Abstract
Cells often move as collective groups during normal embryonic development and wound healing, although the mechanisms governing this type of migration are poorly understood. The Drosophila melanogaster border cells migrate as a cluster during late oogenesis and serve as a powerful in vivo genetic model for collective cell migration. To discover new genes that participate in border cell migration, 64 out of 66 genes that encode PDZ domain-containing proteins were systematically targeted by in vivo RNAi knockdown. The PDZ domain is one of the largest families of protein-protein interaction domains found in eukaryotes. Proteins that contain PDZ domains participate in a variety of biological processes, including signal transduction and establishment of epithelial apical-basal polarity. Targeting PDZ proteins effectively assesses a larger number of genes via the protein complexes and pathways through which these proteins function. par-6, a known regulator of border cell migration, was a positive hit and thus validated the approach. Knockdown of 14 PDZ domain genes disrupted migration with multiple RNAi lines. The candidate genes have diverse predicted cellular functions and are anticipated to provide new insights into the mechanisms that control border cell movement. As a test of this concept, two genes that disrupted migration were characterized in more detail: big bang and the Dlg5 homolog CG6509. We present evidence that Big bang regulates JAK/STAT signaling, whereas Dlg5/CG6509 maintains cluster cohesion. Moreover, these results demonstrate that targeting a selected class of genes by RNAi can uncover novel regulators of collective cell migration.
Keywords: collective migration, PSD95/Dlg/ZO-1 (PDZ) domains, border cells, JAK/STAT, Drosophila
Regulated cell movement is critical for embryonic development, adult wound healing, and normal immune system function. Determining how cells migrate during normal processes can help us better understand how misregulated cell migration contributes to pathologies such as tumor metastasis and inflammation. While some cells migrate singly, others move as small or large groups in a type of migration called collective migration (Friedl and Gilmour 2009). Cells migrate collectively during gastrulation in the embryo and in epithelial sheet migration during wound closure. Notably, this type of group migration has also been observed during tumor invasion and metastasis (Friedl and Gilmour 2009; Yilmaz and Christofori 2010; Friedl et al. 2012). Migrating cells display striking morphological changes induced by dynamic rearrangement of actin filaments and cell-substrate adhesions, which together provide the necessary force for movement (Ridley 2011). Cells migrating collectively further need to coordinate such individual cell motility to precisely modulate cell-cell adhesions and the cytoskeleton among cells in the group (Friedl and Gilmour 2009). Our current understanding of the mechanisms that regulate these and other aspects of collective cell migration in tissues is fairly limited. Therefore, we have turned to a genetically amenable model, the Drosophila border cells, to identify new genes and pathways that control collective cell migration.
Border cells migrate as a cohesive cluster of 6–10 cells during late oogenesis in a highly regulated process (Montell 2003). Border cells are first specified in the anterior follicle cell epithelium at early stage 9. The follicular epithelium is a monolayer of ∼600 cells that surrounds the germline-derived cells of the egg chamber, the basic subunit of the Drosophila ovary. The cytokine-like protein Unpaired (Upd) is secreted from a pair of non-migratory cells, the polar cells, to activate Janus kinase (JAK)/signal transducer activator of transcription (STAT) signaling in the surrounding follicle cells (Silver and Montell 2001; Beccari et al. 2002; Ghiglione et al. 2002; Xi et al. 2003; Silver et al. 2005). Cells expressing the highest levels of active JAK/STAT at the anterior end of the egg chamber become border cells. The border cells form a cluster around the polar cells and subsequently detach from the epithelium. Border cells then migrate over ∼150 µm distance through the germline-derived nurse cell layer to reach the oocyte (Figure 1A). Previous genetic screens identified multiple essential regulators of border cell migration, including the highly conserved steroid hormone receptor and receptor tyrosine kinase (RTK) signaling pathways (Liu and Montell 1999; Bai et al. 2000; Duchek and Rørth 2001; Duchek et al. 2001; Silver and Montell 2001; McDonald et al. 2003; Mathieu et al. 2007). However, none of the screens to date were performed to saturation and, therefore, may have missed critical genes. Moreover, despite the discovery of these and other signaling pathways, in many cases the specific downstream effectors that interpret these signals to produce specific cellular responses in border cells remain unknown.
Figure 1 .
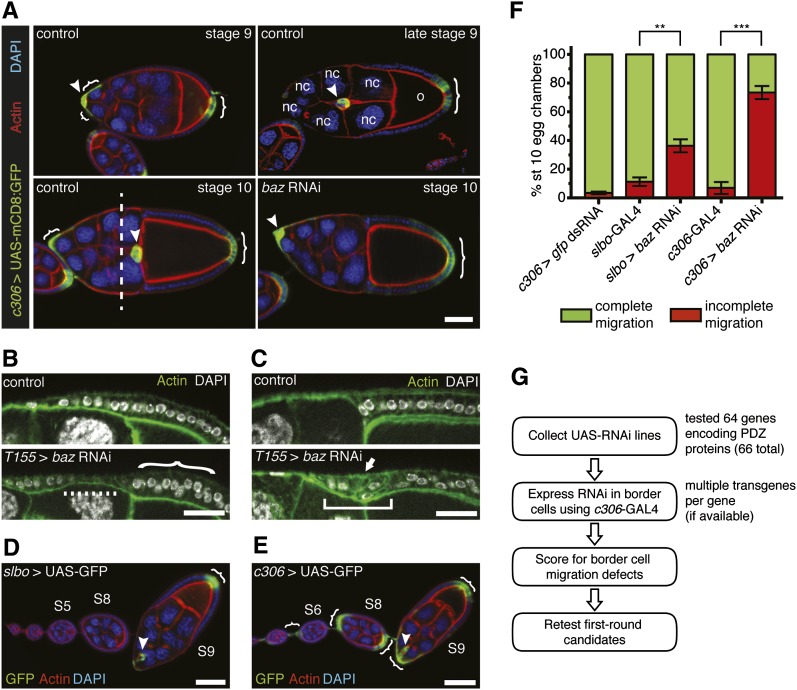
In vivo RNAi knockdown to identify PDZ domain-encoding genes required for border cell migration. (A) Control border cells (arrowheads) migrate between the nurse cells (nc) from stage 9 to 10 of oogenesis to reach the oocyte (o). Border cell clusters that have migrated past the dashed line (stage 10 control) are considered to have completed their migration. Border cells and follicle cells (brackets) express UAS-mCD8:GFP (green) driven by c306-GAL4 in egg chambers at the indicated stages; genotype is c306-GAL4/+; UAS-mCD8:GFP/+. Egg chambers were co-stained for actin (red) and DAPI (blue) to label cell membranes and nuclei, respectively. (Lower right panel) A stage 10 c306-GAL4/+; UAS-mCD8:GFP/UAS-baz RNAi v2914 (baz RNAi) egg chamber in which border cells did not migrate. Scale bar is 20 μm. (B and C) Knockdown of baz in follicle cells (bottom panels) using the follicle cell driver T155-GAL4 disrupts the epithelium compared with control (top panels) at stage 9 (B) and stage 10 (C). Genotypes are T155-GAL4/+ (control) and UAS-baz RNAi/+; +/T155-GAL4. Scale bar is 20 μm. (B) baz RNAi follicle cell layer is thin (dashed line), and some nuclei are misaligned (bracket) compared with control. (C) baz RNAi follicle cells are multilayered (arrow) and fail to retract over the oocyte (square bracket) as in control. (D and E) Ovarioles showing GAL4 expression patterns in border cells (arrowheads) and follicle cells (brackets) as visualized by UAS-mCD8:GFP (green); stages are indicated. Egg chambers were co-stained for actin (red) and DAPI (blue). Scale bar is 50 μm. (D) slbo-GAL4 expression pattern (slbo-GAL4, UAS-mCD8:GFP/+). (E) c306-GAL4 expression pattern (c306-GAL4/+; UAS-mCD8:GFP/+). c306-GAL4 is also expressed in stalk cells, which connect egg chambers within the ovariole. (F) Quantification of migration in stage 10 egg chambers of the indicated genotypes, shown as the percentage with complete (green) or incomplete (red) border cell migration. Error bars represent SEM; n ≥ 50 egg chambers in each of three trials (**P < 0.01; ***P < 0.001; two-tailed unpaired t-test). (G) Outline of the scheme used to survey the role of PDZ genes in border cell migration. Anterior is to the left in this and all subsequent figures.
Correct establishment of cell polarity is critical for the motility of many types of cells, including border cells (Niewiadomska et al. 1999; Pinheiro and Montell 2004; Etienne-Manneville 2008; McDonald et al. 2008). Proteins that regulate epithelial polarity help orient migrating cells and promote motility of both single and collectively migrating cells by organizing the cellular membrane and cytoskeleton (Humbert et al. 2006; Etienne-Manneville 2008; Hidalgo-Carcedo et al. 2011). Moreover, many of these proteins themselves localize in a polarized manner within cells, typically at the cell cortex. Epithelial polarity proteins have also emerged as potential tumor suppressors (Etienne-Manneville 2008; Humbert et al. 2008; Royer and Lu 2011). A large number of polarity proteins implicated in cell migration, such as Par-3, Patj, and Dlg1, contain PSD95/Dlg/ZO-1 (PDZ) domains. The PDZ domain is a small globular module that functions as a protein-protein interaction domain (Harris and Lim 2001; Subbaiah et al. 2011). Specifically, PDZ domains bind to short PDZ-binding motifs (PBM) on target proteins that are mainly, although not exclusively, found at C-termini (Harris and Lim 2001; Subbaiah et al. 2011). PDZ domains can occur alone or as multiple copies and are often found in combination with other protein-protein interaction domains and/or catalytic domains. Proteins with PDZ domains typically mediate the formation of large multi-protein scaffolding complexes that bring molecular components into proximity with each other within the cell (Ranganathan and Ross 1997). PDZ domain-containing proteins regulate signaling, cytoskeletal dynamics, and cell adhesion in addition to polarity, all of which are important for cell motility. Moreover, the polarity PDZ proteins Bazooka (Baz; Par-3 homolog) and Par-6 organize the localization of membrane-associated proteins within the border cell cluster to promote migration (Pinheiro and Montell 2004). This raises the distinct possibility that other, unidentified PDZ domain complexes regulate the polarity and collective migration of border cells.
We sought to identify additional genes that participate in border cell migration using an RNAi knockdown approach. The recent availability of large collections of UAS-RNAi transgenic lines have made it possible to systematically analyze the roles of the majority of genes in the Drosophila genome for specific phenotypes (Dietzl et al. 2007; Ni et al. 2009). These lines are used to knock down gene function in a tissue- and temporal-specific manner using the GAL4/UAS system (Brand and Perrimon 1993; Perrimon et al. 2010). While multiple genome-wide in vivo RNAi screens have been performed (Cronin et al. 2009; Mummery-Widmer et al. 2009; Schnorrer et al. 2010), a substantial number of lines (>12,000) is needed to screen most of the genes in the genome at least once. Moreover, large-scale screens could miss more subtle phenotypes. Several recent studies targeted specific classes of proteins, for example Rab GTPase-activating proteins and kinases, to identify new regulators of cell migration (Simpson et al. 2008; Laflamme et al. 2012). In the present study, we specifically targeted by RNAi knockdown 64 out of 66 genes predicted to encode PDZ domain-containing proteins. We identified 14 high confidence and 17 additional PDZ domain-containing proteins whose knockdown inhibited border cell migration. We provide additional evidence that two genes, big bang and CG6509, regulate specific features of border cells. The genes identified here thus represent a group of conserved signaling pathways and/or intracellular protein complexes that may regulate other types of collectively migrating cells.
