Abstract
The Thai Phase III clinical trial (RV144) showed modest efficacy in preventing HIV-1 acquisition. Plasma collected from HIV-1-uninfected trial participants completing all injections with ALVAC-HIV (vCP1521) prime and AIDSVAX B/E boost were tested for antibody responses against HIV-1 gp120 envelope (Env). Peptide microarray analysis from six HIV-1 subtypes and group M consensus showed that vaccination induced antibody responses to the second variable (V2) loop of gp120 of multiple subtypes. We further evaluated V2 responses by ELISA and surface plasmon resonance using cyclic (Cyc) and linear V2 loop peptides. Thirty-one of 32 vaccine recipients tested (97%) had antibody responses against Cyc V2 at 2 weeks postimmunization with a reciprocal geometric mean titer (GMT) of 1100 (range: 200–3200). The frequency of detecting plasma V2 antibodies declined to 19% at 28 weeks post-last injection (GMT: 110, range: 100–200). Antibody responses targeted the mid-region of the V2 loop that contains conserved epitopes and has the amino acid sequence KQKVHALFYKLDIVPI (HXB2 Numbering sequence 169–184). Valine at position 172 was critical for antibody binding. The frequency of V3 responses at 2 weeks postimmunization was modest (18/32, 56%) with a GMT of 185 (range: 100–800). In contrast, naturally infected HIV-1 individuals had a lower frequency of antibody responses to V2 (10/20, 50%; p=0.003) and a higher frequency of responses to V3 (19/20, 95%), with GMTs of 400 (range: 100–3200) and 3570 (range: 200–12,800), respectively. RV144 vaccination induced antibodies that targeted a region of the V2 loop that contains conserved epitopes. Early HIV-1 transmission events involve V2 loop interactions, raising the possibility that anti-V2 antibodies in RV144 may have contributed to viral inhibition.
Introduction
Vaccination with the heterologous prime-boost vaccine regimen ALVAC-HIV (vCP1521) and AIDSVAXB/E (bivalent rgp120) was associated with a 31.2% reduction in HIV-1 acquisition at 42 months in the modified intent to treat population and provided the first opportunity to study the immune correlates of HIV-1 vaccine-induced protection in humans.1–3 Evaluation of the vaccine regimen in Phase I/II studies showed that this vaccine induced both cellular and humoral immune responses.4,5 Immunogenicity studies have demonstrated the induction of neutralizing antibodies against tier 1 strains of HIV-1, antibody-dependent cell-mediated cytotoxicity (ADCC), antigen-specific lymphoproliferative responses, and CD8+ cytotoxic T lymphocyte (CTL) responses.4–9 Vaccination with recombinant Env proteins alone in AIDSVAX B/E (Vax003 trial) and AIDSVAX B/B (Vax004 trial) generated strong humoral immune responses but did not decrease HIV-1 acquisition in Thai injecting drug users or North American men who have sex with men, respectively.10–12
The role of antibodies in preventing HIV-1 acquisition has been suggested by passive13–16 and active immunization studies in nonhuman primates17,18 as well as by preliminary data in mother–infant transmission studies,19 but has not been confirmed as a general principle of protection in humans. Recently it was found that antibodies against subtype B gp120 V1V2 loops appeared to correlate with decreased risk of infection in RV14420; however, broadly neutralizing antibodies were not seen in that study,21 and neither neutralizing antibodies nor antibody-directed cell-mediated cytotoxicity appeared to correlate with infection risk.20 Nonetheless, several regions of the HIV-1 gp120 glycoprotein contain epitopes that induce antibodies that may play a role in protection. Epitopes found in the second variable (V2) loop have been associated with HIV-1 Env functions that are important in infectivity, CD4 receptor and CCR5/CXCR4 coreceptor usage, escape from antibody neutralization, the formation of Env trimers, and the protection of the coreceptor binding site.22–28 Sequence variation within the V1/V2 loops alters neutralization resistance and viral escape is associated with V2 loop mutations.29–33 Compared to viruses circulating in chronically infected persons, transmitted viruses (subtypes A and C) appear to have shorter V1/V2 regions and fewer N-linked glycosylation sites.30,34–36
