Abstract
Background
Neonatal infections annually claim lives of 1.4 million neonates worldwide. Until now, there is no ideal diagnostic test for detecting sepsis and thus management of possible sepsis cases often depends on clinical algorithm leading to empirical treatment. This often results in unnecessary antibiotic use, which may lead to emergence of antibiotic resistance. Biomarkers have shown great promise in diagnosis of sepsis and guiding appropriate treatment of neonates. In this study, we conducted a literature review of existing biomarkers to analyze their status for use as a point-of-care diagnostic in developing countries.
Methods
PubMed and EMBASE database were searched with keywords, ‘infections’, ‘neonates’, and ‘biomarkers’ to retrieve potentially relevant papers from the period 1980 to 2010. Leading hospitals and manufacturers were communicated to inquire about the cost, laboratory requirements and current standing of biomarkers in clinical use.
Results
The search returned 6407 papers on biomarkers; 65 were selected after applying inclusion and exclusion criteria. Among the studies, C-reactive protein (CRP), procalcitonin (PCT) and interleukin 6 (IL-6) were the most widely studied biomarkers and were considered to be most promising for diagnosing neonatal infections. About 90% of the studies were from developed countries; more than 50% were from Europe.
Conclusions
Extensive work is being performed to find the diagnostic and prognostic value of biomarkers. However, the methodologies and study design are highly variable. Despite numerous research papers on biomarkers, their use in clinical setting is limited to CRP. The methods for detection of biomarkers are far too advanced to be used at the community level where most of the babies are dying. It is important that a harmonized multi-site study is initiated to find a battery of biomarkers for diagnosis of neonatal infections.
Most developing countries have witnessed substantial declines in mortality among children <5 years of age (1,2). In contrast, neonatal mortality has remained relatively constant, with an estimated 3.6 million annual neonatal deaths globally (2-5). Neonatal mortality now accounts for about 40%-50% of under-five child deaths (4-6). More than 90% of these deaths occur in the poorest countries of Asia and Africa (7). Suspected infections, including sepsis, pneumonia and meningitis (hereafter referred to as “infections”) account for an estimated 1.4 million neonatal deaths worldwide every year (5,6).
Low and middle income countries are trying different modalities to achieve MDG4 by 2015. The common intervention is community-based diagnosis of possible sepsis cases, using clinical algorithms and treatments with empirical antibiotics. Highly sensitive algorithms based therapies have performed well in reducing child mortality, irrespective of the antibiotic therapy used (6,8). However, blood culture, as the gold standard for diagnosis, from these algorithm-positive cases yielded bacterial isolates only in 5%-10% of cases. This jeopardized the credibility of the “gold” standard. In recent years, with the advancement of these techniques like real time polymerise chain reaction (RT-PCR) for specific genome and broad range targets, the use of molecular approaches has become common for aetiological diagnosis (9). Although a recent meta-analysis showed that the molecular tests cannot increase the detection frequency of aetiology more than what blood culture already captures (9). Hence it is becoming increasingly important to find a tool to differentiate sick newborns with or without infection, especially to minimize the indiscriminate use of antibiotics. In the last few years, biomarkers, triggered by the host immune system in response to infections, have been targeted as potential indicator for diagnostic and prognostic purposes.
This study was taken up to conduct a structured literature overview on the existing biomarkers for diagnosis of neonatal infections/sepsis and to elucidate their relative potential to be used in resource-poor settings. In addition, the study also investigated the instrumental requirements for detection of biomarkers and the extent of their use in clinical practice.
METHODS
Selection of biomarkers for analysis
After a preliminary examination of the available literature, we consolidated the list of biomarkers for further review. These markers were selected based on the number of papers published on the topic and their potential to be used for diagnosis and prognosis of neonatal infection. Biomarkers included in this analysis are as follows:
Acute phase proteins: C– reactive protein (CRP), procalcitonin (PCT);
Cytokines: interleukin 6 (IL-6), interleukin 8 (IL-8), interferon – gamma (IFN-γ), tumor necrosis factor – alpha (TNF-α);
Cell surface antigens: CD 64, soluble intercellular adhesion molecule (sICAM).
Search strategies
In order to carry out a landscape analysis to identify studies on the diagnostic performance of the aforementioned biomarkers, we searched PubMed and EMBASE bibliography databases. Search strategies for both databases were carefully built to maximize the sensitivity of our search. A combination of text words and subject heading terms specific to each database (MeSH terms for PubMed and EMTREE terms for EMBASE) were used to develop the search strategy (Table 1).
Table 1.
Search strategy, restricted to age (newborn), subject (humans) and time period (January 1980 to April 2010)
| EMBASE: |
