for Arabidopsis. This enabled the analysis of the efficiency of translocation and processing and thus the analysis of the information content of the transit sequence both in vivo and in vitro within the same plant species.
The in vivo import results of the deletion mutants are consistent with the presence of separate, functional domains in the transit sequence of Fd, whereas in vitro the deletion mutants could not be imported in Arabidopsis chloroplasts.
MATERIALS AND METHODS
Reagents
Plasmids pETFD-wt, -323, -342, -361, and -372 contain, respectively, the full-length sequence of the wild-type Fd from Silene pratensis or deletions in the transit sequence of amino acids (starting from the N terminus) 6 to 14, 15 to 25, 26 to 34, and 35 to 42 (Pilon et al., 1995). Plasmids pMOG18 and pMOG23 were described (Sijmons et al., 1990). Antiserum raised against spinach Fd was used (Smeekens et al., 1985). Restriction enzymes, oligonucleotides, sequencing kits, the Cap analog G-(5′)-ppp-(5)-G, and Percoll were from Pharmacia. T7 polymerase transcription kits were from Epicentre Technologies (Madison, WI). Plasmid pET11-d was purchased from Novagen (Madison, WI). Wheat-germ lysate translation kits were from Promega. Goat-anti-rabbit antibody with horseradish peroxidase conjugate were from Bio-Rad. Enhanced chemiluminescence western blotting reagents and all radiochemicals were from Amersham. All other chemicals were of the highest grade available.
Plant Growth Conditions
Arabidopsis was grown in soil in growth chambers with 16 h of light and 8 h of darkness at 22°C and 70% RH.
Chloroplast Isolation from Arabidopsis
To obtain a preparation with maximal import efficiency the procedure of Somerville et al. (1981) was modified. Chloroplasts were isolated from 3- to 4-week-old Arabidopsis ecotype Colombia-0. Typically, 40 g fresh weight of plant material from rosette leaves was used, divided in four portions, and ground in a polytron blender in position 3 for 5 s in 100 mL of ice-cold grinding buffer (330 mm sorbitol, 20 mm tricine/KOH, pH 8.4, 5 mm EGTA, 5 mm EDTA, 10 mm NaCO3, 0, 1% BSA, and 330 mg/L isoascorbate). The suspension was filtered through two layers of Miracloth (Calbiochem) between two layers of cheesecloth, resulting in the crude homogenate. From this point all procedures were done at 0°C to 4°C. The suspension was centrifuged for 5 min at 3000 rpm in an HB-4 rotor (Sorvall) to pellet the crude chloroplast fraction.
The chloroplasts were resuspended in grinding buffer containing 40% (v/v) Percoll. The suspension was layered on top of a step gradient consisting of (bottom to top): 2 mL of 70% (v/v) Percoll/grinding buffer, 4 mL of 50% (v/v) Percoll/grinding buffer, and 4 mL of 40% (v/v) Percoll/grinding buffer in a 15-mL polycarbonate tube. Percoll/grinding buffer was supplemented with 330 mg/L isoascorbate and 132 mg/L glutathione. The gradient was centrifuged for 15 min at 5000 rpm in the HB-4 rotor with the brake off. The (lower) band at the 50% to 70% interface was isolated and collected in a SS34 tube (Sorvall) and diluted with 3 to 4 volumes of grinding buffer. The chloroplasts were centrifuged for 3 min at 3000 rpm in the HB-4 rotor. The pellet was gently resuspended in grinding buffer and collected in a microcentrifuge tube. The intactness of the chloroplasts was assessed by phase-contrast light microscopy (Walker et al., 1987) and found to be better than 90%. The number of chloroplasts was compared with chlorophyll content, as determined by the method of Bruinsma (1961); we found that 1 μg of chlorophyll corresponds to 1.5 × 10−6 chloroplasts. Typically, 150 to 250 μg of chlorophyll was obtained per isolation. Chloroplasts were stored on ice in the dark and used for import assays within 2 h after isolation.
In Vitro Import Experiments
Isolated chloroplasts were centrifuged for 2 min at 2,000 rpm at 4°C in a swing-out centrifuge, and the pellet was resuspended in ice-cold import buffer (330 mm sorbitol, 5 mm Hepes/KOH, pH 8.0, 10 mm NaHCO3, 8 mm MgCl2, and 0.1% BSA). To each sample containing 20 μg of chlorophyll, 1.5 mm DTT (final concentration), 1.2 mg of antipain, and 5 mm ATP (final concentration) were added, making the total volume 100 μL. The samples were kept on ice in the dark for 20 min. Fifty microliters of translation mixture in import buffer containing approximately 200,000 TCA-precipitable cpm was added. The samples were then incubated at 25°C in the light to allow import. After 30 min the samples were placed on ice. From this point all procedures were performed at 4°C under dim-green light. The samples were split into two equal aliquots. One aliquot was treated with 7.5 μL of thermolysin (4 mg/mL thermolysin in import buffer and 10 mm CaCl2) at 4°C for 30 min, after which time EDTA in import buffer was added to a final concentration of 5 mm. To the other aliquot EDTA was added directly. The samples were centrifuged for 2 min at 2,000 rpm in a swing-out centrifuge. The pellet was washed once with grinding buffer. Finally, the chloroplasts were lysed in sample buffer (125 mm Tris/HCl, pH 6.8, 5% SDS [v/w], 10% glycerol [v/v], and 5% β-mercaptoethanol [v/v]), followed by heating to 95°C for 5 min. Analysis by SDS-PAGE and fluorography and quantification were performed essentially as described (Pilon et al., 1990; Knight and Gray, 1995).
