Abstract
The alternative oxidase (AOX) of Neurospora crassa transfers electrons from ubiquinol to oxygen. The enzyme is not expressed under normal conditions. However, when the function of the standard electron transport chain is compromised, AOX is induced, providing cells with a means to continue respiration and growth. Induction of the enzyme represents a form of retrograde regulation because AOX is encoded by a nuclear gene that responds to signals produced from inefficiently functioning mitochondria. To identify genes required for AOX expression, we have screened the N. crassa gene knockout library for strains that are unable to grow in the presence of antimycin A, an inhibitor of complex III of the standard electron transport chain. From the 7800 strains containing knockouts of different genes, we identified 62 strains that have reduced levels of AOX when grown under conditions known to induce the enzyme. Some strains have virtually no AOX, whereas others have only a slight reduction of the protein. A broad range of seemingly unrelated functions are represented in the knockouts. For example, we identified transcription factors, kinases, the mitochondrial import receptor Tom70, three subunits of the COP9 signalosome, a monothiol glutaredoxin, and several hypothetical proteins as being required for wild-type levels of AOX production. Our results suggest that defects in many signaling or metabolic pathways have a negative effect on AOX expression and imply that complex systems control production of the enzyme.
Keywords: alternative oxidase, knockout library, Neurospora crassa, mitochondria
Although the proper formation of mitochondria requires the expression of genes found in both the nuclear and mitochondrial genomes, the vast majority of mitochondrial proteins are encoded in the nucleus. Thus, communication of mitochondrial status to the nucleus must occur to insure that the organelles meet the requirements for growth and development of the organism and to insure that they can properly respond to changing conditions and stress. Communication from the mitochondria to the nucleus that influences the expression of nuclear genes has been termed retrograde regulation (Jazwinski and Kriete 2012; Liu and Butow 2006; Woodson and Chory 2008).
The alternative oxidase (AOX) is a di-iron carboxylate protein that transfers electrons from ubiquinol to molecular oxygen and exists in the mitochondrial inner membrane (Albury et al. 2009; Andersson and Nordlund 1999; Berthold et al. 2000; Berthold and Stenmark 2003). It is found in a variety of organisms, including bacteria, protists, fungi, plants, and animals—but not mammals (McDonald 2008; McDonald and Vanlerberghe 2006). Depending on the organism, expression of AOX can be influenced by developmental signals, tissue specificity, and response to stress (Considine et al. 2001; Djajanegara et al. 2002; Finnegan et al. 1997; Karpova et al. 2002; Nargang and Kennell 2010; Van Aken et al. 2009; Vanlerberghe and McIntosh 1997). In many organisms, AOX occurs at low-to-undetectable levels under normal growth conditions but becomes highly expressed when the standard, cytochrome-mediated, electron transport chain (sETC) is compromised. Thus, because AOX is encoded in the nucleus, it serves as a prime example of a gene that is controlled by retrograde regulation. However, the nature of the retrograde pathway(s) and the factors required to achieve regulation of AOX production are not well understood.
In fungi, a few genes are known that affect AOX production. In Candida albicans there is evidence for the involvement of a histidine kinase in AOX regulation (Huh and Kang 2001). Early work with Neurospora crassa identified the structural gene for AOX as aod-1 whereas another gene, aod-2, was found to be required for the expression of aod-1 (Bertrand et al. 1983; Edwards et al. 1976; Lambowitz et al. 1989; Li et al. 1996). More recently, we used a reporter system and a traditional genetic screen to identify four additional genes, named aod-4, aod-5, aod-6, and aod-7, along with a new allele of aod-2, as required for AOX induction (Descheneau et al. 2005). Thus, these studies defined a minimum of five genes required for proper AOX production in N. crassa. We then demonstrated that aod-2 and aod-5 encode transcription factors (Chae et al. 2007b) of the Zn(II)2Cys6 binuclear cluster (zinc cluster) family (MacPherson et al. 2006). In vitro studies suggested that the proteins form a heterodimer, which binds a specific sequence within the aod-1 promoter region to activate transcription under the appropriate inducing conditions (Chae et al. 2007a,b; Chae and Nargang 2009). Orthologs of AOD2 and AOD5 also are required for AOX production in Podospora anserina (Sellem et al. 2009) and Aspergillus nidulans (Suzuki et al. 2012).
Because the previously described mutant screen was not saturated, it seemed likely that additional genes might also be involved with AOX regulation. However, two factors led us to not simply repeat the screen. First, the screen was designed to detect mutations affecting transcriptional regulation of the aod-1 gene. Additional factors affecting posttranscriptional processes that also may play a role in expression of AOX would not be detected. Second, a gene knockout library for N. crassa (Colot et al. 2006) has been created since our previous screen. Identification of strains affected in their ability to produce AOX in this library would allow direct identification of the genes without the need for mapping and rescue experiments. Here we describe 62 newly identified genes from the N. crassa knockout library that affect the production of AOX to varying extents.
Materials and Methods
Strains and growth of N. crassa
The N. crassa gene knockout library (Colot et al. 2006) was obtained from the Fungal Genetics Stock Center (FGSC) in a series of 96-well microtiter plates holding conidia from individual strains in each well (McCluskey et al. 2010). At the end point of the study described herein, plates 1 through 108 of the library had been examined. Strain 74A-OR23-1A (Nargang lab name NCN251) was used as a control for experiments with the knockout strains.
The tom20RIP sheltered heterokaryon, the methods used to manipulate the heterokaryon, and the control (strain HIV) for experiments with the heterokaryon have been described in detail previously (Harkness et al. 1994a). In summary, the tom20RIP sheltered heterokaryon contains two nutritionally complementing nuclei. One of the nuclei carries a functional tom20 gene, a mutation in the pan-2 gene, and is sensitive to p-fluorophenylalanine (fpa). The other nucleus contains a nonfunctional tom20 allele that was destroyed by repeat-induced point mutation (RIP) mutagenesis. The latter nucleus also carries a mutant allele of the mtr gene that imparts resistance to fpa, as well as a mutation in the his-3 gene. The level of the Tom20 protein can be greatly reduced in the heterokaryon by growth in the presence of fpa plus histidine.
N. crassa cells were grown on solid or in liquid medium according to previously described methods (Davis and De Serres 1970) but using the modified Vogel’s salts developed by Metzenberg (Metzenberg 2004). Medium containing sorbose was used when colonies were desired whereas sucrose containing medium was used when filamentous growth was desired. When needed, inhibitors were added to media at the following final concentrations: antimycin A, 0.5 µg/mL (0.9 µM); chloramphenicol (Cm), 2 mg/mL (6 mM); fpa, 400 µM as described previously (Descheneau et al. 2005; Nargang et al. 1995). At the aforementioned concentration, antimycin A completely inhibits the sETC.
For screening the N. crassa knockout library, a 48-pin inoculation stamp was used that fit precisely into one half of the wells of the 96-well microtiter plates containing the library. Conidia from the library were stamped to Petri plates containing Vogel’s sorbose medium plus antimycin A and to Petri plates without the inhibitor as a control for growth under normal conditions. Petri plates containing antimycin A were examined 5−7 d after inoculation, and those without antimycin were examined 2−3 d after inoculation.
Growth rates in the presence or absence of antimycin A were measured in two ways. In some cases, filamentous growth was monitored by measuring the radius of growth from conidia inoculated in the center of a Petri dish containing Vogel’s sucrose medium at selected time points. In other cases the amount of growth of colonies produced on sorbose containing medium after 2 to 5 d of growth was observed.
Mitochondrial protein import assays
The procedures used to measure the import of mitochondrial precursor proteins have been described previously (Harkness et al. 1994b). In summary, radiolabeled precursors, synthesized by in vitro translation in the presence of [35S]-methionine, were incubated with mitochondria isolated from freshly grown mycelium. After import for specified time periods, proteinase K was added to remove unincorporated precursor proteins. Mitochondria were washed, and mitochondrial proteins were subjected to polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS-PAGE). Proteins were transferred to nitrocellulose and exposed to X-ray film.
Other techniques
Standard procedures were used for gel electrophoresis of proteins (Lämmli 1970), western blot analysis (Good and Crosby 1989), and isolation of mitochondria (Nargang and Rapaport 2007). Measurement of the effect of inhibitors on oxygen consumption in growing cells using an oxygen electrode was as described (Tanton et al. 2003). Potassium cyanide (CN) and salicylhydroxamic acid were used to inhibit the sETC and AOX, respectively. In some figures, irrelevant lanes were removed electronically. Bands on mitochondrial import blots were quantified using Adobe photoshop.