Materials and Methods
Drosophila genetics
All crosses were kept at 25° using standard protocols. c306-GAL4, slbo-GAL4, hsp70-GAL4 (hs-GAL4), T155-GAL4 (Bloomington Stock Center) and tubulin-GAL4 (from A. Page-McCaw) were used to drive UAS-RNAi expression. GAL4 lines were outcrossed to w1118 and used as controls. c96-GAL4, UAS-mCD8:GFP (from A. Zhu) was used to study the bbg expression pattern (Gustafson and Boulianne 1996). The following lines for off-target genes were obtained from the Vienna Drosophila RNAi Center (VDRC) or Harvard Transgenic RNAi Project (TRiP) from the Bloomington Stock Center: UAS-ERR RNAi KK108422 (line v108349, VDRC); UAS-Irbp RNAi (line JF03273, TRiP); and UAS-CG42724 GD4280 RNAi (line v30629, VDRC). Additional fly stocks from the Bloomington Stock Center were two insertions of UAS-CG6509.GFP (http://flybase.org/reports/FBrf0211100.html); PsGefΔ21, FRT19A; PsGefΔ55, FRT19A; UAS-mCD8::GFP; and UAS-GFP dsRNA.
In vivo RNAi knockdown
RNAi lines were obtained from VDRC, TRiP, NIG-Fly, and the Bloomington Stock Center. Supporting Information, Table S1 provides the complete list of RNAi lines. Virgin c306-GAL4; UAS-mCD8:GFP/CyO flies were crossed with males from each UAS-RNAi line. Eight female progeny flies per cross were fattened by feeding with yeast paste for 20 hr at 29° prior to dissection to achieve maximal GAL4/UAS expression. The UAS-baz RNAi v2914 line (VDRC) and the UAS-GFP dsRNA (line 143) (http://flybase.org/reports/FBrf0191479.html) were used as positive and negative controls, respectively. The RNAi lines were tested in batches of 22 lines together with the controls in 24-well plates. Whole ovaries were dissected as described (McDonald and Montell 2005; Prasad et al. 2007). Ovaries were fixed with 4% formaldehyde in potassium phosphate buffer (pH 7.2) for 10 min and washed with potassium phosphate buffer. Fixed ovaries were manually dissociated in 80% glycerol. UAS-mCD8:GFP fluorescence was used to visualize border cells in dissociated ovaries. Analysis of border cell migration was performed with a Zeiss Stereo Discovery V8 epi-fluorescent stereomicroscope. Crosses were set up independently and retested as above to confirm first-round candidates.
Quantitative RT-PCR analysis of gene expression
Virgin hsp70-GAL4 (hs-GAL4) flies were crossed to male UAS-RNAi flies. To express RNAi ubiquitously, adult female progeny were heat shocked for 1 hr, three times a day, at 37° for two days. Ovaries were dissected the following day. RNA was extracted from ovaries or adult female fly carcasses (ovaries removed) using Trizol (Invitrogen). To determine endogenous expression levels, RNA was extracted from 15 to 20 ovary pairs dissected from hs-GAL4/UAS-GFP dsRNA females. The endogenous expression levels of rp49 and tub84B were measured as reference controls (see Table 2 for details). To determine RNAi knockdown, RNA was extracted from 10 to 15 hs-GAL4 > UAS-RNAi female fly carcasses. Note that ovaries were removed because the germline is potentially refractory to long double-stranded hairpin RNA knockdown (Ni et al. 2011). RNA was purified using the Qiagen RNAeasy Kit, followed by cDNA synthesis using the Taqman Reverse Transcription Kit (Applied Biosystems) and 1.5 μg of purified RNA. qRT-PCR was performed using the Roche Lightcycler 480 to run 15 μL reactions containing 0.5 µL cDNA, 0.5 μL 10 µM primer mix, and 7.5 μL of SYBR Green Master Mix (Roche). All qRT-PCR experiments were performed in triplicate on three separate biological samples. In each experiment, UAS-baz RNAi was used as the positive control, and UAS-dsRNA GFP (GFP RNAi) was used as the negative control. RNAi knockdown was calculated using the ΔΔCT method using rp49 gene expression for normalization.
Table 2. Expression levels and RNAi knockdown efficiency as measured by quantitative RT-PCR.
| Gene | Expression Level in the Ovary (CT value ± SD)a,b | RNAi Linec | Percentage Knockdown in Whole Fliesd |
|---|---|---|---|
| Positive candidates | |||
| baz | 25.7 ± 0.350 (L–M) | v2914 | 41*** |
| bbg | 27.4 ± 0.435 (L–M) | v15975 | 89** |
| CG6498 | 26.0 ± 0.640 (L–M) | v35100 | 53* |
| CG6509 | 26.3 ± 0.868 (L–M) | v22496 | 77** |
| Negative candidates | |||
| CG43707 | 34.5 ± 0.885 (L) | v25846 | 40 (ns) |
| CG43955 | 40.1 ± 1.51 (NE) | v103267 | 78** |
| CG9588 | 24.3 ± 0.318 (M–H) | HM05013 | 82*** |
| cnk | 25.5 ± 0.451 (L–M) | HMS00238 | 67*** |
| dlg1 | 25.4 ± 0.575 (L–M) | HMS01521 | 52** |
| dysc | 28.6 ± 0.237 (L–M) | v23278 | 32* |
| Grip | 28.4 ± 0.671 (L–M) | v103551 | 90*** |
| PICK1 | 30.4 ± 0.252 (L) | JF01199 | 21** |
| RHOGAP100F | 31.9 ± 0.874 (L) | HMS00740 | 14 (ns) |
| scrib | 25.8 ± 0.512 (L–M) | v105412 | 50** |
| sif | 31.2 ± 0.0700 (L) | v106832 | 16 (ns) |
| Syn1 | 31.2 ± 0.500 (L) | JF02654 | 32 (ns) |
| vari | 25.7 ± 0.0985 (L–M) | HM05087 | 72*** |
| X11L | 26.0 ± 0.463 (L–M) | v28652 | 55** |
Mean CT value and SD are calculated from three independent qPCR experiments. Expression summary: CT < 20, very high (VH); CT = 20–25, moderate to high (M–H); CT = 25–30, low to moderate (L–M); CT = 30–35, low (L); CT = 35–40, none to low (N–L); CT > 40, not expressed (NE).
For reference, the endogenous expression levels (mean CT value ± SD) of rp49 and tub84b were 17.7 ± 0.112 and 19.1 ± 0.16, respectively.
RNAi lines are from the Vienna Drosophila RNAi Center (prefixed with v) and from the Harvard Transgenic RNAi Project (prefixed with HM, HMS, or JF).
Extent of reduction of target gene expression compared with gfp dsRNA control, calculated using the ΔΔCT method with rp49 expression as reference. The one-tailed unpaired t-test was used to test for significance (ns, P > 0.05; *P = 0.01–0.05; **P = 0.001–0.01; ***P < 0.001).
The following primers were used: baz fwd, CAGGAGCTGCAGATGTCGGATG; baz rev, ctcgtgatcgccatcctccaaaag; bbg fwd, CAATCTCCACACAACGAGCTCCAC; bbg rev, ggagatgccgccaagcttagc; CG43955 fwd, GGCTTTGATAGCTGGGCGAGC; CG43955 rev, gggggccctgaacaagatgaag; CG43707 fwd, GCGGATGGTCGAAACGATATTGCG; CG43707 rev, cttcttgccggatgcattggcg; CG6498 fwd, CCTGCTCCGGAAGATCTCCTATC; CG6498 rev, ctggtaacggagcggtcagttc; CG6509 fwd, CAGCATGATCAGAAGGCGATCCC; CG6509 rev, cacctgcatccgttccagcag; CG9588 fwd, GATGATCGTCTGTCGCGCCAG; CG9588 rev, cgtggaggcgcagatcaacag; cnk fwd, CTCCAGCTGTATGGCCGTATG; cnk rev, ggcctacatcaacatcgccgag; dlg1 fwd, CCCGGCGACAATGGCATCTATG; dlg1 rev, ccagttcgtgcgttacgttctcc; dysc fwd, CTAGGATTGTATCACCGGGTCGC; dysc rev, gcgcgaccagcaaatcgatcatg; Grip fwd, CAGTCCCGACGAGGTGATGAC, Grip rev, cgggactccagtgtgctaaagc; PICK1 fwd, GATTGGCATCAGCATTGGGGGTG; PICK1 rev, cacgctcaccgaattcacagcc; RhoGAP100F fwd, CACGGGCTCAGCGATTTTCGTG; RhoGAP100F rev, cgcacgggtagtgctgaaattgg; rp49 fwd, TACAGGCCCAAGATCGTGAAG; rp49 rev, gacgcactctgttgtcgatacc; scrib fwd, CAATGAAATTGGCCGCCTGCCG; scrib rev, cgaacttgggtatcgggttcgaac; sif fwd, CAAAGTGGCGAGCTGCCCAATC; sif rev, caggttgttgagcagcgaggg; Syn1 fwd, GAATTGGGCAGGGTGCCGTTC; Syn1 rev, ctggaaacggacttcctggcc; tub84B fwd, GGCAAGGAGATCGTCGATCTGG; tub84B rev, gacgctccatcagcagcgag; vari fwd, CTCGTTCACGATGACCATGTCGAAG; vari rev, cataagattcagctccagacgcgc; X11L fwd, GCGTGTTGTTTCGGGCCAGATAC; X11L rev, cagtgctcggctgactttcgc.
Immunostaining and microscopy
Ovarioles were dissected and fixed in 4% formaldehyde in 1× phosphate buffered saline (PBS) with 0.2% v/v Triton X-100 (PBT). Blocking, antibody incubations, and washes were done in PBT with 5 mg/mL BSA (PBT-BSA). The primary antibodies used were: 1:400 mouse anti-alpha-tubulin (DM1A, Sigma); 1:200 rabbit anti-aPKC-zeta (sc-216, Santa Cruz); 1:50 concentrated mouse anti-Dlg1 (4F3, Developmental Studies Hybridoma Bank; DSHB); 1:150 mouse anti-Singed (sn 7c, DSHB); 1:150 rat anti-E-cadherin (DCAD-2, DSHB); 1:10 mouse anti-Fasciclin III (FasIII; 7G10, DSHB); 1:500 rabbit anti-GFP (Life Technologies); 1:1000 rabbit anti-Stat92E (a gift from S. Hou); and 1:500 rabbit anti-Veli (a gift from E. Knust). Secondary antibodies conjugated to Alexa Fluor 488, Alexa Fluor 568, or Alexa Fluor 647 (Life Technologies) were used at 1:400 dilution. Actin was visualized with phalloidin conjugated to Alexa Fluor 568 or Alexa Fluor 647 (Life Technologies) used at 1:400 dilution. DAPI (0.05 μg/mL, Sigma) was used to visualize nuclei. Stained egg chambers were mounted on slides in Aqua-Poly/Mount (Polysciences, Inc.) and imaged with a Zeiss AxioImager Z1 epi-fluorescent compound microscope equipped with the ApoTome system and MRm CCD camera. Either a 20× Plan-Apochromat 0.75 numerical aperture (NA) or a 40× Plan-Neofluar 1.3 NA objective was used. The microscope was controlled by Axiovision 4.8.1 software. For detailed analyses of border cell migration and to verify first-round hits, GAL4/UAS-RNAi crosses were independently set up, and whole ovaries from the adult progeny were fixed and stained for Singed, phalloidin, and DAPI as above. Manually dissociated ovaries were mounted on slides and analyzed as above using the same microscope.