('newborn'/exp OR newborn OR 'newborn'/syn OR 'newborns':ab,ti OR 'neonates':ab,ti OR 'infants':ab,ti) AND ('infection'/exp OR infection OR 'infection'/syn OR 'infections' OR 'sepsis'/exp OR sepsis OR 'sepsis'/syn OR 'bacterial infection'/exp OR 'bacterial infection' OR 'infections':ab,ti OR 'bacterial infection'/syn OR 'bacteremia'/exp OR bacteremia OR 'bacteremia'/syn OR 'septicemia'/exp OR septicemia OR 'septicemia'/syn OR 'systemic inflammatory response syndrome'/syn OR 'systemic inflammatory response syndrome'/exp OR 'systemic inflammatory response syndrome' OR 'meningitis'/exp OR 'meningitis' OR 'meningitis'/syn) AND ('c reactive protein'/exp OR 'c reactive protein' OR 'c reactive protein'/syn OR 'procalcitonin'/exp OR procalcitonin OR 'pct':ab,ti OR 'tumor necrosis factor alpha'/exp OR 'tumor necrosis factor alpha' OR 'tumor necrosis factor alpha'/syn OR 'tnf alpha' OR 'tnf-alpha':ab,ti OR 'gamma interferon'/exp OR 'gamma interferon' OR 'gamma interferon/syn' OR 'ifn-gamma':ab,ti OR 'ifn gamma':ab,ti OR 'intercellular adhesion molecule 1'/exp OR 'intercellular adhesion molecule 1' OR 'intercellular adhesion molecule 1'/syn OR 'icam 1':ab,ti OR 'cd64 antigen'/exp OR 'cd64 antigen' OR 'cd64 antigen'/syn OR 'cd64':ab,ti OR 'interleukin 6'/exp OR 'interleukin 6' OR 'interleukin 6'/syn OR 'il 6':ab,ti OR 'il-6':ab,ti OR 'interleukin 8'/exp OR 'interleukin 8' OR 'interleukin 8'/syn OR 'il 8':ab,ti OR 'il-8':ab,ti) AND ('diagnosis'/exp OR diagnosis OR 'diagnosis'/syn OR 'biological marker'/exp OR 'biological marker' OR 'biological marker'/syn OR markers:ab,ti OR biomarkers:ab,ti OR test:ab,ti OR tests:ab,ti OR indicators:ab,ti) |
| PubMed/Medline: | (neonat* [tw] OR newborn [mh] OR newborn [tw] OR newborns [tw] OR neonate [tw] OR neonates [tw] OR baby [tw] OR babies [tw] OR infant [tw] OR Infants [tw]) AND (“sepsis” [mh] OR sepsis [tw] OR “bacterial Infections” [mh] OR (septic [tw] AND shock [tw]) OR “systemic inflammatory response syndrome” [mh] OR “systemic inflammatory response syndrome” [tw] OR infection [tw] OR infections [tw] OR bacteremia [tw] OR bacteraemia [tw] OR bacteremias [tw] OR bacteraemias [tw] OR septicemia [tw] OR septicemias [tw] OR septicaemia [tw] OR septicaemias [tw] OR bacteremic [tw] OR bacteraemic [tw] OR bacterial [tw] OR viremia [tw] OR viremias [tw] OR viraemia [tw] OR viraemias [tw] OR Viremic [tw] OR viraemic [tw] OR fungemic [tw] OR fungemia [tw] OR fungemias [tw]) AND ((“Diagnosis” [mh] AND (markers [tw] OR marker [tw])) OR markers [tw] OR marker [tw] OR “biological markers” [mh] OR biomarker [tw] OR biomarkers [tw] OR (“sensitivity and specifity” [mh] AND (sensitivity [tw] OR specificity[tw]))) |
The search strategy also adapted individual biomarker specific final queries and ran the search to ensure retrieval of maximum papers. A total of 4868 citations from PubMed and 1539 citations from EMBASE were retrieved. These references were imported into separate libraries using the EndNote software (Thomson Reuters, Philadelphia, PA, USA). The libraries were later merged, and the duplicates were removed. Two reviewers independently screened the titles and abstracts of the retrieved citations to find the articles that were deemed relevant.
Inclusion criteria
For inclusion, the abstract and titles were screened based on the following predetermined criteria: i) the subject population is newborns, ii) the subjects are with culture proved sepsis or suspected infection based on clinical algorithm and iii) the article evaluated any of the proposed biomarkers for diagnosis and/or prognosis of neonatal infections.
The exhaustive search based on the titles and abstracts returned a broad spectrum of infection related studies from which only the cases of sepsis, urinary tract infection, meningitis, pneumonia, respiratory tract infections and umbilical cord infections were considered.
Finally, full text articles with following criteria were included for analysis: i) the age of the newborns ranged from 0 days to 59 days and ii) diagnostic performance of target biomarkers are explored in clinical and/or in culture-confirmed cases of sepsis.
Exclusion criteria
It was challenging to select the relevant articles for this analysis from the large number of papers retrieved (n=6407) based on the above mentioned selection criteria. To make a comprehensive list of appropriate papers, we excluded the articles that dealt with malaria, HIV infection, hepatitis, toxoplasmosis, gestational diabetes, bronchopulmonary dysplasia, antenal and maternofetal studies, in-vitro studies, transplant immunology studies, polymorphisms, necrotizing enterocolitis, foreign languages other than English, letters, comments and editorials and other non-research publication types.
Data extraction
Available full papers were downloaded from PubMed, EMBASE and HINARI sources. Requests for reprints were sent to the authors of the papers which were not available from these sources. Data were extracted and compiled in Excel spreadsheet with the following column headings: Name of Biomarker, Study Title, First Author, Year, Setting, Country, Clinical Characteristics, Sample Size, Age, Specimen source, Method, Cut off, Cost, Sensitivity, Specificity, Positive Predictive Value (PPV), and Negative Predictive Value (NPV).
For a point of reference, we contacted the leading hospitals of several developed and developing countries to learn what tests/biomarkers are currently being used at their clinical settings. We also contacted major diagnostics manufacturers to inquire about the direct costs and laboratory requirements for assaying each of the biomarkers.
RESULTS
Out of 705 potentially relevant papers, 65 were selected for final review after exclusion of 640 papers for the lack of sufficient information, ambiguity in study design and patient characteristics, failure to obtain full-text article or absence of other inclusion criteria (Figure 1).
Figure 1.
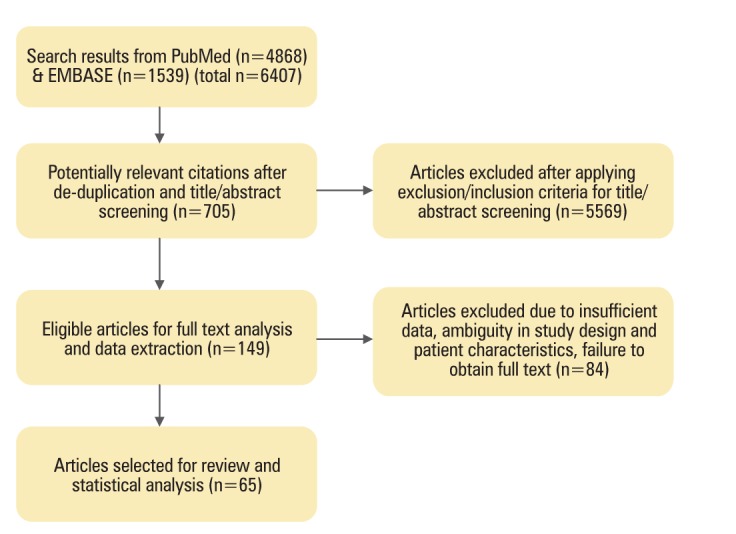
Search strategy and identified articles.