Results
The basis of the screen–strains deficient in AOX are unable to grow in antimycin A
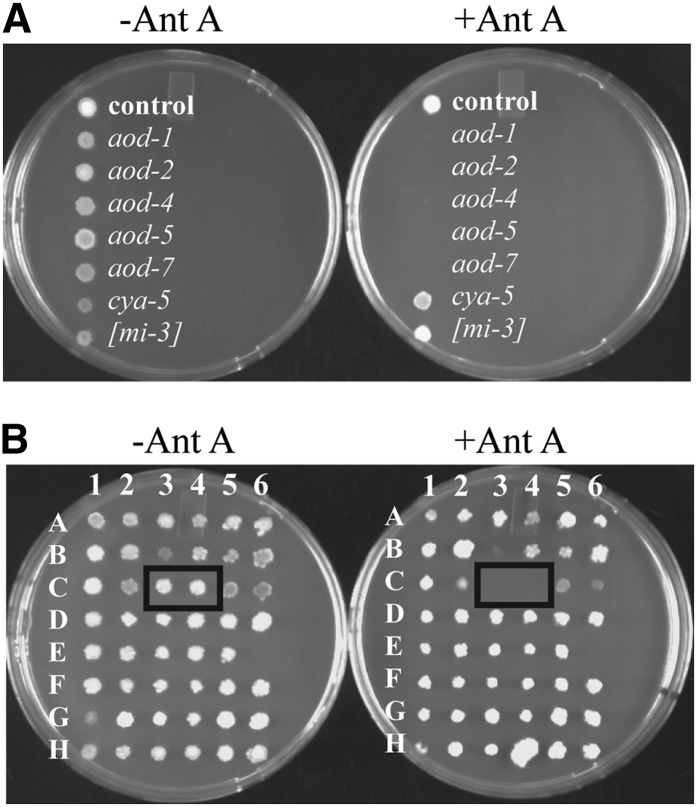
The construction of a library of N. crassa strains containing individual gene knockouts (Colot et al. 2006) maintained on microtiter plates provided a resource to search for genes that might be involved in the production of AOX. When N. crassa is unable to respire through the sETC, it induces AOX, which allows continued growth via this alternative pathway of electron transport. However, if both pathways are blocked, cells are unable to grow. Thus, we reasoned that any of the knockout strains that were unable to grow in the presence of antimycin, which inhibits complex III of the sETC, should also be lacking AOX and the genes affected in such strains would in some way be required for AOX production or function. To test this principle, we set up a microtiter plate containing conidia from several previously described AOX nonproducing strains (Descheneau et al. 2005), including a mutant of the aod-1 gene, which encodes the AOX protein. As controls, we used the parental strain of the knockouts as well as two slow-growing mutants that affected the sETC, cya-5-34 (Nargang et al. 1978), and [mi-3] (Mitchell et al. 1953). A 48-pin stamp was used to transfer conidia from the microtiter plate to Petri dishes containing growth medium with and without antimycin A. Growth of the control and sETC mutant strains in the presence of antimycin A was obvious whereas AOX mutants failed to grow (Figure 1A). As a further test, we stamped out the section of the knockout library known to contain the two different mating type strains with a knockout of the aod-5 gene (plate 1, co-ordinates C3 and C4), which we had previously identified as a transcription factor necessary for the expression of AOX (Chae et al. 2007b). Of the 47 strains that grew on the control plate without antimycin A, only the two containing the aod-5 knockout did not grow in the presence of antimycin A (Figure 1B).
Figure 1 .
Identification of AOX-deficient strains in the knockout library. (A) Conidial suspensions from strains known to lack AOX (aod strains), slow-growing sETC mutants (cya-5 and [mi-3]), and a control wild-type strain (NCN251) were plated on medium with or without antimycin A (+Ant A or –Ant A, respectively). Plates were incubated at 30° for 2 (−Ant A) or 4 (+Ant A) d and then photographed. (B) Plate number 1 of the knockout library contained strains of opposite mating type in which the aod-5 gene had been knocked out. Conidia from this knockout library plate were stamped onto medium with and without antimycin A. The boxes indicate the position of the aod-5 strains at positions C3 and C4.
Examining the knockout library for strains unable to grow on antimycin A
We then proceeded to test the complete library, which consisted of approximately 10,000 strains contained on 108 microtiter plates (96 wells each). Because many of the knocked-out genes are represented in duplicate as both A and a mating type strains, the actual number of knocked out genes screened was approximately 7800. In the initial screening we identified 218 colonies that grew very slowly, or not at all, on medium containing antimycin A while exhibiting obvious growth on medium lacking the drug. The strains represented by these colonies were then inoculated from the library plates into slants and subsequently retested for growth in Petri dishes containing medium with or without antimycin A. In this round of testing, several strains showed relatively good growth in the presence of the drug. The difference in results compared to the original growth observed in the stamped library might be due to low numbers of viable conidia in some wells of the library plates. Other strains were found to grow extremely slowly even on medium that lacked antimycin A. Strains in either of these two categories were not examined further leaving 126 strains with slow, or no growth, in the presence of antimycin A and reasonably robust growth in the absence of the drug.
To continue our screen, we wished to determine the level of AOX that could be produced in each of the remaining 126 mutants. Because the strains were selected based on inability to grow in the presence of antimycin A, we were unable to use the same drug as an AOX inducer for experiments requiring growing cells. Growth in the presence of Cm at 2 mg/mL results in severe but not total inhibition of synthesis of mitochondrially encoded subunits of the respiratory chain and ATP synthase and results in AOX induction in wild-type cells of N. crassa (Li et al. 1996; Tanton et al. 2003). However, we have previously shown that cells unable to produce AOX grow at rates virtually indistinguishable from the wild type in the presence of Cm (Descheneau et al. 2005). We assume that the small amount of mitochondrial protein synthesis that occurs in the presence of Cm is sufficient to support growth of the cells. However, the effects on mitochondria are sufficient to elicit AOX induction.
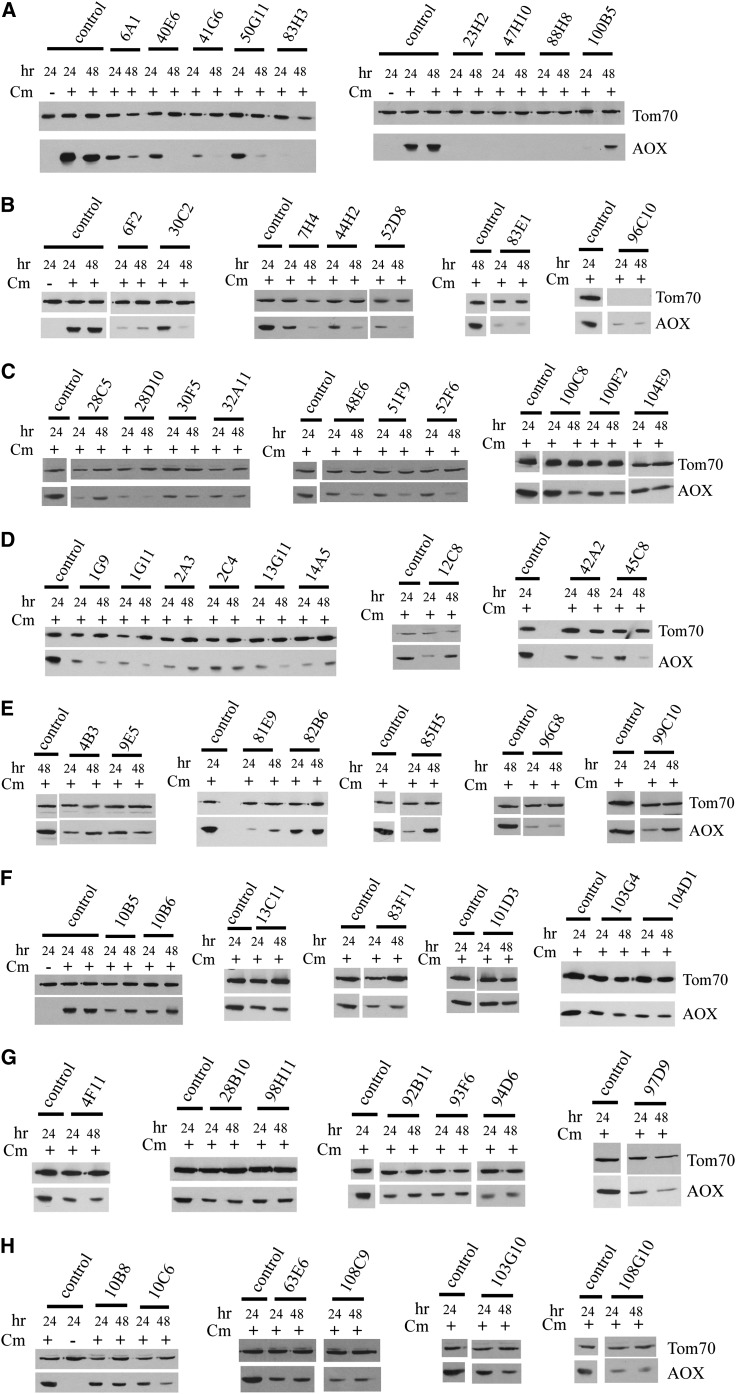
After growth for 24 hr and 48 hr in the presence of Cm, mycelia were harvested and mitochondria were isolated. Western blot analysis of mitochondrial proteins revealed that 62 newly identified strains contained reduced levels of AOX at one or both of the time points (Figure 2). Because the NCU numbers of the predicted proteins at the Neurospora database can change on the basis of more defined annotations, the coordinates of the knockout library 96-well microtiter plates have been used as the primary name for the strains identified. We have used the convention of: number of plate (1−108), followed by the letter of the row (A-H), followed by the number of the column (1−12). Based on a visual assessment of the amount of AOX present on Western blots, we placed each strain into one of three classes (Table 1). Class 1 strains produced virtually no AOX at either 24 hr or 48 hr, or at both time points. An example is strain 23H2 (Figure 2A). Table 1 shows 11 Class 1 strains, but three of these were previously described as AOX-deficient mutants (1C3, aod-5; 96H9, aod-1; 97B1, aod-2). Thus, there are eight new strains that have been identified and assigned to Class 1. The 20 strains in Class 2 exhibited a moderate reduction in the amount of AOX at 24 hr, 48 hr, or both time points. An example is strain 6A1 (Figure 2A). Class 3 contained 34 strains that exhibited some reduction in AOX at one or both of the time points. Strain 4B3 serves as an example (Figure 2E). The NCU number of the predicted proteins and the FGSC strain number of the knockout strain are also shown in Table 1. The BROAD genome sequence website was examined for each gene identified. Known gene functions or characteristic protein domains listed at that website are also included in Table 1. In addition, we have included information regarding the growth rate of the strains in the presence and absence of antimycin A as well as observations on the ability of the strains to form conidiaspores. For unknown reasons, despite the fact that we screened for mutants with poor growth in the presence of antimycin A, a few strains were ultimately found to have good growth in the presence of the drug.
Figure 2 .