Calculation of Stat92E/DAPI intensity ratio
Border cell clusters stained with anti-Stat92E and DAPI were imaged with multiple optical z-sections. A maximum intensity projection image was generated using the Axiovision Extended Focus module. For overlapping nuclei, separate projection images were generated to visually isolate the nuclei. Individual border cell nuclei were first outlined in NIH ImageJ software. The mean DAPI and Stat92E intensities were measured using the “Measure” command in ImageJ. The ratio of Stat92E/DAPI for each nuclei was calculated by dividing the mean Stat92E intensity by the mean DAPI intensity.
Graphs, statistics, and figures
Graphs and statistical analysis were performed in GraphPad Prism 4. The threshold for determining RNAi-induced migration phenotypes was calculated by applying the three-sigma rule on the negative control data (c306-GAL4/+; UAS-mCD8:GFP/UAS-GFP dsRNA). Briefly, the background migration defect was 2.63 ± 2.32% (mean ± SD; data from seven trials, n ≥ 50 egg chambers per trial). The threshold was calculated to be 9.59%, three standard deviation (SD) intervals from the mean. Statistical significance of RNAi knockdown by qRT-PCR was determined using the one-tailed unpaired t-test. In all other cases, the two-tailed unpaired t-test was used. Figures were assembled in Adobe Illustrator CS5. Minor image adjustments (brightness and/or contrast) were done in Axiovision 4.8.1 or Adobe Photoshop CS5. Gene ontology analyses were performed using PANTHER (http://www.pantherdb.org) (Thomas et al. 2003) or AmiGO (http://amigo.geneontology.org) (Ashburner et al. 2000).
Results
RNAi knockdown of PDZ domain-encoding genes in border cells
We first sought to identify all of the Drosophila genes that encode PDZ domain-containing proteins. We used a combination of the InterPro protein signatures (http://www.ebi.ac.uk/interpro/) and FlyBase (http://flybase.org/) databases to identify genes that have at least one PDZ domain. While the human genome encodes more than 250 PDZ domain proteins (Tonikian et al. 2008), Drosophila has 66 PDZ genes (Table S1) (Bilder 2001). Many of these genes have alternatively spliced isoforms, making the total number of PDZ domain proteins slightly higher (Sierralta and Mendoza 2004). We performed a gene ontology analysis to determine the types of proteins that these genes encode along with predicted functions (see Materials and Methods). Drosophila PDZ proteins are annotated predominantly to have protein binding, structural roles and regulation of enzyme activities (Figure S1). The relatively small number of PDZ domain genes in the Drosophila genome makes it a reasonable pool of candidates to test comprehensively for their role in border cell migration.
Knockdown of the multi-PDZ domain protein Baz, which regulates border cell migration, was used as a positive control (Figure 1 and Table S1) (Pinheiro and Montell 2004). We tested different GAL4 drivers to determine the best one for UAS-RNAi knockdown. A ubiquitous GAL4 driver, tubulin-GAL4, was lethal with baz RNAi (line v2914) and therefore was not used. We next tested a follicle cell driver, T155-GAL4, which is expressed early in the germarium in follicle cell precursors followed by expression in all follicle cells starting at stage 9 (Queenan et al. 1997; Liu and Montell 1999). We observed a high proportion of follicle cell defects when baz RNAi was driven by T155-GAL4 (Figure 1, B and C; 33%, n = 136 egg chambers). Regions of the follicle cell epithelium were thin (Figure 1B) and multilayered (Figure 1C). In addition, some follicle cells did not complete their posterior-directed retraction to cover the oocyte at stage 10B (Figure 1C). These results are consistent with the known role for Baz in follicle cell polarity (Cox et al. 2001; Huynh et al. 2001; Abdelilah-Seyfried et al. 2003) and indicates that the RNAi line efficiently knocked down baz function. However, we also observed a large proportion of degenerating egg chambers (33%, n = 136), which precluded us from scoring border cell migration.
We next tested two GAL4 drivers, slbo-GAL4 and c306-GAL4, which are expressed during border cell migration (Figure 1, D and E) (Murphy and Montell 1996; Rørth et al. 1998). We compared the expression patterns by crossing the GAL4 lines to UAS-mCD8:GFP. slbo-GAL4 begins to drive expression at high levels in the newly formed border cells at early stage 9 (Figure 1D). In contrast, c306-GAL4 turns on at earlier stages, beginning around stage 4/5 (Figure 1E). c306-GAL4 is expressed in a larger subset of follicle cells at the anterior end of the egg chamber that includes the presumptive border cells (Figure 1, A and E). Both GAL4 lines are also expressed in a subset of posterior follicle cells (Figure 1, D and E). We compared the border cell migration defects caused by knockdown of baz RNAi (line v2914) using slbo-GAL4 and c306-GAL4. This was scored as the percentage of border cells that migrated (complete) or did not migrate (incomplete) to the oocyte by stage 10 of oogenesis (Figure 1, A and F). We consistently observed stronger inhibition of migration with baz RNAi driven by c306-GAL4 (Figure 1F), possibly because the earlier follicle cell expression allowed time for efficient knockdown of gene function in the presumptive border cells. Therefore, we selected c306-GAL4 for this study (Figure 1G).
We obtained available transgenic UAS-RNAi lines that target each of the PDZ domain-encoding genes from VDRC (Figure 1G and Table S1). A few lines were acquired from two other collections (NIG-Fly and TRiP). Whenever possible, multiple transgenic RNAi lines that target each gene were obtained. RNAi lines can potentially produce off-target effects by non-specific knockdown of other genes (Perrimon et al. 2010). We excluded a few lines that were annotated to have a high number of potential off-target genes (more than 100), but in most cases we were able to test alternative lines. For example, CG43955 had one line (v31686) with 196 predicted off-target genes including taiman, which is required for border cell migration (Bai et al. 2000); we tested the alternative line v103267, which does not have any predicted off-target genes. The only available arc RNAi line (v16826) has 408 predicted off-targets that include taiman and another border cell migration gene, slow border cells (slbo) (Montell et al. 1992), so it was not included in the PDZ gene survey. No lines were available for Mhcl. For the remaining 64 genes, 145 lines were tested for incomplete vs. complete border cell migration (Figure 1, A and G, and Table S1). More than 75% of the genes had multiple RNAi lines, either independent insertions of the same construct (e.g. baz) or independent constructs (e.g. bbg) (Table S1). We knocked down the genes using UAS-RNAi lines driven by c306-GAL4 (see Materials and Methods). baz RNAi was used as a positive control (Figure 1, A and F). RNAi against GFP was used as the negative control and did not significantly disrupt border cell migration (Figure 1F).
The percentage incomplete migration for all of the tested lines is reported in Table S1. The results were classified into two main groups, positive or negative hits. We calculated the minimum threshold for migration defects using our negative control data (see Materials and Methods); migration defects of 9% or fewer of the analyzed egg chambers were considered negative hits. RNAi lines that resulted in more than 9% of egg chambers with border cell migration defects were classified as positive hits. To confirm the first-round positive hits, we retested the strongest RNAi lines for most genes (see Materials and Methods and Figure 1G). Four genes that initially were categorized as positive hits in the first round did not repeat (Table S1). Moreover, upon retesting, some positive lines had stronger migration defects, whereas others were milder; this suggests slight inherent variability of knockdown efficiency. Nonetheless, most RNAi lines when retested exhibited similar strength of migration defects; the variation between trials was generally ≤ 10% (Table S1). Thirty-three genes fell into the negative hit category with the rest being positive hits (Table S1). For the 31 positive gene hits, RNAi knockdown caused migration defects ranging from 10 to 50% of the analyzed egg chambers; none of the lines completely blocked migration. Positive hits were further sorted based on the number of tested lines that had border cell migration defects (Table S1). Genes with all or multiple RNAi lines producing migration defects were designated “high confidence” multiple hits (Table 1) (Booker et al. 2011). A total of 14 positive genes compose the multiple hit category (Table 1) and 17 genes are single hits (Table S1). The positive hit genes have a range of predicted functions, although genes with known or predicted roles in epithelial polarity or cytoskeletal regulation together account for more than half of the hits (Table 1 and Figure S1C).
Table 1. High-confidence PDZ domain-encoding genes in border cell migration identified by RNAi knockdown.
| Gene | Putative Vertebrate Homologa | Other Domains Presentb | No. of Hits / Total Linesc | Known Role in Cell Migrationd |
|---|---|---|---|---|
| bazooka | PARD3 (PAR3) | Oligomerization domain | 2 / 2 | Pinheiro and Montell (2004)e |
| Nakayama et al. (2008)f | ||||
| big bang | PDZD2 | None | 2 / 3 | — |
| CASK ortholog | CASK | Guanylate kinase domain | 2 / 3 | — |
| L27 domain | ||||
| Protein kinase, catalytic domain, inactive | ||||
| SH3 domain | ||||
| CG5921 | Harmonin / USH1C | None | 2 / 2 | — |
| CG6498 | MAST2 | Domain of unknown function | 3 / 3 | — |
| Protein kinase, catalytic domain | ||||
| CG6509 | DLG5 | Guanylate kinase domain | 2 / 3 | Smolen et al. (2010)f |
| Src homology 3 domain | ||||
| Gef26 | RAPGEF2 / PDZ-GEF1 | Cyclic nucleotide-binding domain | 3 / 3 | Huelsmann et al. (2006)e |
| Guanine-nucleotide dissociation stimulator (RasGEF) | ||||
| Ras association domain | ||||
| Ras-like guanine nucleotide exchange factor, N-terminal | ||||
| Lap1 | LRRC7 / ERBB2IP | Leucine-rich repeats | 2 / 2 | — |
| LIMK1 | LIMK1 | LIM zinc-binding domain | 2 / 3 | Zhang et al. (2011)e |
| Protein kinase, catalytic domain | Nishita et al. (2005)f | |||
| par-6 | PARD6 (PAR6) | PB1 domain | 3 / 3 | Pinheiro and Montell (2004)f |
| PatJ | INADL / MPDZ | L27 domain | 2 / 3 | Shin et al. (2007)f |
| Rim | RIMS2 | C2 domain | 2 / 3 | — |
| stardust | MPP5 (PALS1) | Guanylate kinase domain | 2 / 3 | — |
| L27 domain | ||||
| Src homology 3 domain | ||||
| veli | LIN7A / LIN7B / LIN7C | L27 domain | 2 / 2 | — |
Putative homologs were found using NCBI Homologene and UniProt.
