Abstract
The activity of NifA, the transcriptional activator of the nitrogen fixation (nif) gene, is tightly regulated in response to ammonium and oxygen. However, the mechanisms for the regulation of NifA activity are quite different among various nitrogen-fixing bacteria. Unlike the well-studied NifL–NifA regulatory systems in Klebsiella pneumoniae and Azotobacter vinelandii, in Rhodospirillum rubrum NifA is activated by a direct protein–protein interaction with the uridylylated form of GlnB, which in turn causes a conformational change in NifA. We report the identification of several substitutions in the N-terminal GAF domain of R. rubrum NifA that allow NifA to be activated in the absence of GlnB. Presumably these substitutions cause conformational changes in NifA necessary for activation, without interaction with GlnB. We also found that wild-type NifA can be activated in a GlnB-independent manner under certain growth conditions, suggesting that some other effector(s) can also activate NifA. An attempt to use Tn5 mutagenesis to obtain mutants that altered the pool of these presumptive effector(s) failed, though much rarer spontaneous mutations in nifA were detected. This suggests that the necessary alteration of the pool of effector(s) for NifA activation cannot be obtained by knockout mutations.
Introduction
Biological nitrogen fixation is a tightly controlled energy-demanding process. In all studied nitrogen-fixing bacteria, nitrogen fixation is regulated at the transcriptional level, though other levels of regulation are also involved in some bacteria (Dixon & Kahn, 2004; Zhang et al., 2003). Transcription of the nif (nitrogen fixation) operons is activated by a nif-specific transcriptional activator, NifA. However, the mechanism for the regulation of NifA activity is quite different in the various nitrogen-fixing bacteria.
The regulation of NifA is well studied in Klebsiella pneumoniae and occurs at both the transcriptional and post-translational levels. First, the transcription of the nifLA operon in K. pneumoniae is regulated by a general nitrogen regulation (Ntr) system, which involves many gene products, such as PII, NtrB/NtrC and GlnD (Merrick & Edwards, 1995; Minchin et al., 1988; Porter et al., 1995; Reitzer, 2003). glnD encodes a bifunctional, uridylyltrans-ferase/uridylyl-removing enzyme (UTase/UR) that senses the intracellular concentration of glutamine in the cell (Jiang et al., 1998). GlnD reversibly controls the activity of the PII proteins (GlnB and its homologues) by uridylylation or deuridylylation (Adler et al., 1975). PII proteins are some of the most broadly distributed regulatory proteins, and are integrators of signals of nitrogen, carbon and energy status (Arcond´guy et al., 2001; Commichau et al., 2006; Forchhammer, 2004; Leigh & Dodsworth, 2007; Ninfa & Atkinson, 2000; Ninfa & Jiang, 2005; Zhang et al., 2001a, 2006). The products of ntrB and ntrC belong to the family of two-component regulators (Stock et al., 2000). NtrB is a histidine kinase that phosphorylates NtrC under nitrogen-limiting conditions and can also act as a phosphatase to dephosphorylate NtrC under nitrogen-excess conditions (Kamberov et al., 1994). NtrB activity is regulated by PII in response to the carbon/nitrogen balance in the cell (Ninfa & Atkinson, 2000; Ninfa & Jiang, 2005). The phosphory-lated form of NtrC acts as a transcriptional activator of glnA, nifLA, glnK, amtB1 and other operons involved in nitrogen fixation and assimilation (Weiss et al., 1991). Under nitrogen-limiting conditions, the UTase activity of GlnD predominates and it uridylylates PII. The uridylyla-tion of PII prevents its interaction with NtrB. Without interaction with PII the kinase activity of NtrB predominates and it phosphorylates NtrC. The phosphorylated NtrC then activates expression of the nifLA operon in K. pneumoniae. NifA then activates expression of the other nif operons. Under nitrogen-excess conditions, the process is reversed and nif is not expressed. The nifLA operon is also regulated by the Ntr system in Azoarcus sp. BH72, Pantoea agglomerans (formerly called Enterobacter agglomerans) and Pseudomonas stutzeri (Desnoues et al., 2003; Egener et al., 2002; Siddavattam et al., 1995), but this is not the case in Azotobacter vinelandii (Blanco et al., 1993).
Secondly, in both K. pneumoniae and A. vinelandii, NifA activity is post-translationally regulated by NifL. In the presence of ammonium or oxygen, NifA activity is inhibited by NifL through direct protein–protein interaction (Martinez-Argudo et al., 2004b). PII is also involved in the regulation of the NifL–NifA interaction (He et al., 1998; Jack et al., 1999; Little et al., 2000, 2002; Stips et al., 2004), but probably by different mechanisms in these organisms (Little et al., 2000; Reyes-Ramirez et al., 2001; Stips et al., 2004).
In many other nitrogen-fixing bacteria belonging to the α-subgroup Proteobacteria, such as Azospirillum brasilense and Rhodospirillum rubrum, ntrC is not essential for nifA expression, but NifA activity is tightly controlled in response to ammonium (NH4+) (Liang et al., 1992, 1993;Zhang et al., 1995b, 2000). In these bacteria, no NifL homologue has been found, and NifA activity is regulated directly by one of the PII proteins (GlnB). In R. rubrum NifA is activated directly by the uridylylated form of GlnB, and the other PII homologues, GlnK and GlnJ, are unable to activate NifA (Zhang et al., 2001a, 2004). Similarly, in A. brasilense the activation of NifA requires only GlnB, and the other PII homologue (Pz or GlnZ) is not involved (Araújo et al., 2004; de Zamaroczy et al., 1993, 1996; Liang et al., 1992). The detailed mechanism for NifA activation in these organisms is still unknown, but it has been suggested that GlnB binds to the N-terminal GAF domain of NifA to prevent its interaction with the central catalytic domain (Arsene et al., 1996). Consistent with this hypothesis, interaction between NifA and GlnB has been detected in A. brasilense and R. rubrum by the yeast two-hybrid system (Chen et al., 2005; Zhu et al., 2006).
Nitrogenase activity in A. brasilense, R. rubrum and several other nitrogen-fixing bacteria is also tightly regulated at the post-translational level by a reversible mono-ADP ribosy-lation of dinitrogenase reductase (Nordlund & Ludden, 2004; Zhang et al., 1997). Under NH4+ -excess or energy-limiting conditions (such as shifting cells from light to dark for R. rubrum or shifting cells from microaerobic to anaerobic conditions for A. brasilense), dinitrogenase reductase is ADP-ribosylated and thereby inactivated by DRAT (dinitrogenase reductase ADP-ribosyl transferase, the gene product of draT). However, when NH4+ is exhausted or cells are shifted back to light (for R. rubrum) or microaerobic conditions (for A. brasilense), the ADP ribose group can be removed by DRAG (dinitrogenase reductase activating glycohydrolase; the gene product of draG), restoring nitrogenase activity. The activities of DRAT and DRAG are themselves subject to post-translational regulation, in which PII proteins are involved (Huergo et al., 2006, 2007; Klassen et al., 2001, 2005; Wang et al., 2005;Zhang et al., 2001a, b, 2006).
Although no structure of a NifA homologue is available, searching the Pfam protein families database (Finn et al., 2006; Sonnhammer et al., 1998) indicates that it has at least three domains: a GAF domain in the N-terminal region, a central σ54 interaction domain, and a helix–turn–helix (HTH) domain in the C-terminal region (Martinez-Argudo et al., 2004b; Morett & Segovia, 1993; Studholme & Dixon, 2003). GAF domains are ubiquitous small-molecule-binding domains present in cGMP-regulated cyclic nucleotide phosphodiesterases, adenylyl cyclases, the bacterial transcription factor FhlA, and many other signalling and sensory proteins from all three kingdoms of life (Aravind, 1997; Martinez et al., 2002). The GAF domain of A. vinelandii NifA binds α-ketoglutarate (α-KG) and controls its interaction with NifL (Little & Dixon, 2003; Martinez-Argudo et al., 2004a). The central domain of NifA, for σ54 interaction, is homologous with ATPases of the AAA+ family that are associated with diverse cellular activities, and is involved in ATP hydrolysis and interaction with σ54 (Zhang et al., 2002). The HTH motif of the C-terminal domain is involved in DNA binding (Morett et al., 1988).