AOX levels in knockout mutants identified in the screen. The indicated strains were grown for 24 and 48 hr in the presence of Cm. Mycelia were harvested, mitochondria were isolated, and mitochondrial proteins were subjected to SDS-PAGE and Western blot analysis using antibody to AOX, and to Tom70 as a loading control. Results are placed in rows A through H to facilitate reference to results for individual strains. The control (NCN251) was grown in similar fashion. A culture of the control without Cm also was used to demonstrate the lack of AOX without induction by Cm. AOX-deficient strains are shown with a control from the same Western blot. In some cases, irrelevant lanes were removed from the blots and are shown as white strips between lanes.
Table 1. Summary and characterization of knockout strains identified as AOX deficient.
| Knockout Library Grid Number | Mutant Class | Panel of Figure 2 Showing Western | NCU Numbera | FGSC Number | Known or Predicted Protein or Genea (Domain Identified) | Growth Defectb | Growth on Antimycin A for48 hrc | Conidiation Defectd |
|---|---|---|---|---|---|---|---|---|
| 1C3e | 1 | na | 03938.5 | 11227 | aod-5 | 0 | ||
| 23H2 | 1 | A | 08887.5 | 15957 | Hypothetical protein (major facilitator superfamily) | 0 | ||
| 40E6f | 1 | A | 05600.5 | 13805 | Ubiquinone biosynthesis protein (ABC1 family) | 0 | ||
| 12096.5 | Aspartyl aminopeptidase (glycosyl hydrolase family) | |||||||
| 41G6 | 1 | A | 03589.5 | 13923 | Hypothetical protein | Slight | 0 | Slight |
| 47H10 | 1 | A | 00778.5 | 16938 | sed5 vesicle protein (transmembrane adaptor Erv26) | Severe | 0 | Severe |
| 52D8 | 1 | B | 07281.5 | 14469 | Glucose-6-phosphate isomerase (phosphoglucose isomerase) | Severe | 0 | Slight |
| 83H3 | 1 | A | 08365.5 | 18277 | RNA polymerase II mediator complex component Med8 | Slight | 0 | Severe |
| 88H8 | 1 | A | 01542.5 | 19221 | Hypothetical protein (HbrB like domain) | 0 | ||
| 96H9e | 1 | na | 07953.5 | 18947 | aod-1 | |||
| 97B1e | 1 | na | 03352.5 | 19465 | aod-2 | |||
| 100B5 | 1 | A | 08158.5 | 19644 | Dual-specificity phosphatase | Slight | 0 | Slight |
| 1G9 | 2 | D | 00157.5 | 11281 | COP 9 signalosome-1, csn-1 | Slight | 0 | Slight |
| 1G11 | 2 | D | 00467.5 | 11283 | csn-5 | Slight | 0 | Slight |
| 2A3 | 2 | D | 08741.5 | 11299 | Striatin pro 11 (WD domain, G-beta repeat) | 0.7 | Severe | |
| 6A1 | 2 | A | 06799.5 | 11001 | Fungal specific transcription factor, vad-5 | 0 | ||
| 6F2 | 2 | B | 09739.5 | 11062 | Hypothetical protein, ada-7 (Zn(2)-Cys(6) binuclear cluster) | 0.7 | Severe | |
| 7H4 | 2 | B | 03875.5 | 11780 | Chromatin remodeling complex ATPase chain ISW1 (SLIDE, HAND, helicase conserved domain, SNF2 family, type III restriction enzyme, DEAD/DEAH box helicase, class II histone deactylase complex subunits 2 and 3) | 0 | Severe | |
| 12C8 | 2 | D | 01266.5 | 12022 | Phosphoinositide-specific phospholipase C | 0.6 | ||
| 13G11 | 2 | D | 04566.5 | 12420 | Protein kinase SNF1 | 0 | Slight | |
| 14A5 | 2 | D | 03727.5 | 12091 | Hyphal anastomosis-2, ham-2 | 0.4 | Severe | |
| 28C5 | 2 | C | 00007.5 | 16098 | pH response regulator palH, rim-21 | 1.0 | Slight | |
| 28D10 | 2 | C | 00593.5 | 16116 | COP9 signalosome-2, csn-2 | Slight | 0.2 | Slight |
| 30C2 | 2 | B | 06199.5 | 13193 | RNA binding protein Jsn1 (RNA recognition motif) | 1.2 | Severe | |
| 44H2 | 2 | B | 04826.5 | 16834 | Hypothetical protein | 0 | Severe | |
| 45C8 | 2 | D | 03035.5 | 13973 | Hypothetical protein | 0.3 | Slight | |
| 50G11 | 2 | A | 04607.5 | 14316 | Hypothetical protein | 0.3 | Slight | |
| 81E9 | 2 | E | 06084.5 | 20114 | Hypothetical protein (Vps51/Vps67) | 1.6 | Severe | |
| 83E1 | 2 | B | 07881.5 | 18239 | Hypothetical protein | 2.7 | Severe | |
| 85H5 | 2 | E | 09803.5 | 18375 | Thioredoxin (glutaredoxin homolog) | Slight | 0 | Slight |
| 96C10 | 2 | B | 04245.5 | 18888 | Translocase of outer mitochondrial membrane 70, tom70 | 0 | Severe | |
| 96G8g | 2 | E | 06727.5 | 18934 | Spermidine 3, spe-3 (Spermidine synthase) | 2.3 | ||
| 2C4 | 3 | D | 03894.5 | 11324 | Serine/threonine protein kinase ste-20 | 0.2 | ||
| 4B3 | 3 | E | 04062.5 | 11473 | Peroxisome biogenesis factor 20 | 1.1 | ||
| 4F11 | 3 | G | 08480.5 | 11671 | Hsf-type DNA-binding domain-containing protein | 0 | Severe | |
| 9E5 | 3 | E | 08565.5 | 11945 | Hypothetical protein (NACHT domain) | 0.4 | ||
| 10B5 | 3 | F | 09212.5 | 11545 | Serine/threonine protein kinase | 1.7 | ||
| 10B6 | 3 | F | 06563.5 | 11546 | Serine/threonine protein phosphatase PP2A catalytic subunit (calcineurin-like phosphoesterase) | Slight | 0.5 | Severe |
| 10B8 | 3 | H | 07489.5 | 11548 | Phosphatase-Z-like-1, pzl-1 (calcineurin-like phosphoesterase) | 0.4 | Severe | |
| 10C6 | 3 | H | 03853.5 | 11559 | Peptidyl-prolyl cis-trans isomerase (tetratricopeptide repeat) | 2.7 | Slight | |
| 13C11 | 3 | F | 06493.5 | 12370 | Guanine nucleotide-binding protein alpha-1 (ADP ribosylation factor) | 3.3 | ||
| 28B10 | 3 | G | 08137.5 | 16091 | Hypothetical protein | 0 | Slight | |
| 30F5 | 3 | C | 01471.5 | 13235 | Nuclear protein SNF4 (CBS domain) | 0 | ||
| 32A11 | 3 | C | 01955.5 | 13376 | Autophagocytosis protein Aut1 | 1.9 | Slight | |
| 42A2 | 3 | D | 01276.5 | 16557 | N-acetyltransferase 5 | Slight | 0.5 | Slight |
| FR47-like protein | ||||||||
| 48E6g | 3 | C | 04771.5 | 16944 | Fructosyl-amino acid oxidase (FAD dependent oxidoreductase) | 0.5 | Slight | |
| 51F9 | 3 | C | 09530.5 | 14398 | Hypothetical protein | 1.4 | ||
| 52F6 | 3 | C | 08067.5 | 14491 | Osmosensor protein (SH3 domain) | 3.5 | ||
| 63E6 | 3 | H | 03802.5 | 17474 | Trimethyllysine dioxygenase (taurine catabolism dioxygenase) | 2.6 | Slight | |
| 82B6 | 3 | E | 07112.5 | 20169 | Hypothetical protein (pyridine nucleotide-disulphide oxidoreductase) | 1.5 | ||
| 83F11 | 3 | F | 01511.5 | 18261 | Urease accessory protein ureG | Severe | 0.5 | Severe |
| 92B11 | 3 | G | 07579.5 | 19411 | Hypothetical protein (SCA7) | 0.2 | Severe | |
| 93F6 | 3 | G | 03708.5 | 18633 | Hypothetical protein (putative methyl transferase) | 1.8 | ||
| 94D6g | 3 | G | 00712.5 | 18704 | 3-hydroxy-3-methylglutaryl-coenzyme A reductase (sterol sensing domain, patched family) | 2.6 | ||
| 97D9 | 3 | G | 00360.5 | 19497 | NAD-dependent epimerase/dehydratase | 0 | Slight | |
| 98H11 | 3 | G | 10058.5 | 18976 | Phosphoglucomutase 2 | 1.1 | ||
| 99C10 | 3 | E | 09686.5 | 20379 | Clock-controlled gene-8, ccg-8 (transcription factor opl1) | 0 | ||
| 100C8 | 3 | C | 09208.5 | 19659 | Transcription factor SPT8 (WD domain, G-beta repeat) | 0 | ||
| 100F2 | 3 | C | 09527.5 | 20459 | Hypothetical protein | 2.0 | ||
| 101D3 | 3 | F | 09560.5 | 21068 | Superoxide dismutase | 0 | ||
| 103G4 | 3 | F | 09553.5 | 21156 | 3-hydroxybutyryl CoA dehydrogenase | 2.2 | ||
| 103G10g | 3 | H | 08132.5 | 21162 | Alpha-1,3-glucan synthase, Ags2 | Slight | 0.5 | |
| 104D1 | 3 | F | 02973.5 | 21213 | Mitochondrial carrier protein | 0.1 | ||
| 104E9 | 3 | C | 01553.5 | 21233 | Para-hydroxybenzoate-polyprenyltransferase, Coq2 | 0 | ||
| 108C9 | 3 | H | 02979.5 | 21401 | AMP deaminase | 1.8 | Slight | |
| 108G10g | 3 | H | 08279.5 | 21450 | Hypothetical protein | 1.1 |
Information from N. crassa database BROAD Institute (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html).