Protein domains were identified using NCBI Conserved Domains Database and Interpro.
Number of RNAi lines that resulted in a migration defect out of all lines tested.
Cited references describe the Drosophila gene or its homologs.
Pertains to studies in Drosophila.
Pertains to the mammalian homolog.
Validation of candidates
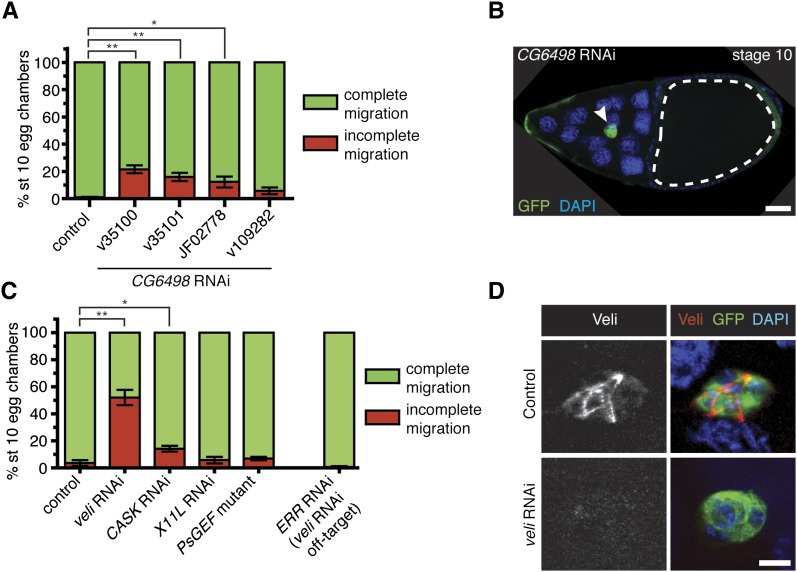
To verify results from the systematic RNAi knockdown of PDZ genes, we performed additional tests for a subset of both positive and negative genes. We first performed a detailed analysis of the migration defects for selected positive first-round genes (see Materials and Methods). CG6498 has high homology to human microtubule-associated serine-threonine kinase 2 (MAST2) (NCBI Homologene; http://www.ncbi.nlm.nih.gov/homologene/); both genes encode proteins with a central serine-threonine kinase domain and a single PDZ domain. Three CG6498 RNAi lines (two different constructs) significantly disrupted migration (Figure 2A; http://flybase.org/reports/FBgn0036511.html). One independent construct, line v109282, did not cause migration defects, possibly due to inefficient knockdown (Figure 2A). In the strongest line, v35100, 21% of the egg chambers had incomplete migration (Figure 2A). Most CG6498 RNAi border cells with a migration defect stopped midway to the oocyte (Figure 2B). Moreover, knockdown of CG6498 driven by the other border cell GAL4, slbo-GAL4, also disrupted border cell migration (Figure S2). We analyzed in more detail the migration defects caused by RNAi lines for two additional multi-hit positive genes, veli and CASK (Figure 2C). Closer examination of RNAi knockdown for both genes revealed migration defects similar to those observed in the first-round analysis (Figure 2C and Table S1). We confirmed that Veli was expressed in border cells using an antibody against Veli protein (Figure 2D). The strongest veli RNAi line (v43094) has a predicted off-target match to estrogen receptor related (ERR). However, RNAi for ERR did not disrupt border cell migration (Figure 2C). Moreover, veli RNAi (v43094) efficiently downregulated Veli levels in border cells (Figure 2D).
Figure 2 .
Confirmation of positive and negative hit genes. (A and C) Quantification of border cell migration at stage 10, shown as the percentage of border cells with complete (green) or incomplete (red) migration in egg chambers expressing RNAi to GFP (control) or the indicated RNAi transgenes driven by c306-GAL4. Error bars represent SEM; n ≥ 50 egg chambers in each of at least three trials (*P < 0.05; **P < 0.01; two-tailed unpaired t-test). (A) Knockdown of CG6498 using multiple transgenes disrupts border cell migration. (B) Representative example of an egg chamber with a border cell migration defect caused by CG6498 RNAi. Genotype is c306-GAL4/+; UAS-mCD8:GFP/UAS-CG6498 RNAi v35100. Border cells (green; arrowhead) stopped a little more than halfway to the oocyte (outlined). DAPI marks nuclei. Scale bar is 20 µm. (C) Border cell migration defects by RNAi knockdown of veli (v43094) and CASK (v34185). Normal border cell migration with RNAi knockdown of X11L (v28652) and in a PsGEF mutant (PsGEFΔ55/PsGEFΔ21). RNAi for ERR (line v108349), the predicted off-target gene for veli RNAi line v43094, did not disrupt border cell migration. (D) Border cells stained with an antibody to Veli. Control border cells (c306-GAL4/+; UAS-mCD8:GFP/+) had detectable Veli (red), which was strongly reduced in veli RNAi border cells (c306-GAL4/+; UAS-mCD8:GFP/UAS-veli RNAi v43094). GFP (green) shows GAL4 expression and DAPI (blue) labels nuclei. Scale bar is 10 µm.
We next confirmed that two genes in the negative hit category did not disrupt border cell migration. X11L RNAi-induced phenotypes were close to the background cutoff migration defects observed in the first-round tests (Table S1). Upon detailed retesting, we verified that knockdown of X11L did not affect border cell migration (Figure 2C). We also obtained two small deletion mutant alleles of the putative Rac guanine-nucleotide exchange factor (GEF) Protostome-specific GEF (PsGEF); these alleles are transcript null, viable, and fertile (Higuchi et al. 2009). In agreement with the RNAi results, egg chambers trans-heterozygous for PsGEFΔ55/PsGEFΔ21 were morphologically normal and did not disrupt border cell migration (Figure 2C).
Although most RNAi lines are expected to produce knockdown of the targeted genes, this has not been tested formally for most individual lines. Therefore, we performed qRT-PCR to ascertain the in vivo knockdown efficiency for selected RNAi lines. We analyzed 18 lines, which target 4 positive genes and 14 negative genes; this encompasses ∼25% of the PDZ genes (Table 2). The tested lines were from multiple collections: the first-generation “GD” and second-generation “KK” long double-stranded hairpin RNA (dsRNA) libraries from VDRC; and the long dsRNA (Valium 1 and 10) and shRNA (Valium 20) libraries from the TRiP collection (http://flybase.org/reports/FBrf0208510.html) (Dietzl et al. 2007; Ni et al. 2009; 2011). To determine whether these genes were expressed during oogenesis, qRT-PCR was used to measure the relative expression levels in wild-type ovarian extracts (Table 2). Most genes were expressed at low or low-to-moderate levels, with the exception of CG43955, which was not expressed. Next, we crossed the 18 RNAi lines to heat shock (hs)-GAL4 and subjected adult flies to heat shock to induce RNAi transgene expression (see Materials and Methods). RNA isolated from whole female flies (ovaries removed) was used to analyze relative levels of transcript in RNAi knockdown flies vs. a non-targeting control (RNAi to GFP). qRT-PCR performed on the resulting cDNA showed that 14 out of 18 RNAi lines achieved statistically significant knockdown of transcripts (Table 2 and Figure S3). Knockdown ranged from mild (21% knockdown by PICK1 RNAi) to strong (90% knockdown by Grip RNAi), with most lines producing more than 40% knockdown. Finally, we compared lines we tested to those identified in two genome-wide in vivo RNAi screens performed to identify genes that regulate Notch signaling or muscle morphogenesis (Mummery-Widmer et al. 2009; Schnorrer et al. 2010). These genome-wide screens found specific phenotypes or lethality with 12 positive hit lines and 9 negative hit lines from our analysis (Table S1). Together, these data confirm specificity for a number of RNAi lines. Moreover, this suggests that the majority of lines tested in this study are expected to reduce relevant transcript levels.
Investigation of two genes, bbg and CG6509, reveals distinct functions in border cells
While the initial analysis of PDZ domain-encoding genes by RNAi knockdown focused on whether border cells completed their migration by the appropriate stage, we wanted to further determine specific function(s) of identified genes in border cells. Earlier studies established that Baz and its partner Par-6 regulate polarity of border cells during detachment from the follicle cell epithelium and subsequent migration (Pinheiro and Montell 2004; McDonald et al. 2008). None of the other high-confidence positive hits from this study have been analyzed previously in border cells, although a few have been found to regulate the migration of other cell types in Drosophila and/or in mammals (Table 1). Two genes, big bang (bbg) and CG6509, were chosen for further tests because they encode different classes of PDZ domain-containing proteins.
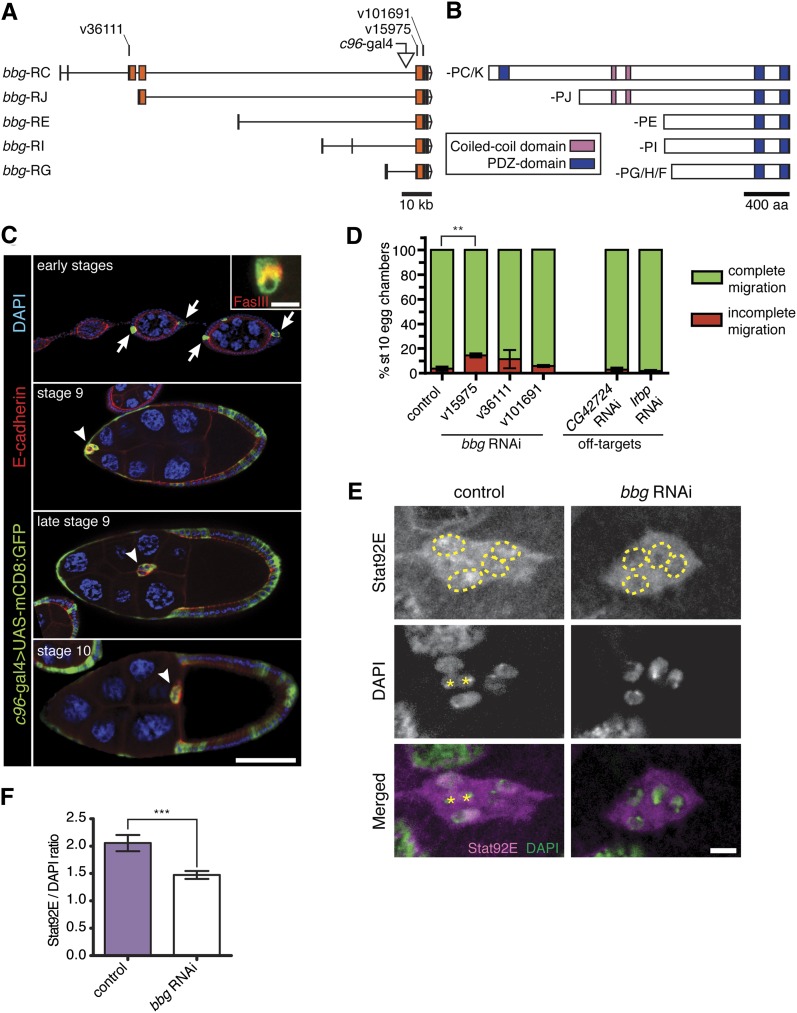
bbg encodes a large multi-PDZ domain protein expressed at various stages of development (Kim et al. 2006). Little is known about the function of bbg in development, except that mutants are mildly bang sensitive (Kim et al. 2006). The bbg gene locus spans over 120 kb and encodes multiple transcripts (5 of the 8 are shown; Figure 3A) (Kim et al. 2006). Bbg protein isoforms are differentiated by the total number of PDZ domains present (three in Bbg-PC/-PK, two in the other isoforms), by variations in the length of the N-terminal region, and the presence of two predicted coiled-coil domains (Figure 3B) (Kim et al. 2006).