To further investigate the mechanisms for NifA activation, we used two different approaches to obtain NifA variants that could be activated in the absence of GlnB. First, we obtained two variants, NifA-M173I and NifA-M173V, that interact better with GlnB in the yeast two-hybrid system. Both NifA variants showed GlnB-independent activity in R. rubrum. Second, a group of NifA variants was obtained as spontaneous Nif+ revertants of a ΔglnB mutant. We also found that wild-type NifA expressed from a multicopy plasmid can be activated in a GlnB-independent manner under certain growth conditions. We then hypothesized that there might be another gene whose elimination might provide the same phenotype, perhaps by altering the pool of small molecular effectors in the cell to allow NifA to be activated in the absence of GlnB. However, we were unable to generate such mutants by Tn5 insertion, though spontaneous mutations were found in nifA. This reinforces the potential for different NifA substitutions to have this phenotype and suggests that knockout mutations cannot significantly alter the level of effector(s) to cause NifA activation.
Methods
Bacterial strains and growth conditions
Escherichia coli was grown in LC medium (similar to Luria–Bertani medium, but with 5 g NaCl l−1). R. rubrum was grown in Supplemental Minimal plus NH4+ (SMN) (rich) medium (Fitzmaurice et al., 1989). Antibiotics were used as necessary at the levels described previously (Zhang et al., 2000).
Whole-cell nitrogenase activity assay
R. rubrum was grown in SMN medium, and then inoculated into MG (a nitrogen-limiting, malate/glutamate minimal medium with glutamate as nitrogen source), MN (minimal medium plus NH4+ ) or MN− (NH4+ -free minimal medium with N2 gas) as described previously (Fitzmaurice et al., 1989; Lehman & Roberts, 1991; Zhang et al., 2001a). Whole-cell nitrogenase activity was monitored by the acetylene reduction assay as described previously (Zhang et al., 1995a).
Random PCR mutagenesis and a yeast two-hybrid selection for nifA mutants
Random mutations in nifA were generated with the GeneMorph II Random Mutagenesis kit (Stratagene) using pUX686 (a pGAD-C1 derivative carrying R. rubrum nifA cloned into the BamHI and PstI sites, AD–NifA fusion) (Zhu et al., 2006) as a template. The mutagenized DNA was digested with BamHI and PstI, and ligated with pGAD-C1. As in our previous report on screening glnB mutants, the mutated plasmids were transformed into Saccharomyces cerevisiae strain pAJ.69 harbouring pUX679 (BD– GlnB fusion) (UY1) by the lithium acetate method (Gietz et al., 1995; Schiestl & Gietz, 1989). The transformants were selected on synthetic defined (SD; a yeast minimal medium) plates lacking leucine, uracil and histidine, and containing 5 mM 3-amino-1,2,4-triazole (3-AT), a competitive inhibitor of the His3 protein (Fields, 1993). Starting with a culture carrying the library of mutated nifA alleles, several hundred colonies appeared on the selection plates, while very few colonies were seen with the unmutagenized control. Plasmids were recovered from some of colonies and nifA was sequenced using the Big Dye Terminator v.3.1 Cycle Sequencing kit (Applied Biosystems) to identify mutations. Some plasmids were then reintroduced into UY1 and shown to support growth on SD plates lacking leucine, uracil and histidine, and containing 1.5 mM 3-AT, thus verifying causality.
Expression of NifA variants in R. rubrum
A 3.8 kb fragment carrying wild-type nifA was subcloned into a suicide vector, pUX19 (Lies, 1994), yielding pUX2021. Two nifA alleles identified in the yeast two-hybrid analysis were reconstructed with pUX2021 by the QuikChange Site-Directed Mutagenesis kit (Stratagene): pUX2022 encodes NifA-M173I and pUX2023 encodes NifA-M173V. These plasmids were transformed into E. coli strain S17-1 (Simon et al., 1983), then conjugated into UR717 (ΔglnB) (Zhang et al., 2000) and UR1325 (ΔglnD) (Zhang et al., 2005). The resulting strains contained both the wild-type nifA allele and a single integrated copy of the wild-type or mutant allele. In the ΔglnB background, these strains were designated UR1739 (the wild-type NifA merodiploid control), UR1740 (NifA-M173I) and UR1741 (NifA-M173V). In the ΔglnD background, they were designated UR1742 (the wild-type NifA merodiploid control), UR1743 (NifA-M173I) and UR1744 (NifA-M173V).
Similarly, wild-type and mutated nifA were also integrated into the chromosome of wild-type (UR2) background, yielding UR2501, UR2502 and UR2503, and into draT (UR213) (Liang et al., 1991) mutant background, yielding UR2327, UR2328 and UR2329.
Tn5 random mutagenesis and identification of the affected loci
pRL27 was used for Tn5 random mutagenesis. It contains a gene (tnp) under the control of the tetA promoter from plasmid RP4 that overproduces Tn5 transposase and a mini-Tn5 element carrying both kanamycin resistance (Kmr) and the origin of replication from plasmid R6K (oriR6K) (Larsen et al., 2002). pRL27 was transferred from E. coli into R. rubrum UR717 (ΔglnB) by bi-parental conjugation, as described previously (Liang et al., 1991). After mating on SMN plates overnight, cells were rinsed from the nitrocellulose filters into SMN containing nalidixic acid (N×) and Km, grown aerobically for 2 days for enrichment of the transconjugants, and then inoculated into MN− (NH4+ -free) medium in gas-tight tubes. After degassing and flushing with N2, cultures were grown anaerobically in the light for 5–10 days until they turned pink or red, indicating that some mutations restored the Nif+ phenotype of the ΔglnB mutant. The cultures were diluted and plated on SMN plates with N× and Km, and single colonies were purified on plates of the same medium. Nitrogenase activity in these mutants was monitored.
To identify the loci affected by the Tn5 insertions in these Nif+ mutants, the total DNA was isolated, digested with BamHI, and then self-ligated. The oriR6K is able to replicate in E. coli strains that contain pir, which allowed the cloning and analysis of the sites of the Tn5 insertions (Larsen et al., 2002).
Protein immunoblotting
A TCA precipitation method was used for rapid protein extraction (Zhang et al., 1993). As described previously (Zhang et al., 1995b), proteins were separated by low cross-linker SDS-PAGE (ratio of acrylamide : bisacrylamide, 172 : 1), electrophoretically transferred onto a nitrocellulose membrane, incubated with polyclonal antibody against A. vinelandii dinitrogenase reductase, and then visualized with horseradish peroxidase. Active dinitrogenase reductase is completely unmodified and migrates as a single band, while inactive dinitrogenase reductase migrates as two bands, since only one subunit of dinitrogenase reductase is ADP-ribosylated, and ADP-ribosylation slows the migration of the modified subunit.
Results
The yeast two-hybrid screen yields NifA variants that have a better interaction with GlnB
In our previous studies, we used the yeast two-hybrid system to detect some variants of GlnB (termed GlnB*) that interact better with NifA than does the wild-type GlnB (Zhu et al., 2006). In R. rubrum, these GlnB* variants activated NifA in the presence of NH4+, whereas NH4+ blocks NifA activity completely in the wild-type. The presence of these GlnB* variants also allowed NifA activity in ΔglnD backgrounds, while the uridylylation of GlnB by GlnD is normally essential for wild-type NifA activation. Our hypothesis is that the role of uridylylation of GlnB is primarily to shift the equilibrium of GlnB from a ‘nitrogen-sufficient’ form toward a ‘nitrogen-deficient’ form, and these GlnB variants apparently shift that equilibrium through direct structural changes (Zhu et al., 2006).