Growth rates were measured by inoculating conidiaspores at the center of Petri dishes (standard 100 mm × 15 mm size) containing Vogel’s medium. Plates were incubated at 30°. The radius of mycelial extension from the inoculation point was measured (in cm) and compared with the wild-type control strain that covers the surface of the plate (radius of growth from the inoculation point of 4.2 cm) in 48 hr. Strains with a “slight” growth defect were defined as having a growth radius of 1.5 to 4.1 cm in 48 hr. A “severe” growth defect was defined as having a growth radius of less than 1.5 cm in 48 hr.
Same as b, but the medium contained Antimycin A. Measurements of growth (in cm) were taken after 48 hr. The control strain grew 1.7 cm in 48 hr on average under these conditions.
Formation of asexual conidiaspores was considered slightly defective if conidiation on slants containing 3 mL of Vogel’s medium in a 1 cm × 10-cm test tube was similar to the control but took longer than the control strain. The defect was considered severe if very few conidia formed, even after extended times.
The aod-1 (Li et al. 1996), aod-2, and aod-5 (Descheneau et al. 2005) genes were all identified during the screen and have been described in detail previously.
Original predicted open reading frame was knocked out but is now known to be divided into two predicted genes.
Strain maintained in the knockout library as a heterokaryon. This implies that the affected gene is essential for viability.
Nature of genes defined by annotations and bioinformatics
The genes identified in the screen represent a broad range of functions. Among the 62 new genes, the two most highly represented groups were 16 hypothetical proteins (9E5, 23H2, 28B10, 41G6, 44H2, 45C8, 50G11, 51F9, 81E9, 82B6, 83E1, 88H8, 92B11, 93F6, 100F2, 108G10) and six kinases or phosphatases (2C4, 10B5, 10B6, 10B8, 13G11, 100B5). If the previously identified aod-2 and aod-5 genes are included, there are also six predicted transcription factors (1C3, 6A1, 6F2, 97B1, 99C10, 100C8). There is no obvious relationship among the predicted functions of the eight new class 1 genes (Table 1) and in most cases it is not obvious how many of the predicted functions might compromise AOX production. One class 1 gene that might be imagined as influencing AOX expression is the dual specificity phosphatase deleted in 100B5, because kinases and phosphatases are well known as important regulatory enzymes. A complication with respect to class 1 strain 40E6 is that the region replaced in this knockout strain actually covers two genes. One of these (NCU05600.5) is listed as a ubiquinone biosynthesis protein with an ABC1 family domain, whereas the other (NCU12096.5) is an aspartyl aminopeptidase. We suspect that the absence of the gene encoding the putative ubiquinone synthesis protein would be the more likely of the two genes to affect AOX production because the reduced form of ubiquinone is the substrate for AOX. NCU05600.5 has recently been shown to contain an atypical serine-threonine kinase domain and the gene has been named stk-33 (Park et al. 2011). The role of the protein in ubiquinone synthesis is not entirely clear, and its closest S. cerevisiae homolog (YPL109c) has not yet been assigned a function. However, the proteins from both species are related to the S. cerevisiae YGL119W protein, also known as Coq8 (Do et al. 2001). The kinase domain of Coq8 is thought to regulate the activity of Coq3, a ubiquinone biosynthetic enzyme (Tauche et al. 2008). However, a coq8 deletion strain has also been shown to have a complex phenotype that includes underproduction of glutathione and sensitivity to oxidative stress (Gazdag et al. 2011). Members of the same family of proteins have also been linked to oxidative stress in Arabidopsis thaliana (Jasinski et al. 2008). It should be noted that a homolog of another ubiquinone synthesis protein, Coq2, was identified as a class 3 mutant (104E9).
Among the 20 class 2 knockouts, it is striking that three encode components of the COP9 signalosome (1G9, 1G11, and 28D10), whereas another two encode predicted transcription factors (6A1, 6F2). The latter two genes were previously identified as vad-5 and ada-7, respectively, in a study of Neurospora transcription factor knockouts (Colot et al. 2006). The class 3 genes represent a broad range of predicted function. Four class 3 genes are predicted to act as kinases or phosphatases.
It was of interest to determine how many of the genes identified in our screen encoded proteins that are known or predicted to be localized to mitochondria. An exhaustive compilation that lists proteins actually identified in purified N. crassa mitochondria by two-dimensional electrophoresis and mass spectrometry, as well as those predicted to be mitochondrial by various in silico analyses has recently been published (Keeping et al. 2011). Each of the NCU numbers identified in our study was compared to the compilation. The matches are shown in Table 2, along with the criteria used in assigning the protein as mitochondrial. Thirteen proteins were found to be known or possible mitochondrial proteins (Table 2).
Table 2. Strains in which the gene knockout is predicted to encode a mitochondrially localized protein.
| Strain | Mutant Class | NCU Number | Localization Criteriaa | Predicted Proteinb |
|---|---|---|---|---|
| 40E6 | 1 | 05600 | A2 | Ubiquinone biosynthesis |
| 96H9 | 1 | 07953 | C1 | Alternative oxidase (AOX) |
| 2A3 | 2 | 08741 | B1 | Striatin pro 11 |
| 96C10 | 2 | 04245 | A1 | Tom70 protein import receptor |
| 96G8 | 2 | 06727 | C1 | Spermidine 3, spermidine synthase |
| 9E5 | 3 | 08565 | B1 | Hypothetical protein |
| 13C11 | 3 | 06493 | D2 | Guanine nucleotide binding protein alpha-1 |
| 28B10 | 3 | 08137 | C1 | Hypothetical [BLAST yeast Mitochondrial ribosomal protein of the small subunit, has similarity to human mitochondrial ribosomal protein MRP-S36] |
| 82B6 | 3 | 07112 | A1 | Hypothetical [HcaD domain conserved many organisms, NAD(FAD) oxidoreductase] |
| 101D3 | 3 | 09560 | A1 | Superoxide dismutase |
| 103G4 | 3 | 09553 | A1 | 3-hydroxybutyryl-CoA dehydrogenase |
| 104D1 | 3 | 02973 | C1 | Mitochondrial carrier protein |
| 104E9 | 3 | 01553 | C1 | Para-hydroxybenzoate-polyprenyltransferase Coq2 |
As defined previously (Keeping et al. 2011). Category A1 proteins were identified in their study from highly purified mitochondria using mass spectrometry. Category A2 proteins were identified by mass spectrometry in other studies or had other evidence of being located in mitochondria. B1 proteins were identified in an earlier study of the mitochondrial proteome (Schmitt et al. 2006) but no other evidence for or against their mitochondrial location exists. C1 proteins were predicted or demonstrated to be mitochondrial by conventional biochemistry. The D2 proteins are listed in MitoP2 Neurospora data, but other evidence that they are not mitochondrial also exists. This refers to a mitochondrial proteome database for several organisms, including Neurospora (http://www.mitop.de/).
Prediction from the N. crassa database. Descriptions in brackets after the first description are taken from Keeping et al. (Keeping et al. 2011).
Time-dependent AOX expression
When wild-type strains are grown for 24 and 48 hr in the presence of Cm, there is typically no difference in the amount of AOX present in their mitochondria (Figure 2, panel A, left and right blots; panel F, left blot). However, several of the strains identified from the KO library showed obvious differences in AOX abundance at the two different times of growth used in the screen. These observations suggest that different pathways of induction might be activated or used after different times of inhibition of the sETC. Although a few class 3 mutants showed this phenotype, it was most obvious in class 1 and 2, where mutants 1G9, 6A1, 7H4, 13G11, 30C2, 40E6, 41G6, 44H2, 50G11, and 52D8 contained more AOX after 24 hr growth in Cm than at 48 hr. In these cases a positive regulatory factor may become depleted or a negative factor may accumulate. Conversely, mutants 12C8, 14A5, 28C5, 81E9, 85H5, 100B5, and 101D5 contained more AOX after 48 hr than 24 hr. In these mutants a positive factor may accumulate or a negative factor might be depleted.
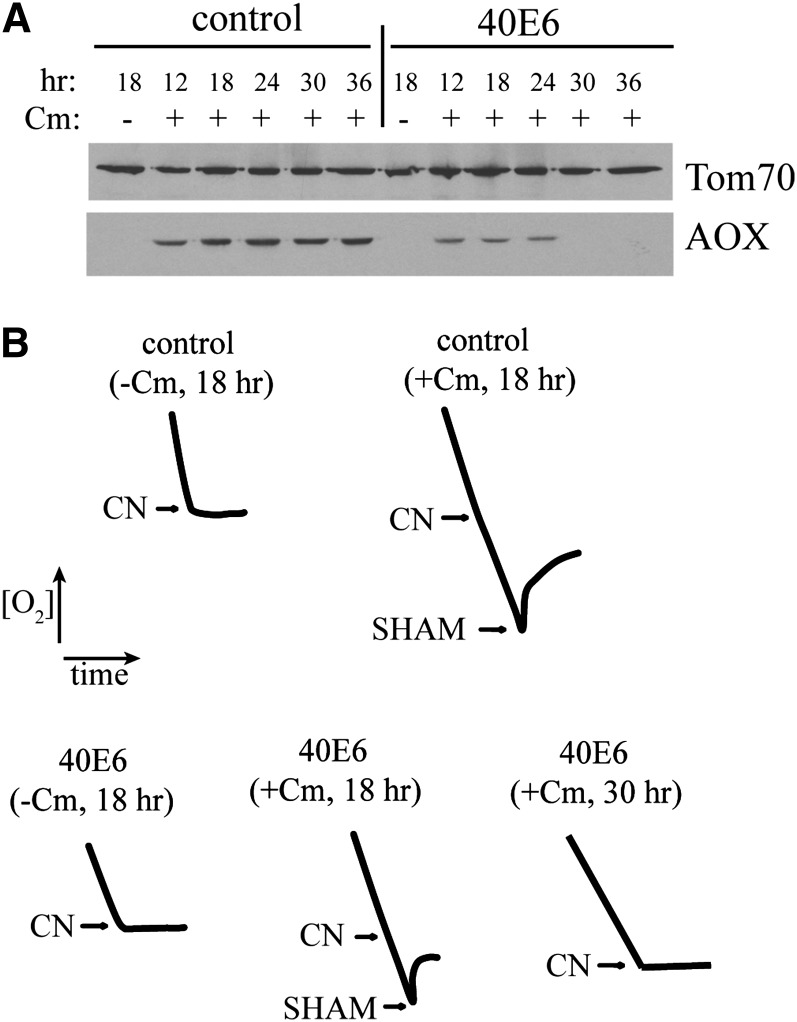
We chose one such class 1 strain (40E6) to determine whether the changes in AOX levels were gradual or sudden. As shown in Figure 3A, the level of AOX in the control strain mitochondria during growth in Cm was consistent over the course of the experiment from 12 to 36 hr. However, for 40E6 grown in Cm, the level of the enzyme was constant from 12 to 24 hr, but then it abruptly disappeared. Furthermore, there was direct correspondence between times that AOX could not be detected by western blot analysis with an absence of the CN-insensitive respiration characteristic of AOX (Figure 3B). It should also be noted that even at the earlier time points, AOX levels are somewhat reduced in 40E6.