Figure 3 .
The multi-PDZ domain protein Big bang regulates nuclear STAT levels in border cells. (A) Schematic diagram of five bbg predicted transcripts (adapted from FlyBase); coding exons are in orange. RNAi target sequences and c96-GAL4 insertion site are indicated. RNAi lines v15975 and v101691 target sequences common to all isoforms. RNAi line v36111 is specific to RC and RK (not shown; differs from RC only in a non-coding exon; see FlyBase) transcripts. (B) Schematic diagram of the eight Bbg protein isoforms, which have either two or three PDZ domains. (C) Egg chambers showing c96-GAL4 expression pattern visualized by UAS-mCD8:GFP (green) at the indicated stages. Egg chambers were co-stained for E-cadherin (red) to mark cell membranes and DAPI (blue) to mark nuclei. Scale bar is 50 µm. (Top panel) c96-GAL4 expression in anterior and posterior polar cells (arrows) at early stages. Inset shows c96-GAL4–positive polar cells (green) co-stained for FasIII (red; scale bar, 5 µm). (Bottom panels) c96-GAL4-driven GFP in border cells (arrowheads) and surrounding follicle cell epithelium during stages 9 and 10. (D) Quantification of border cell migration at stage 10, shown as the percentage of border cells with complete (green) or incomplete (red) migration in egg chambers expressing RNAi to GFP (control) or the indicated RNAi transgenes driven by c306-GAL4. Knockdown of bbg with RNAi line v15975 disrupted border cell migration. bbg RNAi line v36111 had variable effects, and line v101691 did not disrupt migration. RNAi to the predicted off-target genes, CG42724 (line v30629) and Irbp (line JF03273), did not disrupt migration. Error bars represent SEM; n ≥ 50 egg chambers in each of at least three trials (**P = 0.0071; two-tailed unpaired t-test). (E) Reduction of Stat92E levels in border cell nuclei when bbg is knocked down. Stage 9 border cells stained for Stat92E (magenta) and DAPI (green). Stat92E is expressed at higher levels in control border cell nuclei (yellow outline) compared with cytoplasm (c306-GAL4/+; UAS-mCD8:GFP/+). Stat92E is expressed at low levels in bbg RNAi (c306-GAL4/+; UAS-mCD8:GFP/UAS-bbg RNAi v15975) border cell nuclei (outlined). Polar cells (asterisks) were excluded from analyses. Scale bar is 5 μm. (F) Quantification of the fluorescence intensity ratio of STAT nuclear staining to DAPI staining for control (n = 59) or bbg RNAi (n = 88) border cells; genotypes as in (E). At least 16 individual clusters were analyzed. Error bars represent SEM (***P < 0.001; two-tailed unpaired t-test).
bbg is expressed in early oogenesis as well as in discrete patterns in the embryo and larval discs (Gustafson and Boulianne 1996; Kim et al. 2006). We used qRT-PCR to verify that bbg was expressed in ovaries (Table 2). However, its expression during later stages of oogenesis has not been described. bbg was previously identified as the insertion site for the c96-GAL4 enhancer trap line, which has been shown to reliably report the bbg expression pattern (Kim et al. 2006). We analyzed the expression of bbg by crossing c96-GAL4 to UAS-mCD8:GFP (Figure 3C and Figure S4A). c96-GAL4 was restricted to a few follicle cells at the very anterior and posterior ends of the egg chamber at early stages (Figure 3C), in agreement with bbg transcript and protein (Kim et al. 2006). Staining with Fasciclin III (FasIII), which marks the membrane between the pair of polar cells, confirmed that these cells are the anterior and posterior polar cells (Figure 3C). The polar cells later recruit surrounding follicle cells to become border cells at late stage 8 (Silver and Montell 2001; Xi et al. 2003). Starting at stage 8, c96-GAL4-driven GFP expanded to the majority of follicle cells, including border cells (Figure 3C and Figure S4A).
We next confirmed that bbg RNAi-mediated knockdown disrupted border cell migration (Figure 3D). We retested three transgenic RNAi lines that target non-overlapping regions of bbg (Figure 3A). Line v15975 resulted in the strongest migration defects (14% of stage 10 egg chambers), whereas line v36111 had milder and more variable migration defects (Figure 3D). Upon retesting, the third RNAi line against bbg (v101691) did not reliably disrupt migration. Knockdown of bbg using slbo-GAL4 mildly disrupted border cell migration (Figure S2). The first two RNAi lines against bbg each have a predicted off-target gene. However, Irbp RNAi (off-target for v15975) and CG42724 RNAi (off-target for v36111) did not induce migration defects (Figure 3D). Moreover, bbg RNAi line v15975 significantly knocked down bbg levels in vivo (Table 2 and Figure S3).
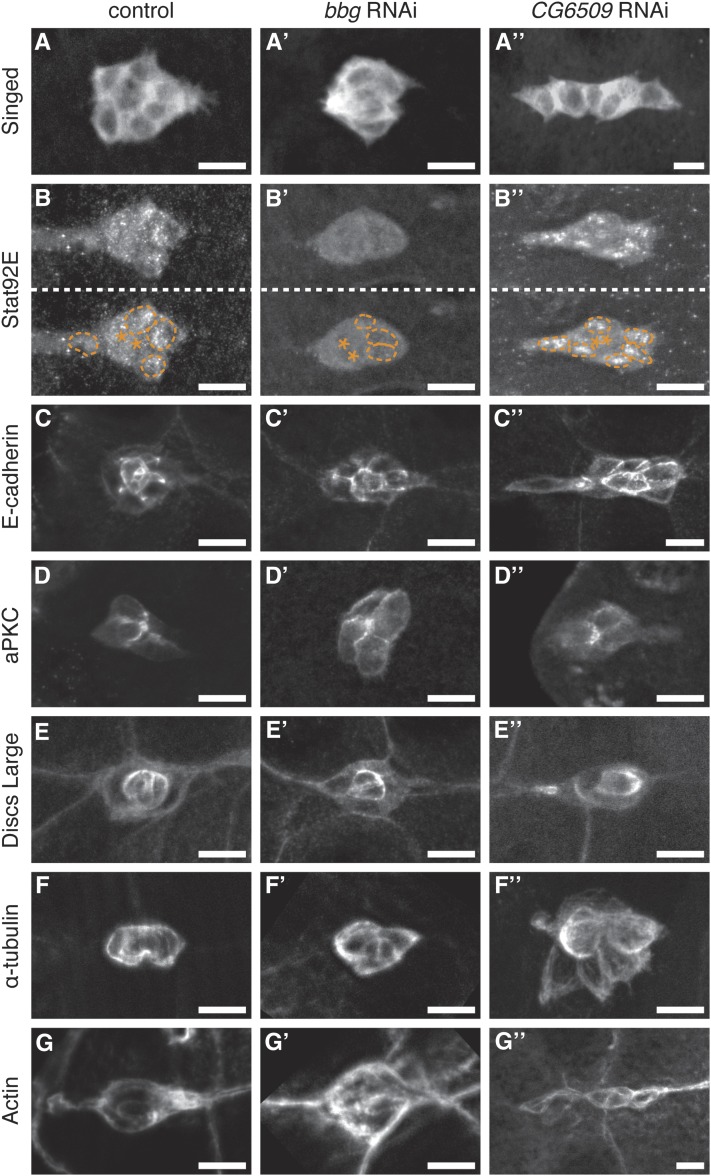
To address whether Bbg regulates a specific aspect of border cell migration, we analyzed the levels and localization of several border cell-enriched proteins (Figure 4). We first analyzed a marker of cell identity, the fascin homolog Singed (Sn). Border cells in which bbg was knocked down by the strongest RNAi line (v15975) had normal levels and localization of Sn compared with control border cells (Figure 4A). The cell adhesion protein E-cadherin and the membrane-associated polarity proteins atypical protein kinase C (aPKC) and discs large 1 (Dlg1) were all localized correctly in bbg RNAi border cells (Figure 4, C–E). Moreover, bbg RNAi did not disrupt F-actin or α-tubulin, indicating no obvious cytoskeletal defects (Figure 4, F and G). Thus, most aspects of border cell differentiation and membrane localization were unchanged when bbg levels were reduced. In contrast, we observed altered Stat92E subcellular localization when bbg was knocked down (Figures 3E and 4B).
Figure 4 .
Markers of cell fate, cell adhesion, polarity, and cytoskeleton in bbg RNAi and CG6509 RNAi border cells. Representative immunofluorescent images of stage 9 control (c306-GAL4/+; UAS-mCD8:GFP/+), bbg RNAi (c306-GAL4/+; UAS-mCD8:GFP/UAS-bbg RNAi v15975), and CG6509 RNAi (c306-GAL4/+; UAS-mCD8:GFP/UAS-CG6509 RNAi v22496) border cells. (A and B) Border cells stained for antibodies to the cell fate markers Singed (A) and Stat92E (B). (A) Singed is enriched in the cytoplasm. (B) Stat92E is enriched in border cell nuclei compared with the cytoplasm. The same cluster is presented with and without border cell nuclei outlined with a dotted line (taken from DAPI staining of nuclei, not shown). Polar cells are marked with an asterisk (*). (C) Border cells stained for the cell adhesion protein E-cadherin, which is high in central polar cells and at the membrane interface between border cells. (D and E) Border cells stained for the cell polarity proteins aPKC (D) and Dlg1 (E). (D) aPKC is an apical cell marker and localizes between border cells; an apical view is shown. (E) Dlg1 is a basolateral cell marker that is enriched in the central polar cells and at lower levels at border cell membranes. (F and G) Border cells stained for the cytoskeletal markers α-tubulin to mark microtubules (F) and phalloidin to label F-actin (G). N ≥ 10 border cell clusters assayed for each genotype. Scale bar is 10 µm.
