Abstract
NANOGP8 is a human (Homo sapiens) retrogene, expressed predominantly in cancer cells where its protein product is tumorigenic. It arose through retrotransposition from its parent gene, NANOG, which is expressed predominantly in embryonic stem cells. Based on identification of fixed and polymorphic variants in a genetically diverse set of human NANOG and NANOGP8 sequences, we estimated the evolutionary origin of NANOGP8 at approximately 0.9 to 2.5 million years ago, more recent than previously estimated. We also discovered that NANOGP8 arose from a derived variant allele of NANOG containing a 22-nucleotide pair deletion in the 3′ UTR, which has remained polymorphic in modern humans. Evidence from our experiments indicates that NANOGP8 is fixed in modern humans even though its parent allele is polymorphic. The presence of NANOGP8-specific sequences in Neanderthal reads provided definitive evidence that NANOGP8 is also present in the Neanderthal genome. Some variants between the reference sequences of NANOG and NANOGP8 utilized in cancer research to distinguish RT-PCR products are polymorphic within NANOG or NANOGP8 and thus are not universally reliable as distinguishing features. NANOGP8 was inserted in reverse orientation into the LTR region of an SVA retroelement that arose in a human-chimpanzee-gorilla common ancestor after divergence of the orangutan ancestral lineage. Transcription factor binding sites within and beyond this LTR may promote expression of NANOGP8 in cancer cells, although current evidence is inferential. The fact that NANOGP8 is a human-specific retro-oncogene may partially explain the higher genetic predisposition for cancer in humans compared with other primates.
Keywords: pseudogene evolution, retroelement, SVA element, cancer, human diversity
The human (Homo sapiens) genome contains nearly three million copies of retroelements—segments of DNA derived from reverse transcription of RNA—constituting more than 40% of the genome (International Human Genome Sequencing Consortium 2001). These retroelements are classified as long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), LTR elements, and retropseudogenes (also known as processed pseudogenes). The number of genomic retroelements has been increasing over evolutionary time through retrotransposition, by which an existing segment of DNA is transcribed in cells of the germline, the resulting RNA is reverse transcribed, and the derived DNA is inserted into the genome. From that point forward, the retrotransposed element at its new location in the DNA is heritable and may eventually become fixed in the genome of a species, while its parent sequence remains at its original site in the genome.
Retroelements are especially abundant in human and great ape genomes, due in part to retrotranspositional bursts that predate the evolutionary divergence of their ancestral lineages. As a consequence, more than 99% of retroelements in the human and chimpanzee (Pan troglodytes) genomes are orthologous, although recent retrotranspositional activity has generated thousands of lineage-specific retroelements in these two species (Chimpanzee Sequencing and Analysis Consortium 2005).
Some full-length LINEs are autonomous retroelements, containing functional genes that encode all of the products necessary for their own retrotransposition. These products do not act solely on LINEs but may retrotranspose RNA molecules transcribed in the germline from various segments of DNA—including LINEs, SINEs, LTR elements, and genes—progressively increasing the number of retroelements in the genome. Over evolutionary time, as retroelements become fixed in the genome of a species, those located in or near a gene may exert a functional role in expression of that gene (Wang et al. 2005).
More than 8000 retropseudogenes in the human genome are derived from protein-encoding parent genes (Zhang et al. 2003) and can be readily identified by their similarity to the mRNAs encoded by those genes—they lack the gene’s introns and often have a remnant of a 3′ poly(A) tail—and by short target-site duplications (TSDs) at their borders. A significant number of genes have generated retropseudogene families consisting of multiple copies derived from the same parent gene. For example, 22.8% of all retropseudogenes in the human genome are members of ribosomal protein pseudogene families, each with numerous members. Protein-encoding cellular housekeeping genes, such as CYCS and GADPH, have likewise generated large pseudogene families (Zhang et al. 2003; Liu et al. 2009).
Most retropseudogenes have undergone extensive mutational decay over evolutionary time, acquiring mutations that render them non-functional, such as large deletions and insertions (including retroelement insertions), frameshifts, and premature termination codons. Such mutations accumulate because retropseudogenes typically fail to function as genes from the time of their origin and are thus free from constraints of purifying selection. Some evolutionarily recent retropseudogenes lack any disabling mutations but are nonetheless transcriptionally quiescent because they are removed from the promoter and transcription-enhancing sequences of their parent gene. A relatively small number of retropseudogenes can be classified as retrogenes because they are transcribed and translated into functional proteins, often in different cell types and under different transcriptional controls than their parent gene (Rohozinski et al. 2012; Ishiguro et al. 2012; Ma et al. 2011; Snider et al. 2010).
The NANOG gene encodes a homeobox transcription factor that plays an essential role in maintaining pluripotency in vertebrate embryonic stem cells (Mitsui et al. 2003; Chambers et al. 2003; Chambers et al. 2007; Wang et al. 2006; Tay et al. 2008). Its pseudogene family in the human genome consists of 11 members, named NANOGP1 through NANOGP11, NANOGP1 being a single duplication pseudogene, and NANOGP2 through NANOGP11, 10 retropseudogenes (Booth and Holland 2004). Booth and Holland (2004) determined the relative evolutionary ages of the NANOG pseudogenes based on substitutional divergence from the NANOG parent gene and concluded that NANOGP8 is the most recent. Fairbanks and Maughan (2006) reported that the human and chimpanzee genomes share all NANOG pseudogenes at orthologous chromosomal positions except for NANOGP8, which is absent from the chimpanzee genome. NANOGP8 therefore arose in the hominin ancestral lineage after the hominin-panin (human-chimpanzee) ancestral divergence and is a lineage-specific retropseudogene unique to humans among extant species.
In addition to its presence in embryonic stem cells, the NANOG protein is present in cancer stem cells (CSCs) in a variety of human cancers. Two distinct but related NANOG proteins may be active in CSCs, one encoded by NANOG and the other by NANOGP8, which is transcriptionally active as a retrogene in several types of cancer cells (Zhang et al. 2006; Jeter et al. 2009, 2011; Zbinden et al. 2010; Ambady et al. 2010; Eberle et al., 2010; Zhang et al. 2010, Ma et al. 2011, 2012; Ishiguro et al. 2012; Uchino et al. 2012; Ibrahim et al. 2012). Although both the NANOG gene and the NANOGP8 retrogene may be expressed in CSCs, experimental evidence suggests that CSCs preferentially express NANOGP8 and that the NANOGP8 protein promotes tumorigenesis more readily than NANOG (Jeter et al. 2009; Zbinden et al. 2010; Jeter et al. 2011; Uchino et al. 2012). Importantly, knockdown of NANOG/NANOGP8 mRNA in cancer cells inhibits tumorigenesis and clonogenic growth of breast, colon, prostate, and gastrointestinal cancer cell lines (Jeter et al. 2009; Jeter et al. 2011; Uchino et al. 2012). These discoveries confirm a key role for NANOGP8 in tumorigenesis and suggest that suppression of NANOGP8 gene expression or protein activity may potentially be developed as a treatment for cancer.
The cancer rate in humans exceeds that of wild and captive great apes, and this difference is probably due in part to genetic differences (McClure 1973; Beniashvili 1989; Waters et al. 1998; Puente et al. 2006). In an attempt to determine the genetic basis of this discrepancy, Puente et al. (2006) examined 333 genes known to influence the incidence and progression of cancer, comparing them in the human and chimpanzee genomes. They determined that all of these genes are present at orthologous chromosomal positions, contain open reading frames, and are highly conserved in both species. NANOGP8, however, had not been identified as a cancer-promoting retrogene at the time of their study and was not included in it. Several retrogenes are known to influence the incidence and progression of cancer (Rohozinski et al. 2012; Young et al. 2010; Liu et al. 2009). However, to date, NANOGP8 is the only known cancer-promoting retrogene that is exclusive to humans and thus may be partially responsible for the higher predisposition for cancer in humans compared with other primates.