Plasmid Constructions
Standard procedures were used for restriction enzyme digestions, analysis of restriction sites, ligations, and transformation of Escherichia coli. The NcoI-BamHI fragments from plasmids pETFD-wt, -323, -342, -361, and -372 containing pre-Fd were cloned in pMOG18 (Sijmons et al., 1990) under the control of the constitutively active cauliflower mosaic virus 35S promoter and enhancer, in front of a nopaline synthase termination signal to allow expression in plants, resulting in plasmids pWA1, -2, -3, -4, and -5. The EcoRI-HindIII fragments were subcloned in the binary vector pMOG23 (Sijmons et al., 1990), resulting in plasmids pC1, -2, -3, -4, and -5 to enable plant transformation. The sequence of the inserts in pC1–5 was confirmed by dideoxy sequencing.
Plant Transformation
Constructs pC1–5 were transformed to Agrobacterium tumefaciens strain LBA-4404 containing helper plasmid pTiAch5 (Hoekema et al., 1983), using a freeze-thaw method as described (An et al., 1988). The intactness of the transformed binary vectors in A. tumefaciens was determined by plasmid isolation and restriction mapping as described (An et al., 1988). Arabidopsis seeds ecotype C24 were used in a root-explant transformation procedure according to the method of Valvekens (1988). Plants were regenerated from kanamycin-resistant callus. Calli transformed with the different constructs regenerated with comparable efficiencies, although the number of transgenic lines varied per construct; for each construct 14 to 39 independent transgenic lines were regenerated and self-crossed to generate the seeds for the F1 line. The F1 plants were tested for the presence of S. pratensis pre-Fd by immunoblotting and were self-crossed. These plants were used in Figure 3. F2 plants were germinated on Murashige and Skoog medium containing 125 mg/L kanamycin to select homozygous lines. Homozygous F2 lines were used in Figures 4 and 5. Integration of the T-DNA was determined by Southern blotting.
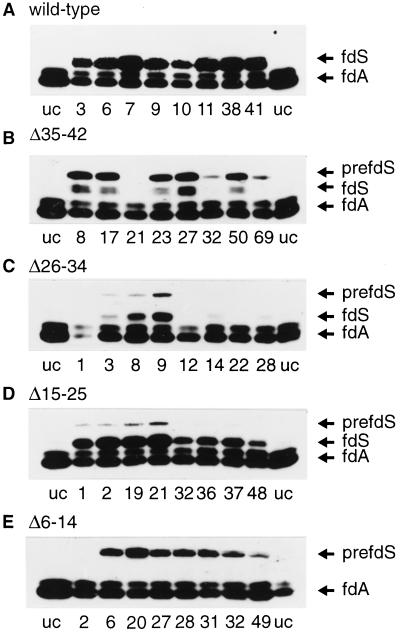
Figure 3.
Immunoblot of total plant extracts from 1-week-old seedlings. Transgenic lines were randomly selected. An equal amount of total protein (10 μg) was loaded per lane. The numbers indicate the number of the independent transgenic plant lines. uc, Untransformed control plant extracts from untransformed Arabidopsis line C24; PrefdS, preFd from S. pratensis; fdS, mature Fd from S. pratensis; fdA, endogenous Arabidopsis mature Fd.
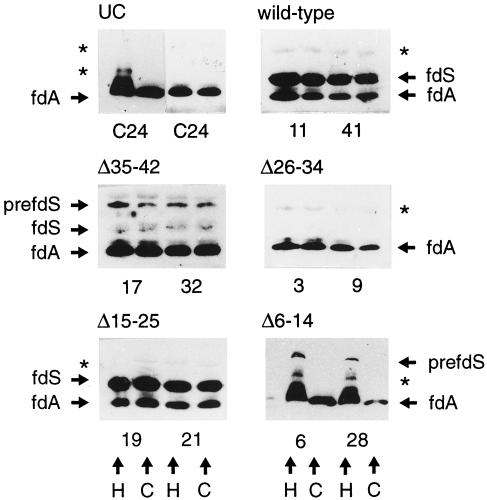
Figure 4.
Immunoblot of fractionated plant extracts. Equal amounts of chlorophyll (1 μg) from the crude homogenate and the isolated chloroplasts were loaded in each lane. The numbers indicate the number of the transgenic plant line. H, Crude homogenate; C, isolated chloroplasts; UC, untransformed control plant extracts from untransformed Arabidopsis line C24; prefdS, pre-Fd from S. pratensis; fdS, mature from S. pratensis; fdA, endogenous Arabidopsis mature Fd. Asterisks indicate unspecific bands that were detected in the untransformed control plants.