JAK/STAT signaling specifies border cell fate, recruits border cells to form a cluster, and promotes their motility (Silver and Montell 2001; Beccari et al. 2002; Silver et al. 2005). Nuclear STAT localization reflects high levels of JAK/STAT signal activation (Vinkemeier 2004). Stat92E (the Drosophila STAT homolog) becomes enriched in border cell nuclei as they are specified in the epithelium and is maintained throughout their migration (Silver et al. 2005). Control migrating border cells have visibly higher nuclear Stat92E compared with the cytoplasm (Figure 3E). However, bbg RNAi reduced the levels of nuclear Stat92E in most border cells (Figure 3E). To quantitate this effect, we measured the ratio of nuclear Stat92E to DAPI staining in stage 9 migrating border cells (see Materials and Methods). The ratio of nuclear STAT to DAPI signal was reduced from ∼2.0 in control border cells to ∼1.5 in bbg RNAi border cells (Figure 3F). Nonetheless, high nuclear Stat92E was observed in premigratory bbg RNAi border cells (Figure S4B). This result suggests that nuclear Stat92E was initially normal in bbg RNAi border cells but that it was not maintained adequately after border cells began to migrate. bbg RNAi border cell clusters contain a similar number of cells compared with control clusters (Figure S4C). Thus, Bbg functions after border cells are specified and recruited into the cluster to maintain optimal STAT levels during border cell migration.
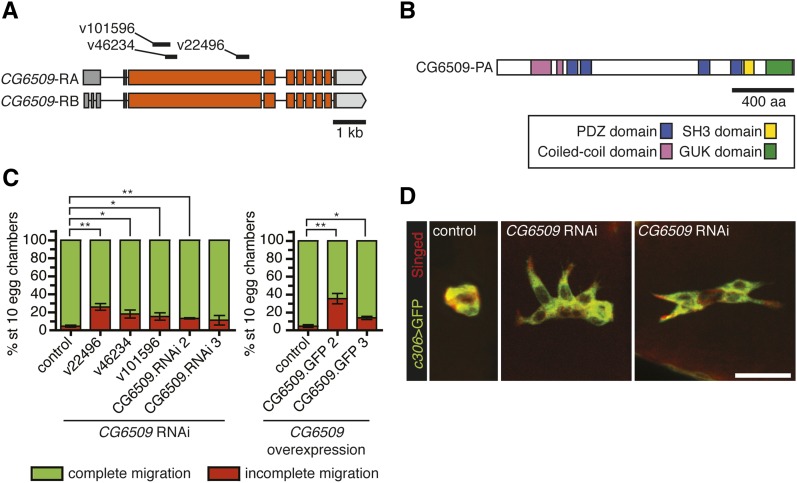
The second gene we analyzed in more detail, CG6509, encodes a member of the membrane-associated guanylate kinase (MAGUK) proteins (Figure 5, A and B). The domain architecture of CG6509, a combination of PDZ, SH3, and guanylate kinase (GUK) homology domains, is characteristic of members of the MAGUK family of scaffolding proteins (Oliva et al. 2012). Four other MAGUK-encoding genes were identified as positive hits: the multi-hit genes CASK ortholog (CASK) and stardust (sdt) and the single-hit genes menage a trois (metro) and polychaetoid (pyd). We verified that knockdown of CG6509 with multiple RNAi lines disrupted border cell migration (Figure 5C). Although the RNAi line v22496 initially fell below the migration defect cutoff, upon retesting, it inhibited border cell migration in 26% of stage 10 egg chambers (Figure 5C and Table S1). Moreover, line v22496 produced significant knockdown of CG6509 transcript levels (Table 2 and Figure S3). Knockdown of CG6509 using the border cell-specific slbo-GAL4 mildly delayed migration, confirming a requirement in border cells (Figure S2). Three CG6509 RNAi lines (v22496, v46234, and v101596) do not have predicted off-target genes, further indicating that the phenotypes are specific to CG6509 knockdown.
Figure 5 .
The MAGUK family member CG6509 regulates border cell cluster morphology. (A) Schematic of the CG6509 transcripts, which differ only in the 5′ non-coding exons (adapted from FlyBase); coding exons in orange. RNAi target sequences are indicated. (B) Schematic of CG6509 protein showing the conserved domains. (C) Quantification of border cell migration at stage 10, shown as the percentage of border cells with complete (green) or incomplete (red) migration in egg chambers expressing multiple RNAi lines or overexpression of full-length UAS-CG6509 (different insertions of same transgene) driven by c306-GAL4. Error bars represent SEM; n ≥ 50 egg chambers in each of at least three trials (*P < 0.05; **P < 0.01; two-tailed unpaired t-test). (D) Stage 9 border cells stained for GFP (green) and Singed (red) to reveal border cell cluster morphology. Representative example of a control (c306-GAL4/+; UAS-mCD8:GFP/+) border cell cluster. Two examples of CG6509 RNAi (c306-GAL4/+; UAS-mCD8:GFP/UAS-CG6509 RNAi v22496) border cells in which the cluster is partially dissociated (middle panel) or elongated (right panel). Scale bar is 20 μm.
Analysis of border cell markers in CG6509 RNAi border cells, similar to that performed for bbg (see above), did not reveal obvious changes compared with control (Figure 4). Nonetheless, RNAi knockdown of CG6509 markedly affected the morphology of border cell clusters (Figures 4 and 5D). Control clusters are generally round and fairly compact (96%; n = 21). In contrast, 44% of CG6509 RNAi border cell clusters (n = 36) no longer had a compact shape and, instead, were dissociated or elongated (Figure 5D). Finally, overexpression of CG6509 in border cells also disrupted their migration; the strongest UAS-CG6509 line disrupted migration in 35% of stage 10 egg chambers (Figure 5C). These data together indicate that having proper levels of CG6509 is important for normal border cell migration and cohesion of the cluster.
Discussion
RNAi knockdown of specific classes of genes identify regulators of border cell migration
The advantage of using RNAi to test a selected class of genes, like the one performed here, is the rapid identification of genes involved in a particular process such as border cell migration. The less labor-intensive nature of this approach ensures that even those genes whose knockdown results in incompletely penetrant phenotypes are detected. Recently, this method was used to identify Evi5 as a new GTPase-activating protein for Rab11 in border cell migration (Laflamme et al. 2012). Moreover, targeted RNAi knockdown of microtubule-associated proteins demonstrated a requirement for the Lis-1 complex in border cells (Yang et al. 2012). In this study, we chose to systematically target 64 genes that encode PDZ domain-containing proteins because of their known functions in processes critical for cell migration, such as cell polarity, adhesion, and signaling. The majority of these PDZ genes have not been examined for functions in cell migration in any organism. The 14 genes designated as high-confidence hits likely represent new members of protein complexes required for border cell migration. Importantly, several positive genes, the multi-hit genes baz and par-6 and the single-hit gene dishevelled (dsh), were previously identified as regulators of border cell migration (Pinheiro and Montell 2004; Bastock and Strutt 2007). Most genes in the positive class, which includes 17 additional genes with one phenotypic RNAi line, have direct or putative mammalian homologs. The results of this study thus provide a list of PDZ genes whose roles in cell migration and motility are predicted to be conserved.
Knockdown of positive hit PDZ genes resulted in mild to moderate migration defects, with most border cells able to detach from the epithelium and migrate partway to the oocyte. These observations suggest that RNAi for these genes resulted in partially penetrant phenotypes, either due to partial knockdown of gene function or because the gene is not completely essential for full border cell motility. Incomplete knockdown could occur if the RNAi transgene is not expressed at the right time or at strong enough levels (Perrimon et al. 2010; Booker et al. 2011). To overcome this potential problem, the c306-GAL4 driver was used because it is expressed early in follicle cells and maintained in the migrating border cells. Whenever possible, multiple independent insertion lines and/or constructs were tested to minimize the potential issue of inefficient RNAi constructs. Multiple independent hits increase the likelihood that the migration defects caused by RNAi are specific. Our assessment of RNAi efficiency by qRT-PCR confirms that 75% of the tested RNAi lines effectively knocked down the relevant targeted transcript. While knockdown efficiency ranged from 20 to 90%, most RNAi lines decreased transcript levels by 40% or more. Furthermore, our results indicate that, at least for baz, partial knockdown (∼40% reduction of transcripts) significantly disrupted border cell migration.
The relatively mild nature of the phenotypes alternatively suggests that these genes have support or partially redundant roles in border cell migration. A striking example of this is the two receptor tyrosine kinases, the epidermal growth factor receptor (EGFR) and the PDGF/VEGF receptor related (PVR), that guide border cells to the oocyte in response to secreted growth factors (Duchek and Rørth 2001; McDonald et al. 2006). Loss of either receptor alone has modest effects, but simultaneous loss of both receptors severely inhibits posterior-directed migration (Duchek and Rørth 2001; Duchek et al. 2001; McDonald et al. 2003). This contrasts with other genes, such as slbo, that are required for early border cell fate and whose loss completely inhibits migration (Montell et al. 1992). It remains to be seen whether the genes identified in this study play partially redundant roles. Many of the genes identified lack classical mutant alleles, and therefore, RNAi is the most direct method to assess their functions at present. Once loss-of-function alleles are tested and/or created, the mutant results can be compared with the RNAi knockdown results. In the future, it will also be important to identify the cellular and membrane-associated proteins to which these PDZ domain proteins bind. The results from this study accordingly present a collection of candidates to search for PDZ-interacting substrates in border cells and other migratory cells.
Epithelial polarity and cytoskeletal-associated genes are highly represented hits
A key group of genes identified here are those involved in epithelial cell polarity. Significantly, these and other epithelial polarity proteins are required for mammalian cell motility and have been implicated in tumor invasion and metastasis (Etienne-Manneville 2008; Hidalgo-Carcedo et al. 2011; Subbaiah et al. 2011; Martin-Belmonte and Perez-Moreno 2012). Border cells retain many epithelial characteristics during migration, including polarized localization of Par-6 and Baz and upregulation of E-cadherin (Niewiadomska et al. 1999; Pinheiro and Montell 2004). Nine of the positive-hit genes (baz, CASK, dsh, Lap1, par-6, Patj, pyd, sdt, and veli) regulate apical-basal polarity to establish distinct membrane domains (Ashburner et al. 2000; Guillemot et al. 2008; Martin-Belmonte and Perez-Moreno 2012). This raises the possibility that these polarity proteins regulate the localization of junctional proteins in border cells to organize and promote migration, similar to the known functions of baz and par-6 (Pinheiro and Montell 2004; Llense and Martín-Blanco 2008). Many of the proteins in the polarity group form known multi-protein complexes. Par-6 and Baz form a complex with the serine-threonine kinase aPKC in some contexts, and Sdt, Patj, and Veli form another complex with the transmembrane protein Crumbs (Martin-Belmonte and Perez-Moreno 2012). The identification of multiple members of these complexes in this study indicates that specific complexes function in border cell migration and confirms the sensitivity of this approach.
Notably, most of the polarity genes with phenotypes encode proteins that are associated with apical junctions of epithelial cells, for example, the Baz and Crumbs complexes, rather than basolateral junctions (Laprise and Tepass 2011; Martin-Belmonte and Perez-Moreno 2012). Two basolateral polarity complex proteins that contain PDZ domains, Dlg1 and Scribbled (Scrib), regulate mammalian epithelial cell migration (Dow and Humbert 2007). Moreover, Dlg1 is highly expressed in follicle cells and border cells (Szafranski and Goode 2004). Surprisingly, dlg1 or scrib RNAi did not disrupt border cell migration even though their transcript levels were significantly knocked down. These proteins suppress cell invasion in ovarian follicle cells (Goode and Perrimon 1997; Szafranski and Goode 2007) and in a model of tumor invasion (Pagliarini and Xu 2003), thus they may have a different role in border cells. Indeed, loss of dlg1 depolarizes the follicle cell epithelia, induces uncontrolled invasion, and may even stimulate border cell motility (Goode and Perrimon 1997; Szafranski and Goode 2004). Therefore, the activity of Dlg1, and possibly Scrib, likely needs to be downregulated in border cells to allow their detachment and migration. We previously found that the basolateral protein Par-1 is required for detachment of border cells from the follicle cell epithelium and their subsequent motility (McDonald et al. 2008). Therefore, border cells may use a different set of basolateral polarity proteins for migration compared with other types of epithelial cells.