The V2 loop contains a putative α4β7 integrin binding motif (LDI/V) at amino acid (aa) residues 179–181 (HBX2 numbering system) that was shown to interact with the gut mucosal homing receptor.23,24 Under physiological conditions, the α4β7 receptor facilitates the migration of lymphocytes to the gut and some have postulated that HIV-1 exploits this interaction to infect lymphocytes localized at mucosal tissues.23,24,37,38 However, further studies are needed to confirm the role of α4β7 in viral transmission with unliganded gp120 trimers.39
Broadly neutralizing antibodies that interact with quaternary neutralizing epitopes (QNE) of trimeric native Env spikes have been shown to interact with conserved aa embedded within the variable (V) loops of gp120 including the V2 loop.40–47 Furthermore, antibodies found in individuals that exhibit potent cross-clade neutralizing activity have been shown to target conserved regions of the V1, V2, and V3 loops, providing evidence that V2 contributes to the formation of QNE that can induce potent cross-clade neutralizing antibodies.25,43 The structure of scaffolded V1/V2 complexed with the broadly neutralizing monoclonal antibody PG9 has been resolved.47 This potent antibody binds to glycans and aa sequences within the V1/V2 regions. Antibodies against the V2 loop target several conserved residues that are sensitive to glycosylation patterns and changes within the loop alter the efficiency by which antibodies neutralize HIV-1.26,30,41,48
T cell peptide epitope mapping studies with peripheral blood mononuclear cells (PBMCs) from HIV-1-uninfected RV144 vaccine recipients using the interferon-gamma ELISpot assay demonstrated preferential CD4+ T cell responses to the V2 loop region compared to other regions of gp120.49 Antibody epitope mapping studies were undertaken to assess whether there was a similar preferential targeting of regions within gp120.
In this study we report that the RV144 vaccine regimen generated antibodies to the V2 region of gp120. Using cyclic as well as linear peptides, we demonstrate that the vaccine-induced antibodies recognize the mid-region of the V2 loop. A role for these antibodies in protection from HIV acquisition was recently suggested by a genetic sieve analysis of subjects who enrolled in RV144 and became HIV-1 infected.50
Materials and Methods
Study subjects for phase III Thai clinical trial (RV144)
The design and findings of the RV144 Phase III clinical trial have been published previously.1 In this study we used three sets of RV144 plasma samples (sets A, C, and Z) that were randomly generated by the Statistical Center for HIV/AIDS Research Prevention (SCHARP), Seattle, WA, and the EMMES Corporation, Rockville, MD from volunteers who received all immunizations according to schedule and were HIV seronegative at the end of the study (Table 1). It was not possible to use the same sample set for all studies due to the limited volumes of sample available. However, all plasma separations were performed at the same centralized laboratory. Each plasma set consisted of uninfected subjects in the per-protocol population with ratios of 4:1 vaccine:placebo for both male and female subjects. Simple random sampling was used within each of the four strata. For peptide microarray and biotinylated linear peptide analyses, plasma samples from 80 vaccine and 20 placebo recipients, designated plasma set A and set C, were used, respectively (Table 1). For cyclic (Cyc) peptide analysis, plasma samples from 32 vaccine and 8 placebo recipients, designated plasma set Z, were used (Table 1). RV144 plasma samples from three different time points were tested: baseline or prevaccination (visit 1), 2 weeks (visit 8), and 28 weeks (visit 9) post-final inoculation. The RV144 trial was conducted under a protocol approved by IRBs from the Thai Ministry of Public Health, Mahidol University, the Scientific Subcommittee of the National AIDS Committee, Thailand, the Royal Thai Army, Thailand, and the U.S. Army Medical Research and Materiel Command, United States (http://clinicaltrials.gov number NTC00223080).
Table 1.
Definition of Plasma Sets from RV144 Subjects and the Assays for Which They Were Used
| Plasma set | No. vaccine/No. placebo recipients | Assay |
|---|---|---|
| A | 80/20 | Peptide microarray |
| C | 80/20 | Biotinylated linear peptide analysis |
| Z | 32/8 | Whole gp120 ELISA cyclic peptide analysis: ELISA, surface plasmon resonance |
Plasma sets used for the analysis of the V2 loop and gp120 responses by microarray, ELISA, and Biacore.
HIV-1 naturally infected individuals
Plasma samples from Thai blood donors infected with HIV-1 circulating recombinant form (CRF) 01_AE (WRAIR protocol #1617) were used in ELISA and Biacore assays with Cyc V2 peptides.