We supposed that it might be possible to find GlnB-independent (and therefore NH4+ -constitutive) NifA variants using a similar approach. That is, we reasoned that if binding of GlnB–UMP to NifA stabilizes the transcriptionally active form of NifA, then NifA variants that are equilibrium-shifted toward that form for structural reasons might have a higher affinity for GlnB–UMP and/or GlnB. Because wild-type NifA has a very low affinity for the non-UMP form of GlnB in the yeast system (Zhu et al., 2006), we supposed that NifA variants with improved affinity for GlnB should have a better interaction in the yeast.
We therefore mutagenized nifA in the yeast expression vector by error-prone PCR, and screened for NifA variants with improved interaction with GlnB. We initially tried 30 cycles of PCR with the GeneMorph II Random Mutagenesis kit from Stratagene. The mutagenized plasmid pool was transferred into a yeast strain producing wild-type GlnB (UY1), and selection for growth on minimal medium (SD) plates lacking leucine, uracil and histidine, but containing 5 mM 3-AT, a competitive inhibitor of His3 protein, was performed (Zhu et al., 2006). Several clones were purified, and plasmids from these yeast strains were isolated and the nifA genes were sequenced. Eight out of nine plasmids had multiple substitutions in nifA, though two were siblings (first screening in Table 1). Intriguingly, four of six non-identical clones had substitutions at Met-173, suggestive of causation. Because of the high mutation frequency, we remutagenized nifA with the following modifications: increasing the amount of template by fivefold and decreasing the number of PCR cycles from 30 to 12. Twenty-four plasmids containing the nifA alleles were isolated from yeast strains that grew well on the selection medium, and 19 plasmids were shown to be causative of the phenotype after they were reintroduced into UY1. Again, M173I and M173V substitutions predominated among non-identical plasmids (second screening in Table 1). We then reconstructed single M173I and M173V substitutions in the yeast plasmid and verified that these were both able to support growth of yeast on selective medium.
Table 1. Location of substitution(s) in nifA.
| Plasmid | Location of residues (substitutions) in NifA* |
|---|---|
| First screening: | |
| pMNA-2-1 | M173I (ATG→ATT), I196F, I217I, L418L, T516M |
| pMNA-5-1 | No mutation |
| pMNA-11-1 and 19-1 | M173V (ATG→GTG), D126Y, K208R, E413E |
| pMNA-12-1 | M173V (ATG→GTG), V48G, T64S, R156H, P282T, E324A, R438H, G524D |
| pMNA-14-5 and 23-4 | L41I, V96I, V132M, S241P, A360A, A376V, V473A |
| pMNA-24-1 | M173I (ATG→ATA), P116L, A482V, M428L |
| pMNA-26-4 | D126D, P146P |
| Second screening: | |
| pMNA-2-2 | V96A, S183R |
| pMNA-3-2 | M173V (ATG→GTG), S16P, A376A |
| pMNA-4-1 and 7-1 | M173I (ATG→ATT), A200A, R481F |
| pMNA-8-6 | M173I (ATG→ATC), Y380Y |
| pMNA-9-2 | M173I (ATG→ATT), V75A, K275K |
| pMNA-10-1 | S32T, L486P |
| pMNA-12-2 | E321N, F329F |
| pMNA-14-4 and 26-3 | M173V (ATG→GTG), S22S, A243T, P511T |
| pMNA-15-4 and 16-4 | M173I (ATG→ATA) |
| pMNA-17-5 | M173I (ATG→ATA), T370A |
| pMNA-18-2 | M173V (ATG→GTG), G126N, P207L |
| pMNA-20-3 | M173I (ATG→ATT), Y522C |
| pMNA-24-2 | M173V (ATG→GTG) |
| pMNA-27-1 | M173I (ATG→ATT), K591N |
| pMNA-28-1 | M173I (ATG→ATT), V86A |
| pMNA-30-1 | M173I (ATGAATC), Y380Y, G556V |
The substitutions and codon changes at the M173 residue are shown in bold type. Base substitutions that did not change the encoded amino acid are also indicated (e.g. I217I).
NifA-M173V and NifA-M173I variants are active in R. rubrum mutants lacking GlnB-UMP or GlnB
The above results show that NifA-M173V and NifA-M173I interact better with GlnB in yeast than wild-type. We then tested the physiological effect of their presence at normal levels in R. rubrum. Appropriate mutations were constructed and cloned into a pUX19 suicide vector, yielding pUX2022 (NifA-M173I) and pUX2023 (NifA-M173V). These plasmids, together with pUX2021 carrying wild-type nifA, were integrated into ΔglnB (UR717) or ΔglnD (UR1325) backgrounds. The nitrogenase activity in these strains is shown in Table 2 under nif-derepressing conditions in MG (a nitrogen-limiting, minimal medium with glutamate as nitrogen source). In both ΔglnB and ΔglnD backgrounds, the altered NifA variants caused substantial nitrogenase activity, while little NifA activation was seen with wild-type NifA. This indicates that these substitutions not only allow NifA to interact better with GlnB in yeast, but also cause NifA to be activated independently of the GlnB uridylylation state (based on the ΔglnD result) and in fact of any GlnB interaction at all (based on the ΔglnB result). This result is consistent with the original hypothesis of the conformational change in NifA, though it obviously does not prove it. For unknown reasons these NifA variants showed higher nitrogenase activity in the ΔglnB mutant background than in the ΔglnD mutant background. One simple explanation is that the NifA activity in these variants can be slightly inhibited by interaction with unmodified GlnB, since these NifA variants were shown to interact well with unmodified GlnB in yeast.
Table 2. Nitrogenase activity in the ΔglnB or the ΔglnD mutant with wild-type or altered nifA integrated into the chromosome when grown in MG (nitrogen-limiting) medium.
| UR strain | nifA integrated into DglnB or DglnD background | Nitrogenase activity* |
|---|---|---|
| UR1739 | Wild-type nifA integrated into ΔglnB mutant (UR717) | <10 |
| UR1740 | nifA11 (encoding NifA-M173I) integrated into ΔglnB mutant (UR717) | 470 |
| UR1741 | nifA12 (encoding NifA-M173V) integrated into ΔglnB mutant (UR717) | 450 |
| UR1742 | Wild-type nifA integrated into ΔglnD mutant (UR1325) | <10 |
| UR1743 | nifA11 (encoding NifA-M173I) integrated into ΔglnD mutant (UR1325) | 200 |
| UR1744 | nifA12 (encoding NifA-M173V) integrated into ΔglnD mutant (UR1325) | 230 |
Each unit of nitrogenase activity is expressed as nanomoles ethylene produced per hour per millilitre of cells at an OD600 of 1. Each activity value is from at least five replicate assays from different individually grown cultures. SD, 5–15 %.
Nitrogenase activity is still regulated by NH4+ in these NifA variants
As mentioned in the Introduction, nitrogenase activity in R. rubrum is regulated post-translationally by the DRAT– DRAG regulatory system: DRAT modifies dinitrogenase reductase to inactivate nitrogenase activity, while DRAG removes the covalently modified ADP ribose group from dinitrogenase and restores its activity. A screen/selection for mutants of Rhodopseudomonas palustris has been reported recently in which several mutants were found that produced hydrogen in the presence of NH4+ , and four of these had mutations in nifA (Rey et al., 2007). Interestingly, the nitrogenase activity in these nifA mutants is no longer regulated by NH4+ , even though Rhodopseudomonas palustris appears to have draTG homologues (Rey et al., 2007). We investigated whether substitutions in our NifA variants also altered the DRAT– DRAG regulation in R. rubrum. We found that although mutants with NifA-M173I and M173V variants possessed GlnB-independent NifA activity, and NifA activity was no longer regulated by NH4+ , the nitrogenase activity in these mutants was still regulated by NH4+ . As shown in Table 3, when grown in MG (nitrogen-limiting) medium, strains carrying wild-type nifA (UR2501), NifA-M173I (UR2502) or NifA-M173V (UR2503) showed high nitrogenase activity, similar to that seen in UR2 (wild-type). However, little or very low nitrogenase activity was seen in these strains when they were grown in MN (minimal medium plus NH4+ ). To test the hypothesis that nitrogenase activity in UR2502 and UR2503 was tightly regulated by the DRAT–DRAG regulatory system, we examined the behaviour of these NifA variants in a draT background. This draT mutant (UR213) lacks the DRAT activity that ADP-ribosylates dinitrogenase reductase, so nitrogenase activity is no longer regulated in response to either darkness or addition of NH4+ (Liang et al., 1991). draT mutants with NifA-M173I or M173V (UR2328 or UR2329) showed high nitrogenase activity in both MG- and MN-grown cultures, while draT mutants with wild-type NifA (UR2327) showed little nitrogenase activity in MN-grown cultures, since in this strain NifA activity is still regulated by NH4+.