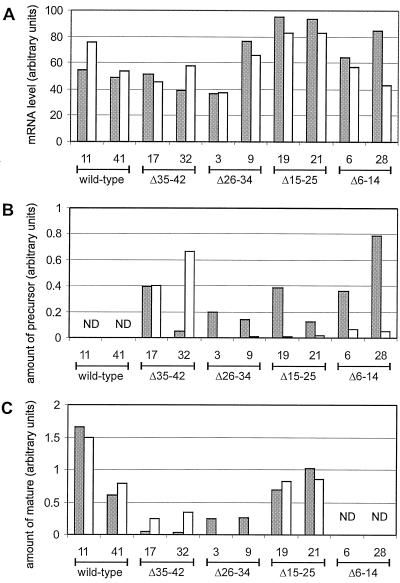
Figure 5.
Quantitative analysis of the expression of S. pratensis Fd. A, Quantitative analysis of a northern blot of total RNA from 1-week-old (gray bars) or 3-week-old (white bars) seedlings. B and C, Quantitative analysis of an immunoblot of total plant extracts of 1-week-old (gray bars) or 3-week-old (white bars) seedlings of precursor- and mature-sized proteins, respectively. The values are the averages of three experiments. ND, Not detectable.
Preparation of Plant Extracts
Plant leaf material, 0.1 to 0.5 g fresh weight, was ground on ice in a microcentrifuge tube in extraction buffer (100 mm NaCl, 50 mm Tris/HCl, pH 7.5, 0, 5% [v/v] Triton X-100, 10 mm β-mercaptoethanol, and 1 mm PMSF). The extract was centrifuged for 5 min in an Eppendorf centrifuge to pellet the insoluble fraction. The soluble fraction was mixed with sample buffer, and 10 μg of protein was applied to SDS-PAGE and immunoblotting was performed essentially as described (Pilon et al., 1990), except that blots were developed with enhanced chemiluminescence reagent. Blots were quantified with a personal densiometer (model SI, Molecular Dynamics, Sunnyvale, CA). All values were averaged over two experiments and corrected for the amount of endogenous Arabidopsis Fd that was detected and correlated to a range of known amounts of purified pre-Fd present on the same blot.
Northern and Southern Blotting
Total RNA was isolated and applied to northern blotting for the quantitative data in Figure 5, as described (Quaedvlieg et al., 1995). DNA was precipitated out of the remaining solution, digested with HindIII, and applied to Southern blotting. Blots were probed with a random-primed BamHI-BstEII fragment from pETFD-wt corresponding to the mature sequence of S. pratensis pre-Fd or an 18S rRNA (Pruitt and Meyerowitz, 1986) probe, which corrected for loading differences. Bands were quantified with a phosphor imager (model SI, Molecular Dynamics).
General Methods
Published methods were used for SDS-PAGE (Laemmli, 1970), protein assays (Bradford, 1976), and in vitro transcription and translation (Pilon et al., 1995).
RESULTS
Fd Precursors
In this study the information content of the Fd transit sequence was analyzed both in vivo and in vitro. The transit sequence of the S. pratensis Fd wild-type precursor and the four deletion mutant precursors Δ6–14, Δ15–25, Δ26–34, and Δ35–42 are given in Figure 1.
Figure 1.
Schematic representation of the transit sequence of S. pratensis Fd. Deletions are indicated with dashes and substitutions are indicated in bold. The arrow represents the processing site.
In Vitro Protein Uptake Assay with Arabidopsis Chloroplasts
It has been reported that, in general, plastids from younger tissues are most active for import (Dahlin and Cline, 1991). Pea chloroplasts are usually isolated from 10- to 12-d-old seedlings. The small size of Arabidopsis, however, made it necessary to use 3- to 4-week-old seedlings to obtain reproducible yields and intactness. Relative to pea, Arabidopsis chloroplasts appeared to have a higher buoyant density. Arabidopsis chloroplasts were pelleted through a Percoll solution of 60% (v/v), whereas pea chloroplasts could only be pelleted through a concentration of 40% (v/v) (not shown). Typically, our preparations contained more than 90% intact chloroplasts. Arabidopsis chloroplasts were more fragile compared with pea chloroplasts, which forced us to reduce the number of manipulations in subsequent import assays (see Methods).
Conditions for import of wild-type S. pratensis pre-Fd into Arabidopsis chloroplasts were optimized. Upon incubation in the presence of ATP, the wheat-germ-synthesized, radiolabeled, wild-type precursor was detected as a mature-sized protein in the chloroplast pellet. This mature-sized protein was located inside of the chloroplasts, since it was protected against externally added protease thermolysin (see Fig. 2). The optimal Mg-ATP concentration was found to be 5 mm (not shown). Magnesium, bicarbonate, and potassium phosphate were reported to influence photosynthetic activity of Arabidopsis chloroplasts (Somerville et al., 1981). To investigate the effect of these salts on import, their concentration in the import assay was varied and the import efficiency was assayed (not shown). The optimal MgCl2 concentration was 8 mm. The addition of 10 mm NaHCO3 raised the efficiency of import, whereas the addition of 10 mm KH2PO4 had no effect. Under the optimized conditions the amount of added precursor that was fully translocated into Arabidopsis chloroplasts varied between 20% and 30% (see Fig. 2), which is comparable to the import efficiency of this precursor with pea chloroplasts.