Figure 3 .
Temporal changes in AOX expression. (A) Mycelium was harvested from cultures of strain 40E6 and the control strain (NCN251) after growth in the presence or absence of Cm for the indicated times. Mitochondria were isolated and subjected to SDS-PAGE and Western blot analysis using the antibodies indicated on the right. (B) Immediately before harvesting the cultures used in (A), 2 mL of the culture was removed and analyzed in a respirometer to determine the effect of inhibitors of the sETC (CN) or the AOX (salicylhydroxamic acid; SHAM) on oxygen consumption. The tracings show the decrease in O2 concentration over time. Arrows indicate the point at which the indicated inhibitors were added.
Tom70 acts as the major receptor for the AOX precursor protein
Among the genes identified as affecting AOX production, tom70 was somewhat unexpected (strain 96C10). Tom70 is a protein of the outer mitochondrial membrane TOM (translocase of the outer mitochondrial membrane) holo-complex (Ahting et al. 1999; Dekker et al. 1998). The TOM complex is responsible for the import of cytosolically synthesized mitochondrial preproteins into the organelle. Tom70, along with another protein of the complex, Tom20, are the major receptors for mitochondrial precursor proteins in the cytosol (Endo and Yamano 2010; Rapaport and Nargang 2004). Tom70 is chiefly involved in the recognition of hydrophobic proteins of the inner membrane that carry internal targeting signals and multiple membrane spanning domains such as AAC, the ATP/ADP carrier. Tom20 is best known for recognizing matrix-destined precursors carrying an N-terminal targeting signal which is removed from precursor proteins once they reach the matrix. However, Tom20 also has a degree of overlapping specificity for precursors recognized by Tom70 (Brix et al. 1999; Söllner et al. 1990; Steger et al. 1990). Therefore, such proteins are still imported in mutants lacking Tom70, though with reduced efficiency (Grad et al. 1999; Steger et al. 1990). Because N. crassa AOX carries a classic cleavable N-terminal targeting signal (Li et al. 1996), it was expected to be imported into mitochondria using Tom20 as its major receptor. However, a few other proteins with cleavable targeting signals have been shown to depend on Tom70 for their import (see Discussion).
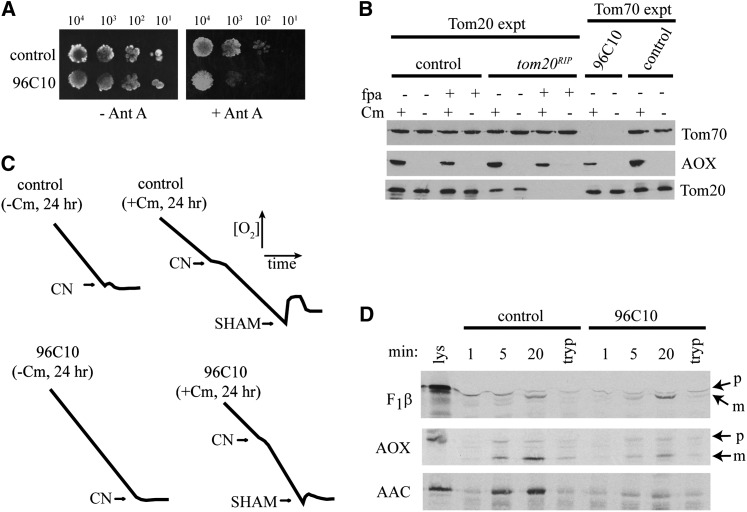
To validate the effects of the tom70 knockout on AOX, we investigated the phenotype of the strain in more detail. Although we saw no growth of the strain on antimycin A containing medium after 48 hr (Table 1), slow growth was detectable after four d (Figure 4A). We then examined the steady-state levels of AOX protein in mitochondria lacking Tom70 or depleted for Tom20. Because Tom20 is an essential protein (Harkness et al. 1994a), we could not study a knockout strain. Therefore, we produced mitochondria with greatly reduced levels of Tom20 from a previously constructed tom20RIP sheltered heterokaryon (Harkness et al. 1994a). Growth of the heterokaryon in the presence of fpa results in numerical superiority of the nucleus that is devoid of a functional tom20 gene resulting in a reduction of the Tom20 protein in the mitochondria of the culture (See Methods). We isolated mitochondria from cultures of the tom70 knockout strain (96C10), a Tom20-depleted strain, and appropriate controls. All strains were grown both in the presence and absence of Cm. Mitochondrial proteins were subjected to SDS-PAGE and analyzed on Western blots for the presence of various mitochondrial proteins. Growth of the tom20RIP sheltered heterokaryon in the presence of fpa resulted in mitochondria virtually devoid of Tom20, while Tom70 levels were unaffected (Figure 4B, Tom20 expt, tom20RIP lanes). Levels of AOX in these mitochondria after growth in the presence of fpa and Cm were reduced compared with AOX in tom20RIP cultures grown without fpa. However, the levels of AOX in the tom20RIP cultures are virtually identical to the control cells grown under the corresponding conditions (Figure 4B, Tom20 expt +fpa +Cm lanes for control and tom20RIP).
Figure 4 .
Tom70 is the major receptor for AOX import into mitochondria. (A) The indicated number of conidia from the control (NCN251) and strain 96C10 were spotted onto medium without (−Ant A) or with (+Ant A) antimycin A. Plates were photographed after 2 d (−Ant A) or 4 d (+Ant A) of growth. (B) The indicated strains were grown in liquid medium containing inhibitors as shown. Mitochondria were isolated and subjected to SDS-PAGE followed by Western blot analysis. Antibodies used are indicated on the right. Controls were strain HIV for the Tom20 experiment and NCN251 for the Tom70 experiment. (C) Respiration measurements were taken as in Figure 3B. (D) Mitochondria isolated from the control (NCN251) and 96C10 were incubated with radiolabeled precursor proteins of F1β, AOX, and AAC. After the indicated times of incubation, mitochondria were treated with proteinase K to remove unincorporated precursor proteins. Mitochondria were then washed and subjected to SDS-PAGE. The gel was transferred to nitrocellulose and exposed to x-ray film. Lys, 33% of the radiolabeled lysate used in each lane; p, precursor form of the protein; m, mature form of the protein after removal of the targeting signal; tryp, mitochondria were treated with trypsin to remove surface receptors before the addition of labeled precursor to demonstrate that the import observed in other lanes was dependent on mitochondrial surface receptors.
Thus, fpa appears to have an unknown effect in reducing AOX levels, but the presence or absence of Tom20 does not result in changes in AOX levels relative to the control. On the other hand, mitochondria isolated from the tom70 knockout strain have levels of Tom20 similar to the control strain, but have much reduced AOX levels compared with the relevant control (Figure 4B, Tom70 expt, +Cm lanes), consistent with the results in Figure 2B for 96C10. Respiration measurements on tom70 knockout cells grown in the presence of Cm showed that the reduced amount of AOX in the cells was still sufficient to allow CN-insensitive respiration to occur (Figure 4C). We assume that the reduced level of AOX in the mitochondria of tom70 knockout cells grown in Cm is sufficient to allow slow growth in the presence of antimycin A as seen in Figure 4A.
We also directly measured import of radiolabeled precursor proteins into isolated mitochondria with reduced levels of Tom70 (Figure 4D). AOX import was obviously reduced in mitochondria lacking Tom70, as was import of AAC a protein known to depend on Tom70 for its import (Ryan et al. 1999; Söllner et al. 1990; Young et al. 2003b). In contrast, import of the precursor of the β subunit of the F1 portion of the mitochondrial ATP synthase (F1β), a protein that contains a cleavable N-terminal targeting signal, occurred at a slightly higher level in the mitochondria lacking Tom70. For two quantified experiments, the percentage of import into the mitochondria lacking Tom70 was 100% and 123% of the control for F1β; 39% and 47% for AAC; and 35% and 48% for AOX. The simplest interpretation of these data is that Tom70 is the major receptor involved in the import of the AOX precursor. However, we cannot formally eliminate the possibility that another unknown factor that is decreased in mitochondria lacking Tom70 may play a role in AOX import.
Discussion
This study has identified many genes that are required to achieve full expression of AOX under conditions that induce the enzyme in N. crassa. Absence of many of the genes has only minor effects on AOX (class 3), whereas loss of others has a more pronounced effect (classes 1 and 2). Many of the knockout strains identified in our screen exhibit reduced growth rates and/or reduced ability to form conidia. Thus, such genes are not likely to be specific for an AOX regulatory pathway. Loss of these genes may affect several functions that include AOX expression. More information on AOX expression at the transcriptional and posttranscriptional levels, as well as the assembly of the enzyme into the mitochondria inner membrane, will be required to understand the effects in each case.