The other major group of genes identified in this study encodes proteins with known or predicted roles in cytoskeletal regulation. This is consistent with established roles for the actin cytoskeleton and microtubules in migrating cells (Kaverina and Straube 2011; Ridley 2011). Like most migrating cells, border cells normally extend and retract actin-rich cellular protrusions that provide traction for migration and help them sense directional guidance cues (Murphy and Montell 1996; Fulga and Rørth 2002; Prasad and Montell 2007). Several microarray screens identified an enrichment of cytoskeletal-associated proteins in border cells compared with non-migratory cells (Borghese et al. 2006; Wang et al. 2006). Moreover, regulators of actin and microtubules promote the formation of dynamic border cell protrusions (Zhang et al. 2011; Kim et al. 2011; Yang et al. 2012). The cytoskeletal regulator Lim Kinase 1 (LIMK1), which encodes a serine-threonine kinase with two LIM domains in addition to a single PDZ domain, was a multi-hit gene identified by our study. LIMK1 functions downstream of the Rac GTPase to regulate actin dynamics through the actin-regulatory protein cofilin (Bernard 2007). Moreover, LIMK1 mildly rescues the border cell migration defects caused by inactivation of Rac (Zhang et al. 2011). Our results demonstrate that LIMK1 itself is required for border cell migration. However, more work is needed to determine the extent to which LIMK1 functions primarily through Rac in border cells, as has been proposed (Zhang et al. 2011), or has any additional functions. Two Rac-GEFs, myoblast city and elmo (Ced-12), are required for border cell migration (Bianco et al. 2007; Geisbrecht et al. 2008). In contrast, we found that another putative Drosophila Rac-GEF (Higuchi et al. 2009), PsGEF, is not required for border cell migration. These results highlight the complex roles of Rac-effector proteins in specific cell and tissue contexts. Further investigation of the cytoskeletal-associated genes identified in this study is anticipated to provide new insights into the regulation of border cell motility.
Roles of Bbg and CG6509 in cell migration
Although the targeted RNAi survey of PDZ gene function was designed to focus only on the extent of border cell migration, studies with bbg and CG6509 revealed genes that regulate distinct features of border cells. Our results indicate that Bbg regulates levels of active Stat92E within migrating border cells. Activation of the JAK/STAT pathway in the follicle cells surrounding the polar cells is the first step in the specification of border cell fate and recruitment of cells to form the border cell cluster (Silver and Montell 2001). Subsequently, JAK/STAT signaling is actively maintained during migration (Silver et al. 2005). It was unclear from previous studies what mechanisms control nuclear Stat92E levels in border cells, although both active transport of Upd ligand mRNA and endocytosis appear to be important (Silver et al. 2005; Van de Bor et al. 2011). Despite the relatively mild migration defect caused by bbg knockdown, partially migrated bbg RNAi border cells exhibited reduced nuclear Stat92E and presumably reduced JAK/STAT activation. Moreover, STAT levels were unaffected in border cells prior to migration. Thus, Bbg is a new regulator of JAK/STAT signaling that upregulates and/or maintains nuclear Stat92E levels in migrating border cells. As the protein interaction partners of Bbg have yet to be identified, the mechanism for Bbg-mediated regulation of STAT activity remains to be elucidated.
Border cells migrate as a morphologically distinct and interconnected group. Knockdown of CG6509 in border cells disrupted this cluster organization in addition to delaying their migration. This suggests that CG6509 helps keep border cells together in a collective cluster. The predicted mammalian homolog of CG6509, Dlg5, has been implicated in regulating cell migration (Smolen et al. 2010) and epithelial polarity (Nechiporuk et al. 2007). From mouse knockout studies, Dlg5 was proposed to maintain cell polarity through trafficking of cadherin-catenin complexes and stabilization of adherens junctions (Nechiporuk et al. 2007). The cluster morphology defects we observed with CG6509 knockdown are consistent with defects in cell polarity and/or cell-cell adhesion. Nonetheless, we did not observe gross alterations in the levels or localization of E-cadherin and polarity proteins in CG6509 RNAi border cells; however, we cannot rule out the possibility of subtle changes in these proteins and/or residual CG6509 gene function. The disorganized cluster phenotypes produced by knockdown of CG6509 resemble those caused by loss of JNK activity (Llense and Martín-Blanco 2008; Melani et al. 2008). JNK signaling promotes border cell cluster cohesion through regulation of cell polarity proteins such as Baz and cell-cell adhesion via Integrins and E-cadherin (Llense and Martín-Blanco 2008). Further investigation will be needed to determine whether CG6509 regulates cell-cell contacts within the border cell cluster and whether it functions downstream of or in parallel to JNK signaling.
Supplementary Material
Acknowledgments
We thank the Vienna Drosophila RNAi Center, NIG-Fly Stock Center, Harvard Transgenic RNAi Project, Bloomington Stock Center, Developmental Studies Hybridoma Bank (University of Iowa), E. Knust, A. Page-McCaw, S. Hou, K. O’Connor-Giles, and A. Zhu for fly stocks and antibodies. Special thanks to A. Burtscher for technical assistance and K. O’Connor-Giles for helpful discussions. This work was supported by National Institutes of Health (NIH)/National Institute of Child Health and Human Development training grant T32 HD-007104 to G.A. and NIH/National Institute of General Medical Sciences R01 GM-078526 and American Recovery and Reinvestment Act (ARRA) funds through this grant to J.A.M.
Footnotes
Communicating editor: K. S. McKim
Literature Cited
- Abdelilah-Seyfried S., Cox D. N., Jan Y. N., 2003. Bazooka is a permissive factor for the invasive behavior of discs large tumor cells in Drosophila ovarian follicular epithelia. Development 130: 1927–1935 [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., et al. , 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J., Uehara Y., Montell D. J., 2000. Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell 103: 1047–1058 [DOI] [PubMed] [Google Scholar]
- Bastock R., Strutt D., 2007. The planar polarity pathway promotes coordinated cell migration during Drosophila oogenesis. Development 134: 3055–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beccari S., Teixeira L., Rørth P., 2002. The JAK/STAT pathway is required for border cell migration during Drosophila oogenesis. Mech. Dev. 111: 115–123 [DOI] [PubMed] [Google Scholar]
- Bernard O., 2007. Lim kinases, regulators of actin dynamics. Int. J. Biochem. Cell Biol. 39: 1071–1076 [DOI] [PubMed] [Google Scholar]
- Bianco A., Poukkula M., Cliffe A., Mathieu J., Luque C. M., et al. , 2007. Two distinct modes of guidance signalling during collective migration of border cells. Nature 448: 362–365 [DOI] [PubMed] [Google Scholar]
- Bilder D., 2001. PDZ proteins and polarity: functions from the fly. Trends Genet. 17: 511–519 [DOI] [PubMed] [Google Scholar]
- Booker M., Samsonova A. A., Kwon Y., Flockhart I., Mohr S. E., et al. , 2011. False negative rates in Drosophila cell-based RNAi screens: a case study. BMC Genomics 12: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghese L., Fletcher G., Mathieu J., Atzberger A., Eades W. C., et al. , 2006. Systematic analysis of the transcriptional switch inducing migration of border cells. Dev. Cell 10: 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Cox D. N., Seyfried S. A., Jan L. Y., Jan Y. N., 2001. Bazooka and atypical protein kinase C are required to regulate oocyte differentiation in the Drosophila ovary. Proc. Natl. Acad. Sci. USA 98: 14475–14480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin S. J. F., Nehme N. T., Limmer S., Liegeois S., Pospisilik J. A., et al. , 2009. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325: 340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156 [DOI] [PubMed] [Google Scholar]
- Dow L. E., Humbert P. O., 2007. Polarity regulators and the control of epithelial architecture, cell migration, and tumorigenesis. Int. Rev. Cytol. 262: 253–302 [DOI] [PubMed] [Google Scholar]
- Duchek P., Rørth P., 2001. Guidance of cell migration by EGF receptor signaling during Drosophila oogenesis. Science 291: 131–133 [DOI] [PubMed] [Google Scholar]
- Duchek P., Somogyi K., Jékely G., Beccari S., Rørth P., 2001. Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell 107: 17–26 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., 2008. Polarity proteins in migration and invasion. Oncogene 27: 6970–6980 [DOI] [PubMed] [Google Scholar]
- Friedl P., Gilmour D., 2009. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10: 445–457 [DOI] [PubMed] [Google Scholar]
- Friedl P., Locker J., Sahai E., Segall J. E., 2012. Classifying collective cancer cell invasion. Nat. Cell Biol. 14: 777–783 [DOI] [PubMed] [Google Scholar]
- Fulga T. A., Rørth P., 2002. Invasive cell migration is initiated by guided growth of long cellular extensions. Nat. Cell Biol. 4: 715–719 [DOI] [PubMed] [Google Scholar]
- Geisbrecht E. R., Haralalka S., Swanson S. K., Florens L., Washburn M. P., et al. , 2008. Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev. Biol. 314: 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione C., Devergne O., Georgenthum E., Carballès F., Médioni C., et al. , 2002. The Drosophila cytokine receptor Domeless controls border cell migration and epithelial polarization during oogenesis. Development 129: 5437–5447 [DOI] [PubMed] [Google Scholar]
- Goode S., Perrimon N., 1997. Inhibition of patterned cell shape change and cell invasion by Discs large during Drosophila oogenesis. Genes Dev. 11: 2532–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot L., Paschoud S., Pulimeno P., Foglia A., Citi S., 2008. The cytoplasmic plaque of tight junctions: a scaffolding and signalling center. Biochim. Biophys. Acta 1778: 601–613 [DOI] [PubMed] [Google Scholar]
- Gustafson K., Boulianne G. L., 1996. Distinct expression patterns detected within individual tissues by the GAL4 enhancer trap technique. Genome 39: 174–182 [DOI] [PubMed] [Google Scholar]
- Harris B. Z., Lim W. A., 2001. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 114: 3219–3231 [DOI] [PubMed] [Google Scholar]
- Hidalgo-Carcedo C., Hooper S., Chaudhry S. I., Williamson P., Harrington K., et al. , 2011. Collective cell migration requires suppression of actomyosin at cell-cell contacts mediated by DDR1 and the cell polarity regulators Par3 and Par6. Nat. Cell Biol. 13: 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi N., Kohno K., Kadowaki T., 2009. Specific retention of the protostome-specific PsGEF may parallel with the evolution of mushroom bodies in insect and lophotrochozoan brains. BMC Biol. 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsmann S. S., Hepper C. C., Marchese D. D., Knöll C. C., Reuter R. R., 2006. The PDZ-GEF dizzy regulates cell shape of migrating macrophages via Rap1 and integrins in the Drosophila embryo. Development 133: 2915–2924 [DOI] [PubMed] [Google Scholar]