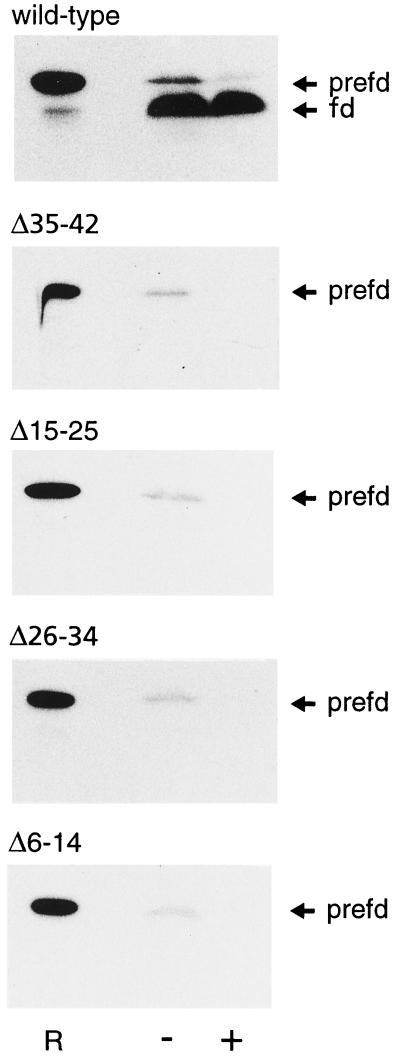
Figure 2.
Fluorogram of the in vitro import of pre-Fd (prefd) and deletion mutants in Arabidopsis chloroplasts. Lane R, 5% of the added precursor was loaded as a reference sample; lane +, a protease treatment with thermolysin; lane –, no protease treatment.
Using the optimized conditions the import of the four deletion mutants Δ6–14, Δ15–25, Δ26–34, and Δ35–42 was compared with the import of the full-length precursor. None of the wheat-germ-synthesized, radiolabeled deletion mutants could be imported by the Arabidopsis chloroplasts (see Fig. 2). For all of the deletion mutants some precursor protein was associated with the chloroplast pellet but it was not resistant to the protease thermolysin. These data are in contrast to the previously obtained in vitro import data with the pea chloroplasts that import deletion mutants Δ26–34 and Δ35–42 with a low efficiency (Pilon et al., 1995). As a control, these experiments with pea chloroplasts were reproduced with the same translation mixtures that were used for the Arabidopsis assays (not shown).
The Expression of S. pratensis Fd Forms in Transgenic Plants
To study the in vivo import capacities of the deletion mutants of S. pratensis Fd, transgenic Arabidopsis plant lines were generated (see Methods) that stably expressed the wild-type S. pratensis Fd and the four deletion mutants.
As conversion to the mature size is catalyzed by a specific enzyme thought to be present only in the chloroplast, processing can be used as a first indication of localization. Therefore, the form in which the S. pratensis Fd is present in the transgenic plants was analyzed. Eight independent lines per construct were randomly chosen and protein extracts were made from leaves of 1-week-old seedlings, resolved by SDS-PAGE, and immunoblotted (see Fig. 3). The endogenous mature Arabidopsis Fd could be detected as a double band (see Fig. 3, lane uc). The location of the S. pratensis Fd-precursor- and mature-sized protein was determined from reference standards (not shown). In plants transformed with the S. pratensis full-length construct, only a band corresponding to the mature-sized S. pratensis Fd was detected next to the endogenous Fd (see Fig. 3A). No precursor-sized Fd was present, suggesting that the protein was efficiently imported and processed by the chloroplast. The expression levels of the S. pratensis Fd gene were variable, as expected when analyzing different transgenic plant lines. Figure 3B shows the results with transgenic plants expressing deletion mutant Δ35–42, with the deletion close to the processing site. Not all lines showed enough expression of the mutant gene to allow detection of the S. pratensis Fd. In the remaining lines expressing deletion mutant Δ35–42, the S. pratensis Fd could be detected both as a precursor-sized band and as two approximately mature-sized processed bands. The large amount of precursor visible indicates that import, processing, or both are less efficient compared with the wild-type precursor.
The results with deletion mutant Δ26–34 are shown in Figure 3C. Most transgenic lines did not show significant levels of S. pratensis Fd. This could be an indication that the precursor is not stable, perhaps because its import competence is affected or because the construct is poorly expressed. In some lines, however, S. pratensis Fd could be detected both as a precursor- and as a mature-sized band, whereas in other lines, only a faint, mature-sized band could be detected. Figure 3D shows the immunoblot of deletion mutant Δ15–25. In all tested lines the mature-sized S. pratensis Fd could be detected, indicating import and processing. In some lines small amounts of precursor-sized protein could be detected. Since no precursor-sized protein could be detected in plants expressing the wild-type S. pratensis Fd, import of deletion mutant Δ15–25 appears to be slightly affected. Figure 3E shows the deletion mutant Δ6–14. In lines expressing this construct only precursor-sized S. pratensis Fd could be detected. Since the deletion in this construct is far removed from the processing site, this lack of processing strongly suggests that the precursor was not translocated into the chloroplast in vivo.