In the research reported here, we document the evolutionary history of NANOGP8, distinguish polymorphic from fixed variants between NANOGP8 and NANOG, and use a comparative genomic approach to identify a series of retrotranspositional and mutational events that allowed NANOGP8 to evolve as a human-specific retro-oncogene.
Materials and Methods
DNA samples
Human DNA samples were obtained from the Coriell Institute for Medical Research (Camden, NJ). Samples were from the Human Variation Panel, including 110 individuals in the SNP500Cancer panel (Packer et al. 2004) and 9 individuals in the Africans South of the Sahara panel.
PCR amplification and identification of amplified fragments
PCR amplification of DNA fragments specific to NANOG or NANOGP8 from genomic DNA is complicated by the exceptionally high similarity between NANOG and NANOGP8, and their similarity to other NANOG pseudogenes in the human genome. These sequence similarities substantially limit identification of appropriate primer-binding sites because all but a few sites result in co-amplification of unintended non-target fragments from one or more paralogous sequences, often the same size as the target fragment. To preclude unintended amplification, we selected primer pairs that target sites unique to NANOG and/or NANOGP8, excluding all other NANOG pseudogenes. In some cases, primers pairs co-amplified fragments from both NANOG and NANOGP8, but fragments were differentiated by size due to the presence of introns in fragments amplified from NANOG and their absence in those amplified from NANOGP8. Predicted fragment sizes, primer targeting, and preclusion of unintended amplification were verified by Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast).
Table 1 lists the primers used for PCR amplifications from genomic DNA, their target sites, primer pairings, and the sources and sizes of amplified fragments for these pairings. All primers were manufactured by Integrated DNA Technologies (Coralville, IA). PCR amplification was accomplished using Qiagen HotStarTaq Plus Master Mix (Valencia, CA) according to the manufacturer’s recommendations. The amplification protocol consisted of an initial denaturation step of 5 min at 95°, followed by 35 cycles of amplification consisting of 30 s denaturation at 94°, 30 s for primer annealing at 62°, and from 0.5 to 2 min of extension at 72°, depending on the expected amplification product size (1 min/1 kb). PCR products were separated on 1% agarose gels run in 0.5X TBE and visualized by ethidium bromide staining and UV transillumination. Special care was taken during PCR setup to avoid extraneous human DNA contamination, including the use of clean rooms and DNA-free reagents and tubes. Controls lacking template DNA were included with each experiment.
Table 1. Primers for PCR amplification and sequencing, primer binding sites, primer pairings, and amplified fragment sizes and sites.
| Primer | Binding Sitea | |
|---|---|---|
| F1: 5′CAAAGCACATCTTGCCAGGA3′ | c.–212 to c.–193 (5′ insertion site of NANOGP8) | |
| F2: 5′GGCCGAAGAATAGCAATGGTGTGACG3′ | c.473 to c.498 (exon 3 in NANOG and NANOGP8) | |
| F3: 5′CTCCAGTCACAGACAGTTCTGGTTGTCC3′ | c.502–64C to c.502–37C (intron 3 in NANOG) | |
| F4: 5′GAATAGCAATGGTGTGACGCAGAAGG3′ | c.477 to c.496 (splice site for exons 3 and 4 in NANOGP8) | |
| F5: 5′GGACAGCCCTGATTCTTCCACCAG3′ | c.189 to c.212 (second exon in NANOGP8, internal primer for sequencing cloned DNA) | |
| R1: 5′GGTTATTAAAATGTCTTTTCTAGGCAGGGCGC3′ | c.*512 to c.*543 (3′ boundary of Alu element in 3′ UTR of NANOG and NANOGP8) | |
| R2: 5′CTTATCTATAGCCAGAGACGGCAGCC3′ | c.*546 to c.*551 and c.*574 to c.*593 (22 nucleotide-pair deletion in 3′ UTR of NANOG and NANOGP8) | |
| R3: 5′GCTTCTATCAATGTTGTCCTTAGC3′ | c.*550 to c.*573 (ancestral, non-deletion site in NANOG 3′ UTR) | |
| R4: 5′CCATACTCCACCCTCCATGAG3′ | c.*140 to c.*160 (3′ UTR in NANOGP8, internal primer for sequencing cloned DNA) | |
| Primer Pair | Fragment Size | Fragment Sitea |
| F1/R1 | 1681 | c.–212 to c.*543 from NANOGP8 |
| F2/R1 | 1132 | c.473 to c.*543 from NANOG |
| 997 | c.473 to c.*543 from NANOGP8 | |
| F2/R2 | 1157 | c.473 to c.*593 from NANOG deletion allele |
| 1025 | c.473 to c.*593 from NANOGP8 | |
| F2/R3 | 1154 | c.473 to c.*573 from NANOG allele without deletion |
| F3/R1 | 1029 | c.502–64C to c.*543 from NANOG |
| F4/R1 | 990 | c.477 to c.*543 from NANOGP8 |
Nucleotides are numbered in accordance with Nomenclature for the Description of Sequence Variants of the Human Genome Variation Society (http://www.hgvs.org/mutnomen), as follows: The symbol “c.” refers to coding sequence. Numbering in the reading frame begins at the first nucleotide of the initiation codon and ends with the final nucleotide of the termination codon. Nucleotides in the 5′ UTR are numbered in reverse, denoted with a negative sign (–), with the nucleotide preceding the first nucleotide of the reading frame designated as –1. Nucleotides in the 3′ UTR are numbered consecutively, denoted with an asterisk (*), with the first nucleotide beyond the final nucleotide of the reading frame designated as *1.
DNA sequencing
A 1681 bp PCR fragment consisting of 78% of the complete sequence of NANOGP8, including the entire 5′ UTR and reading frame and the 3′ UTR between the termination codon and an Alu element insertion, was amplified with primer pair F1/R1 (Table 1) from 10 geographically diverse individuals selected from the Coriell SNP500Cancer and Africans South of the Sahara panels (supporting information, File S1). PCR fragments of the target size amplified from these 10 individuals were cloned using the pGEM-T Easy Vector System II (Promega, Madison, WI). Recombinant clones were identified by standard blue/white screening methods with IPTG and X-Gal. Plasmid DNA from each selected recombinant clone was purified using GenElute plasmid miniprep Kit (Sigma, St. Louis, MO). Isolated plasmid DNA was sequenced bidirectionally using standard M13 (F/R) primers and internal primers F4 and R5 (Table 1).
A 1029 bp PCR product specific to NANOG, amplified with primer pair F3/R1 (Table 1), and a 990 bp PCR product specific to NANOGP8, amplified with primer pair F4/R1, were purified using a standard ExoSAP (exonuclease I/shrimp alkaline phosphatase) protocol and single-pass sequenced directly from PCR products using the F3 primer for NANOG and the F4 primer for NANOGP8.
DNA sequencing, assembly, and alignment
DNA sequencing was performed at the Brigham Young University DNA Sequencing Center (Provo, UT) using standard ABI Prism Taq dye-terminator cycle-sequencing methodology. DNA sequences were analyzed and assembled using Geneious software (Biomatters, Auckland, New Zealand), and aligned with either megablast (http://www.ncbi.nlm.nih.gov) or MEGA 5.05 for Mac OS X (http://www.megasoftware.net/megamac.php) (Tamura et al. 2011).