Table 3. Nitrogenase activity in R. rubrum wild-type or draT mutant with wild-type or altered nifA integrated into the chromosome when grown in MG (nitrogen-limiting) and MN (NH4+-excess) media.
| Strain | nifA integrated into wild-type or draT background | Nitrogenase activity* | |
|---|---|---|---|
|
| |||
| MG (nitrogen-limiting) | MN (NH4+ -excess) | ||
| UR2 | Wild-type | 800 | <10 |
| UR2501 | Wild-type with wild-type nifA | 750 | <10 |
| UR2502 | Wild-type with nifA11 (encoding NifA-M173I) | 660 | 30 |
| UR2503 | Wild-type with nifA12 (encoding NifA-M173V) | 770 | 30 |
| UR2327 | draT mutant with wild-type nifA | 610 | <10 |
| UR2328 | draT mutant with nifA11 (encoding NifA-M173I) | 660 | 450 |
| UR2329 | draT mutant with nifA12 (encoding NifA-M173V) | 500 | 540 |
Units of nitrogenase activity are expressed as in Table 2. Each activity value is from at least five replicate assays from different individually grown cultures. SD, 5–15 %.
The protein level and the modification status of dinitrogenase reductase are completely consistent with the nitrogenase activity seen in these mutants in Table 3. As shown in Fig. 1, when grown in MN (NH4+ -excess) medium, little dinitrogenase reductase accumulated in UR2, UR2501, and UR2327, while high levels of dinitrogenase reductase accumulated in mutants with the NifA-M173I or M173V variant (UR2502, UR2503, UR2328 and UR2329). The difference is that the dinitrogenase reductase was modified in UR2502 and UR2503, but was completely unmodified in UR2328 and UR2329, since these two strains lack DRAT. There is also a third band between the unmodified band (indicated as U) and the modified band (indicated as M). Since it is absent under NH4+ -excess conditions and can cross-react with the antibody against A. vinelandii dinitrogenase reductase, it might be the alternative (Fe-only) dinitrogenase reductase or another unmodified form of dinitrogenase reductase. When grown in MG medium, all strains had similar amounts of unmodified dinitrogenase reductase (data not shown). These results indicate that the activity of NifA-M173I or NifA-M173V is no longer regulated by NH4+ , although nitrogenase activity in these variants is still regulated by NH4+ through the DRAT/DRAG system.
Fig. 1.
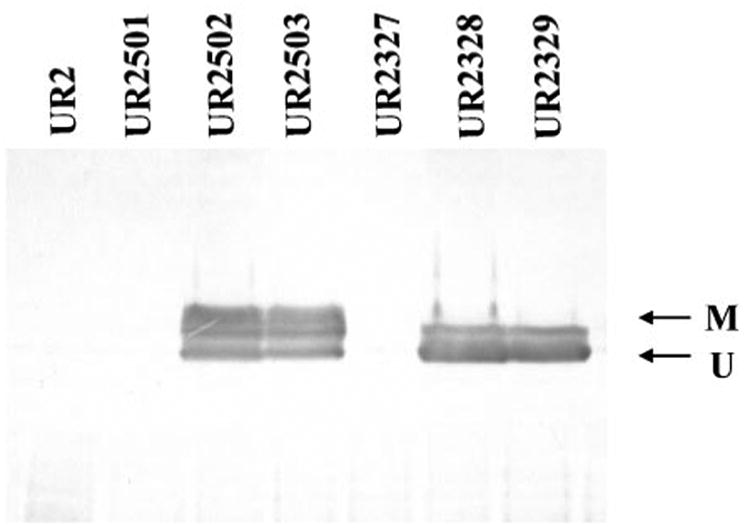
Immunoblot of the dinitrogenase reductase in UR2 (wild-type), UR2501 (wild-type NifA in wild-type background), UR2502 (NifA-M173I in wild-type background), UR2503 (NifA-M173V in wild-type background), UR2327 (wild-type NifA in draT mutant background), UR2328 (NifA-M173I in draT mutant background) and UR2329 (NifA-M173V in draT mutant background) when cells were grown in MN (NH4+ -excess) medium. Proteins were extracted quickly with TCA, separated by SDS-PAGE, and immunoblotted with antibody against A. vinelandii dinitrogenase reductase. As described in Methods, active dinitrogenase reductase is unmodified and migrates as a single band (indicated by U), while inactive dinitrogenase reductase migrates as two bands with the upper band representing the modified subunit (indicated by M). A third band between the unmodified and the modified bands might be an alternative dinitrogenase reductase or another unmodified form of dinitrogenase reductase.
We noticed a high nitrogenase activity in UR2328 and UR2329 (draT mutants with NifA-M173I and NifA-M173V variants) when grown MN medium. This seems to be inconsistent with the hypothesis that we proposed above that NifA activity in these variants can be slightly inhibited by interaction with unmodified GlnB, since GlnB should be unmodified in MN-grown cells. We suggest two possible explanations. (i) Cell growth and nitrogenase activity vary in different media. However, we were unable to perform the best comparison of nitrogenase activity in glnB draT and glnD draT mutants with these NifA* variants in MN medium, since glnD mutants fail to grow in MN medium (Zhang et al., 2005). (ii) Alternatively, the protein level of GlnB is much lower in MN-grown cells than in MG-grown cells, since one of its promoters is repressed under N-excess conditions (Cheng et al., 1999). This lower level of GlnB may be insufficient to inhibit NifA activity in MN-grown cells.
Wild-type NifA can be activated in the absence of GlnB under N2-fixing conditions when expressed from a multicopy plasmid
As we reported previously, GlnB is essential for NifA activation and little nitrogenase activity is seen in a ΔglnB3 mutant (UR717) in MG (nitrogen-limiting) medium (Zhang et al., 2000) (Table 4). When wild-type nifA was expressed from a multicopy plasmid in the ΔglnB3 mutant background, a low level of nitrogenase activity was detected in UR744 when it was grown in MG medium (Zhang et al., 2000) (Table 4). This result is consistent with the equilibrium hypothesis: when elevated amounts of NifA are present due to the multicopy plasmid, higher levels of NifA (inactive form) should accumulate, and the equilibrium should produce a small population of the active form of NifA, leading to some low level of nitrogenase activity. Surprisingly, we found dramatically different nitrogenase activity in these strains when they were grown in MN− (NH4+ -free) medium with N2 gas. As shown in Table 4, a ΔglnB3 mutant (UR717) failed to grow in MN− medium, and little nitrogenase activity was detected. However, UR744 (ΔglnB3 mutant with multicopy nifA) grew reasonably well and had a moderate nitrogenase activity in MN− medium, similar to that seen in the wild-type control (UR741). For unknown reasons, nitrogenase activity in MN−-grown cultures of the wild type strains was always lower than that in MG-grown cultures, as has been reported previously in both wild-type (UR2) and other strains (Zhang et al., 2001a, 2005). These results support the notion that in the absence of GlnB, some other effector(s) (either proteins or small molecules) can also affect NifA activation.
Table 4. Nitrogenase activity in R. rubrum strains containing single or multiple copies of R. rubrum nifA when grown in MG (nitrogen-limiting) or MN− (NH4+ -free) media.
| Strain | Plasmid (gene) | Nitrogenase activity* | |
|---|---|---|---|
|
| |||
| MG (nitrogen-limiting)† | MN− (NH4+-free)‡ | ||
| UR2 (wild-type) | None (single copy of R. rubrum nifA) | 800 | 400 |
| UR717 (ΔglnB3) | None (single copy of R. rubrum nifA) | <10 | <10 |
| UR741 (wild-type) | pYPZ239 (multicopy of R. rubrum nifA) | 650 | 300 |
| UR744 (ΔglnB3) | pYPZ239 (multicopy of R. rubrum nifA) | 50 | 280 |
Units of nitrogenase activity are expressed as in Table 2.