Prevalence of research on biomarkers
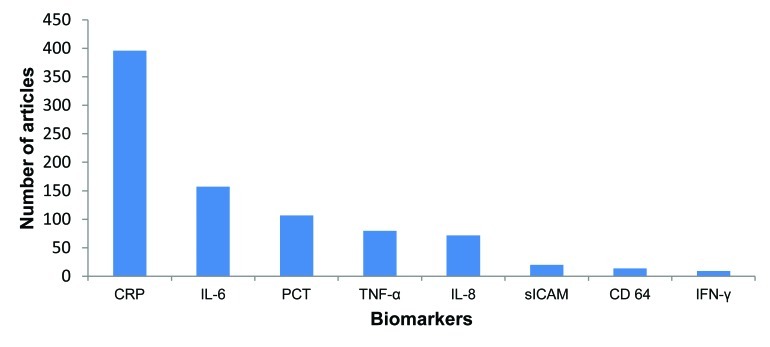
Review of relevant papers, published during January1980 to April 2010, revealed that CRP was the most extensively studied biomarker (n=396), followed by IL-6 (n=157), PCT (n=107), TNF-alpha (n=80), IL-8 (n=72), sICAM (n=20), CD64 (n=14), IFN-γ (n=9) (Figure 2). However, even the less frequent markers showed promise in respect of their ability to differentiate the sepsis from non-sepsis cases.
Figure 2.
Distribution of studies according to biomarkers studies. CRP – C-reactive protein, IL – interleukin, TNF – tumor necrosis factor, sICAM (soluble intercellular adhesion molecule, IFN – interferon.
Heterogeneity of the studies
Biomarker research studies widely differed by study groups in respect to their inclusion criteria for patients, case definition, test methodologies and cut off values for markers (Table 2). The range of cut off value was as wide as 0.2 to 95 mg/mL for CRP, 0.34 to 100 ng/mL for PCT, 3.6 to 500 pg/mL for IL-6; and 1 to 1000 pg/mL for IL-8. Accordingly, sensitivity and specificity of the tests also varied widely among the studies. For convenience of interpretation of sensitivity and specificity, we divided the studies in smaller subgroups based on their cut off values used by the study groups (Table 2).
Table 2.
Sensitivity, specificity and cut-off values of biomarkers in reviewed studies*
| Biomarker | Cut-off range | Cut-off sub-ranges | Percentage of papers | Sensitivity ranges (%) | Specificity ranges (%) | Cut-off | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|
|
CRP |
0.2–95 mg/L |
0.2–10
11–30
31–95 |
70
15
15 |
41–96
33–56
23–87 |
72–100
74–96
48–98 |
mean=17.1
median=10 |
mean=66.53
median=69 (IQR=26.3) |
mean=86.14
median=90 (IQR=13.9) |
|
PCT |
0.34 –100 ng/mL |
0.34–1.0
2–10
11–100 |
48
39
13 |
58–100
59–95
21–95 |
50–100
50–100
87–100 |
mean=8.92
median=1.17 |
mean=77.93
median=8 (IQR=14.8) |
mean=81.84
median=82.5 (IQR=16.6) |
|
IL-6 |
3.6–500 pg/mL |
2–10
11–30
31–100
101–500 |
15
34
31
20 |
88–96
61–90
57–100
74–97 |
66–89
56–90
43–100
70–100 |
mean=76.49
median=30 |
mean=77.87
median=80 (IQR=29.9) |
mean=78.61
median=78 (IQR=18.9) |
|
IL-8 |
1–1000 pg/mL |
0.6–100
101–1000 |
74
26 |
34–92
36–92 |
52–96
65–96 |
mean=220.53
median=70 |
mean=72.48
median=80 (IQR=15.5) |
mean=80.57
median=82 (IQR=21.7) |
|
CD64 |
different units used |
– |
– |
79–100 |
81–96.8 |
not analyzable |
mean=82.42
median=92 (IQR=17) |
mean=82.79
median=88 (IQR=15) |
|
sICAM |
250–300 µg/L |
– |
– |
78–80 |
61–90 |
mean=275
median=275 |
mean=79
median=79 (IQR=1) |
mean=75.5
median=75.5 (IQR=14.5) |
| TNF-α | 1.7–70 pg/mL | – | – | 54–100 | 43–96.6 | mean=18.94 median=7.5 | mean=78.72 median=80.4 (IQR=22.7) | mean=81.4 median=93 (IQR=14.9) |
CRP – C-reactive protein, IL – interleukin, TNF – tumor necrosis factor, sICAM (soluble intercellular adhesion molecule, IFN – interferon, IQR – interquartile range.
*Reported in refs 13-77.
Characteristics of biomarkers
The mean cut-off point of CRP was 17 mg/L, with 66% sensitivity and 86% specificity. PCT appeared to be a more relevant marker than CRP for diagnosing bacterial sepsis at earlier stages, with the mean sensitivity of 77.93%, specificity of 81.84%, and a cut-off of 8.92 ng/mL. For IL-6, the mean sensitivity at “zero hour” was 77.87%, specificity was 78.61%, and the mean cut- off value was 76.49 pg/mL (Table 2). The mean value of cut-off for IL-8 was 220.53 pg/mL, sensitivity 72.48% and specificity 80.57% (Table 2).
CRP and PCT have been extensively studied and compared for their efficacy to diagnose sepsis cases in young infants. Overall, the studies reported that the optimum sensitivity and specificity for CRP was obtained during the window of 24-48 hours after the onset of symptoms. On the other hand, PCT was sensitive enough to detect the cases much earlier than CRP. However, some studies also suggested that serial measurements of CRP over a period of 2-3 days after onset clinical symptom, using varying cut-off values, improved the diagnostic performance of CRP (10,11).
CD64 demonstrated the mean sensitivity of 92% and a specificity of 82.79% during the first 24 hours of infection. sICAM yielded the mean sensitivity of 79% and specificity of 75.5%. Finally, the mean sensitivity of TNF-α was 78.72%, and the specificity was 81.4% (Table 2). Unfortunately, we could not find any analyzable data for IFN-γ from any of the relevant studies.
Minimum laboratory requirements and cost analysis
According to retrieved studies, the major techniques for detecting biomarkers were immunoassays, and cell sorting for CD64. The immunoassays were usually accompanied by a variety of readers to quantify the level of specific markers. Most of the studies used enzyme-linked immunosorbent assay (ELISA) readers for quantification along with other tests like immunoturbidimetric and nephelometric assays. These methods, except ELISA, were rarely available at the resource-poor settings.
The detection method for CRP as the extensively studied and widely used marker was also immunoassay, usually using an ELISA reader. In resource-poor settings, qualitative or semiquantitative latex agglutination test is also used for the detection of CRP. Other immunoassays are only available at the tertiary level and/or private commercial facilities in low income countries. Some studies used immunoluminometry and chemiluminescence for the detection of PCT. Immunochromatographic tests (ICT) was also reported for PCT in a few studies, but these were still at the developmental stage and only at research level (12). Flow cytometry was invariably used by all the studies for detection of cell surface antigen CD64 and the literature revealed very little information about the cost associated with these techniques (13-77).