Nucleotide numbering and DNA variant nomenclature
Nucleotide numbering and DNA-variant designations were assigned in accordance with Nomenclature for the Description of Sequence Variants of the Human Genome Variation Society (http://www.hgvs.org/mutnomen), with the following two clarifications: (1) Guidelines specify that the largest transcript of a gene should be used. Accordingly, we used the coding sequence of the alternate reference assembly of NANOG with introns removed (AC_000144.1 [gi 157704453]: 7755619.0.7762294). To maintain consistent numbering in NANOG and NANOGP8, all nucleotides in NANOGP8 were numbered according to their alignment with the inferred mRNA sequence in the alternate reference assembly for NANOG. Variations in numbering are relevant only to the 3′ UTR where deletions and insertions are present in some sequences. (2) To accurately reflect mutational evolution, all substitution variants within NANOG or NANOGP8, or between the two, are designated with the ancestral nucleotide first, followed by the derived nucleotide (e.g. G > A), regardless of which nucleotide is present in any particular reference sequence.
Results and Discussion
The origin of NANOGP8 is evolutionarily more recent than previously estimated
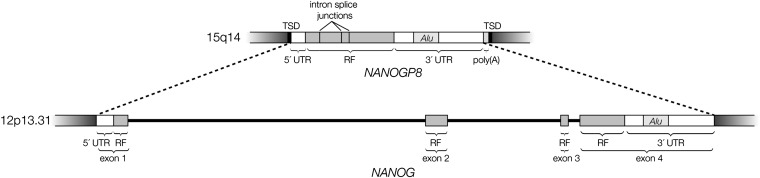
NANOGP8 resides on the long arm of human chromosome 15 in a relatively gene-free region (15q14). As depicted in Figure 1, NANOGP8 is bordered on both ends by a target-site duplication of 15 nucleotide pairs, and it lacks all three introns that are present in NANOG. It has a 5′ UTR of 197 nucleotide pairs, an intact reading frame of 918 nucleotide pairs, a 3′ UTR of 967 nucleotide pairs, including an Alu element, and a poly(A) tail in the current human primary and alternate reference assemblies (NC_000015.9 [gi 224589806]: c35375427 . . 35377509, AC_000147.1 [gi 157709134]: c12222739 . . 12220659).
Figure 1 .
Comparison of NANOGP8 with its parent gene NANOG. 3′ UTR, 3′ untranslated region; 5′ UTR, 5′ untranslated region; Alu, Alu element in 3′ UTR; RF, reading frame; TSD, target site duplication.
As Booth and Holland (2004) pointed out, the Alu element in the 3′ UTR of NANOGP8 is also present in the NANOG parent gene but is absent from all other human NANOG pseudogenes, evidence of a relatively recent evolutionary origin for this Alu element and a more recent origin for NANOGP8. Fairbanks and Maughan (2006) found this Alu element in the chimpanzee NANOG gene and also found it to be absent in all chimpanzee NANOG pseudogenes. Using the current human NANOG mRNA reference sequence as a query (NM_024865.2 [gi: 153945815]), we searched the Sumatran orangutan (Pongo abelii) and rhesus macaque (Macaca mulatta) assembled genomes and the gorilla (Gorilla gorilla) whole-genome shotgun sequences for this Alu element, and we found it to be present in the gorilla NANOG gene but absent from the orthologous site in the orangutan and rhesus macaque NANOG genes. It therefore was inserted into NANOG in the human-chimpanzee-gorilla common ancestral lineage between 8 and 16 million years ago, after the divergence of the orangutan ancestral lineage (Locke et al. 2011). Because this Alu element is present exclusively in NANOG and NANOGP8 and absent in all other NANOG pseudogenes, we used its 3′-insertion site as a primer-binding site (primer R1, Table 1) in several of our PCR experiments to exclude unintended amplification from other NANOG pseudogenes.
NANOGP8 is highly similar to its parent NANOG gene’s mRNA. Booth and Holland (2004), Jeter et al. (2009), and Uchino et al. (2012) aligned NANOG and NANOGP8 reference sequences within the reading frame and identified variants between them to estimate the evolutionary age of NANOGP8 or to distinguish NANOG and NANOGP8 RT-PCR products in cancer cells. Of the eight reading-frame variants between NANOG and NANOGP8 identified in these studies, only two (c.144G > A and c.759G > C) were common to all three studies, indicative of modern polymorphisms in NANOG or NANOGP8. Identification of modern polymorphisms is important for molecular-clock analysis because their inclusion as presumably fixed substitutions could overestimate the age of NANOGP8. Moreover, reliance on variants between reference sequences to distinguish NANOG and NANOGP8 RT-PCR products in cancer research may result in misidentification of these products if a variant is a modern polymorphism within either NANOG or NANOGP8.
To initially identify modern polymorphisms within NANOG, we aligned the primary and alternate reference assemblies for this gene throughout the full length of the transcribed region, excluding introns (primary reference assembly NC_000012.11 [gi 22458903]: 7941995 . . 7948655, alternate reference assembly AC_000144.1 [gi 157704453]: 7755619 . . 7762294). We found a surprisingly high number of variants between these two reference sequences, suggesting a high degree of modern polymorphism within NANOG: 16 substituted and 27 deleted nucleotides within the 2115 nucleotides of the mRNA. The degree of variation between these two NANOG reference sequences within the reading frame is equal to the presumably fixed variation previously reported between NANOG and NANOGP8 in the same region (Booth and Holland 2004; Jeter et al. 2009), calling into question the accuracy of the previous age estimate of 5.2 million years ago for the origin of NANOGP8 (Booth and Holland 2004).
Six of the substitutions we identified between the NANOG reference sequences are in the reading frame: c.165C > T, c.246G > T, c.276G > A, c.363C > T, c.531C > T, and c.798C > T (Table 2). All six have been identified as modern polymorphisms in the current NCBI dbSNP Report for NANOG (http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=79923), with relatively high minor allele frequencies (MAF) ranging from 0.1836 to 0.3863 (rs4294629, rs2889551, rs4354764, rs4438116, rs4012939, and rs4012937). All six are in the third nucleotide of their respective codons, and only one is non-synonymous (c.246T > G p.Asn82Lys, rs2889551). Booth and Holland (2004) experimentally confirmed this latter substitution variant as a modern polymorphism in NANOG through direct sequencing. The remaining 10 substitutions are in the 3′ UTR, as are all 27 deletions. Of those deletions, 22 are contiguous as a single deleted segment downstream of the 3′ border of the Alu element (c.*552_*573del), and 4 are variations within poly-T mononucleotide repeats [c.*184T(12–15) and c.*223T(18–19)]. The remaining deletion is a single nucleotide within the 3′ UTR Alu element (c.*496del).
Table 2. Variants between NANOG and NANOGP8 sequences detected by comparison of current primary and alternate reference assemblies and sequences we obtained experimentally.
| Variant in Coding DNAa | Variant in Proteina | NANOG | NANOGP8 |
|---|---|---|---|
| –135T > C | — | T | T/Cb |
| 47C > A | Ala16Glu | C | C/A |
| 126T > C | = | T | T/C |
| 144G > A | = | G | A |
| 165T > C | = | T/C | T |
| 190G > T | Asp64Tyr | G | G/Tb |
| 246T > G | Asn82Lys | T/G | T |
| 276G > A | = | G/A | G |
| 363C > T | = | C/T | C |
| 531C > T | = | C/T | C |
| 552A > T | = | A | A/Tb |
| 629C > T | Thr210Ile | C | C/Tb |
| 754A > C | Met252Leu | A | A/Cb |
| 759G > C | Gln253His | G | C |
| 798C > T | = | C/T | C |
| 916–917del | Ter309fs | TG | TG/delb |
| *7G > A | — | G | G/Ab |
| *44G > A | — | G | G/Ab |
| *184T(12–17) | — | poly(T) | poly(T) |
| *223T(17–20) | — | poly(T) | poly(T) |
| *243G > A | — | G/A | G |
| *310T > C | — | T/C | T |
| *313C > G | — | C | C/G |
| *315C > T | — | C | C/T |
| *413G > A | — | G/A | G |
| *467G > A | — | G | G/A |
| *496del | — | T/del | T |
| *512G > A | — | G | G/A |
| *552–*573del | — | =/del | del |
| *606T > G | — | T | G |
| *663A > G | — | A/G | A |
| *802C > T | — | C/T | C |
| *843C > T | — | C/T | C |
| *875G > A | — | G/A | A |
| *910C > A | — | C/A | C |
| *956A > G | — | A/G | A |
Polymorphic variants are indicated with a forward slash separating the ancestral and derived variants, with the ancestral variant indicated first. All variants are polymorphic in either NANOG or NANOGP8 except for three, 144G > A, 759G > C, and *606T > G, indicated in boldface.