Recombinant proteins and peptides
CHO-expressed recombinant gp120 proteins derived from MN and A244 were kindly provided by Dr. Marc Gurwith, Global Solutions for Infectious Disease, San Francisco, CA. HIV-1 CRF01_AE 92TH023 gp120 protein was expressed in 293T cells and purified using Galanthus nivalis lectin columns.
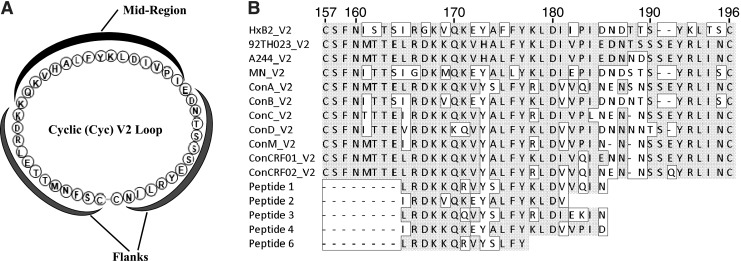
Peptide microarrays and Cyc peptides were synthesized by JPT Peptide Technologies. Peptides were cyclized by disulfide bond formation (Fig. 1A) and the purity was determined to be greater than 90% by high-pressure liquid chromatography and mass spectrometry. The aa sequences of Cyc V2 and V3 peptides were based on vCP1521 Env glycoprotein of HIV-1 CRF01 AE (92TH023 strain) GenBank accession number EF553537.1 (Fig. 1). Cyc V2 peptides of varying lengths as well as those with scrambled mid-region (Scr MR) or scrambled flanking regions (Scr Fl) were synthesized with or without biotin at the amino terminus of the peptide. HIV-1 strains 92TH023 and A244 have identical V2 loop mid-regions (Fig. 1B). Cyc V3 peptide was not biotinylated. Cyclic nonbiotinylated peptides were used in all ELISAs and biotinylated peptides were used for all Biacore binding studies. The sequences of Cyc peptides are shown in Table 2. The aa sequences of the scrambled regions of the mid-region of Cyc V2 Scr MR and the flanking regions of Cyc V2 Scr Fl are shown in bold (Table 2). The integrin binding motif, LDI, is underlined.
FIG. 1.
Graphic representation of the cyclic V2 loop and alignment of V2 loop amino acid sequences. (A) Amino acid sequence of the cyclic (Cyc) V2 loop of the HIV-1 CRF01_AE 92TH023 strain. The flanks and mid-region are labeled. (B) Alignment of V2 loop amino acid sequences. Sequences that vary from 92TH023 are boxed and the first (157) and last (196) amino acids of the V2 are shown on top of the alignment. Numbering is based on the HXB2 strain. Cyclic V2 peptides were synthesized based on clone 92TH023. Peptides for microarray analysis were based on consensus (Con) sequences and peptides 1–6 represent linear N-linked biotinylated peptides.
Table 2.
Cyclic and Linear Peptides Used in the Study
| Cyclic peptides based on 92TH023 strain | HXB2 amino acid numbering | Amino acid sequence |
|---|---|---|
| Cyc V2 (42 aa) | 157–196 | CSFNMTTELRDKKQKVHALFYKLDIVPIEDNTSSSEYRLINC |
| Cyc V2 (25aa) | 173–193 | CHALFYKLDIVPIEDNTSSSEYRLC |
| Cyc V2 (16aa) | 176–189 | CFYKLDIVPIEDNTSC |
| Cyc V2 Scr FI | 157–196 | CENLTDKMFTSRKQKVHALFYKLDIVPISESRLDETNYNISC |
| Cyc V2 Scr MR | 157–196 | CSFNMTTELRDKQVLFKDIHKIVKPLYAEDNTSSSEYRLINC |
| Cyc V3 (35aa) | 296–331 | CTRPSNNTRTSINIGPGQVFYRTGDIIGDIRKAYC |
| Linear peptides strain specificity | HXB2 amino acid numbering | Amino acid sequence |
|---|---|---|
| Peptide 1 | 165–185 | LRDKKQRVYSLFYKLDVVQIN |
| Subtype A | ||
| Peptide 2 | 165–185 | IRDKVQKEYALFYKLDVVPID |
| Subtype B | ||
| Peptide 3 | 165–185 | LRDKKQQVYSLFYRLDIEKIN |
| Subtype A | ||
| Peptide 4 | 165–185 | IRDKKQKEYALFYKLDVVPID |
| Subtype A and B | ||
| Peptide 6 | 165–178 | LRDKKQRVYSLFYK |
| Subtype A | ||
The amino acid sequence of cyclic V2 and V3 loop peptides is based on HIV-1 CRF01_AE strain 92TH023. Scrambled regions are shown in bold and the integrin binding motif LDI/V is underlined. Linear biotinylated peptide sequences are based on the sequences of HIV-1 strains deposited at the LANL databse, except for peptide 4, which is a consensus V2 loop sequence. The specific strains pertaining to peptides 1–3 are listed in Materials and Methods. Peptide 6 is the 14 amino acid N-terminal fragment of peptide 1.