Global distribution of biomarker research
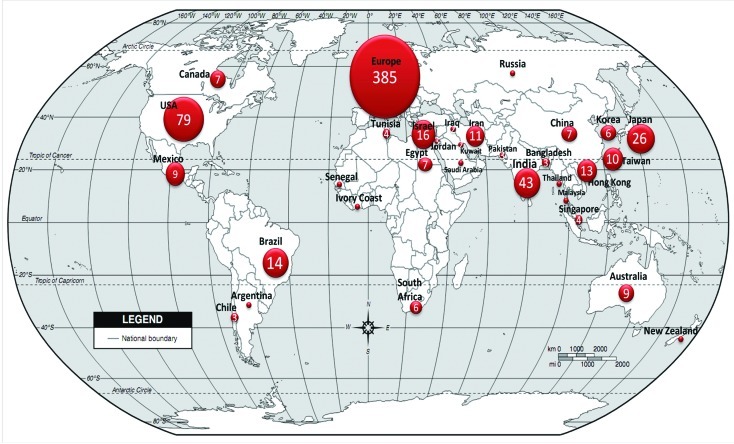
We tried to establish the geographical distribution of biomarker research based on retrieved studies. Research on biomarkers was mostly confined to the developed countries with a large share of the total numbers of articles (385/705, 55%) from Europe, followed by North America (95/705, 13%). In contrast, only 9.5% (68/705) papers were from South Asia and Africa (Figure 3); the regions where more than a third of neonatal deaths occur.
Figure 3.
Geographical distribution of published research on biomarkers in the last three decades (January 1980 to April 2010).
DISCUSSION
Biomarkers may have great potentials for diagnosis of neonatal sepsis and have been studied for more than two decades. The frontiers of research in biomarkers as diagnostic tools for detecting neonatal sepsis have progressed considerably over the years and persist in advancing novel technologies. This review suggests that many newer biomarkers have come into play, and thorough investigations on them are in progress.
Among the numerous biomarkers in the field of neonatal sepsis diagnosis, this review identified 8 predominant markers, as determined by number of publications: CRP, PCT, IL-6, IL-8, IFN-γ, TNF-α, CD64 and sICAM. Of these, CRP was the most widely used diagnostic and prognostic marker. Despite its limitation due to late appearance and persistence for relatively longer period (76), it was used as the standard marker to measure the potential and efficacy of newer biomarkers. Among other markers, PCT has come up as more promising, with the comparative advantage of early detection in sepsis and quick reduction in its levels in response to appropriate therapy (72). PCT also has the additional advantage of being specifically responsive to bacterial infections and not viral (73). On the other hand, IFN-γ seems to be particularly responsive to viral infection at very early stage of infection (74).
Based on the available data about the detection time, the markers could be classified into three groups; early phase (IL-6, IL-8, CD64, sICAM, TNF-α and IFN-γ), mid phase (PCT) and late phase (CRP). The unique dynamics of appearance and disappearance of specific markers would be useful for possible multiplexing to capture the neonatal infection cases irrespective of their disease status.
Other biomarkers have also showed promise, and most of them revealed potential for detection of sepsis at very early stage of the disease. IL-6 demonstrated a high potential with the ability to detect the cases at very early stage of infection and monitor the appropriateness of therapy, based on its characteristic early appearance and short half life (75). Newer markers like sICAM and CD64 also have the potential to detect sepsis cases at very early stage of disease, with high sensitivity but compromised specificity (57-62).
With all possible and definite potentials of biomarkers, none of them is currently in use for patient care, except CRP. The review identified several reasons for this slow transition of biomarkers from the research laboratories to their real-life use in clinical care. The main cause of this hindrance is the heterogeneity between the research protocols used by different groups. The study designs are heterogenic with respect to cut off values used to define positivity, which sometimes varied by about 100 folds (Table 2). In some studies different threshold levels were used for same biomarker, based on the duration of illness at the time of collection of blood (10,11). This is an impractical approach to be implemented at any clinical setting.
Defining “zero hour” is an important parameter to characterize the biomarkers as early or late infection detectors. However, this definition varied from study to study as it was mostly decided based on the blood collection time, and only few studies considered the first onset of illness. The requirement for early onset diagnosis is not usually relevant for low and middle income countries where care seeking behaviour is poor, and thus the babies are brought to the hospital when the disease process has already progressed to a severe state.
We also observed that case definition for sepsis differed from study to study. In majority of the studies, sepsis was defined based on clinical algorithm, which also varied from study to study. Furthermore, there were studies where only culture-proved cases were considered as sepsis. This is a challenging issue: if we consider all clinically suspected sepsis cases as true, we run the risk of diluting true sepsis cases; if not, then we are possibly missing out the actual infections which are not captured by blood culture.
Additionally, >90% of biomarker studies were from developed countries, and >50% were specifically from Europe. Therefore, there is almost no data from developing countries where the populations are different with respect to their exposure to microbes, aetiology of infection, nutritional status, time for care-seeking behaviour, and other factors.
In conclusion, biomarker research has many limitations, its progress has slowed down and research results are far from reaching the population where biomarkers are needed most.
Several steps are needed to facilitate the uptake of biomarkers as tools to diagnose neonatal infections in the developing countries; i) a multi-country and multi-site study using a harmonized protocol to detect the most promising biomarkers, ii) formulation of their use in single and/or multiplex format, iii) development of point care device and their trial in the facility level and iv) validation of point of care device in large population based sites of multiple countries.
Acknowledgment
We thank Maksuda Islam for administrative support. We also acknowledge support from the principal grant holder, Prof. Igor Rudan from the University of Edinburgh, for financial support and technical guidance.
Funding: Bill and Melinda Gates Foundation.
Ethical approval: Not required.
Authorship declaration: MSI and SKS conceived and designed the study and developed the protocol. MMe JKM, RM, MMo and SKS conducted the literature review and analysis. Mme and SKS participated in writing and revision of the manuscript.
Competing interests: All authors have completed the Unified Competing Interest form at www.icmje.org/coi_disclosure.pdf (available on request from the corresponding author) and declare support from Bill and Melinda Gates Foundation for the submitted work. The authors declare no financial relationships with any organizations that might have an interest in the submitted work in the previous 3 years; and no other relationships or activities that could appear to have influenced the submitted work.