In accordance with human genetic nomenclature guidelines for designating DNA variants in genes (http://www.hgvs.org/mutnomen), nucleotides in the reading frame are numbered relative to the first nucleotide in the ATG initiation codon; positions preceded by a negative (–) sign are in the 5′ UTR and are numbered in reverse relative to the first nucleotide in the ATG initiation codon; and positions indicated with an asterisk (*) are in the 3′ UTR relative to the first nucleotide beyond the termination codon. The symbol “=” denotes that a nucleotide substitution has no effect on the protein (i.e. a synonymous variant in the reading frame).
Variants at these sites were not present in the comparison of primary and alternate reference assemblies of NANOGP8 or in other publications. However, we observed polymorphism in at least two individuals for each of these sites in the sequences we obtained.
We also aligned the sequences for NANOGP8 from the current primary and alternate reference assemblies (primary reference assembly NC_000015.9 [gi 224589806]: c35375427 . . 35377509, alternate reference assembly AC_000147.1 [gi 157709134]: c12222739 . . 12220659). The number of variants between these NANOGP8 sequences was considerably fewer than in NANOG: four substitution variants, all in the 3′ UTR (c.*75C > T, c.*313C > G, c.*315C > T, and c.*512G > A), and variation in one of the two poly(T) mononucleotide repeats of the 3′ UTR [c.*223T(18–20)].
Alignment of the human NANOG and NANOGP8 reference sequences with other NANOG pseudogenes, with the NANOG gene in the assembled chimpanzee, orangutan, and rhesus macaque genomes, and with the gorilla NANOG sequence from a WGS contig (gi 269709276) allowed us to classify all substitution variants within and between NANOG and NANOGP8 as ancestral or derived. We were unable to determine which mononucleotide repeat polymorphisms are ancestral in the two poly(T) segments in the 3′ UTR because they vary in repeat number within and among species, as expected for mononucleotide repeats. A 22-nucleotide pair deletion in the 3′ UTR (c.*552_*573del) is present only in NANOG and NANOGP8 and is exclusive to humans; it thus is derived. With the exception of this deletion and the poly(T) mononucleotide repeats in the 3′ UTR, NANOGP8 carries the ancestral nucleotides for all variants between the two reference sequences of NANOG, and likewise, NANOG carries the ancestral sequence for all variants between the two reference sequences of NANOGP8. From these observations, we concluded that the ancestral nucleotides for all substitution variants between the two reference sequences of NANOG and between the two reference sequences of NANOGP8 were present in the parent allele of NANOG at the time of NANOGP8’s origin.
On the basis of this conclusion, we merged the primary and alternate reference sequences for NANOG by converting all substitution variants between the two to the ancestral form and did the same for NANOGP8, and then we aligned these converted NANOG and NANOGP8 sequences. This comparison reduced the number of potentially fixed substitution variants between NANOG and NANOGP8 to four: three within the reading frame (c.47C > A, c.144G > A, and c.759G > C) and one in the 3′ UTR (*606T > G). In all four cases, NANOG carries the ancestral nucleotide, and the derived variant is in NANOGP8 (Table 2). This analysis, therefore, allowed us to reconstruct the sequence of the NANOG parent allele at the time of NANOGP8’s origin, permitting a more accurate dating of this origin.
To identify additional modern polymorphisms in NANOGP8 and to obtain further evidence of potentially fixed variants, we used primer pair F1/R1 to amplify 78% of the complete sequence of NANOGP8, including the entire 5′ UTR and reading frame and the first 511 nucleotides of the 3′ UTR, from 10 geographically diverse individuals. Then we cloned these PCR fragments and obtained high-quality sequences of the complete clones (GenBank accession nos. JX104830–JX104848, supporting information, File S1). To more extensively examine a particular region in the fourth exon of NANOG and NANOGP8, which contains an important variant (c.759G > C p.Gln253His, described in detail later), we amplified a 1029 nucleotide-pair fragment from exon 4 of NANOG with primer pair F3/R1 (Table 1) and a 990 nucleotide-pair fragment from the same region of NANOGP8 with primer pair F4/R1 (Table 1) from 94 geographically diverse individuals. We then conducted single-pass sequencing of these PCR products, obtaining approximately 590 nucleotides of reliable sequence from the 5′ end of exon 4 into the first poly(T) segment of the 3′ UTR (GenBank accession nos. JX104849–JX104942 for NANOGP8, and JX104943–JX105036 for NANOG, File S2 and File S3).
We found that the c.47C > A variant is polymorphic and apparently rare in NANOGP8 (see File S1), as suggested by Jeter et al. (2009), reducing the number of evidently fixed variants between NANOG and NANOGP8 to three: c.144G > A, c.759G > C, and c.*606T > G. The latter of these variants lies near the end of the 3′ UTR, outside of the region we sequenced, and thus, we could not confirm whether it is fixed or polymorphic. All evidence of its fixation is derived from four NANOGP8 sequences that include the entire 3′ UTR in the NCBI nucleotide database; no other NANOGP8 sequences in the NCBI nucleotide database contain this region. We therefore have focused our analysis on the two evidently fixed variants within the reading frame: c.144G > A and c.759G > C.
Table 2 summarizes all variants we found between NANOG and NANOGP8 by aligning different reference sequences and in the sequences we obtained experimentally, identifying those variants confirmed to be modern polymorphisms in either NANOG or NANOGP8. All variants between the NANOG and NANOGP8 reading frames identified in previous publications (Booth and Holland 2004; Zhang et al. 2006; Jeter et al. 2009; Uchino et al. 2012) were evident in the sequences we obtained, all but two of them modern polymorphisms.
Of the two evidently fixed variants between NANOG and NANOGP8 within the reading frame, one is synonymous (c.144G > A), and one is non-synonymous (c.759G > C p.Glu253His). Sequencing of PCR products from exon 4 of NANOG and NANOGP8 in DNA samples from 94 geographically diverse individuals demonstrated that the ancestral G at c.759 in NANOG is homozygous in all individuals tested and that the derived C at this site in NANOGP8 is homozygous in all individuals tested, strong evidence that this variant is an ancient mutation in NANOGP8 and a fixed variant that reliably distinguishes NANOG and NANOGP8. Therefore, the proteins encoded by NANOG and NANOGP8 differ by only a single fixed amino acid substitution, although modern polymorphisms in both NANOG and NANOGP8 alter other amino acids in some individuals.
Presumably we could have utilized SNP databases for NANOG to identify additional polymorphisms. However, our analysis of variants between NANOG and NANOGP8 suggests that SNP databases for NANOG are prone to error due to misassignment of short-read sequences, a consequence of the high similarity of paralogous sequences in NANOG and its pseudogenes. For example, the current NCBI dbSNP report for NANOG has incorrectly assigned the two evidently fixed variants in NANOGP8 to NANOG as modern polymorphisms (rs2377097 for c.144G > A, and rs4012938 for c.759G > C), as well as several other variants that, according to our review of sequences, belong to NANOGP8 or other NANOG pseudogenes.