Although the AOX precursor protein contains a cleavable N-terminal targeting signal, its efficient import appears to be dependent on Tom70. Tom70 was previously shown to enhance the import of a few precursors containing N-terminal cleavable targeting signals (Hachiya et al. 1995; Hines et al. 1990; Hines and Schatz 1993; Schlossmann et al. 1994). A detailed investigation of precursors of this class revealed that their presequences were recognized by Tom20. However, the mature portions of these precursor proteins were prone to aggregation, which could be prevented by association with Tom70 (Yamamoto et al. 2009). It was suggested that the requirement for Tom70 is related to its known function as a docking site for cytosolic chaperone proteins (Fan et al. 2011; Young et al. 2003b), which would likely be associated with aggregation prone precursor proteins. Furthermore, a chaperone activity of Tom70 toward at least some precursors was also identified (Yamamoto et al. 2009). Thus, the AOX with its hydrophobic domains that interact with the inner mitochondrial membrane (Andersson and Nordlund 1999; Berthold et al. 2000; Berthold and Stenmark 2003) fits nicely into this model of Tom70 function.
Six genes were identified that are predicted to be transcription factors. Two of these, AOD2 and AOD5, were previously characterized as required for AOX production. However, it is now clear that these proteins are involved in the transcription of other genes as well. It has been shown in P. anserina that the orthologs of AOD2 and AOD5 (RSE-2 and RSE-3, respectively) are involved in transcription of AOX but also play a role in transcription of genes involved in gluconeogenesis (Sellem et al. 2009). Similarly, the A. nidulans orthologs (AcuM and AcuK, respectively) are involved in gluconeogenesis and AOX regulation (Hynes et al. 2007; Suzuki et al. 2012). Interestingly, the Aspergillus fumigatus orthologs also are involved in iron uptake (Liu et al. 2010). The other transcription factors identified here, may also have multiple roles, including possible contributions to AOX transcription.
We also identified six genes that encode putative kinases or phosphatases. Phosphorylation/dephosphorylation events are well known to be associated with effects on protein function and gene expression. Perhaps the most interesting gene in this category is the Snf1 kinase, which is disrupted in strain 13G11. The S. cerevisiae Snf1 kinase and its mammalian ortholog, AMPK, are known to play roles in sensing cellular energy levels (Hardie 2007; Turcotte et al. 2010; Young et al. 2003a). When activated by conditions resulting in reduced ATP levels, the kinase acts to phosphorylate transcription factors involved in expression of genes for processes such as gluconeogenesis and the glyoxylate cycle. In fact, the Snf1 kinase is involved in phosphorylation of Rds2 (Soontorngun et al. 2007), the Saccharomyces cerevisiae ortholog of N. crassa AOD2 (Chae et al. 2007b). Rds2 is involved in gluconeogenesis and increased Snf1-dependent phosphorylation of the protein is observed during growth in ethanol as compared to growth in glucose. Binding of Rds2 at the promoter for the FBP1 gene encoding fructose-1,6-bisphophatase, was dependent on Snf1 activity (Soontorngun et al. 2007). Interestingly, the Snf4 gene (30F5) that encodes a homolog of a regulatory factor of the S. cerevisiae Snf1 kinase complex also was identified in our screen as a class 3 mutant. Thus, it will be of interest to determine if phosphorylation of AOD2 by N. crassa SNF1 plays a role in activation of the protein as a transcription factor of the aod-1 gene. For A. nidulans, it was recently suggested that malate may be a ligand that binds to the PAS domains of either AcuM (AOD2) and/or AcuK (AOD5) to activate the heterodimeric transcription factor (Suzuki et al. 2012). Because deletion of the snf1 gene in N. crassa does not completely eliminate AOX expression, it is conceivable that both phosphorylation by SNF1 and binding of malate may be involved in AOD2 activation.
Reactive oxygen species (ROS) are thought to play a role in mitochondrial biogenesis and in eliciting stress responses in many systems (Bae et al. 2011; Collins et al. 2012; Leister 2012; Rhoads and Vanlerberghe 2004; Yoboue and Devin 2012). Increased levels of ROS are known to be associated with increased levels of AOX in certain fungi such as C. albicans (Huh and Kang 2001), Magnoporthe grisea (Yukioka et al. 1998), and A. fumigatus (Magnani et al. 2007). On the other hand, this does not appear to be the case for A. nidulans (Pusztahelyi et al. 2011; Suzuki et al. 2012) or P. anserina (Borghouts et al. 2001). Our screen detected only two genes with an obvious relationship to cellular redox conditions. Superoxide dismutase, which converts superoxide to molecular oxygen and hydrogen peroxide, is inactivated in the class 3 strain 101D3, which has a minor AOX reduction after 48 hr growth in Cm. The specific protein (NCU09560.5) is predicted to encode a version of the enzyme that uses manganese as a cofactor, a hallmark of mitochondrial versions of the enzyme (Holley et al. 2010) and the protein was identified as a Neurospora mitochondrial protein in a proteomic study (Keeping et al. 2011). Because there is only a slight reduction of AOX in the strain after growth in Cm, it would appear that the signaling mechanism for AOX transcription is largely unaffected. Given that AOX is present in this strain during growth in Cm, it is not obvious why it does not grow in the presence of antimycin A (Table 1). One possibility is that the signaling mechanism for AOX production is different for growth in antimycin A vs. Cm. Another possibility is that the increased ROS that is likely produced upon antimycin A inhibition proves toxic to cells in the absence of the mitochondrial superoxide dismutase.
The gene removed in the class 2 mutant 85H5, is listed at the genome database as a thioredoxin that also has a glutaredoxin domain (NCU09803.5). BLAST searches against the S. cerevisae proteome demonstrated that NCU09803.5 is most highly related to the monothiol glutaredoxins, Grx3 and Grx4 (supporting information, Table S1). These yeast proteins, as well as NCU09803.5, contain a thioredoxin-like domain at their N-terminus, but also the CGFS motif that defines monothiol glutaredoxins (Herrero and De La Torre-Ruiz 2007; Lillig et al. 2008; Meyer et al. 2009). The latter domain replaces the CXXC motif found in classical thioredoxins and dithiol glutaredoxins.
A search of the N. crassa database identified a GRX5 protein that represents a mitochondrial monothiol glutaredoxin and revealed that NCU09803.5 is the only non-mitochondrial form of the protein in the organism (see Table S1). This is similar to Schizosaccharomyces pombe, which contains only a single Grx3/Grx4 ortholog, called Grx4 (Chung et al. 2005). These proteins are known to play important roles in various aspects of iron metabolism, including iron sensing, influencing the localization and activity of iron responsive transcription factors, and general intracellular iron trafficking (Lill et al. 2012; Rouhier et al. 2010). With respect to our study, it is of interest to note that depletion of both Grx3 and Grx4 in S. cerevisiea has been shown to result in severe reduction in activity of di-iron enzymes in both the cytosol and mitochondria (Mühlenhoff et al. 2010). Thus, the defect in expression of AOX, a di-iron carboxylate enzyme, seems likely to be related to iron metabolism in the 85H5 strain. The observation that 85H5 does not grow in the presence of antimycin A even though substantial levels of AOX accumulate after 48 hr growth in Cm may reflect an accumulation of AOX protein without inserted iron.
Three class 2 mutants encode components of the COP9 signalosome (CSN). The CSN is a highly conserved complex in eukaryotes consisting of eight subunits in higher organisms, with some variation in the number of subunits in lower eukaryotes. The CSN is best known for its role in the regulation of protein degradation, but other activities, including possible direct roles in gene expression and regulation of transcription factors, have been associated with the complex or its subunits (reviewed in Chamovitz 2009, Wei and Deng 2003, and Wei et al. 2008). In N. crassa, study of a csn-2 disruption strain revealed a role for CSN in the regulation of the circadian clock (He et al. 2005). An in depth study of the N. crassa CSN complex revealed that it contains seven different subunits: CSN1 through CSN6 plus CSN7a (Wang et al. 2010). With the exception of CSN3, cells lacking any of the seven subunits had pleiotropic phenotypes, including defects in proteosome regulation, growth, conidiation, and circadian rhythms. Our screen identified knockouts of the csn-1, csn-2, and csn-5 genes as having effects on AOX production (Table 1). At the time we examined the library the csn-3 and csn-4 gene knockouts also were present but were not identified in the screen. Interestingly, an A. nidulans mutant lacking CSN5 (CsnE) was found to exhibit a complex phenotype that included alterations in redox regulation (Nahlik et al. 2010). Furthermore, an interaction between CSN5 (Jab-1) and the antioxidant protein thioredoxin that affects the activity of the AP-1 transcription factor was seen in human cells (Hwang et al. 2004). Thus, the effects observed on AOX expression in the csn-5 knockout in our study may be the result of altered redox regulation. It will be interesting to determine whether the effect of the csn gene knockouts on AOX abundance in N. crassa is due to an effect on transcription or a posttranscriptional process. There is considerable evidence in other systems suggesting that the CSN2 and CSN5 proteins have roles as individual proteins in gene expression, unrelated to their roles in the complex (Chamovitz 2009; Wei et al. 2008).
Our study has revealed several new genes that affect AOX production. It is valid to ask whether we have found any genes that should be named aod (AOX deficient). Because our previously named aod-2 and aod-5 genes were thought to be AOX-specific transcription factors but are now known to be involved in gluconeogenesis as well (described previously), it may be difficult to assign aod to any gene in the absence of further data regarding the exact function of the gene products. The requirement for many different genes to achieve the full level of AOX in mitochondria suggests that complex pathways of expression exist for the enzyme. Closer examination of the mutants identified here will help to elucidate the nature of these mechanisms.