References
- 1.Thaver D, Zaidi AK. Burden of neonatal infections in developing countries: a review of evidence from community-based studies. Pediatr Infect Dis J. 2009;28:S3–9. doi: 10.1097/INF.0b013e3181958755. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad OB, Lopez AD, Inoue M.The decline in child mortality: a reappraisal. Bull World Health Organ 2000781175–1191.[REMOVED HYPERLINK FIELD] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryce J, Boschi-Pinto C, Shibuya K, Black RE. WHO estimates of the causes of death in children. Lancet. 2005;365:1147–1152. doi: 10.1016/S0140-6736(05)71877-8. [DOI] [PubMed] [Google Scholar]
- 4.Lawn JE, Cousens S, Zupan J, Lancet Neonatal Survival Steering Team 4 million neonatal deaths: When? Where? Why? Lancet. 2005;365:891–900. doi: 10.1016/S0140-6736(05)71048-5. [DOI] [PubMed] [Google Scholar]
- 5.Black RE, Cousens S, Johnson HL, Lawn JE, Rudan I, Bassani DG, et al. Child Health Epidemiology Reference Group of WHO and UNICEF Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet. 2010;375:1969–1987. doi: 10.1016/S0140-6736(10)60549-1. [DOI] [PubMed] [Google Scholar]
- 6.Baqui AH, El-Arifeen S, Darmstadt GL, Ahmed S, Williams EK, Seraji HR, et al. Projahnmo Study Group Effect of community-based newborn-care intervention package implemented through two service-delivery strategies in Sylhet district, Bangladesh: a cluster-randomized controlled trial. Lancet. 2008;371:1936–1944. doi: 10.1016/S0140-6736(08)60835-1. [DOI] [PubMed] [Google Scholar]
- 7.Qazi SA, Stoll BJ. Neonatal sepsis a major global public health challenge. Pediatr Infect DisJ. 2009;28(Suppl1):S1–S2. doi: 10.1097/INF.0b013e31819587a9. [DOI] [PubMed] [Google Scholar]
- 8.Bang AT, Bang RA, Reddy MH, Baitule SB, Deshmukh MD, Paul VK. de C Marshal TF. Simple clinical criteria to identify sepsis or pneumonia in neonates in the community needing treatment or referral. Pediatr Infect Dis J. 2005;24:335–341. doi: 10.1097/01.inf.0000157094.43609.17. [DOI] [PubMed] [Google Scholar]
- 9.Pammi M, Flores A, Leeflang M, Versalovic J. Molecular assays in the diagnosis of neonatal sepsis: A systematic review and meta-analysis. Pediatrics. 2011;128:e973. doi: 10.1542/peds.2011-1208. [DOI] [PubMed] [Google Scholar]
- 10.Pourcyrous M, Bada HS, Korones SB, Baselski V, Wong SP. Significance of serial C-reactive protein responses in neonatal infection and other disorders. Pediatrics. 1993;92:431–435. [PubMed] [Google Scholar]
- 11.Benitz WE, Han MY, Madan A, Ramachandra P. Serial serum C-reactive protein levels in the diagnosis of neonatal infection. Pediatrics. 1998;102:E41. doi: 10.1542/peds.102.4.e41. [DOI] [PubMed] [Google Scholar]
- 12.Carrol ED, Thomson AP, Hart CA. Procalcitonin as a marker of sepsis. Int J Antimicrob Agents. 2002;20:1–9. doi: 10.1016/S0924-8579(02)00047-X. [DOI] [PubMed] [Google Scholar]
- 13.Resch B, Gusenleitner W, Muller WD. Procalcitonin and interleukin-6 in the diagnosis of early-onset sepsis of the neonate. Acta Paediatr. 2003;92:243–245. doi: 10.1111/j.1651-2227.2003.tb00534.x. [DOI] [PubMed] [Google Scholar]
- 14.Roman J, Fernandez F, Velasco F, Rojas R, Roldan MR, Torres A. Serum TNF levels in neonatal sepsis and septic shock. Acta Paediatr. 1993;82:352–354. doi: 10.1111/j.1651-2227.1993.tb12695.x. [DOI] [PubMed] [Google Scholar]
- 15.Bilavsky E, Yarden-Bilavsky H, Ashkenazi S, Amir J. C-reactive protein as a marker of serious bacterial infections in hospitalized febrile infants. Acta Paediatr. 2009;98:1776–1780. doi: 10.1111/j.1651-2227.2009.01469.x. [DOI] [PubMed] [Google Scholar]
- 16.Chiesa C, Pellegrini G, Panero A, Osborn JF, Signore F, Assumma M, et al. C-reactive protein, interleukin-6, and procalcitonin in the immediate postnatal period: influence of illness severity, risk status, antenatal and perinatal complications, and infection. Clin Chem. 2003;49:60–68. doi: 10.1373/49.1.60. [DOI] [PubMed] [Google Scholar]
- 17.Dollner H, Vatten L, Austgulen R. Early diagnostic markers for neonatal sepsis: comparing C-reactive protein, interleukin-6, soluble tumour necrosis factor receptors and soluble adhesion molecules. J Clin Epidemiol. 2001;54:1251–1257. doi: 10.1016/S0895-4356(01)00400-0. [DOI] [PubMed] [Google Scholar]
- 18.Garland SM, Bowman ED. Reappraisal of C-reactive protein as a screening tool for neonatal sepsis. Pathology. 2003;35:240–243. doi: 10.1080/0031302031000123227. [DOI] [PubMed] [Google Scholar]
- 19.Kocabas E, Sarikçioglu A, Aksaray N, Seydaoglu G, Seyhun Y, Yaman A. Role of procalcitonin, C-reactive protein, interleukin-6, interleukin-8 and tumor necrosis factor-alpha in the diagnosis of neonatal sepsis. Turk J Pediatr. 2007;49:7–20. [PubMed] [Google Scholar]
- 20.Hansen AB, Verder H, Staun-Olsen P. Soluble intercellular adhesion molecule and C-reactive protein as early markers of infection in newborns. J Perinat Med. 2000;28:97–103. doi: 10.1515/JPM.2000.012. [DOI] [PubMed] [Google Scholar]
- 21.Palmer A, Carlin JB, Freihorst J, Gatchalian S, Muhe L, Mulholland K, et al. WHO Young Infant Study Group The use of CRP for diagnosing infections in young infants < 3 months of age in developing countries. Ann Trop Paediatr. 2004;24:205–212. doi: 10.1179/027249304225018948. [DOI] [PubMed] [Google Scholar]