Two independent lines of evidence from our analysis allowed us to estimate the evolutionary age of NANOGP8. First, evidence that NANOGP8 is present in the Neanderthal genome (which we will discuss momentarily) and evidence published by Fairbanks and Maughan (2006) that it is absent from the chimpanzee genome indicate that it must have originated before the human-Neanderthal divergence and after the hominin-panin divergence. This observation allowed us to establish lower and upper boundaries for its origin, independent of any variants it carries relative to its parent gene. The lower boundary is the human-Neanderthal divergence, which Green et al. (2008) estimated as 520,000 to 800,000 years ago, based on mitochondrial genome analysis, whereas Green et al. (2010) estimated it as between 270,000 and 440,000 years ago, based on nuclear genome-wide analysis. The upper boundary is the hominin-panin divergence, which was recently estimated as 5.5–7 million years ago, based on a synthesis of genomic and fossil data (Scally et al., 2012).
Second, inferred substitution rates for human retropseudogenes can be used to estimate the origin of NANOGP8 by identifying fixed variants in NANOGP8 relative to the sequence of the NANOG parent allele at the time of NANOGP8’s origin. Such an estimate is based on the assumption that substitutions in NANOGP8 are selectively neutral and that fixed variants accumulate in NANOGP8 at the same rate as in other autosomal human retropseudogenes. Although there is strong evidence that NANOGP8 is a retrogene expressed in cancer cells, there is no evidence in our data or in previous publications to contradict the assumption that variants in NANOGP8 are selectively neutral. Therefore, in line with Booth and Holland’s (2004) assumption, we have treated NANOGP8 as a neutrally evolving retropseudogene for estimation of its evolutionary age.
According to a recent review (Keightly 2012), the most rigorous analysis of autosomal substitution rates in human retropseudogenes was that conducted by Nachman and Crowell (2000), which Booth and Holland (2004) combined with results of Martinez-Arias et al. (2001) to derive a rate of 1.25 × 10−9 substitutions fixed per site per year. Utilizing this rate, Booth and Holland (2004) estimated the origin of NANOGP8 as 5.2 million years ago (six variants across 915 sites in the reading frame, excluding the termination codon). Our analysis reduced the number of evidently fixed variants to two (c.144G > A and c.759G > C) and extended the sequenced region to 1622 nucleotides, which, based on this substitution rate, reduces the estimated origin of NANOGP8 to approximately one million years ago.
Nachman and Crowell (2000) and Keightly (2012) pointed out that inferred substitution rates for human retropseudogenes are dependent on assumptions regarding the ancestral effective population size and the time of the hominin-panin divergence. By varying these parameters, Nachman and Crowell (2000) listed several possible rates, ranging from 0.65 × 10−9 to 1.35 × 10−9 per year, and Keightly (2012) suggested two rates: 1.1 × 10−9 per year and 0.5 × 10−9 per year. The former of these is similar to the rate proposed by Booth and Holland (2004), and the latter is consistent with a genomic mutation rate of (0.5–0.6) × 10−9 per year for modern humans, as summarized by Scally et al. (2012). If we apply the extremes of the rates proposed by Keightly (2012) and Nachman and Crowell (2000) as a range (1.35 × 10−9 to 0.5 × 10−9 per year), the origin of NANOGP8 falls between 0.9 and 2.5 million years ago, still considerably less than previously estimated.
A derived 22 nucleotide-pair deletion is polymorphic in NANOG but monomorphic in NANOGP8
NANOGP8 contains a derived 22 nucleotide-pair deletion in the 3′ UTR (c.*552_*573del). Jeter et al. (2009, 2011) considered this deletion to be a variant that distinguishes NANOGP8 from NANOG based on the human genome primary reference assembly available at the time, and they relied on it as a primer-binding site to differentially amplify NANOG and NANOGP8 qRT-PCR products from cancer cells, as did Ma et al. (2011, 2012) and Ibrahim et al. (2012). However, in the current human genome primary reference assembly, both NANOG and NANOGP8 contain this deletion, indicating that it is not a universally reliable distinguishing feature. The NANOG sequence in the alternate reference assembly does not carry this deletion, evidence that the deletion is polymorphic in NANOG among modern humans, as noted by Zbinden et al. (2010).
The presence of this deletion in NANOGP8 suggests that the deletion arose in NANOG prior to the origin of NANOGP8 and that NANOGP8 then arose from the NANOG allele containing the deletion. Our observation that this deletion is derived and polymorphic in modern humans offers further evidence that NANOGP8’s origin must be relatively recent because a more ancient deletion in the NANOG parent allele should have become fixed or lost. Initially, this observation that NANOGP8’s parent allele is derived and polymorphic suggested to us that the presence/absence of NANOGP8 might also be polymorphic among modern humans.
To test this possibility, we utilized primer pair F2/R1 to simultaneously amplify different-sized fragments from NANOG and NANOGP8 (Table 1) to screen for the possible presence/absence of NANOGP8 in 119 geographically diverse individuals from the entire Coriell SNP500Cancer and Africans South of the Sahara panels. This experiment, as well as subsequent experiments with primer pairs F2/R2 and F4/R2 (Table 1), demonstrated that all individuals in both panels uniformly carry NANOGP8, evidence that NANOGP8 originated in Africa and was fixed there prior to the migrations approximately 60,000 years ago that founded the first Homo sapiens populations outside of Africa.
We screened all individuals in these two panels for the presence/absence of the 22 nucleotide-pair deletion (c.*552_*573del) in both NANOG and NANOGP8 with primer pairs F2/R2 and F2/R3 (Table 1). The deletion was uniformly present in NANOGP8 in all individuals, but both the ancestral (=) and deletion (*552–*573del) alleles in NANOG were highly polymorphic and distributed in populations throughout the world, with no evident geographic pattern for their distribution (Table 3).
Table 3. Distribution of the ancestral (=) and deletion (*552–*573del) alleles in the 3′ UTR of NANOG among 119 geographically diverse individuals.
| Population | Number of Individuals Tested | Homozygous for Ancestral (=) Allelea | Heterozygous | Homozygous for Deletion (*552–*573del) Alleleb |
|---|---|---|---|---|
| Africans South of the Sahara | 9 | 1 | 6 | 2 |
| Biaka Pygmy | 4 | 1 | 2 | 1 |
| Mbuti Pygmy | 5 | 2 | 1 | 2 |
| African-American | 14 | 2 | 8 | 4 |
| Druze | 5 | 5 | 0 | 0 |
| Indo-Pakistani | 3 | 3 | 0 | 0 |
| Russian Krasnodar | 3 | 2 | 1 | 0 |
| Ami | 5 | 0 | 2 | 3 |
| Chinese | 5 | 1 | 4 | 0 |
| Japanese | 4 | 0 | 3 | 1 |
| Southeast Asian | 3 | 0 | 2 | 1 |
| Pacific | 4 | 3 | 1 | 0 |
| South American | 7 | 4 | 2 | 1 |
| Mexican | 8 | 4 | 4 | 0 |
| Mexican-American | 9 | 2 | 4 | 3 |
| Puerto Rican | 8 | 1 | 2 | 5 |
| CEPH-Utah | 22 | 12 | 10 | 0 |
| Unidentified | 1 | 0 | 1 | 0 |
| Total | 119 | 43 | 53 | 23 |
Frequency of the ancestral (=) sequence = 0.5840.
Frequency of deletion (*552–*573del) = 0.4160.
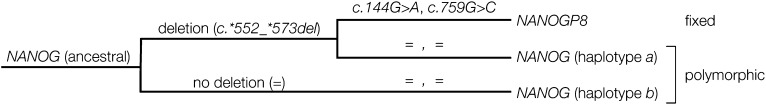
We found strong evidence in sequences we obtained and in genomic, EST, and SNP databases of two highly prevalent haplotypes of NANOG with worldwide distribution, as well as two intragenic recombinant haplotypes in a limited number of individuals, as detailed in File S3 and File S4. These two haplotypes differ by five substitution variants in the reading frame and by the *552–*573del deletion. This evidence also suggests that divergence of these two haplotypes predates the origin of NANOGP8. The haplotype derived from the parent allele of NANOGP8 carries the derived deletion *552–*573del and is in the current primary human genome assembly. The other haplotype is in the current alternate genome assembly. Figure 2 summarizes the evolutionary relationships of NANOGP8 with these two haplotypes of NANOG, portraying the relative times of occurrence for the two principal variants that are evidently fixed in NANOGP8 (c.144G > A and c.759G > C), and deletion *552–*573del in NANOGP8 and its parent allele. In Figure 2, we have named the two haplotypes in the primary and alternate assemblies, respectively, a and b to coincide with their designations as alleles a and b by Zbinden et al. (2010).