Peptides representing the more sequence-conserved segment in the first two-thirds of the V2 loop were selected from the set of all recorded V2 sequences in the Los Alamos National Laboratory (LANL) database in order to maximize predicted functional diversity. The sequences of the linear V2 peptides are shown in Table 2, and the integrin binding motif in each of the linear peptides is underlined. Polar and charged amino acids mediate most chemical functions of proteins such as solvent interactions (surface accessibility), binding, posttranslational modification, and catalysis. Peptide 1 was selected as the V2 loop mid-region from a strain with the most charged and polar aa among circulating strains from subtype A, strain QB585.2102M.Ev1v5.C. Peptide 3 was selected as the V2 loop mid-region sequence found in circulating strains with the most polar and charged aa and also exhibiting the most common V2 length of 39 aa from subtype A, strain 01TZA341. Peptide 2 represents the sequence most commonly found in circulating subtype B strains recorded in the LANL database as of May 2011, strain 878v3_2475. Peptide 4 represents the consensus sequences for V2 in the region covered by peptides 1–3. Peptide 6 contains the N-terminal 14 aa of peptide 1. Peptides were biotinylated at the N-terminus during synthesis and were purchased from Genemed Biotechnologies, Inc., South San Francisco, CA.
Reagents
Carboxymethylcellulose 5 (CM5) chips, streptavidin (SA) chips, and the amine coupling kit were purchased from GE Healthcare, Piscataway, NJ. Lysozyme, Costar Spin-X centrifuge tube cellulose acetate filters (0.22 μm pore size), Dulbecco's phosphate-buffered saline (D-PBS), Tween 20, and Thimerosal were purchased from Sigma-Aldrich, St. Louis, MO. Affinity purified sheep antihuman IgG (gamma chain specific) antibodies were purchased from The Binding Site, Birmingham, UK. Skim milk was from Applichem, St. Louis, MO. ABTS peroxidase substrate solution was from KPL, Gaithersburg, MD. Horseradish peroxidase-conjugated goat antihuman IgG antibodies were from Bethyl Laboratories, Montgomery, TX. Strepta Well plates were from Roche, Mannheim, Germany. Alkaline phosphatase-conjugated goat antihuman IgG was purchased from SouthernBiotech, Birmingham, AL. Anti-IgG Cy5 antibody was from Jackson ImmunoResearch Laboratories, West Grove, PA. Superblock T20 PBS blocking buffer, Immulon 2HB U bottom, and Immulon 4HBX flat ELISA plates were purchased from Thermo Fisher Scientific, Rockford, IL.
Peptide microarray
Each array consisted of 1423 tiled Env peptides arranged in three identical subarrays, allowing for triplicate evaluation of the entire set of peptides. Peptide sequences covered the entire gp160 subtype consensus sequences from six HIV-1 group M subtypes (A, B, C, D, CRF01, and CRF02) and a consensus group M gp160, Con-S,51 for a total of 1423 peptides (15-mers overlapping by 12 aa). We used consensus sequences rather than the vaccine strains, as we wished to focus on cross-reactive responses rather than type-specific responses to the vaccine immunogens. The peptides were generated using an alignment of the seven consensus sequences with LANLs PeptGen tool (www.hiv.lanl.gov), so that peptides remain in register throughout the Env despite insertions and deletions, and identical peptides found in more than one subtype were represented only once. Microarray development was performed using the HS4800 Pro Hybridization Station (Tecan, Männedorf, Switzerland). All arrays were blocked with Superblock T20 PBS blocking buffer for 0.5 h at 30°C, followed by a 2-h incubation at 30°C with heat-inactivated plasma diluted 1:100 in Superblock T20. Arrays were then incubated for 45 min at 30°C with a secondary antibody, 1.5 mg/ml anti-IgG Cy5 antibody diluted 1:1000 with Superblock T20. Washes between all steps were with PBS containing 0.1% Tween. Arrays were scanned at a wavelength of 635 nm using an Axon Genepix 4300 Scanner (Molecular Devices, Sunnyvale, CA) at a PMT setting of 600, 50% laser power. Images were analyzed using Genepix Pro 7 software (Molecular Devices). Spot intensities were corrected for prevaccine results.