- Humbert P. O., Dow L. E., Russell S. M., 2006. The Scribble and Par complexes in polarity and migration: friends or foes? Trends Cell Biol. 16: 622–630 [DOI] [PubMed] [Google Scholar]
- Humbert P. O., Grzeschik N. A., Brumby A. M., Galea R., Elsum I., et al. , 2008. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene 27: 6888–6907 [DOI] [PubMed] [Google Scholar]
- Huynh J. R., Petronczki M., Knoblich J. A., St Johnston D., 2001. Bazooka and PAR-6 are required with PAR-1 for the maintenance of oocyte fate in Drosophila. Curr. Biol. 11: 901–906 [DOI] [PubMed] [Google Scholar]
- Kaverina I., Straube A., 2011. Regulation of cell migration by dynamic microtubules. Semin. Cell Dev. Biol. 22: 968–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Cho A., Yin H., Schafer D. A., Mouneimne G., et al. , 2011. Psidin, a conserved protein that regulates protrusion dynamics and cell migration. Genes Dev. 25: 730–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y., Renihan M. K., Boulianne G. L., 2006. Characterization of big bang, a novel gene encoding for PDZ domain-containing proteins that are dynamically expressed throughout Drosophila development. Gene Expr. Patterns 6: 504–518 [DOI] [PubMed] [Google Scholar]
- Laflamme C., Assaker G., Ramel D., Dorn J. F., She D., et al. , 2012. Evi5 promotes collective cell migration through its Rab-GAP activity. J. Cell Biol. 198: 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprise P., Tepass U., 2011. Novel insights into epithelial polarity proteins in Drosophila. Trends Cell Biol. 21: 401–408 [DOI] [PubMed] [Google Scholar]
- Liu Y., Montell D. J., 1999. Identification of mutations that cause cell migration defects in mosaic clones. Development 126: 1869–1878 [DOI] [PubMed] [Google Scholar]
- Llense F., Martín-Blanco E., 2008. JNK signaling controls border cell cluster integrity and collective cell migration. Curr. Biol. 18: 538–544 [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F., Perez-Moreno M., 2012. Epithelial cell polarity, stem cells and cancer. Nat. Rev. Cancer 12: 23–38 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Sung H.-H., Pugieux C., Soetaert J., Rørth P., 2007. A sensitized PiggyBac-based screen for regulators of border cell migration in Drosophila. Genetics 176: 1579–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. A., Montell D. J., 2005. Analysis of cell migration using Drosophila as a model system. Methods Mol. Biol. 294: 175–202 [DOI] [PubMed] [Google Scholar]
- McDonald J. A., Pinheiro E. M., Montell D. J., 2003. PVF1, a PDGF/VEGF homolog, is sufficient to guide border cells and interacts genetically with Taiman. Development 130: 3469–3478 [DOI] [PubMed] [Google Scholar]
- McDonald J. A., Pinheiro E. M., Kadlec L., Schupbach T., Montell D. J., 2006. Multiple EGFR ligands participate in guiding migrating border cells. Dev. Biol. 296: 94–103 [DOI] [PubMed] [Google Scholar]
- McDonald J. A., Khodyakova A., Aranjuez G., Dudley C., Montell D. J., 2008. PAR-1 kinase regulates epithelial detachment and directional protrusion of migrating border cells. Curr. Biol. 18: 1659–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melani M., Simpson K. J., Brugge J. S., Montell D., 2008. Regulation of cell adhesion and collective cell migration by hindsight and its human homolog RREB1. Curr. Biol. 18: 532–537 [DOI] [PubMed] [Google Scholar]
- Montell D. J., 2003. Border-cell migration: the race is on. Nat. Rev. Mol. Cell Biol. 4: 13–24 [DOI] [PubMed] [Google Scholar]
- Montell D. J., Rørth P., Spradling A. C., 1992. slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell 71: 51–62 [DOI] [PubMed] [Google Scholar]
- Mummery-Widmer J. L., Yamazaki M., Stoeger T., Novatchkova M., Bhalerao S., et al. , 2009. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature 458: 987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A. M., Montell D. J., 1996. Cell type-specific roles for Cdc42, Rac, and RhoL in Drosophila oogenesis. J. Cell Biol. 133: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M., Goto T. M., Sugimoto M., Nishimura T., Shinagawa T., et al. , 2008. Rho-kinase phosphorylates PAR-3 and disrupts PAR complex formation. Dev. Cell 14: 205–215 [DOI] [PubMed] [Google Scholar]
- Nechiporuk T., Fernandez T. E., Vasioukhin V., 2007. Failure of epithelial tube maintenance causes hydrocephalus and renal cysts in Dlg5−/− mice. Dev. Cell 13: 338–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.-Q., Liu L.-P., Binari R., Hardy R., Shim H.-S., et al. , 2009. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182: 1089–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.-Q., Zhou R., Czech B., Liu L.-P., Holderbaum L., et al. , 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8: 405–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomska P., Godt D., Tepass U., 1999. DE-Cadherin is required for intercellular motility during Drosophila oogenesis. J. Cell Biol. 144: 533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishita M., Tomizawa C., Yamamoto M., Horita Y., Ohashi K., et al. , 2005. Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J. Cell Biol. 171: 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva C., Escobedo P., Astorga C., Molina C., Sierralta J., 2012. Role of the MAGUK protein family in synapse formation and function. Dev. Neurobiol. 72: 57–72 [DOI] [PubMed] [Google Scholar]
- Pagliarini R. A., Xu T., 2003. A genetic screen in Drosophila for metastatic behavior. Science 302: 1227–1231 [DOI] [PubMed] [Google Scholar]
- Perrimon N., Ni J.-Q., Perkins L., 2010. In vivo RNAi: today and tomorrow. Cold Spring Harb. Perspect. Biol. 2: a003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro E. M., Montell D. J., 2004. Requirement for Par-6 and Bazooka in Drosophila border cell migration. Development 131: 5243–5251 [DOI] [PubMed] [Google Scholar]
- Prasad M., Montell D. J., 2007. Cellular and molecular mechanisms of border cell migration analyzed using time-lapse live-cell imaging. Dev. Cell 12: 997–1005 [DOI] [PubMed] [Google Scholar]
- Prasad M., Jang A. C.-C., Starz-Gaiano M., Melani M., Montell D. J., 2007. A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nat. Protoc. 2: 2467–2473 [DOI] [PubMed] [Google Scholar]
- Queenan A. M., Ghabrial A., Schüpbach T., 1997. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development 124: 3871–3880 [DOI] [PubMed] [Google Scholar]
- Ranganathan R., Ross E. M., 1997. PDZ domain proteins: scaffolds for signaling complexes. Curr. Biol. 7: R770–R773 [DOI] [PubMed] [Google Scholar]
- Ridley A. J., 2011. Life at the leading edge. Cell 145: 1012–1022 [DOI] [PubMed] [Google Scholar]
- Rørth P., Szabo K., Bailey A., Laverty T., Rehm J., et al. , 1998. Systematic gain-of-function genetics in Drosophila. Development 125: 1049–1057 [DOI] [PubMed] [Google Scholar]
- Royer C., Lu X., 2011. Epithelial cell polarity: a major gatekeeper against cancer? Cell Death Differ. 18: 1470–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer F., Schönbauer C., Langer C. C. H., Dietzl G., Novatchkova M., et al. , 2010. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature 464: 287–291 [DOI] [PubMed] [Google Scholar]
- Shin K., Wang Q., Margolis B., 2007. PATJ regulates directional migration of mammalian epithelial cells. EMBO Rep. 8: 158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierralta J., Mendoza C., 2004. PDZ-containing proteins: alternative splicing as a source of functional diversity. Brain Res. Brain Res. Rev. 47: 105–115 [DOI] [PubMed] [Google Scholar]
- Silver D. L., Montell D. J., 2001. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell 107: 831–841 [DOI] [PubMed] [Google Scholar]
- Silver D. L., Geisbrecht E. R., Montell D. J., 2005. Requirement for JAK/STAT signaling throughout border cell migration in Drosophila. Development 132: 3483–3492 [DOI] [PubMed] [Google Scholar]
- Simpson K. J., Selfors L. M., Bui J., Reynolds A., Leake D., et al. , 2008. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat. Cell Biol. 10: 1027–1038 [DOI] [PubMed] [Google Scholar]
- Smolen G. A., Zhang J., Zubrowski M. J., Edelman E. J., Luo B., et al. , 2010. A genome-wide RNAi screen identifies multiple RSK-dependent regulators of cell migration. Genes Dev. 24: 2654–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah V. K., Kranjec C., Thomas M., Banks L., 2011. PDZ domains: the building blocks regulating tumorigenesis. Biochem. J. 439: 195–205 [DOI] [PubMed] [Google Scholar]
- Szafranski P., Goode S., 2004. A Fasciclin 2 morphogenetic switch organizes epithelial cell cluster polarity and motility. Development 131: 2023–2036 [DOI] [PubMed] [Google Scholar]
- Szafranski P., Goode S., 2007. Basolateral junctions are sufficient to suppress epithelial invasion during Drosophila oogenesis. Dev. Dyn. 236: 364–373 [DOI] [PubMed] [Google Scholar]
- Thomas P. D., Kejariwal A., Campbell M. J., Mi H., Diemer K., et al. , 2003. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 31: 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonikian R., Zhang Y., Sazinsky S. L., Currell B., Yeh J.-H., et al. , 2008. A specificity map for the PDZ domain family. PLoS Biol. 6: e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Bor V., Zimniak G., Cerezo D., Schaub S., Noselli S., 2011. Asymmetric localisation of cytokine mRNA is essential for JAK/STAT activation during cell invasiveness. Development 138: 1383–1393 [DOI] [PubMed] [Google Scholar]
- Vinkemeier U., 2004. Getting the message across, STAT! Design principles of a molecular signaling circuit. J. Cell Biol. 167: 197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Bo J., Bridges T., Dugan K. D., Pan T.-C., et al. , 2006. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev. Cell 10: 483–495 [DOI] [PubMed] [Google Scholar]
- Xi R., McGregor J. R., Harrison D. A., 2003. A gradient of JAK pathway activity patterns the anterior-posterior axis of the follicular epithelium. Dev. Cell 4: 167–177 [DOI] [PubMed] [Google Scholar]
- Yang N., Inaki M., Cliffe A., Rørth P., 2012. Microtubules and Lis-1/NudE/Dynein regulate invasive cell-on-cell migration in Drosophila. PLoS ONE 7: e40632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz M., Christofori G., 2010. Mechanisms of motility in metastasizing cells. Mol. Cancer Res. 8: 629–642 [DOI] [PubMed] [Google Scholar]
- Zhang L., Luo J., Wan P., Wu J., Laski F., et al. , 2011. Regulation of cofilin phosphorylation and asymmetry in collective cell migration during morphogenesis. Development 138: 455–464 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.