Localization of S. pratensis Fd
Processing is one indication of import by the chloroplast. However, to determine the intracellular localization of the newly introduced Fd, chloroplasts were isolated from the transgenic plants. For these experiments two independent lines with good expression of the S. pratensis Fd (see Fig. 3) were selected for each construct. Because very young Arabidopsis plants are too small to allow reproducible isolation of intact chloroplasts, 3-week-old seedlings were used for these experiments, the same age as the seedlings used in the in vitro experiments. Equal amounts of chloroplasts, as determined by chlorophyll content, were applied to immunoblotting both as crude homogenate and as purified intact chloroplasts (see “Methods and Materials”). The results are shown in Figure 4. The untransformed control plants only showed the endogenous Arabidopsis Fd that resides in the chloroplast, indicating that the method of determining the localization is suitable. Two unspecific bands marked with an asterisk were detected in the untransformed controls. These bands could also be detected in some samples from the transgenic plant lines. In transgenic plants expressing the wild-type pre-Fd from S. pratensis, mature-sized Fd was detected in all samples with approximately the same intensity in the crude homogenate and the isolated chloroplasts. With deletion mutant Δ35–42, a precursor and a mature-sized band were detected both in the crude homogenate and in the isolated chloroplasts. With deletion mutant Δ26–34, no S. pratensis Fd could be detected in the crude homogenate or in the isolated chloroplasts. With deletion mutant Δ15–25 mature-sized S. pratensis Fd was detected with the same intensity in both fractions. In transgenic plants expressing the deletion mutant, Δ6–14 precursor-sized S. pratensis Fd could only be detected in the crude homogenate but not in the chloroplast fraction.
In summary, the newly introduced S. pratensis mature-sized Fd from either the wild type, Δ15–25, or Δ35–42 colocalized with the endogenous Arabidopsis Fd inside the chloroplast, indicating translocation of these precursors. The precursor-sized S. pratensis Fd from deletion mutant Δ35–42 was localized or attached to the chloroplast. In contrast, the precursor-sized band from deletion mutant Δ6–14 did not localize in the chloroplasts, indicating that it was not translocated or bound to the surface. For deletion mutant Δ26–34 the localization of the S. pratensis Fd could not be determined, since no protein could be detected in these samples from older plants.
Expression of the S. pratensis Fd Transgene
To determine whether the amounts of S. pratensis Fd in plant homogenates from 3-week-old (Fig. 4) and 1-week-old seedlings (Fig. 3) were correlated with transcript levels of the introduced S. pratensis Fd gene, total RNA was isolated and applied to a northern blot. The filter was incubated with a probe that hybridizes to the mRNA of the mature part of S. pratensis Fd and quantified (see Fig. 5A). All lines showed a significant detectable expression of the S. pratensis Fd gene, whereas in untransformed plants, no expression could be detected (not shown). The expression of the S. pratensis Fd gene was comparable after 1 or 3 weeks at the mRNA level. Only in line 28 from plants expressing deletion mutant Δ6–14 was the expression lower in 3-week-old seedlings. This indicates that the difference in import pattern of the precursors was not caused by a difference in transgene expression. The absence of the mutant Δ26–34 S. pratensis Fd protein in 3-week-old seedlings was not caused by the lack of expression of the S. pratensis Fd gene and therefore had to originate from posttranscriptional processes.
To further investigate the finding that in older plants the S. pratensis Fd protein could not be detected from deletion mutant Δ26–34 (see Fig. 4), whereas it could be detected in 1-week-old seedlings (see Fig. 3), the levels of S. pratensis Fd protein were quantified. Plant extracts were prepared from 1- and 3-week-old seedlings for quantitative immunoblotting. Figure 5, B and C, shows the amount of S. pratensis precursor and mature protein that could be detected in 1- or 3-week-old seedlings. In plants expressing the wild-type construct, no precursor-sized protein could be detected, whereas the amount of mature-sized protein was the highest. The amount of mutant (Δ6–14, Δ15–25, and Δ26–34) precursor-sized Fd decreased with age, except in plants expressing deletion mutant Δ35–42, from which the precursor-sized Fd was localized to the chloroplast, whereas the amount of mature-sized Fd from deletion mutants Δ15–25 and Δ35–42 after 1 or 3 weeks was comparable (see Fig. 5C). From deletion mutant Δ6–14 no mature-sized protein could be detected.
DISCUSSION
In this study the topogenic information of a chloroplast transit sequence was analyzed both in vitro and in vivo in the same plant species, Arabidopsis. In vitro the full-length S. pratensis Fd precursor was imported in an ATP-dependent fashion by isolated Arabidopsis chloroplasts. In vivo the full-length precursor was imported, and the protein was processed to the mature size and localized in the chloroplast in transgenic Arabidopsis. Four mutants with deletions in adjacent regions of the transit sequence were analyzed for their in vitro and in vivo import competence. In vitro none of these mutant precursors was imported by Arabidopsis chloroplasts. In contrast, in transgenic plants expressing these mutant precursor constructs, more subtle effects were observed. Full loss of import was observed for deletion mutant Δ6–14; in this case, only precursor-sized S. pratensis Fd was detected and it was not localized in the chloroplast. In plants expressing deletion mutants Δ15–25 and Δ26–34, both mature- and precursor-sized proteins were detected. The mature-sized protein of deletion mutant Δ15–25 was localized in the chloroplast, whereas from deletion mutant Δ26–34, the mutant precursor could not be localized. The main effect of deleting amino acids 35 to 42 was on processing. Both precursor-sized and processed forms were observed with Δ35–42 and both were localized in the chloroplasts.