Nitrogenase activities of these strains in MG medium are similar to those recorded previously (Zhang et al., 2001a).
UR717 grew very slowly in MN− medium, with a final OD600 of 0.3 after 2 days of growth, while UR2 and UR741 had a final OD600 of ∼3. UR744 grew slightly slower in MN− medium, with a final OD600 of 1.7.
Tn5 random mutagenesis of ΔglnB mutant and screening for mutants with high nitrogenase activity in this background
As shown above, there is GlnB-independent NifA activation, which is affected in some way by nitrogen status. To identify the effector(s) involved in NifA regulation, we performed Tn5 mutagenesis on a ΔglnB mutant and sought mutants that would allow NifA to be activated in a GlnB-independent manner. Our reasoning was that some Tn5 insertions might alter the pools of the effector molecules, and such mutants would provide useful information about these effector(s).
After Tn5 mutagenesis and enrichment in MN− medium, many mutants displayed a Nif+ phenotype in a ΔglnB mutant background. The sites of the Tn5 insertion in about 50 strains were identified by sequencing, and several strains showed identical loci of the Tn5 insertion in ORFs named Rru_A3654/3555, A2353, A1638, A1681, A0934 and B0039 in the R. rubrum genome, indicating that these are siblings (Table 5). When selected Tn5 insertions were subcloned into a suicide vector and moved back into the ΔglnB background, the strains remained Nif−. Thus, the Tn5 insertions were not causative of the Nif+ phenotype.
Table 5. Loci of Tn5 insertion and substitutions in nifA in R. rubrum mutants.
| Tn5 insertion locus* | ORF* | Substitution in nifA |
|---|---|---|
| 4194344 (9)† | Between Rru_A3654 and Rru_A3654 | A243T (2)‡ |
| 40371 (3) | Rru_ B0039 | M173I (2) |
| 2738176 (5) | Rru_ A2353 | L184R (2) |
| 1929051 (3) | Rru_ A1638 | M173I |
| 1974362 (3) | Rru_A1681 | G36E |
| 1113948 (21) | Rru_ A0934 | M173I |
| 3073513 (1) | Rru_A2643 | G36E |
| 2087876 (1) | Rru_ A1795 | No mutation in nifA |
| 3405657 (1) | Rru_ A2956 | M173I |
| 3047404 (1) | Rru_A2619 | M173L |
| 3533303 (1) | Rru_ A3067 | M173V |
| 3336134 (1) | Rru_ A2892 | T38P |
| 1264071 (1) | Rru_ A1073 | T38P |
| 184684 (1) | Rru_A0158 | M173V |
The Tn5 insertion loci and its ORF are based on the R. rubrum genomic sequence at http://www.ncbi.nlm.nih.gov/sites/entrez?db=genome&cmd=Retrieve&dopt=Overview&list_uids=19059.
The numbers in parentheses indicate the number of strains that were sequenced and had identical loci of Tn5 insertion.
The numbers in parentheses indicate the number of strains in which nifA was sequenced.
Because our PCR mutagenesis of nifA revealed GlnB-independent variants, we reasoned that some of these Nif+ mutants found after Tn5 mutagenesis might also have spontaneous mutations in nifA. We amplified and sequenced nifA from 17 Nif+ strains obtained by Tn5 mutagenesis. As shown in Table 5, all strains but one had mutations in nifA, and some had the identical M173I or M173V substitution mentioned above. To confirm that these nifA mutations were causative of the GlnB-independence, we reconstructed several single substitutions in nifA, integrated them into the chromosome of a ΔglnB mutant and measured the nitrogenase activity. As shown in Table 6, these new NifA variants with M173L, L184R, A243T, G36E and T38P substitutions caused moderate nitrogenase activity in a ΔglnB mutant background, similar to that seen in NifA-M173I and M73V variants. All of these NifA variants have substitutions in the N terminus, strongly suggesting the important role of this GAF domain in the regulation of NifA activity.
Table 6. Nitrogenase activity in R. rubrum ΔglnB mutants with NifA variants when grown in MG (nitrogen-limiting) medium.
| Strain | nifA integrated into ΔglnB mutant (UR717) | Nitrogenase activity* |
|---|---|---|
| UR2 | Wild-type | 800 |
| UR1739 | Wild-type nifA integrated into ΔglnB mutant | <10 |
| UR1740 | nifA11 (encoding NifA-M173I) integrated into ΔglnB mutant | 470 |
| UR1741 | nifA12 (encoding NifA-M173V) integrated into ΔglnB mutant | 450 |
| UR11742 | nifA23 (encoding NifA-M173L) integrated into ΔglnB mutant | 450 |
| UR11743 | nifA24 (encoding NifA-L184R) integrated into ΔglnB mutant | 400 |
| UR11744 | nifA25 (encoding NifA-A243T) integrated into ΔglnB mutant | 500 |
| UR11745 | nifA26 (encoding NifA-G36E) integrated into ΔglnB mutant | 220 |
| UR11746 | nifA27 (encoding NifA-T38P) integrated into ΔglnB mutant | 310 |
Units of nitrogenase activity are expressed as in Table 2.
Discussion
We previously proposed a model in which GlnB has at least two forms: a ‘nitrogen-sufficient’ form and a ‘nitrogen-deficient’ form. The uridylylation of GlnB is primarily to shift the equilibrium of GlnB from a nitrogen-sufficient form to a nitrogen-deficient form (Zhu et al., 2006). Similar to GlnB, NifA appears to exist in at least two forms, dependent on the nitrogen status of the cell. Under nitrogen-excess conditions GlnB is deuridylylated, and this unmodified form of GlnB is probably unable to interact well with NifA under normal physiological conditions. Under nitrogen-limiting conditions, GlnB is uridylylated and this modified GlnB might have a much better affinity for NifA. The interaction with GlnB–UMP causes a conformational change in NifA, which activates its activity.
We report here that several substitutions in the N-terminal GAF domain of NifA, derived using two independent methods, allow NifA to be activated in the absence of GlnB. We believe that these substitutions also cause a conforma-tional change in NifA, similar to that caused by the interaction with GlnB–UMP. The NifA-M173I or M173V variants interact strongly with GlnB in yeast. Because GlnB is presumably unmodified in yeast, this indicates that con-formational change in the T-loop of GlnB by uridylylation is not necessary for its interaction with these NifA variants. Because the inhibition of NifA activity by NH4+ is mediated by PII and these NifA variants showed GlnB-independent activity, the activity of these NifA variants is also no longer subject to regulation in response to NH4+ (Table 3). However, because of the DRAT–DRAG regulatory system, the nitrogenase activity in these NifA variants is still regulated by NH4+ , and high nitrogenase activity was only seen in draT mutants with altered nifA under nitrogen-excess (MN) conditions (Table 3). This is quite different from a recent report that nitrogenase activity in some Rhodopseudomonas palustris nifA mutants is not regulated by NH4+, even though Rhodopseudomonas palustris appears to have draTG homologues (Rey et al., 2007). However, the DRAT–DRAG regulatory system has not been studied in Rhodopseudomonas palustris, and it is not known if it is actually functional.