Figure 2 .
The evolutionary relationship of NANOGP8 with the two major haplotypes of NANOG, as distinguished by evidently fixed reading-frame variants c.144G > A and c.759G > C, and the 22-nucleotide 3′ UTR deletion c.*552_*573del.
The presence of modern polymorphic variants in NANOGP8 and between and within the two major haplotypes of NANOG has important implications for research on NANOGP8 expression in cancer cells. All haplotypes of NANOG can be readily distinguished from NANOGP8 in genomic DNA by the presence of introns in NANOG and the absence of introns in NANOGP8. Also, the 5′ insertion boundary of NANOGP8 reliably distinguishes it from NANOG. We utilized these features to unambiguously distinguish NANOG and NANOGP8 in our experiments. However, because RT-PCR products from NANOG and NANOGP8 are derived from mRNAs, they lack these distinguishing features. Several published studies have relied on variants between NANOG and NANOGP8 in reference sequences for primer design or for RFLPs to distinguish their RT-PCR products in cancer cells (Zhang et al. 2006; Jeter et al. 2009, 2011; Ambady et al. 2010; Zbinden et al. 2010; Eberle et al. 2010; Zhang et al. 2010; Ma et al. 2011, 2012; Uchino et al. 2012; Ibrahim et al. 2012). However, some of these variants, according to our data, are modern polymorphisms and, therefore, are unreliable for making these distinctions. Based on a detailed comparison of our data with data from these studies, as described in File S5, we determined that there is sufficient collective evidence from reliable fixed variants in sequenced RT-PCR products to confirm that NANOGP8 is expressed in several different types of cancer cells and that its protein is the predominant form of NANOG in these cancer cells (Jeter et al. 2009, 2011; Ibrahim et al. 2012). However, some distinctions in previous studies of NANOG and NANOGP8 RT-PCR products, and some conclusions regarding the presence or absence of NANOGP8 expression in cancer cells, were based on unreliable polymorphic variants and may have been incorrect.
NANOGP8 is present in the Neanderthal genome
We also examined the Neanderthal genome in the region on chromosome 15 orthologous to the insertion site in the human genome of NANOGP8 to determine whether NANOGP8 is present in that genome. (By way of information, annotation of the current Neanderthal assembly is incorrect for NANOG and NANOGP8: NANOG on chromosome 12 is misidentified as NANOGP8, and NANOGP8 on chromosome 15 is not identified.) Neanderthal sequences consist of short-read sequences aligned to the human reference assembly on the basis of sequence similarity. Because of the extremely high similarity of NANOG and NANOGP8, short-read sequences assigned to NANOGP8 in the Neanderthal genome assembly may in fact be derived from NANOG or another NANOG pseudogene, or vice versa. Therefore, we searched for Neanderthal short-read sequences spanning sites that are unique to NANOGP8 and capable of distinguishing it from the NANOG parent gene and other NANOG pseudogenes. The most reliable and unambiguous of such sites is the NANOGP8 5′-insertion boundary, which is unique to NANOGP8. We found two relatively long Neanderthal reads spanning this 5′-insertion boundary, one 78 and the other 57 nucleotides in length. We used these reads as queries for BLAST searches of the human genome and found no sites other than this insertion boundary in the human genome with significant similarity to the full lengths of each of these reads. Therefore, these two reads alone provide conclusive evidence that NANOGP8 is present in the Neanderthal genome.
Also informative as confirmatory evidence are the exon splice sites in NANOGP8, which differ from NANOG and NANOGP1 by their absence of introns, but may be similar to the paralogous splice sites in other NANOG retropseudogenes. A single read of 36 nucleotides spanning the splice site between exons 1 and 2 is assigned to NANOGP8 in the Neanderthal genome assembly. However, this read has equal similarity to the paralogous site in NANOGP9 and thus cannot be conclusively assigned to NANOGP8. A single read of 37 nucleotides spanning the splice site between exons 3 and 4 is identical only to the sequence at this site in NANOGP8. It differs by a single nucleotide from the paralogous site in NANOGP7 and by two nucleotides from this site in NANOGP4. One of the derived variants in NANOG and NANOGP8 that differs from NANOGP7 and NANOGP4 is a synonymous substitution variant (c.498T > C) that mutated in the parent NANOG gene after the origin of NANOGP7 and NANOGP4 (the most recent NANOG pseudogene before NANOGP8), prior to the hominin-panin divergence. In the human genome, it is present only in NANOG, NANOGP1, and NANOGP8, and the Neanderthal read containing it is distinguishable from NANOG and NANOGP1 because it lacks the intron at the splice site between exons 3 and 4 in NANOG and NANOGP1. Therefore, this read is correctly assigned to NANOGP8 in the Neanderthal genome.
The c.759G > C p.Glu253His substitution variant in NANOGP8 (which, according to the sequences we obtained, is unique to and evidently fixed in NANOGP8) is present in all three reads assigned to this region in the Neanderthal genome. Therefore, these reads are correctly assigned to NANOGP8, and offer evidence that this mutation predates divergence the Neanderthal and modern-human ancestral lineages.
In summary, the presence of two reads spanning the 5′-insertion boundary of NANOGP8, the presence of a NANOGP8-specific read spanning the splice site between exons 3 and 4, and the presence of three reads with the NANOGP8-specific c.759G > C p.Glu253His substitution variant collectively offer definitive evidence that NANOGP8 is present in the Neanderthal genome.
NANOGP8 is embedded in an SVA retroelement that may promote its transcription in cancer cells
SVA (SINE-R–VNTR–Alu) elements constitute a family of related composite retroelements composed of five segments: (1) a CCCCTC hexamer repeat on the 5′ end; (2) a segment composed of two truncated Alu elements in reverse orientation relative to the rest of the element; (3) a variable tandem nucleotide repeat (VNTR) segment; (4) a SINE-R segment derived from human endogenous retrovirus-K10 (HERV-K10), consisting of portions of the HERV-K10 env gene and the long terminal repeat (LTR); and (5) a poly(A) tail on the 3′ end of the element (Wang et al. 2005). They constitute the youngest class of retroelements in humans, having evolved about 13.5 million years ago exclusively in the human-great ape ancestral lineage shortly before divergence of the orangutan ancestral lineage from the human-chimpanzee-gorilla common ancestral lineage (Wang et al. 2005). Consequently, SVA elements are present exclusively in the genomes of humans and great apes. In the human genome, these elements fall into six subfamilies, named SVA_A through SVA_F. The SVA_A family, the most ancient, is found in the human, chimpanzee, gorilla, and orangutan genomes; it is retrotranspositionally quiescent in modern humans and great apes (Wang et al. 2005).