S. pratensis Fd Forms in Transgenic Plants
The amount of precursor-sized protein that could be detected in 3-week-old plants expressing deletion mutants Δ6–14, Δ15–25, and Δ26–34 decreased compared with 1-week-old seedlings. Since the expression on mRNA level was comparable, this can be caused by a difference in translation efficiency, a decreased precursor stability, or an altered import properties of the chloroplast. This does not affect the accumulation of mature-sized wild-type S. pratensis Fd, but it affects the accumulation of mature-sized Fd of deletion mutant Δ26–34.
Difference between in Vitro and in Vivo Import
There is a striking discrepancy between the in vitro and in vivo import properties of the deletion mutants in Arabidopsis chloroplasts. Deletion mutants Δ15–25 and Δ35–42 were imported by the chloroplast in 3-week-old-seedlings in vivo, but not in vitro by chloroplasts isolated from seedlings of the same age. Mature-sized protein from deletion mutant Δ26–34 could only be detected in 1-week-old seedlings in vivo, no import was observed in 3-week-old seedlings either in vitro or in vivo. The lack of import of the mutants in vitro results most likely from both intrinsic properties of these precursor proteins, which reduces their ability to be imported, and the relatively low translocation efficiency of an in vitro import system. In vivo protein accumulation in chloroplasts occurs constantly and the organelle is maintained in its native cellular environment in transgenic plants. For stable precursors in vivo import can be less efficient and still not limit mature-sized protein accumulation.
The finding that the information deduced from the in vitro import system does not fully reflect the in vivo situation was also observed in the unicellular algae Chlamydomonas reinhardtii (Lawrence and Kindle, 1997; Kindle and Lawrence, 1998). Also in transgenic tobacco plants, when analyzing the intraorganellar sorting of thylakoid proteins (de Boer et al., 1991) or the sorting of a chloroplast envelope inner membrane protein (de Castro Silva-Filho et al., 1997), the in vitro system did not reflect the in vivo situation. However, these studies addressed the problem of proper sorting within the organelle, whereas in this paper, targeting and import into the chloroplast were studied.
Difference between Pea and Arabidopsis Chloroplasts
Pea and Arabidopsis chloroplasts differed in their capacity to import two of the deletion mutants in vitro. Deletion mutants Δ26–34 and Δ35–42 could be imported by isolated pea chloroplasts, although with a reduced efficiency (Pilon et al., 1995). No detectable import occurred into Arabidopsis chloroplasts. In all of the experiments a Fd precursor from S. pratensis was used for import in pea and Arabidopsis chloroplasts, which might also affect the import efficiency. Differences in import capacities might partly be explained by the difference in age of the plants used to isolate the chloroplasts for the in vitro import assays. For practical reasons the Arabidopsis plants were older than the pea plants at the time of chloroplast isolations. Previously, Dahlin and Cline (1991) showed that older plastids have a reduced import capacity in vitro. However, our isolated Arabidopsis chloroplasts, despite their age, were still capable of importing the wild-type precursor. Thus, it appears that Arabidopsis chloroplasts are more stringent than pea chloroplasts in their requirement for the transit sequence structure
Domain Structure of the Transit Sequence of Fd
The N-terminal uncharged region of the Fd transit sequence (residues 6–14) contains essential information for chloroplast targeting. This region was also found to interact with lipids of the chloroplast envelope (Pilon et al., 1995). Surprisingly, this region is the least conserved part of the Fd transit sequence. The only features shared by other Fd transit sequences in this region are the hydrophobic nature of these amino acids and the lack of charged residues.
The central region of the transit sequence (residues 15–34) is more tolerant to deletions. The two mutations analyzed in this region, Δ15–25 and Δ26–34, still allow import in vivo, although at a reduced efficiency. In vitro the deletion of amino acids 15 to 25 still allowed initial recognition by the pea chloroplasts but no import (Pilon et al., 1995). Therefore, the central region seems to be involved in envelope translocation after initial recognition. This region of the transit sequence is rich in Ala and hydroxylated amino acids and contains several helix-breaking residues that are conserved among Fd transit sequences. It can be hypothesized that this region contains spacer-like elements that allow proper positioning of specific parts of the transit sequence. In 3-week-old seedlings the reduced import efficiency of deletion mutant Δ26–34 affected the accumulation of mature-sized Fd, indicating that this region becomes more important in older plants.
The C-terminal region of the transit sequence is the most conserved part of the Fd transit sequence and is required for proper processing in vivo. The previously performed in vitro assays with pea chloroplasts indicated that the C terminus also contained a region needed for envelope recognition (Pilon et al., 1995). Our results obtained for deletion mutant Δ35–42 show that import and processing are not strictly coupled in vivo.