Supplementary Material
Acknowledgments
We are grateful to the FGSC for supplying the strains used in this work. We are pleased to acknowledge use of materials generated by P01 GM068087 “Functional Analysis of a Model Filamentous Fungus.” C.R. received scholarship support from the Deutscher Akademischer Austausch Dienst of Germany as an exchange student. S.C. and N.E. were supported by undergraduate summer research scholarships from the Natural Sciences and Engineering Research Council of Canada (NSERC). This work was supported by an NSERC Discovery Grant to FEN.
Footnotes
Communicating editor: R. B. Brem
Literature Cited
- Ahting U., Thun C., Hegerl R., Typke D., Nargang F. E., et al. , 1999. The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 147: 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albury M. S., Elliott C., Moore A. L., 2009. Towards a structural elucidation of the alternative oxidase in plants. Physiol. Plant. 137: 316–327 [DOI] [PubMed] [Google Scholar]
- Andersson M. E., Nordlund P., 1999. A revised model of the active site of alternative oxidase. FEBS Lett. 449: 17–22 [DOI] [PubMed] [Google Scholar]
- Bae Y. S., Oh H., Rhee S. G., Yoo Y. D., 2011. Regulation of reactive oxygen species generation in cell signaling. Mol. Cells 32: 491–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold D. A., Stenmark P., 2003. Membrane-bound diiron carboxylate proteins. Annu. Rev. Plant Biol. 54: 497–517 [DOI] [PubMed] [Google Scholar]
- Berthold D. A., Andersson M. E., Nordlund P., 2000. New insight into the structure and function of the alternative oxidase. Biochim. Biophys. Acta 1460: 241–254 [DOI] [PubMed] [Google Scholar]
- Bertrand H., Argan A., Szakacs N. A., 1983. Genetic control of the biogenesis of cyanide insensitive respiration in Neurospora crassa, pp. 495–507 in Mitochondria 1983, edited by Schweyen R. J., Wolf K., Kaudewitz F. Walter de Gruyter Co., Berlin [Google Scholar]
- Borghouts C., Werner A., Elthon T. E., Osiewacz H. D., 2001. Copper-modulated gene expression and senescence in the filamentous fungus Podospora anserina. Mol. Cell. Biol. 21: 390–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brix J., Rudiger S., Bukau B., Schneider-Mergener J., Pfanner N., 1999. Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J. Biol. Chem. 274: 16522–16530 [DOI] [PubMed] [Google Scholar]
- Chae M. S., Nargang F. E., 2009. Investigation of regulatory factors required for alternative oxidase production in Neurospora crassa. Physiol. Plant. 137: 407–418 [DOI] [PubMed] [Google Scholar]
- Chae M. S., Lin C. C., Kessler K. E., Nargang C. E., Tanton L. L., et al. , 2007a Identification of an alternative oxidase induction motif in the promoter region of the aod-1 gene in Neurospora crassa. Genetics 175: 1597–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae M. S., Nargang C. E., Cleary I. A., Lin C. C., Todd A. T., et al. , 2007b Two zinc cluster transcription factors control induction of alternative oxidase in Neurospora crassa. Genetics 177: 1997–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamovitz D. A., 2009. Revisiting the COP9 signalosome as a transcriptional regulator. EMBO Rep. 10: 352–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W. H., Kim K. D., Roe J. H., 2005. Localization and function of three monothiol glutaredoxins in Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 330: 604–610 [DOI] [PubMed] [Google Scholar]
- Collins Y., Chouchani E. T., James A. M., Menger K. E., Cocheme H. M., et al. , 2012. Mitochondrial redox signalling at a glance. J. Cell Sci. 125: 801–806 [DOI] [PubMed] [Google Scholar]
- Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C., et al. , 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Considine M. J., Daley D. O., Whelan J., 2001. The expression of alternative oxidase and uncoupling protein during fruit ripening in mango. Plant Physiol. 126: 1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. H., De Serres F. J., 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 17: 79–143 [Google Scholar]
- Dekker P. J. T., Ryan M. T., Brix J., Müller H., Hönlinger A., et al. , 1998. Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol. Cell. Biol. 18: 6515–6524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descheneau A. T., Cleary I. A., Nargang F. E., 2005. Genetic evidence for a regulatory pathway controlling alternative oxidase production in Neurospora crassa. Genetics 169: 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djajanegara I., Finnegan P. M., Mathieu C., McCabe T., Whelan J., et al. , 2002. Regulation of alternative oxidase gene expression in soybean. Plant Mol. Biol. 50: 735–742 [DOI] [PubMed] [Google Scholar]
- Do T. Q., Hsu A. Y., Jonassen T., Lee P. T., Clarke C. F., 2001. A defect in coenzyme Q biosynthesis is responsible for the respiratory deficiency in Saccharomyces cerevisiae abc1 mutants. J. Biol. Chem. 276: 18161–18168 [DOI] [PubMed] [Google Scholar]
- Edwards D. L., Chalmers J. H., Jr, Guzik H. J., Warden J. T., 1976. Assembly of the cyanide-insensitive respiratory pathway in Neurospora crassa, pp. 865–872 in Genetics and Biogenesis of Chloroplasts and Mitochondria, edited by Bucher T. H., Neupert W., Sebald W., Werner S. Elsevier/North-Holland Biomedical Press, Amsterdam [Google Scholar]
- Endo T., Yamano K., 2010. Transport of proteins across or into the mitochondrial outer membrane. Biochim. Biophys. Acta 1803: 706–714 [DOI] [PubMed] [Google Scholar]
- Fan A. C., Kozlov G., Hoegl A., Marcellus R. C., Wong M. J., et al. , 2011. Interaction between the human mitochondrial import receptors Tom20 and Tom70 in vitro suggests a chaperone displacement mechanism. J. Biol. Chem. 286: 32208–32219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan P. M., Whelan J., Millar A. H., Zhang Q., Smith M. K., et al. , 1997. Differential expression of the multigene family encoding soybean mitochondrial alternative oxidase. Plant Physiol. 114: 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdag Z., Fujs S., Koszegi B., Kalman N., Papp G., et al. , 2011. The abc1-/coq8- respiratory-deficient mutant of Schizosaccharomyces pombe suffers from glutathione underproduction and hyperaccumulates Cd2+. Folia Microbiol. (Praha) 56: 353–359 [DOI] [PubMed] [Google Scholar]
- Good A. G., Crosby W. L., 1989. Anaerobic induction of alanine aminotransferase in barley root tissue. Plant Physiol. 90: 1305–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad L., Descheneau A., Neupert W., Lill R., Nargang F., 1999. Inactivation of the Neurospora crassa mitochondrial outer membrane protein TOM70 by repeat-induced point mutation (RIP) causes defects in mitochondrial protein import and morphology. Curr. Genet. 36: 137–146 [DOI] [PubMed] [Google Scholar]
- Hachiya N., Mihara K., Suda K., Horst M., Schatz G., et al. , 1995. Reconstitution of the initial steps of mitochondrial protein import. Nature 376: 705–709 [DOI] [PubMed] [Google Scholar]
- Hardie D. G., 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8: 774–785 [DOI] [PubMed] [Google Scholar]
- Harkness T. A., Metzenberg R. L., Schneider H., Lill R., Neupert W., et al. , 1994a Inactivation of the Neurospora crassa gene encoding the mitochondrial protein import receptor MOM19 by the technique of “sheltered RIP.” Genetics 136: 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkness T. A., Nargang F. E., Van Der Klei I., Neupert W., Lill R., 1994b A crucial role of the mitochondrial protein import receptor MOM19 for the biogenesis of mitochondria. J. Cell Biol. 124: 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Cheng P., Liu Y., 2005. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 19: 1518–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E., De La Torre-Ruiz M. A., 2007. Monothiol glutaredoxins: a common domain for multiple functions. Cell. Mol. Life Sci. 64: 1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines V., Schatz G., 1993. Precursor binding to yeast mitochondria. J. Biol. Chem. 268: 449–454 [PubMed] [Google Scholar]
- Hines V., Brandt A., Griffiths G., Horstmann H., Brütsch H., et al. , 1990. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 9: 3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley A. K., Dhar S. K., St Clair D. K., 2010. Manganese superoxide dismutase vs. p53: regulation of mitochondrial ROS. Mitochondrion 10: 649–661 [DOI] [PubMed] [Google Scholar]
- Huh W., Kang S., 2001. Characterization of the gene family encoding alternative oxidase from Candida albicans. Biochem. J. 356: 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C. Y., Ryu Y. S., Chung M. S., Kim K. D., Park S. S., et al. , 2004. Thioredoxin modulates activator protein 1 (AP-1) activity and p27Kip1 degradation through direct interaction with Jab1. Oncogene 23: 8868–8875 [DOI] [PubMed] [Google Scholar]
- Hynes M. J., Szewczyk E., Murray S. L., Suzuki Y., Davis M. A., et al. , 2007. Transcriptional control of gluconeogenesis in Aspergillus nidulans. Genetics 176: 139–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski M., Sudre D., Schansker G., Schellenberg M., Constant S., et al. , 2008. AtOSA1, a member of the Abc1-like family, as a new factor in cadmium and oxidative stress response. Plant Physiol. 147: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski S. M., Kriete A., 2012. The yeast retrograde response as a model of intracellular signaling of mitochondrial dysfunction. Front Physiol 3: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova O. V., Kuzmin E. V., Elthon T. E., Newton K. J., 2002. Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 14: 3271–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeping A., Deabreu D., Dibernardo M., Collins R. A., 2011. Gel-based mass spectrometric and computational approaches to the mitochondrial proteome of Neurospora. Fungal Genet. Biol. 48: 526–536 [DOI] [PubMed] [Google Scholar]