- 22.Joram N, Boscher C, Denizot S, Loubersac V, Winer N, Roze JC, et al. Umbilical cord blood procalcitonin and C reactive protein concentrations as markers for early diagnosis of very early onset neonatal infection. Arch Dis Child Fetal Neonatal Ed. 2006;91:F65–66. doi: 10.1136/adc.2005.074245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon BH, Romero R, Shim JY, Shim SS, Kim CJ, Jun JK. C-reactive protein in umbilical cord blood: a simple and widely available clinical method to assess the risk of amniotic fluid infection and funisitis. J Matern Fetal Neonatal Med. 2003;14:85–90. doi: 10.1080/jmf.14.2.85.90. [DOI] [PubMed] [Google Scholar]
- 24.Mathers NJ, Pohlandt F. Diagnostic audit of C-reactive protein in neonatal sepsis. Eur J Pediatr. 1987;18:147–151. doi: 10.1007/BF02343221. [DOI] [PubMed] [Google Scholar]
- 25.Khassawneh M, Hayajneh WA, Kofahi H, Khader Y, Amarin Z, Daoud A. Diagnostic markers for neonatal sepsis: comparing C-reactive protein, interleukin-6 and immunoglobulin M. Scand J Immunol. 2007;65:171–175. doi: 10.1111/j.1365-3083.2006.01878.x. [DOI] [PubMed] [Google Scholar]
- 26.Laborada G, Rego M, Jain A, Guliano M, Stavola J, Ballabh P, et al. Diagnostic value of cytokines and C-reactive protein in the first 24 hours of neonatal sepsis. Am J Perinatol. 2003;20:491–501. doi: 10.1055/s-2003-45382. [DOI] [PubMed] [Google Scholar]
- 27.Manucha V, Rusia U, Sikka M, Faridi MM, Madan N. Utility of haematological parameters and C-reactive protein in the detection of neonatal sepsis. J Paediatr Child Health. 2002;38:459–464. doi: 10.1046/j.1440-1754.2002.00018.x. [DOI] [PubMed] [Google Scholar]
- 28.Marcus N, Mor M, Amir L, Mimouni M, Waisman Y. The quick-read C-reactive protein test for the prediction of bacterial gastroenteritis in the pediatric emergency department. Pediatr Emerg Care. 2007;23:634–637. doi: 10.1097/PEC.0b013e31814a6a52. [DOI] [PubMed] [Google Scholar]
- 29.Sakha K, Husseini MB, Seyyedsadri N. The role of the procalcitonin in diagnosis of neonatal sepsis and correlation between procalcitonin and C-reactive protein in these patients. Pak J Biol Sci. 2008;11:1785–1790. doi: 10.3923/pjbs.2008.1785.1790. [DOI] [PubMed] [Google Scholar]
- 30.Guibourdenche J, Bedu A, Petzold L, Marchand M, Mariani-Kurdjian P, Hurtaud-Roux MF, et al. Biochemical markers of neonatal sepsis: value of procalcitonin in the emergency setting. Ann Clin Biochem. 2002;39:130–135. doi: 10.1258/0004563021901874. [DOI] [PubMed] [Google Scholar]
- 31.Vazzalwar R, Pina-Rodrigues E, Puppala BL, Angst DB, Schweig L. Procalcitonin as a screening test for late-onset sepsis in preterm very low birth weight infants. J Perinatol. 2005;25:397–402. doi: 10.1038/sj.jp.7211296. [DOI] [PubMed] [Google Scholar]
- 32.López Sastre JB, Pérez Solís D, Roqués Serradilla V, Fernández Colomer B, Coto Cotallo GD, Krauel Vidal X, et al. Grupo de Hospitales Castrillo Procalcitonin is not sufficiently reliable to be the sole marker of neonatal sepsis of nosocomial origin screening. BMC Pediatr. 2006;6:16. doi: 10.1038/sj.jp.7211296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spada S, Cuccu A, Mussap M, Testa M, Puddu M, Pisu C, et al. Reliability of procalcitonin in neonatology. Experience in 59 preterm newborns. J Matern Fetal Neonatal Med. 2009;22(Suppl 3):96–101. doi: 10.1080/14767050903195450. [DOI] [PubMed] [Google Scholar]
- 34.Ballot DE, Perovic O, Galpin J, Cooper PA.Serum procalcitonin as an early marker of neonatal sepsis. S Afr Med J 200494851–854.[REMOVED HYPERLINK FIELD] [PubMed] [Google Scholar]
- 35.Turner D, Hammerman C, Rudensky B, Schlesinger Y, Schimmel MS. The role of procalcitonin as a predictor of nosocomial sepsis in preterm infants. Acta Paediatr. 2006;95:1571–1576. doi: 10.1080/08035250600767811. [DOI] [PubMed] [Google Scholar]
- 36.Isidor B, Caillaux G, Gilquin V, Loubersac V, Caillon J, Roze JC, et al. The use of procalcitonin in the diagnosis of late-onset infection in neonatal intensive care unit patients. Scand J Infect Dis. 2007;39:1063–1066. doi: 10.1080/00365540701466181. [DOI] [PubMed] [Google Scholar]
- 37.Boo NY, Nor Azlina AA, Rohana J. Usefulness of a semi-quantitative procalcitonin test kit for early diagnosis of neonatal sepsis. Singapore Med J. 2008;49:204–8. [PubMed] [Google Scholar]
- 38.López Sastre JB, Solís DP, Serradilla VR, Colomer BF, Cotallo GD. Grupo de Hospitales Castrillo. Evaluation of procalcitonin for diagnosis of neonatal sepsis of vertical transmission. BMC Pediatr. 2007;7:9. doi: 10.1186/1471-2431-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santuz P, Soffiati M, Dorizzi RM, Benedetti M, Zaglia F, Biban P. Procalcitonin for the diagnosis of early-onset neonatal sepsis: a multilevel probabilistic approach. Clin Biochem. 2008;41:1150–1155. doi: 10.1016/j.clinbiochem.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 40.Bender L, Thaarup J, Varming K, Krarup H, Ellermann-Eriksen S, Ebbesen F. Early and late markers for the detection of early-onset neonatal sepsis. Dan Med Bull. 2008;55:219–223. [PubMed] [Google Scholar]