The observations made on the transgenic plants are consistent with a structure of the Fd transit sequence consisting of at least three functional domains. This provides an in vivo basis for the previously proposed domain hypothesis that was based on biophysical characterizations and in vitro assays with pea chloroplasts (Pilon et al., 1995). The N-terminal region mediates the initial reactions of the import process, whereas the C-terminal region is important for processing. It is possible that the central region provides flexibility between the N- and C-terminal region, resulting in the overall efficiency of the import process.
Given the proposed roles of the domains of the transit sequence in the import process it will be interesting to investigate the interaction between parts of the transit sequence and known proteins involved in chloroplast import. The possibility of analyzing translocation in vivo would provide an important tool with which to validate results obtained in vitro. Because Arabidopsis is genetically accessible and suitable for a mutant analysis, this system will be exploited further in the future.
ACKNOWLEDGMENT
Vectors pMOG18 en pMOG23 were a kind gift from Mogen International (Leiden, The Netherlands).
Footnotes
This work is part of the joint program “Chloroplast Protein Import” in collaboration with Prof. B. de Kruijff and Prof. W. Vredenberg, and was supported by a Netherlands Organization for Scientific Research/Earth and Life Sciences grant.
LITERATURE CITED
- An G, Ebert PR, Mitra A, Ha SB. Binary vectors. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. pp. A3/1–19. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bruinsma J. A comment on the spectrophotometric determination of chlorophyll. Biochim Biophys Acta. 1961;52:576–578. doi: 10.1016/0006-3002(61)90418-8. [DOI] [PubMed] [Google Scholar]
- Cline K, Werner-Washburne M, Lubben TH, Keegstra K. Precursors to two nuclear-encoded chloroplast proteins bind to the outer envelope membrane before being imported into chloroplasts. J Biol Chem. 1985;260:3691–3696. [PubMed] [Google Scholar]
- Dahlin C, Cline K. Developmental regulation of the plastid protein import apparatus. Plant Cell. 1991;3:1131–1140. doi: 10.1105/tpc.3.10.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer D, Bakker H, Lever A, Bouma T, Salentijn E, Weisbeek P. Protein targeting towards the thylakoid lumen of chloroplasts: proper localization of fusion proteins is only observed in vivo. Embo J. 1991;10:2765–2772. doi: 10.1002/j.1460-2075.1991.tb07825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer D, Weisbeek PJ. Chloroplast protein topogenesis: import, sorting and assembly. Biochim Biophys Acta. 1991;1071:221–253. doi: 10.1016/0304-4157(91)90015-o. [DOI] [PubMed] [Google Scholar]
- de Castro Silva-Filho M, Wieers M, Flugge UI, Chaumont F, Boutry M. Different in vitro and in vivo targeting properties of the transit peptide of a chloroplast envelope inner membrane protein. J Biol Chem. 1997;272:15264–15269. doi: 10.1074/jbc.272.24.15264. [DOI] [PubMed] [Google Scholar]
- Gray JC, Row PE. Protein translocation across chloroplast envelope membranes. Trends Cell Biol. 1995;5:243–247. doi: 10.1016/s0962-8924(00)89018-2. [DOI] [PubMed] [Google Scholar]
- Hageman J, Baecke C, Ebskamp M, Pilon M, Smeekens S, Weisbeek P. Protein import into and sorting inside the chloroplast are independent processes. Plant Cell. 1990;2:479–494. doi: 10.1105/tpc.2.5.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Muckel E, Heemeyer F, Vonheijne G, Soll J. A receptor component of the chloroplast protein translocation machinery. Science. 1994;266:1989–1992. doi: 10.1126/science.7801125. [DOI] [PubMed] [Google Scholar]
- Hoekema A, Hirsch PR, Hooykaas P, Schilperoort RA. A binary vector strategy based on seperation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature. 1983;303:179–180. [Google Scholar]
- Kessler F, Blobel G. Interaction of the protein import and folding machineries in the chloroplast. Proc Natl Acad Sci USA. 1996;93:7684–7689. doi: 10.1073/pnas.93.15.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F, Blobel G, Patel HA, Schnell DJ. Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science. 1994;266:1035–1039. doi: 10.1126/science.7973656. [DOI] [PubMed] [Google Scholar]
- Kindle KL, Lawrence SD. Transit peptide mutations that impair in vitro and in vivo chloroplast protein import do not affect accumulation of the gamma-subunit of chloroplast ATPase. Plant Physiol. 1998;116:1179–1190. doi: 10.1104/pp.116.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Gray JC. The N terminal hydrophobic region of the mature phosphate translocator is sufficient for targeting to the chloroplast inner envelope membrane. Plant Cell. 1995;7:1421–1432. doi: 10.1105/tpc.7.9.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawrence S, Kindle K. Alterations in the Chlamydomonas plastocyanin transit peptide have distinct effects on in vitro import and in vivo protein accumulation. J Biol Chem. 1997;272:20357–20363. doi: 10.1074/jbc.272.33.20357. [DOI] [PubMed] [Google Scholar]