- Lambowitz A. M., Sabourin J. R., Bertand H., Nickels R., McIntosh L., 1989. Immunological identification of the alternative oxidase of Neurospora crassa mitochondria. Mol. Cell. Biol. 9: 1362–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämmli U. K., 1970. Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Leister D., 2012. Retrograde signaling in plants: from simple to complex scenarios. Front Plant Sci 3: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Ritzel R. G., Mclean L. T. T., McIntosh L., Ko T., et al. , 1996. Cloning and analysis of the alternative oxidase of Neurospora crassa. Genetics 142: 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R., Hoffmann B., Molik S., Pierik A. J., Rietzschel N., et al. , 2012. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim. Biophys. Acta. 1823: 1491–1508 [DOI] [PubMed] [Google Scholar]
- Lillig C. H., Berndt C., Holmgren A., 2008. Glutaredoxin systems. Biochim. Biophys. Acta 1780: 1304–1317 [DOI] [PubMed] [Google Scholar]
- Liu Z., Butow R. A., 2006. Mitochondrial retrograde regulation. Annu. Rev. Genet. 40: 159–185 [DOI] [PubMed] [Google Scholar]
- Liu H., Gravelat F. N., Chiang L. Y., Chen D., Vanier G., et al. , 2010. Aspergillus fumigatus AcuM regulates both iron acquisition and gluconeogenesis. Mol. Microbiol. 78: 1038–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson S., Larochelle M., Turcotte B., 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70: 583–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani T., Soriani F. M., Martins V. P., Nascimento A. M., Tudella V. G., et al. , 2007. Cloning and functional expression of the mitochondrial alternative oxidase of Aspergillus fumigatus and its induction by oxidative stress. FEMS Microbiol. Lett. 271: 230–238 [DOI] [PubMed] [Google Scholar]
- McCluskey K., Wiest A., Plamann M., 2010. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J. Biosci. 35: 119–126 [DOI] [PubMed] [Google Scholar]
- McDonald A. E., 2008. Alternative oxidase: an inter-kingdom perspective on the function and regulation of this broadly distributed “cyanide-resistant” terminal oxidase. Funct. Plant Biol. 35: 535–552 [DOI] [PubMed] [Google Scholar]
- McDonald A. E., Vanlerberghe G. C., 2006. Origins, evolutionary history, and taxonomic distribution of alternative oxidase and plastoquinol terminal oxidase. Comp. Biochem. Physiol. Part D 1: 357–364 [DOI] [PubMed] [Google Scholar]
- Metzenberg R., 2004. Bird medium: an alternative to Vogel medium. Fungal Genet. Newsl. 51: 19–20 [Google Scholar]
- Meyer Y., Buchanan B. B., Vignols F., Reichheld J. P., 2009. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu. Rev. Genet. 43: 335–367 [DOI] [PubMed] [Google Scholar]
- Mitchell M. B., Mitchell H. K., Tissieres A., 1953. Mendelian and non-Mendelian factors affecting the cytochrome system in Neurospora Crassa. Proc. Natl. Acad. Sci. USA 39: 606–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenhoff U., Molik S., Godoy J. R., Uzarska M. A., Richter N., et al. , 2010. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 12: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahlik K., Dumkow M., Bayram O., Helmstaedt K., Busch S., et al. , 2010. The COP9 signalosome mediates transcriptional and metabolic response to hormones, oxidative stress protection and cell wall rearrangement during fungal development. Mol. Microbiol. 78: 964–979 [DOI] [PubMed] [Google Scholar]
- Nargang F. E., Kennell J. C., 2010. Mitochondria and respiration, pp. 155–178 in Cellular and Molecular Biology of Filamentous Fungi, edited by Borkovich K. A., Ebbole D. J. ASM Press, Washington, DC [Google Scholar]
- Nargang F. E., Rapaport D., 2007. Neurospora crassa as a model organism for mitochondrial biogenesis, pp. 107–123 in Mitochondria. Practical Protocols, edited by Leister D. L., Herrmann J. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- Nargang F. E., Bertrand H., Werner S., 1978. A nuclear mutant of Neurospora crassa lacking subunit 1 of cytochrome c oxidase. J. Biol. Chem. 253: 6364–6369 [PubMed] [Google Scholar]
- Nargang F. E., Künkele K.-P., Mayer A., Ritzel R. G., Neupert W., et al. , 1995. “Sheltered disruption” of Neurospora crassa MOM22, an essential component of the mitochondrial protein import complex. EMBO J. 14: 1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G., Servin J. A., Turner G. E., Altamirano L., Colot H. V., et al. , 2011. Global analysis of serine-threonine protein kinase genes in Neurospora crassa. Eukaryot. Cell 10: 1553–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusztahelyi T., Klement E., Szajli E., Klem J., Miskei M., et al. , 2011. Comparison of transcriptional and translational changes caused by long-term menadione exposure in Aspergillus nidulans. Fungal Genet. Biol. 48: 92–103 [DOI] [PubMed] [Google Scholar]
- Rapaport D., Nargang F. E., 2004 Mitochondrial biogenesis: protein import into and across the outer membrane, pp. 37–58 in Topics in Current Genetics, Vol. 8. Biogenesis of Mitochondria and Associated Diseases, edited by M. Bauer and C. Koehler. Springer-Verlag, Berlin Heidelberg. [Google Scholar]
- Rhoads D. M., Vanlerberghe G. C., 2004. Mitochondria-nucleus interactions: evidence for mitochondrial retrograde communication in plant cells, pp. 83–106 in Plant Mitochondria: From Genome to Function, edited by Day D. A., Millar A. H., Whelan J. Kluwer Academic Publishers, Great Britain [Google Scholar]
- Rouhier N., Couturier J., Johnson M. K., Jacquot J. P., 2010. Glutaredoxins: roles in iron homeostasis. Trends Biochem. Sci. 35: 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M. T., Muller H., Pfanner N., 1999. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem. 274: 20619–20627 [DOI] [PubMed] [Google Scholar]
- Schlossmann J., Dietmeier K., Pfanner N., Neupert W., 1994. Specific recognition of mitochondrial preproteins by the cytosolic domain of the import receptor MOM72. J. Biol. Chem. 269: 11893–11901 [PubMed] [Google Scholar]
- Schmitt S., Prokisch H., Schlunck T., Camp II D. G., Ahting U., et al. , 2006. Proteome analysis of mitochondrial outer membrane from Neurospora crassa. Proteomics 6: 72–80 [DOI] [PubMed] [Google Scholar]
- Sellem C. H., Bovier E., Lorin S., Sainsard-Chanet A., 2009. Mutations in two zinc cluster proteins activate alternative respiratory and gluconeogenic pathways and restore senescence in long-lived respiratory mutants of Podospora anserina. Genetics 182: 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T., Pfaller R., Griffiths G., Pfanner N., Neupert W., 1990. A mitochondrial import receptor for the ADP/ATP carrier. Cell 62: 107–115 [DOI] [PubMed] [Google Scholar]
- Soontorngun N., Larochelle M., Drouin S., Robert F., Turcotte B., 2007. Regulation of gluconeogenesis in Saccharomyces cerevisiae is mediated by activator and repressor functions of Rds2. Mol. Cell. Biol. 27: 7895–7905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger H. F., Söllner T., Kiebler M., Dietmeier K. A., Trülzsch K. S., et al. , 1990. Import of ADP/ATP carrier into mitochondria: two receptors act in parallel. J. Cell Biol. 111: 2353–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Murray S. L., Wong K. H., Davis M. A., Hynes M. J., 2012. Reprogramming of carbon metabolism by the transcriptional activators AcuK and AcuM in Aspergillus nidulans. Mol. Microbiol. 84: 942–964 [DOI] [PubMed] [Google Scholar]
- Tanton L. L., Nargang C. E., Kessler K. E., Li Q., Nargang F. E., 2003. Alternative oxidase expression in Neurospora crassa. Fungal Genet. Biol. 39: 176–190 [DOI] [PubMed] [Google Scholar]
- Tauche A., Krause-Buchholz U., Rodel G., 2008. Ubiquinone biosynthesis in Saccharomyces cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res. 8: 1263–1275 [DOI] [PubMed] [Google Scholar]
- Turcotte B., Liang X. B., Robert F., Soontorngun N., 2010. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 10: 2–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O., Giraud E., Clifton R., Whelan J., 2009. Alternative oxidase: a target and regulator of stress responses. Physiol. Plant. 137: 354–361 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe G. C., McIntosh L., 1997. Alternative oxidase: from gene to function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 703–734 [DOI] [PubMed] [Google Scholar]
- Wang J., Hu Q., Chen H., Zhou Z., Li W., et al. , 2010. Role of individual subunits of the Neurospora crassa CSN complex in regulation of deneddylation and stability of cullin proteins. PLoS Genet. 6: e1001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N., Deng X. W., 2003. The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 19: 261–286 [DOI] [PubMed] [Google Scholar]
- Wei N., Serino G., Deng X. W., 2008. The COP9 signalosome: more than a protease. Trends Biochem. Sci. 33: 592–600 [DOI] [PubMed] [Google Scholar]
- Woodson J. D., Chory J., 2008. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Fukui K., Takahashi H., Kitamura S., Shiota T., et al. , 2009. Roles of Tom70 in import of presequence-containing mitochondrial proteins. J. Biol. Chem. 284: 31635–31646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoboue E. D., Devin A., 2012. Reactive oxygen species-mediated control of mitochondrial biogenesis. Int. J. Cell Biol. 2012: 403870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E. T., Dombek K. M., Tachibana C., Ideker T., 2003a Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem. 278: 26146–26158 [DOI] [PubMed] [Google Scholar]
- Young J. C., Hoogenraad N. J., Hartl F. U., 2003b Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112: 41–50 [DOI] [PubMed] [Google Scholar]
- Yukioka H., Inagaki S., Tanaka R., Katoh K., Miki N., et al. , 1998. Transcriptional activation of the alternative oxidase gene of the fungus Magnaporthe grisea by a respiratory-inhibiting fungicide and hydrogen peroxide. Biochim. Biophys. Acta 1442: 161–169 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.