- 41.Verboon-Maciolek MA, Thijsen SF, Hemels MA, Menses M, van Loon AM, Krediet TG, et al. Inflammatory mediators for the diagnosis and treatment of sepsis in early infancy. Pediatr Res. 2006;59:457–461. doi: 10.1203/01.pdr.0000200808.35368.57. [DOI] [PubMed] [Google Scholar]
- 42.Nishimaki S, Sato M, An H, Shima Y, Akaike T, Yokoyama U, et al. Comparison of markers for fetal inflammatory response syndrome: fetal blood interleukin-6 and neonatal urinary beta(2)-microglobulin. J Obstet Gynaecol Res. 2009;35:472–476. doi: 10.1111/j.1447-0756.2008.00988.x. [DOI] [PubMed] [Google Scholar]
- 43.Silveira RC, Procianoy RS. Evaluation of interleukin-6, tumour necrosis factor-alpha and interleukin-1beta for early diagnosis of neonatal sepsis. Acta Paediatr. 1999;88:647–650. doi: 10.1080/08035259950169314. [DOI] [PubMed] [Google Scholar]
- 44.Hatzidaki E, Gourgiotis D, Manoura A, Korakaki E, Bossios A, Galanakis E, et al. Interleukin-6 in preterm premature rupture of membranes as an indicator of neonatal outcome. Acta Obstet Gynecol Scand. 2005;84:632–638. doi: 10.1111/j.0001-6349.2005.00747.x. [DOI] [PubMed] [Google Scholar]
- 45.Santana Reyes C, García-Muńoz F, Reyes D, González G, Dominguez C, Domenech E. Role of cytokines (interleukin-1beta, 6, 8, tumour necrosis factor-alpha, and soluble receptor of interleukin-2) and C-reactive protein in the diagnosis of neonatal sepsis. Acta Paediatr. 2003;92:221–227. doi: 10.1111/j.1651-2227.2003.tb00530.x. [DOI] [PubMed] [Google Scholar]
- 46.Magudumana MO, Ballot DE, Cooper PA, Trusler J, Cory BJ, Viljoen E, et al. Serial interleukin 6 measurements in the early diagnosis of neonatal sepsis. J Trop Pediatr. 2000;46:267–271. doi: 10.1093/tropej/46.5.267. [DOI] [PubMed] [Google Scholar]
- 47.Kurt AN, Aygun AD, Godekmerdan A, Kurt A, Dogan Y, Yilmaz E. Serum IL-1beta, IL-6, IL-8, and TNF-alpha levels in early diagnosis and management of neonatal sepsis. Mediators Inflamm. 2007;2007:31397. doi: 10.1093/tropej/46.5.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoon BH, Romero R, Park JS, Kim M, Oh SY, Kim CJ, et al. The relationship among inflammatory lesions of the umbilical cord (funisitis), umbilical cord plasma interleukin 6 concentration, amniotic fluid infection, and neonatal sepsis. Am J Obstet Gynecol. 2000;183:1124–1129. doi: 10.1067/mob.2000.109035. [DOI] [PubMed] [Google Scholar]
- 49.Tasci Y, Dilbaz B, Uzmez Onal B, Caliskan E, Dilbaz S, Doganci L, et al. The value of cord blood interleukin-6 levels for predicting chorioamnionitis, funisitis and neonatal infection in term premature rupture of membranes. Eur J Obstet Gynecol Reprod Biol. 2006;128:34–39. doi: 10.1016/j.ejogrb.2005.11.049. [DOI] [PubMed] [Google Scholar]
- 50.Weeks JW, Reynolds L, Taylor D, Lewis J, Wan T, Gall SA. Umbilical cord blood interleukin-6 levels and neonatal morbidity. Obstet Gynecol. 1997;90:815–818. doi: 10.1016/S0029-7844(97)00421-3. [DOI] [PubMed] [Google Scholar]
- 51.Kashlan F, Smulian J, Shen-Schwarz S, Anwar M, Hiatt M, Hegyi T. Umbilical vein interleukin 6 and tumor necrosis factor alpha plasma concentrations in the very preterm infant. Pediatr Infect Dis J. 2000;19:238–243. doi: 10.1097/00006454-200003000-00013. [DOI] [PubMed] [Google Scholar]
- 52.Krueger M, Nauck MS, Sang S, Hentschel R, Wieland H, Berner R. Cord blood levels of interleukin-6 and interleukin-8 for the immediate diagnosis of early-onset infection in premature infants. Biol Neonate. 2001;80:118–123. doi: 10.1159/000047130. [DOI] [PubMed] [Google Scholar]
- 53.Naccasha N, Hinson R, Montag A, Ismail M, Bentz L, Mittendorf R. Association between funisitis and elevated interleukin-6 in cord blood. Obstet Gynecol. 2001;97:220–224. doi: 10.1016/S0029-7844(00)01149-2. [DOI] [PubMed] [Google Scholar]
- 54.Källman J, Ekholm L, Eriksson M, Malmström B, Schollin J. Contribution of interleukin-6 in distinguishing between mild respiratory disease and neonatal sepsis in the newborn infant. Acta Paediatr. 1999;88:880–884. doi: 10.1111/j.1651-2227.1999.tb00065.x. [DOI] [PubMed] [Google Scholar]
- 55.de Bont ES, Martens A, van Raan J, Samson G, Fetter WP, Okken A, et al. Diagnostic value of plasma levels of tumor necrosis factor alpha (TNF alpha) and interleukin-6 (IL-6) in newborns with sepsis. Acta Paediatr. 1994;83:696–699. doi: 10.1111/j.1651-2227.1994.tb13121.x. [DOI] [PubMed] [Google Scholar]
- 56.Doellner H, Arntzen KJ, Haereid PE, Aag S, Austgulen R. Interleukin-6 concentrations in neonates evaluated for sepsis. J Pediatr. 1998;132:295–9. doi: 10.1016/S0022-3476(98)70448-2. [DOI] [PubMed] [Google Scholar]
- 57.Adib M, Ostadi V, Navaei F, Saheb Fosoul F, Oreizi F, Shokouhi R, et al. Evaluation of CD11b expression on peripheral blood neutrophils for early detection of neonatal sepsis. Iran J Immunol. 2006;3:9–14. [PubMed] [Google Scholar]
- 58.Layseca-Espinosa E, Pérez-González LF, Torres-Montes A, Baranda L, de la Fuente H, Rosenstein Y, et al. Expression of CD64 as a potential marker of neonatal sepsis. Pediatr Allergy Immunol. 2002;13:319–327. doi: 10.1034/j.1399-3038.2002.01064.x. [DOI] [PubMed] [Google Scholar]