- Lubeck J, Soll J, Akita M, Nielsen E, Keegstra K. Topology of Iep110, a component of the chloroplastic protein import machinery present in the inner envelope membrane. EMBO J. 1996;15:4230–4238. [PMC free article] [PubMed] [Google Scholar]
- Oblong JE, Lamppa GK. Identification of 2 structurally related proteins involved in proteolytic processing of precursors targeted to the chloroplast. EMBO J. 1992;11:4401–4409. doi: 10.1002/j.1460-2075.1992.tb05540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SE, Keegstra K. Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell. 1994;6:93–105. doi: 10.1105/tpc.6.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, de Boer D, Knols SL, Koppelman MH, van der Graaf RM, de Kruijff B, Weisbeek PJ. Expression in Escherichia coli and purification of a translocation-competent precursor of the chloroplast protein ferredoxin. J Biol Chem. 1990;265:3358–3361. [PubMed] [Google Scholar]
- Pilon M, de Kruijff B, Weisbeek PJ. New insights into the import mechanism of the ferredoxin precursor into chloroplasts. J Biol Chem. 1992a;267:2548–2556. [PubMed] [Google Scholar]
- Pilon M, Rietveld AG, Weisbeek PJ, de Kruijff B. Secondary structure and folding of a functional chloroplast precursor protein. J Biol Chem. 1992b;267:19907–19913. [PubMed] [Google Scholar]
- Pilon M, Wienk H, Sips W, Deswaaf M, Talboom I, Vanthof R, Dekortekool G, Demel R, Weisbeek PJ, de Kruijff B. Functional domains of the ferredoxin transit sequence involved in chloroplast import. J Biol Chem. 1995;270:3882–3893. doi: 10.1074/jbc.270.8.3882. [DOI] [PubMed] [Google Scholar]
- Pruitt RE, Meyerowitz EM. Characterization of the genome of Arabidopsis thaliana. J Mol Biol. 1986;187:169–183. doi: 10.1016/0022-2836(86)90226-3. [DOI] [PubMed] [Google Scholar]
- Quaedvlieg N, Dockx J, Rook F, Weisbeek P, Smeekens S. The homeobox gene ATH1 of Arabidopsis is derepressed in the photomorphogenic mutants cop1 and det1. Plant Cell. 1995;7:117–129. doi: 10.1105/tpc.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C, Ellis RJ. Transport of proteins into chloroplasts: the precursor of small subunit of ribulose bisphosphate carboxylase is processed to the mature size in two steps. Eur J Biochem. 1984;142:343–346. doi: 10.1111/j.1432-1033.1984.tb08292.x. [DOI] [PubMed] [Google Scholar]
- Schnell DJ. Shedding light on the chloroplast protein import machinery. Cell. 1995;83:521–524. doi: 10.1016/0092-8674(95)90090-x. [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Kessler F, Blobel G. Isolation of components of the chloroplast protein import machinery. Science. 1994;266:1007–1012. doi: 10.1126/science.7973649. [DOI] [PubMed] [Google Scholar]
- Sijmons PC, Dekker BM, Schrammeijer B, Verwoerd TC, van den Elzen PJ, Hoekema A. Production of correctly processed human serum albumin in transgenic plants. Biotechnology. 1990;8:217–221. doi: 10.1038/nbt0390-217. [DOI] [PubMed] [Google Scholar]
- Smeekens S, Bauerle C, Hageman J, Keegstra K, Weisbeek P. The role of the transit peptide in the routing of precursors toward different chloroplast compartments. Cell. 1986;46:365–375. doi: 10.1016/0092-8674(86)90657-4. [DOI] [PubMed] [Google Scholar]
- Smeekens S, van Binsbergen J, Weisbeek P. The plant ferredoxin precursor: nucleotide sequence of a full length cDNA clone. Nucleic Acids Res. 1985;13:3179–3194. doi: 10.1093/nar/13.9.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S, van Steeg H, Bauerle C, Weisbeek P. Import into chloroplasts of a yeast mitochondrial protein directed by ferredoxin and plastocyanin transit peptides. Plant Mol Biol. 1987;9:377–388. doi: 10.1007/BF00014912. [DOI] [PubMed] [Google Scholar]
- Somerville CR, Somerville SC, Ogren WL. Isolation of photosynthetically active protoplasts and chloroplasts from Arabidopsis thaliana. Plant Sci Lett. 1981;21:89–96. [Google Scholar]
- Tranel PJ, Froehlich J, Goyal A, Keegstra K. A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J. 1995;14:2436–2446. doi: 10.1002/j.1460-2075.1995.tb07241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Montagu MV, Lijsbettens MV. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandervere PS, Bennett TM, Oblong JE, Lamppa GK. A chloroplast processing enzyme involved in precursor maturation shares a zinc binding motif with a recently recognized family of metalloendopeptidases. Proc Natl Acad Sci USA. 1995;92:7177–7181. doi: 10.1073/pnas.92.16.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waegemann K, Soll J. Characterization of the protein import apparatus in isolated outer envelopes of chloroplasts. Plant J. 1991;1:149–158. [Google Scholar]
- Walker D, Zoran G, Cerovic Z, Robinson S. Isolation of intact chloroplasts: general principles and criteria of integrity. Methods Enzymol. 1987;148:145–157. [Google Scholar]
- Wu CB, Seibert FS, Ko K. Identification of chloroplast envelope proteins in close physical proximity to a partially translocated chimeric precursor protein. J Biol Chem. 1994;269:32264–32271. [PubMed] [Google Scholar]