Another important observation that we report here is that some other effector(s) could also activate NifA activity in a GlnB-independent manner. When cells were grown in MN− (NH4+ -free) medium, UR744 (ΔglnB mutant with a multicopy R. rubrum nifA) showed a nitrogenase activity similar to that seen in UR741 (wild-type with a multicopy R. rubrum nifA) (Table 4). One possibility is that some effector(s) could also interact with and activate NifA, similar to the role of GlnB–UMP interaction. The difference in NifA activation between MG- and MN−-growing cells suggests a dramatic change in the pools of some effector(s) under these different growth conditions. It is unknown whether the effector is a small molecule or protein. As mentioned in the Introduction, small molecules, such as ATP, ADP and α-KG, apparently play important roles in the regulation of PII function in E. coli, R. rubrum and other bacteria and archaea (Dodsworth & Leigh, 2006; Dodsworth et al., 2005; Forchhammer, 2004; Jiang & Ninfa, 2007; Johansson & Nordlund, 1997; Jonsson & Nordlund, 2007; Ninfa & Jiang, 2005; Wolfe et al., 2007; Zhu et al., 2006). These small molecules or other unidentified effector(s) might also be able to bind R. rubrum NifA to modulate its activity directly. It has been reported that α-KG binds the N-terminal GAF domain of A. vinelandii NifA to prevent its inactivation by NifL (Little & Dixon, 2003). NifA belongs to the AAA+ superfamily of ATPases (Zhang et al., 2002), so ATP is also a candidate effector. We also noticed previously that NH4+ addition causes a more rapid loss of nitrogenase activity in MN−-grown cells than in MG-grown cells (Zhang et al., 2000, 2005). These results suggest that changes in pools of these effector(s) might affect the DRAT–DRAG regulatory system, which is also regulated by PII (Huergo et al., 2006, 2007;Zhang et al., 2001b, 2003). Our unsuccessful attempt to use Tn5 to find other genes whose elimination allowed NifA to be activated in the absence of GlnB suggests that knockout mutations cannot alter the level of effector(s) significantly to cause the activation of NifA.
Previous studies indicated that the N-terminal GAF domain of NifA in A. brasilense and Herbaspirillum seropedicae plays an important role in regulation. The N-terminal GAF domain of A. brasilense NifA interacts directly with GlnB, as detected using a yeast two-hybrid system (Chen et al., 2005). We also found interaction between the N-terminal GAF domain of R. rubrum NifA and GlnB by this method (data not shown). Deletion of this GAF domain in A. brasilense and H. seropedicae NifA altered NifA regulation, and the truncated NifA of A. brasilense showed significant NifA activity in glnB mutant backgrounds that prevent wild-type NifA activation (Arsène et al., 1999). The activity of truncated NifA of H. seropedicae was no longer regulated by NH4+ (Souza et al., 1999). Several tyrosine residues in the N-terminal region of A. brasilense NifA have also been suggested to be involved in NifA regulation (Arsène et al., 1999; Chen et al., 2005). However, R. rubrum NifA with a Y29F substitution (Y29 is the only conserved tyrosine residue in the N-terminal domain of R. rubrum NifA) was inactive in ΔglnB and ΔglnD backgrounds (data not shown), indicating that this residue is unimportant in R. rubrum NifA regulation. When we constructed five different truncations of the GAF domain in R. rubrum NifA (residues Δ1–57, Δ1–106, ΔN– 172, Δ2–243, and Δ2–220) NifA activity was completely abolished, but we cannot rule out the possibility of indirect effects on protein stability (data not shown).
In summary, we have identified several substitution mutations in the GAF domain of R. rubrum NifA, allowing NifA to be activated in the absence of GlnB. The activities of two NifA variants, NifA-M173I and NifA-M173V, are no longer responsive to NHz4 . With the addition of the mutation in draT, which abolishes the post-translational regulation of nitrogenase activity, nifA draT double mutants were able to fix nitrogen and produce hydrogen under NH4+-excess conditions, and thus would be potentially useful for the production of biohydrogen.
Acknowledgments
This work was supported by the College of Agricultural and Life Sciences, University of Wisconsin–Madison, and National Institute of General Medical Sciences (NIGMS) grant GM65891 to G.P.R. and the grant from Chinese National Basic Research Program 2001CB108904 to J. L. We thank Philip James in Betty Craig's laboratory (University of Wisconsin–Madison) for the yeast strains and plasmids for the yeast two-hybrid system, and William W. Metcalf (University of Illinois–Urbana-Champaign) for generously providing the strain and plasmid for Tn5 mutagenesis. We also thank David Wolfe for helpful criticisms on the manuscript.
Abbreviations
- 3-AT
3-amino-1,2,4-triazole
- α-KG
α-ketoglutarate
References
- Adler SP, Purich D, Stadtman ER. Cascade control of Escherichia coli glutamine synthetase. Properties of the PII regulatory protein and the uridylyltransferase-uridylyl-removing enzyme. J Biol Chem. 1975;250:6264–6272. [PubMed] [Google Scholar]
- Araújo LM, Monteiro RA, Souza EM, Steffens MB, Rigo LU, Pedrosa FO, Chubatsu LS. GlnB is specifically required for Azospirillum brasilense NifA activity in Escherichia coli. Res Microbiol. 2004;155:491–495. doi: 10.1016/j.resmic.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Aravind L. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- Arcondéguy T, Jack R, Merrick M. PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol Mol Biol Rev. 2001;65:80–105. doi: 10.1128/MMBR.65.1.80-105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsene F, Kaminski PA, Elmerich C. Modulation of NifA activity by PII in Azospirillum brasilense: evidence for a regulatory role of the NifA N-terminal domain. J Bacteriol. 1996;178:4830–4838. doi: 10.1128/jb.178.16.4830-4838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsène F, Kaminski PA, Elmerich C. Control of Azospirillum brasilense NifA activity by PII: effect of replacing Tyr residues of the NifA N-terminal domain on NifA activity. FEMS Microbiol Lett. 1999;179:339–343. doi: 10.1111/j.1574-6968.1999.tb08747.x. [DOI] [PubMed] [Google Scholar]
- Blanco G, Drummond M, Woodley P, Kennedy C. Sequence and molecular analysis of the nifL gene of Azotobacter vinelandii. Mol Microbiol. 1993;9:869–879. doi: 10.1111/j.1365-2958.1993.tb01745.x. [DOI] [PubMed] [Google Scholar]
- Chen S, Liu L, Zhou X, Elmerich C, Li JL. Functional analysis of the GAF domain of NifA in Azospirillum brasilense: effects of Tyr→Phe mutations on NifA and its interaction with GlnB. Mol Genet Genomics. 2005;273:415–422. doi: 10.1007/s00438-005-1146-5. [DOI] [PubMed] [Google Scholar]
- Cheng J, Johansson M, Nordlund S. Expression of PII and glutamine synthetase is regulated by PII, the ntrBC products, and processing of the glnBA mRNA in Rhodospirillum rubrum. J Bacteriol. 1999;181:6530–6534. doi: 10.1128/jb.181.20.6530-6534.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau FM, Forchhammer K, Stülke J. Regulatory links between carbon and nitrogen metabolism. Curr Opin Microbiol. 2006;9:167–172. doi: 10.1016/j.mib.2006.01.001. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M, Paquelin A, Elmerich C. Functional organization of the glnB-glnA cluster of Azospirillum brasilense. J Bacteriol. 1993;175:2507–2515. doi: 10.1128/jb.175.9.2507-2515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zamaroczy M, Paquelin A, Peltre G, Forchhammer K, Elmerich C. Coexistence of two structurally similar but functionally different PII proteins in Azospirillum brasilense. J Bacteriol. 1996;178:4143–4149. doi: 10.1128/jb.178.14.4143-4149.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoues N, Lin M, Guo X, Ma L, Carreno-Lopez R, Elmerich C. Nitrogen fixation genetics and regulation in a Pseudomonas stutzeri strain associated with rice. Microbiology. 2003;149:2251–2262. doi: 10.1099/mic.0.26270-0. [DOI] [PubMed] [Google Scholar]