- 59.Ng PC, Li K, Wong RP, Chui KM, Wong E, Fok TF. Neutrophil CD64 expression: a sensitive diagnostic marker for late-onset nosocomial infection in very low birthweight infants. Pediatr Res. 2002;51:296–303. doi: 10.1203/00006450-200203000-00006. [DOI] [PubMed] [Google Scholar]
- 60.Ng PC, Li G, Chui KM, Chu WC, Li K, Wong RP, et al. Neutrophil CD64 is a sensitive diagnostic marker for early-onset neonatal infection. Pediatr Res. 2004;56:796–803. doi: 10.1203/01.PDR.0000142586.47798.5E. [DOI] [PubMed] [Google Scholar]
- 61.Zeitoun AA, Gad SS, Attia FM, Abu Maziad AS, Bell EF. Evaluation of neutrophilic CD64, interleukin 10 and procalcitonin as diagnostic markers of early- and late-onset neonatal sepsis. Scand J Infect Dis. 2010;42:299–305. doi: 10.3109/00365540903449832. [DOI] [PubMed] [Google Scholar]
- 62.Bhandari V, Wang C, Rinder C, Rinder H. Hematologic profile of sepsis in neonates: neutrophil CD64 as a diagnostic marker. Pediatrics. 2008;121:129–134. doi: 10.1542/peds.2007-1308. [DOI] [PubMed] [Google Scholar]
- 63.Franz AR, Steinbach G, Kron M, Pohlandt F. Interleukin-8: a valuable tool to restrict antibiotic therapy in newborn infants. Acta Paediatr. 2001;90:1025–1032. doi: 10.1111/j.1651-2227.2001.tb01359.x. [DOI] [PubMed] [Google Scholar]
- 64.Franz AR, Kron M, Pohlandt F, Steinbach G. Comparison of procalcitonin with interleukin 8, C-reactive protein and differential white blood cell count for the early diagnosis of bacterial infections in newborn infants. Pediatr Infect Dis J. 1999;18:666–671. doi: 10.1097/00006454-199908000-00003. [DOI] [PubMed] [Google Scholar]
- 65.Franz AR, Sieber S, Pohlandt F, Kron M, Steinbach G. Whole blood interleukin 8 and plasma interleukin 8 levels in newborn infants with suspected bacterial infection. Acta Paediatr. 2004;93:648–653. doi: 10.1111/j.1651-2227.2004.tb02991.x. [DOI] [PubMed] [Google Scholar]
- 66.Franz AR, Bauer K, Schalk A, Garland SM, Bowman ED, Rex K, et al. International IL-8 Study Group. Measurement of interleukin 8 in combination with C-reactive protein reduced unnecessary antibiotic therapy in newborn infants: a multicenter, randomized, controlled trial. Pediatrics. 2004;114:1–8. doi: 10.1542/peds.114.1.1. [DOI] [PubMed] [Google Scholar]
- 67.Franz AR, Steinbach G, Kron M, Pohlandt F. Reduction of unnecessary antibiotic therapy in newborn infants using interleukin-8 and C-reactive protein as markers of bacterial infections. Pediatrics. 1999;104:447–453. doi: 10.1542/peds.104.3.447. [DOI] [PubMed] [Google Scholar]
- 68.Santana C, Guindeo MC, González G, García-Muńoz F, Saavedra P, Doménech E. Cord blood levels of cytokines as predictors of early neonatal sepsis. Acta Paediatr. 2001;90:1176–1181. doi: 10.1111/j.1651-2227.2001.tb03250.x. [DOI] [PubMed] [Google Scholar]
- 69.Bentlin MR, de Souza Rugolo LM, Júnior AR, Hashimoto M, Lyra JC. Is urine interleukin-8 level a reliable laboratory test for diagnosing late onset sepsis in premature infants? J Trop Pediatr. 2007;53:403–408. doi: 10.1093/tropej/fmm054. [DOI] [PubMed] [Google Scholar]
- 70.Bonac B, Derganc M, Wraber B, Hojker S. Interleukin-8 and procalcitonin in early diagnosis of early severe bacterial infection in critically ill neonates. Pflugers Arch. 2000;440(5) Suppl:R72–74. doi: 10.1007/s004240000011. [DOI] [PubMed] [Google Scholar]
- 71.Austgulen R, Arntzen KJ, Haereid PE, Aag S, Dollner H. Infections in neonates delivered at term are associated with increased serum levels of ICAM-1 and E-selectin. Acta Paediatr. 1997;86:274–280. doi: 10.1111/j.1651-2227.1997.tb08889.x. [DOI] [PubMed] [Google Scholar]
- 72.Athhan F, Akagündüz B, Genel F, Bak M, Can D. Procalcitonin: a marker of neonatal sepsis. J Trop Pediatr. 2002;48:10–14. doi: 10.1093/tropej/48.1.10. [DOI] [PubMed] [Google Scholar]
- 73.Delévaux I, André M, Colombier M, Albuisson E, Meylheuc F, Bégue RJ, et al. Can procalcitonin measurement help in differentiating between bacterial infection and other kinds of inflammatory processes? Ann Rheum Dis. 2003;62:337–340. doi: 10.1136/ard.62.4.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ng PC, Li K, Wong RPO, Chui K, Wong E, Li G, et al. Proinflammatory and anti-inflammatory cytokine responses in preterm infants with systemic infections. Arch Dis Child Fetal Neonatal Ed. 2003;88:209–213. doi: 10.1136/fn.88.3.F209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buck C, Bundschu J, Gallati H, Bartmann P, Pohlandt F. Interleukin-6: a sensitive parameter for the early diagnosis of neonatal bacterial infection. Pediatrics. 1994;93:54–58. [PubMed] [Google Scholar]
- 76.Ng PC. Diagnostic markers of infection in neonates. Arch Dis Child Fetal Neonatal Ed. 2004;89:F229–235. doi: 10.1136/adc.2002.023838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ng PC, Cheng SH, Chui KM, Fok TF, Wong MY, Wong W, et al. Diagnosis of late onset neonatal sepsis with cytokines, adhesion molecule, and C-reactive protein in preterm very low birthweight infants. Arch Dis Child Fetal Neonatal Ed. 1997;77:F221–F227. doi: 10.1136/fn.77.3.F221. [DOI] [PMC free article] [PubMed] [Google Scholar]