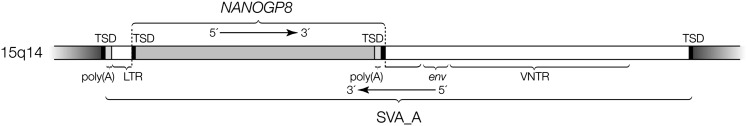
We aligned the genomic region of human chromosome 15 consisting of NANOGP8 plus 4000 nucleotides of flanking DNA sequence on both ends with orthologous sequences in the most recent genomic assemblies of common chimpanzee, Sumatran orangutan, and rhesus macaque. We also aligned two chromosome 15 contigs from gorilla whole-genome shotgun (WGS) sequences with portions of this region (gi 269664462 and gi 269664460). These alignments showed that NANOGP8 is embedded in reverse orientation in the LTR region of an SVA_A retroelement in the human genome (Figure 3). This SVA_A retroelement is present in the human, chimpanzee, and gorilla genomes, but it is absent at the orthologous site in the orangutan and rhesus macaque genomes, indicating that it was inserted into chromosome 15 in the common ancestral lineage of humans, chimpanzees, and gorillas after divergence of the orangutan ancestral lineage. The evolutionary history of NANOGP8 and its genomic context in the human genome, therefore, consists of insertion of an SVA_A element into the chromosome 15 homeolog in the human-chimpanzee-gorilla common ancestral lineage during a period between 8 and 16 million years ago (Locke et al. 2011), followed by insertion of NANOGP8 into this SVA element exclusively in the human ancestral lineage approximately 0.9 to 2.5 million years ago, according to our estimate.
Figure 3 .
Insertion of NANOGP8 into the LTR region of an SVA_A retroelement. env, envelope gene of a HERV-K endogenous retrovirus; LTR, long terminal repeat of a HERV-K endogenous retrovirus; poly(A), poly(A) tail; TSD, target site duplication; VNTR, variable nucleotide tandem repeat region.
The core promoter elements of the LTR region of this SVA_A element reside in a 215 nucleotide-pair segment upstream of the 5′ border of NANOGP8, albeit in reverse orientation relative to NANOGP8. LTRs are known to possess promoter and enhancer activities in germline and cancer cells, although little is known about the specific promoter activity of SVA LTRs. Wang et al. (2005) suggested that SVA-LTR promoter elements may regulate transcription of genes residing near them and that transcription of SVA elements may extend beyond their borders as evidenced by transduced segments of flanking DNA carried by approximately 10% of retrotransposed SVA elements in the human genome.
The LTR in the common ancestor of all SVA elements was evolutionarily derived from a HERV-K LTR sequence, and the promoter activity of HERV-K LTRs has been extensively documented through experimentation (Fuchs et al. 2011). The LTR region upstream of the NANOGP8 5′ border has no recognizable TATA box and is in reverse orientation relative to NANOGP8. However, three experimental observations of HERV-K LTR promoters suggest that this SVA_A LTR may be capable of promoting transcription of NANOGP8 in cancer cells. First, HERV-K genes are transcriptionally repressed in somatic cells but may gain transcriptional activity in germline and cancer cells (Cohen et al. 2009; Fuchs et al. 2011). Second, although the SVA_A LTR is in reverse orientation relative to NANOGP8, HERV-K promoters are capable of promoting antisense and bidirectional transcription (Dunn et al. 2006, Gogvadze et al. 2009). Third, HERV-K LTRs do not rely on canonical promoter sequences (such as the TATA box) but rather on transcription-factor binding sites, specifically Sp1 and Sp3 binding sites, shown experimentally to promote transcription in HERV-K LTRs (Fuchs et al. 2011).
We aligned the SVA LTR region upstream of NANOGP8 with the HERV-K LTR core promoter sequences identified by Fuchs et al. (2011) and found that an Sp1/Sp3 binding site, named GC-box3, shown experimentally to be essential and functional as a transcription activator in a HERV-K LTR (see Figure 4 in Fuchs et al. 2011), is fully conserved at nucleotides –396 through –401 relative to the initiation codon of NANOGP8, albeit in reverse orientation. Moreover, computational analysis of the SVA LTR with TESS (Schug 2003) identified a potential Sp1 binding site at nucleotides –255 through –264 relative to the initiation codon, also in reverse orientation. Thus, the presence of inferred transcription-factor binding sites in this SVA_A LTR sequence and the position of NANOGP8 relative to this sequence suggest a potential role for this LTR in promoting NANOGP8’s transcription in cancer cells. Transcription regulation may also extend to sequences upstream of the insertion border of the SVA element. Computational analysis with TFSEARCH (Heinemeyer et al. 1998) identified 95 potential transcription-factor binding sites within 1000 nucleotide pairs upstream of the NANOGP8 initiation codon. This evidence is entirely inferential; the actual promoter elements and transcription factor binding sites that influence NANOGP8 transcriptional activity are best determined experimentally, and the appropriate experiments are underway (Jeter et al. 2011).
Conclusions
The principal conclusions of our research are as follows: NANOGP8 is a human-specific retro-oncogene that arose approximately 0.9 to 2.5 million years ago in a common ancestor of humans and Neanderthals. Our evidence strongly indicates that it is fixed in modern humans, whereas its NANOG parent allele containing a derived deletion was polymorphic at the time of its origin and has remained polymorphic in one of two major haplotypes for NANOG in modern humans. The endogenous retroviral promoter elements present in an SVA LTR may promote transcription of NANOGP8 in cancer cells, although current evidence supporting this proposition is inferential rather than experimental. The SVA_A element that carries NANOGP8 in humans originated in a common ancestor of humans, chimpanzees, and gorillas after the divergence of the orangutan ancestral lineage.
Our observations of modern polymorphisms in NANOG and NANOGP8 underscore the unreliability of variants between reference sequences for accurate experimental distinction of NANOGP8 from NANOG RT-PCR products, particularly in studies of gene expression in cancer cells. Our observations further indicate that adequate screening in genetically diverse populations is essential to confirm which variants are modern polymorphisms and which are evidently fixed. Such screening is especially valuable if the variant may play a functional role in the protein or in gene expression. Furthermore, short-read sequences may be unreliable for polymorphism screening due to a high likelihood of incorrect assignment when a pseudogene or retrogene is highly similar to its parent gene, as NANOGP8 is to NANOG.
The c.759G > C p.Glu253His variant in NANOGP8 is of particular importance because it encodes the only fixed difference between the NANOG and NANOGP8 proteins. The mutant amino acid residue lies outside of the homeodomain, and its direct effect on the NANOGP8 protein’s biochemical function is unknown. Jeter et al. (2011) reported that most biological activities of the NANOG and NANOGP8 proteins are similar in cancer cells. However, their observations also suggest some enhanced tumorigenic functions for the NANOGP8 protein compared with the NANOG protein, possibly a consequence of this single amino acid substitution.
The major functional difference between NANOG and NANOGP8 is at the level of gene expression. NANOG is expressed predominantly in embryonic stem cells where NANOGP8 is apparently quiescent. By contrast, as Jeter et al. (2011) pointed out, “NANOGP8 is the predominant ‘isoform’ [of the protein] expressed in cancer cells and may, therefore, have evolved new functions distinct from those of NANOG1 in ESCs [embryonic stem cells]” (p. 11).
The recent evolution of NANOGP8 as a human-specific retro-oncogene is highly relevant for cancer research because NANOGP8 is absent in non-human species used as models for cancer. This situation may be a contributing factor to the delay in discovery of NANOGP8 as a retro-oncogene, and it highlights the value of evolutionary and comparative genomic research to identify other potential oncogenes that may be lineage-specific to humans or have human-specific effects. Finally, the recent evolution of NANOGP8 exclusively in the human ancestral lineage may partially explain some uniquely human aspects of cancer, including the higher predisposition for cancer in humans compared with other primates (Puente et al. 2006).
Supplementary Material
Acknowledgments
The authors wish to acknowledge Haagen Klaus for helpful comments on the manuscript and two anonymous reviewers for their comments. This research was funded by grants from Utah Valley University (to A.D.F. and T.H.O.) and from Brigham Young University (to P.J.M.).