- Dixon R, Kahn D. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol. 2004;2:621–631. doi: 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- Dodsworth JA, Leigh JA. Regulation of nitrogenase by 2-oxoglutarate-reversible, direct binding of a PII-like nitrogen sensor protein to dinitrogenase. Proc Natl Acad Sci U S A. 2006;103:9779–9784. doi: 10.1073/pnas.0602278103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodsworth JA, Cady NC, Leigh JA. 2-Oxoglutarate and the PII homologues NifI1 and NifI2 regulate nitrogenase activity in cell extracts of Methanococcus maripaludis. Mol Microbiol. 2005;56:1527–1538. doi: 10.1111/j.1365-2958.2005.04621.x. [DOI] [PubMed] [Google Scholar]
- Egener T, Sarkar A, Martin DE, Reinhold-Hurek B. Identification of a NifL-like protein in a diazotroph of the β-subgroup of the Proteobacteria, Azoarcus sp. strain BH72. Microbiology. 2002;148:3203–3212. doi: 10.1099/00221287-148-10-3203. [DOI] [PubMed] [Google Scholar]
- Fields S. The two-hybrid system to detect protein–protein interactions. Methods: A Companion to Meth Enzymol. 1993;5:116–124. [Google Scholar]
- Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzmaurice WP, Saari LL, Lowery RG, Ludden PW, Roberts GP. Genes coding for the reversible ADP-ribosylation system of dinitrogenase reductase from Rhodospirillum rubrum. Mol Gen Genet. 1989;218:340–347. doi: 10.1007/BF00331287. [DOI] [PubMed] [Google Scholar]
- Forchhammer K. Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol Rev. 2004;28:319–333. doi: 10.1016/j.femsre.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- He L, Soupene E, Ninfa A, Kustu S. Physiological role for the GlnK protein of enteric bacteria: relief of NifL inhibition under nitrogen-limiting conditions. J Bacteriol. 1998;180:6661–6667. doi: 10.1128/jb.180.24.6661-6667.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huergo LF, Chubatsu LS, Souza EM, Pedrosa FO, Steffens MBR, Merrick M. Interactions between PII proteins and the nitrogenase regulatory enzymes DraT and DraG in Azospirillum brasilense. FEBS Lett. 2006;580:5232–5236. doi: 10.1016/j.febslet.2006.08.054. [DOI] [PubMed] [Google Scholar]
- Huergo LF, Merrick M, Pedrosa FO, Chubatsu LS, Araujo MS, Souza EM. Ternary complex formation between AmtB, GlnZ and the nitrogenase regulatory enzyme DraG reveals a novel facet of nitrogen regulation in bacteria. Mol Microbiol. 2007;66:1523–1535. doi: 10.1111/j.1365-2958.2007.06016.x. [DOI] [PubMed] [Google Scholar]
- Jack R, De Zamaroczy M, Merrick M. The signal transduction protein GlnK is required for NifL-dependent nitrogen control of nif gene expression in Klebsiella pneumoniae. J Bacteriol. 1999;181:1156–1162. doi: 10.1128/jb.181.4.1156-1162.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Ninfa AJ. Escherichia coli PII signal transduction protein controlling nitrogen assimilation acts as a sensor of adenylate energy charge in vitro. Biochemistry. 2007;46:12979–12996. doi: 10.1021/bi701062t. [DOI] [PubMed] [Google Scholar]
- Jiang P, Peliska JA, Ninfa AJ. Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry. 1998;37:12782–12794. doi: 10.1021/bi980667m. [DOI] [PubMed] [Google Scholar]
- Johansson M, Nordlund S. Uridylylation of the PII protein in the photosynthetic bacterium Rhodospirillum rubrum. J Bacteriol. 1997;179:4190–4194. doi: 10.1128/jb.179.13.4190-4194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson A, Nordlund S. In vitro studies of the uridylylation of the three PII protein paralogs from Rhodospirillum rubrum: the transferase activity of R. rubrum GlnD is regulated by α-ketoglutarate and divalent cations but not by glutamine. J Bacteriol. 2007;189:3471–3478. doi: 10.1128/JB.01704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamberov ES, Atkinson MR, Chandran P, Ninfa AJ. Effect of mutations in Escherichia coli glnL (ntrB), encoding nitrogen regulator II (NRII or NtrB), on the phosphatase activity involved in bacterial nitrogen regulation. J Biol Chem. 1994;269:28294–28299. [PubMed] [Google Scholar]
- Klassen G, de Souza EM, Yates MG, Rigo LU, Inaba J, Pedrosa FO. Control of nitrogenase reactivation by the GlnZ protein in Azospirillum brasilense. J Bacteriol. 2001;183:6710–6713. doi: 10.1128/JB.183.22.6710-6713.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen G, Souza EM, Yates MG, Rigo LU, Costa RM, Inaba J, Pedrosa FO. Nitrogenase switch-off by ammonium ions in Azospirillum brasilense requires the GlnB nitrogen signal-transducing protein. Appl Environ Microbiol. 2005;71:5637–5641. doi: 10.1128/AEM.71.9.5637-5641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen RA, Wilson MM, Guss AM, Metcalf WW. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol. 2002;178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- Lehman LJ, Roberts GP. Identification of an alternative nitrogenase system in Rhodospirillum rubrum. J Bacteriol. 1991;173:5705–5711. doi: 10.1128/jb.173.18.5705-5711.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JA, Dodsworth JA. Nitrogen regulation in bacteria and archaea. Annu Rev Microbiol. 2007;61:349–377. doi: 10.1146/annurev.micro.61.080706.093409. [DOI] [PubMed] [Google Scholar]
- Liang JH, Nielsen GM, Lies DP, Burris RH, Roberts GP, Ludden PW. Mutations in the draT and draG genes of Rhodospirillum rubrum result in loss of regulation of nitrogenase by reversible ADP-ribosylation. J Bacteriol. 1991;173:6903–6909. doi: 10.1128/jb.173.21.6903-6909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YY, de Zamaroczy M, Arséne F, Paquelin A, Elmerich C. Regulation of nitrogen fixation in Azospirillum brasilense Sp7: involvement of nifA, glnA and glnB gene products. FEMS Microbiol Lett. 1992;79:113–119. doi: 10.1111/j.1574-6968.1992.tb14028.x. [DOI] [PubMed] [Google Scholar]
- Liang YY, Arséne F, Elmerich C. Characterization of the ntrBC genes of Azospirillum brasilense Sp7: their involvement in the regulation of nitrogenase synthesis and activity. Mol Gen Genet. 1993;240:188–196. doi: 10.1007/BF00277056. [DOI] [PubMed] [Google Scholar]
- Lies DP. Genetic manipulation and the overexpression analysis of posttranslational nitrogen fixation regulation in Rhodospirillum rubrum PhD thesis. University of Wisconsin; Madison: 1994. [Google Scholar]
- Little R, Dixon R. The amino-terminal GAF domain of Azotobacter vinelandii NifA binds 2-oxoglutarate to resist inhibition by NifL under nitrogen-limiting conditions. J Biol Chem. 2003;278:28711–28718. doi: 10.1074/jbc.M301992200. [DOI] [PubMed] [Google Scholar]
- Little R, Reyes-Ramirez F, Zhang Y, van Heeswijk WC, Dixon R. Signal transduction to the Azotobacter vinelandii NIFL–NIFA regulatory system is influenced directly by interaction with 2-oxoglutarate and the PII regulatory protein. EMBO J. 2000;19:6041–6050. doi: 10.1093/emboj/19.22.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R, Colombo V, Leech A, Dixon R. Direct interaction of the NifL regulatory protein with the GlnK signal transducer enables the Azotobacter vinelandii NifL–NifA regulatory system to respond to conditions replete for nitrogen. J Biol Chem. 2002;277:15472–15481. doi: 10.1074/jbc.M112262200. [DOI] [PubMed] [Google Scholar]
- Martinez SE, Beavo JA, Hol WJG. GAF domains: two-billion-year-old molecular switches that bind cyclic nucleotides. Mol Interv. 2002;2:317–323. doi: 10.1124/mi.2.5.317. [DOI] [PubMed] [Google Scholar]
- Martinez-Argudo I, Little R, Dixon R. Role of the amino-terminal GAF domain of the NifA activator in controlling the response to the antiactivator protein NifL. Mol Microbiol. 2004a;52:1731–1744. doi: 10.1111/j.1365-2958.2004.04089.x. [DOI] [PubMed] [Google Scholar]