Footnotes
Communicating editor: B. S. Yandell
Literature Cited
- Ambady S., Malcuit C., Kashpur O., Kole D., Holmes W. F., et al. , 2010. Expression of NANOG and NANOGP8 in a variety of undifferentiated and differentiated human cells. Int. J. Dev. Biol. 54: 1743–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniashvili D. S., 1989. An overview of world literature on spontaneous tumors in nonhuman primates. J. Med. Primatol. 18: 423–437 [PubMed] [Google Scholar]
- Booth H. A. F., Holland P. W. H., 2004. Eleven daughters of NANOG. Genomics 84: 229–238 [DOI] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., et al. , 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113: 643–655 [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., et al. , 2007. Nanog safeguards pluripotency and mediates germline development. Nature 450: 1230–1234 [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium, 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87 [DOI] [PubMed] [Google Scholar]
- Cohen C. J., Lock W. M., Mager D. L., 2009. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448: 105–114 [DOI] [PubMed] [Google Scholar]
- Dunn C. A., Romanish M. T., Gutierrez L. E., van de Lagemaat L. N., Mager D. L., 2006. Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene 366: 335–342 [DOI] [PubMed] [Google Scholar]
- Eberle I., Pless B., Braun M., Dingermann T., Marschalek R., 2010. Transcriptional properties of human NANOG1 and NANOG2 in acute leukemic cells. Nucleic Acids Res. 38: 5384–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbanks D. J., Maughan P. J., 2006. Evolution of the NANOG pseudogene family in the human and chimpanzee genomes. BMC Evol. Biol. 6: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs N. V., Kraft M., Tondera C., Hanschmann K. M., Löwer J., et al. , 2011. Expression of the human endogenous retrovirus (HERV) group HML-2/HERV-K does not depend on canonical promoter elements but is regulated by transcription factors Sp1 and Sp3. J. Virol. 85: 3436–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogvadze E., Stukacheva E., Buzdin A., Sverdlov E., 2009. Human-specific modulation of transcriptional activity provided by endogenous retroviral insertions. J. Virol. 83: 6098–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. E., Malaspinas A. S., Krause J., Briggs A. W., Johnson P. L. F., et al. , 2008. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell 134: 416–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. E., Krause J., Briggs A. W., Maricic T., Stenzel U., et al. , 2010. A draft sequence of the Neanderthal genome. Science 328: 710–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer T., Wingender E., Reuter I., Hermjakob H., Kel A. E., et al. , 1998. Databases on transcriptional regulation: TRANSDAC, TRRD, and COMPEL. Nucleic Acids Res. 26: 364–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim E. E., Babaei-Jadidi R., Saadeddin A., Spencer-Deen B., Hossaini S., et al. , 2012. Embryonic NANOG activity defines colorectal cancer stem cells and modulated through AP1- and TCF-dependent mechanisms. Stem Cells 30: 2076–2087 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium, 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Ishiguro T., Sato A., Ohata H., Sakai H., Nakagama H., et al. , 2012. Differential expression of Nanog1 and Nanogp8 in colon cancer cells. Biochem. Biophys. Res. Commun. 418: 199–204 [DOI] [PubMed] [Google Scholar]
- Jeter C. R., Badeaux M., Choy G., Chandra D., Patrawala L., et al. , 2009. Functional evidence that the self-renewal gene NANOG regulates human tumor development. Stem Cells 27: 993–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeter C. R., Liu B., Liu X., Chen X., Liu C., et al. , 2011. NANOG promotes cancer stem cell characteristics and prostate cancer resistance to androgen deprivation. Oncogene 30: 3833–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightly P. D., 2012. Rates and fitness consequences of new mutations in humans. Genetics 190: 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. F., Bera T. K., Liu L. J., Pastan I., 2009. A primate-specific POTE-actin fusion protein plays a role in apoptosis. Apoptosis 14: 1237–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke D. P., Hillier L. W., Warren W. C., Worley K. C., Nazareth L. V., 2011. Comparative and demographic analysis of orang-utan genomes. Nature 469: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Liang D., Liu J., Axcrona K., Kvalheim G., et al. , 2011. Prostate cancer cell lines under hypoxia exhibit greater stem-like properties. PLoS ONE 6: 329170. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ma Y., Liang D., Liu J., Axcrona K., Kvalheim G., et al. , 2012. Synergistic effect of SCF and G-SCF on stem-like properties in prostate cancer cell lines. Tumour Biol. 33: 967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Arias R., Calafell F., Mateu E., Comas D., Andrés A., et al. , 2001. Sequence variability of a human pseudogene. Genome Res. 11: 1071–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure H. M., 1973. Tumors in nonhuman primates: observations during a six year period in the Yerkes primate center colony. Am. J. Phys. Anthropol. 38: 425–429 [DOI] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., et al. , 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642 [DOI] [PubMed] [Google Scholar]
- Nachman M. W., Crowell S. L., 2000. Estimate of the mutation rate per nucleotide in human. Genetics 156: 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer B. R., Yeager M., Staats B., Welch R., Crenshaw A., et al. , 2004. SNP500Cancer: a public resource for sequence validation and assay development for genetic variation in candidate genes. Nucleic Acids Res. 32: D528–D532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente X. S., Velasco G., Gutiérrez-Fernández A., Bertranpetit J., King M. C., et al. , 2006. Comparative analysis of cancer genes in the human and chimpanzee genomes. BMC Genomics 7: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohozinski J., Edwards C. L., Anderson M. L., 2012. Does expression of the retrogene UTP14c in the ovary pre-dispose women to ovarian cancer? Med. Hypotheses 78: 446–449 [DOI] [PubMed] [Google Scholar]
- Scally A., Dutheil J. Y., Hillier L. W., Jordan G. E., Goodhead I., et al. , 2012. Insights into hominid evolution from the gorilla genome sequence. Nature 483: 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J., 2003 Using TESS to predict transcription factor binding sites in DNA sequence, Unit 2.6 in Current Protocols in Bioinformatics, edited by A. Baxevanis. J. Wiley & Sons, Hoboken, NJ. [DOI] [PubMed] [Google Scholar]
- Snider L., Geng L. N., Lemmers R. J., Kyba M., Ware C. B., et al. , 2010. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 6: e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y., Zhang J., Thomson A. M., Lim B., Rigoutsos I., 2008. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455: 1124–1128 [DOI] [PubMed] [Google Scholar]
- Uchino K., Hirano G., Hirahashi M., Isobe T., Shirakawa T., et al. , 2012. Human Nanog pseudogene8 promotes the proliferation of gastrointestinal cancer cells. Exp. Cell Res. 318: 1799–1807 [DOI] [PubMed] [Google Scholar]
- Wang H., Xing J., Grover D., Hedges D. J., Han K., et al. , 2005. SVA elements: a hominid-specific retroposon family. J. Mol. Biol. 354: 994–1007 [DOI] [PubMed] [Google Scholar]
- Wang J., Rao S., Chu J., Shen X., Levasseur D. N., et al. , 2006. A protein interaction network for pluripotency of embryonic stem cells. Nature 444: 364–368 [DOI] [PubMed] [Google Scholar]
- Waters D. J., Sakr W. A., Hayden D. W., Lang C. M., McKinney L., et al. , 1998. Workgroup 4: spontaneous prostate carcinoma in dogs and nonhuman primates. Prostate 36: 64–67 [DOI] [PubMed] [Google Scholar]
- Young J., Ménétrey J., Goud B., 2010. RABC is a retrogene that encodes a centrosomal protein involved in cell cycle progression. J. Mol. Biol. 397: 69–88 [DOI] [PubMed] [Google Scholar]
- Zbinden M., Duquet A., Lorente-Trigos A., Ngwabyt S. B., Borges I., et al. , 2010. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 29: 2659–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang X., Li M., Han J., Chen B., et al. , 2006. NANOGP8 is a retrogene expressed in cancers. FEBS J. 273: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Zhang J., Wang X., Chen B., Xiao Z., Li W., et al. , 2010. The human pluripotency gene NANOG/NANOGP8 is expressed in gastric cancer and associated with tumor development. Oncol. Lett. 1: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Harrison P. M., Liu Y., Gerstein M., 2003. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 13: 2541–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.