- Martinez-Argudo I, Little R, Shearer N, Johnson P, Dixon R. The NifL–NifA system: a multidomain transcriptional regulatory complex that integrates environmental signals. J Bacteriol. 2004b;186:601–610. doi: 10.1128/JB.186.3.601-610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick MJ, Edwards RA. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin SD, Austin S, Dixon RA. The role of activator binding sites in transcriptional control of the divergently transcribed nifF and nifLA promoters from Klebsiella pneumoniae. Mol Microbiol. 1988;2:433–442. doi: 10.1111/j.1365-2958.1988.tb00049.x. [DOI] [PubMed] [Google Scholar]
- Morett E, Segovia L. The σ54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morett E, Cannon W, Buck M. The DNA-binding domain of the transcriptional activator protein NifA resides in its carboxy terminus, recognises the upstream activator sequences of nif promoters and can be separated from the positive control function of NifA. Nucleic Acids Res. 1988;16:11469–11488. doi: 10.1093/nar/16.24.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa AJ, Atkinson MR. PII signal transduction proteins. Trends Microbiol. 2000;8:172–179. doi: 10.1016/s0966-842x(00)01709-1. [DOI] [PubMed] [Google Scholar]
- Ninfa AJ, Jiang P. PII signal transduction proteins: sensors of α-ketoglutarate that regulate nitrogen metabolism. Curr Opin Microbiol. 2005;8:168–173. doi: 10.1016/j.mib.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Nordlund S, Ludden PW. Post-translational regulation of nitrogenase in photosynthetic bacteria. In: Klipp W, Masepohl B, Gallon JR, Newton WE, editors. Genetics and Regulation of Nitrogen Fixation in Free-Living Bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2004. pp. 175–196. [Google Scholar]
- Porter SC, North AK, Kustu S. Mechanism of transcriptional activation by NtrC. In: Hoch JA, Silhavy TJ, editors. Two-Component Signal Transduction. Washington, DC: American Society for Microbiology; 1995. pp. 147–158. [Google Scholar]
- Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- Rey FE, Heiniger EK, Harwood CS. Redirection of metabolism for biological hydrogen production. Appl Environ Microbiol. 2007;73:1665–1671. doi: 10.1128/AEM.02565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Ramirez F, Little R, Dixon R. Role of Escherichia coli nitrogen regulatory genes in the nitrogen response of the Azotobacter vinelandii NifL–NifA complex. J Bacteriol. 2001;183:3076–3082. doi: 10.1128/JB.183.10.3076-3082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Siddavattam D, Steibl HD, Kreutzer R, Klingmuller W. Regulation of nif gene expression in Enterobacter agglomerans: nucleotide sequence of the nifLA operon and influence of temperature and ammonium on its transcription. Mol Gen Genet. 1995;249:629–636. doi: 10.1007/BF00418032. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer UB, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- Sonnhammer ELL, Eddy SR, Birney E, Bateman A, Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza EM, Pedrosa FO, Drummond M, Rigo LU, Yates MG. Control of Herbaspirillum seropedicae NifA activity by ammonium ions and oxygen. J Bacteriol. 1999;181:681–684. doi: 10.1128/jb.181.2.681-684.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stips J, Thummer R, Neumann M, Schmitz RA. GlnK effects complex formation between NifA and NifL in Klebsiella pneumoniae. Eur J Biochem. 2004;271:3379–3388. doi: 10.1111/j.1432-1033.2004.04272.x. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Studholme DJ, Dixon R. Domain architectures of σ54-dependent transcriptional activators. J Bacteriol. 2003;185:1757–1767. doi: 10.1128/JB.185.6.1757-1767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Franke CC, Nordlund S, Norén A. Reversible membrane association of dinitrogenase reductase activating glycohydro-lase in the regulation of nitrogenase activity in Rhodospirillum rubrum; dependence on GlnJ and AmtB1. FEMS Microbiol Lett. 2005;253:273–279. doi: 10.1016/j.femsle.2005.09.049. [DOI] [PubMed] [Google Scholar]
- Weiss DS, Batut J, Klose KE, Keener J, Kustu S. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- Wolfe DM, Zhang Y, Roberts GP. Specificity and regulation of interaction between the PII and AmtB1 proteins in Rhodospirillum rubrum. J Bacteriol. 2007;189:6861–6869. doi: 10.1128/JB.00759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Burris RH, Ludden PW, Roberts GP. Posttranslational regulation of nitrogenase activity by anaerobiosis and ammonium in Azospirillum brasilense. J Bacteriol. 1993;175:6781–6788. doi: 10.1128/jb.175.21.6781-6788.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Burris RH, Ludden PW, Roberts GP. Comparison studies of dinitrogenase reductase ADP-ribosyl transferase/dinitrogenase reductase activating glycohydrolase regulatory systems in Rhodospirillum rubrum and Azospirillum brasilense. J Bacteriol. 1995a;177:2354–2359. doi: 10.1128/jb.177.9.2354-2359.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cummings AD, Burris RH, Ludden PW, Roberts GP. Effect of an ntrBC mutation on the posttranslational regulation of nitrogenase activity in Rhodospirillum rubrum. J Bacteriol. 1995b;177:5322–5326. doi: 10.1128/jb.177.18.5322-5326.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Burris RH, Ludden PW, Roberts GP. Regulation of nitrogen fixation in Azospirillum brasilense. FEMS Microbiol Lett. 1997;152:195–204. doi: 10.1111/j.1574-6968.1997.tb10428.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Pohlmann EL, Ludden PW, Roberts GP. Mutagenesis and functional characterization of the glnB, glnA, and nifA genes from the photosynthetic bacterium Rhodospirillum rubrum. J Bacteriol. 2000;182:983–992. doi: 10.1128/jb.182.4.983-992.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pohlmann EL, Ludden PW, Roberts GP. Functional characterization of three GlnB homologs in the photosynthetic bacterium Rhodospirillum rubrum: roles in sensing ammonium and energy status. J Bacteriol. 2001a;183:6159–6168. doi: 10.1128/JB.183.21.6159-6168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pohlmann EL, Halbleib CM, Ludden PW, Roberts GP. Effect of PII and its homolog GlnK on reversible ADP-ribosylation of dinitrogenase reductase by heterologous expression of the Rhodospirillum rubrum dinitrogenase reductase ADP-ribosyl transferase-dinitrogenase reductase-activating glycohydrolase regulatory system in Klebsiella pneumoniae. J Bacteriol. 2001b;183:1610–1620. doi: 10.1128/JB.183.5.1610-1620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chaney M, Wigneshweraraj SR, Schumacher J, Bordes P, Cannon W, Buck M. Mechanochemical ATPases and transcriptional activation. Mol Microbiol. 2002;45:895–903. doi: 10.1046/j.1365-2958.2002.03065.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Pohlmann EL, Ludden PW, Roberts GP. Regulation of nitrogen fixation by multiple PII homologs in the photosynthetic bacterium Rhodospirillum rubrum. Symbiosis. 2003;35:85–100. [Google Scholar]
- Zhang Y, Pohlmann EL, Roberts GP. Identification of critical residues in GlnB for its activation of NifA activity in the photosynthetic bacterium Rhodospirillum rubrum. Proc Natl Acad Sci U S A. 2004;101:2782–2787. doi: 10.1073/pnas.0306763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pohlmann EL, Roberts GP. GlnD is essential for NifA activation, NtrB/NtrC-regulated gene expression, and posttranslational regulation of nitrogenase activity in the photosynthetic, nitrogen-fixing bacterium Rhodospirillum rubrum. J Bacteriol. 2005;187:1254–1265. doi: 10.1128/JB.187.4.1254-1265.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wolfe DM, Pohlmann EL, Conrad MC, Roberts GP. Effect of AmtB homologs on the posttranslational regulation of nitrogenase activity in response to ammonium and energy signals in Rhodospirillum rubrum. Microbiology. 2006;152:2075–2089. doi: 10.1099/mic.0.28903-0. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Conrad MC, Zhang Y, Roberts GP. Identification of Rhodospirillum rubrum GlnB variants that are altered in their ability to interact with different targets in response to nitrogen-status signals. J Bacteriol. 2006;188:1866–1874. doi: 10.1128/JB.188.5.1866-1874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