Peptide microarray spot intensities were extracted from the raw images using the GenePix software, and peptide intensities were calculated as follows: (1) Spot intensities were defined as foreground median intensity minus background median intensity. (2) Resulting intensities were log2 transformed and centered by subtracting the average (log2) intensities of the empty (control) spots on the slide. (3) For each unique peptide, a single intensity was defined as the median of the triplicate intensities on the slide for that peptide. To remove effects unrelated to true peptide-binding activity, such as nonspecific binding of primary antibody in the sample, array normalization was performed in an attempt to remove such effects. Nonspecific binding may be related to physicochemical properties of the peptide, we used the z-scales to correct for nonspecific binding biases.52 To arrive at a summary z-scale score for each peptide, the z-scale values of the individual aa comprising that peptide were summed. Here, the normalization was done in two stages: (1) a regression model for the probe intensities was derived from their overall z-scores and (2) each probe was normalized by subtracting its predicted intensity (representing the bias) from the observed intensity. A linear model with t4 distributed errors was used, which provides more robustness against probe outliers as described previously.53 This normalization was performed on each array separately. Once the data were normalized, the probe intensities were smoothed and a score was calculated for each peptide that was used to detect binding regions. Based on the assumption that an antibody binds a region of approximately 9 aa or more, normalized peptide intensities were smoothed using a sliding window mean statistics of size 9 aa, i.e., peptides with HXB2 positions within 9 aa of one another were grouped together. This smoothing step can be made subtype specific (e.g., for subtype A) or across all subtypes to borrow strength across multiple peptides. For baseline correction, samples were corrected by subtracting, for each sample, the peptide intensities measured prevaccination for that sample.
Once scores have been calculated and corrected for baseline, the score cutoff to call positive peptides can be set arbitrarily, determined based on simple validation, or based on a false detection rate (FDR) cutoff. Here the placebo group from the RV144 data was used to estimate the FDR and set at an appropriate threshold. Peptides with log2-fold change greater than 1.1 (with respect to baseline) were called positive, which led to an FDR of approximately 5%.
ELISA for cyclic peptides and recombinant gp120
Briefly, U-bottom 2HB plates were coated with either 1 μg/ml of cyclic peptide or with 3 μg/ml of recombinant gp120 in D-PBS at 4°C overnight. Wells were washed three times with wash buffer (PBS, 0.1% Tween 20, and 0.01% Thimerosal, pH 7.4) using Microplate Washer ELX405, Bio Tek, Winooski, VT, and blocked with blocking buffer (D-PBS, 5% skim milk) for 2 h at room temperature. Plasma was initially diluted 1:100 in blocking buffer and then serial 2-fold dilutions were performed and added to wells for 2 h at room temperature. Wells were washed five times with wash buffer and HRP-conjugated goat antihuman IgG (1:25,000) was added to wells for 1 h at room temperature. Plates were washed five times with wash buffer, 100 μl/well of substrate was added, and color was allowed to develop at room temperature for 1 h in the dark. Plates were read at A405 nm using an ELISA reader Spectramax 340 PC, Molecular Devices. The data are expressed as end point titers, with the titers being defined as the reciprocal of the highest dilution that yielded an absorbance value above 2.5 times the background value (wells that did not contain recombinant protein or peptides).
Biotinylated linear peptide ELISA
Briefly, StreptaWell plates (Roche) were coated with 1 μg/ml biotinylated linear V2 peptides for 1.5 h at 37°C and then washed six times with PBS containing 0.05% Tween-20, pH 7.4. Plates were incubated for 1.5 h at 37°C with RV144 plasma diluted 1:100 in RPMI media containing 15% fetal bovine serum. The plates were washed six times and alkaline phosphatase-conjugated goat antihuman IgG (1:2,000) was added for 1.5 h at 37°C. After washing, 10% diethanolamine substrate was added for 30 min to develop color, and the plates were read at A405 nm. All reagents were added in a volume of 50 μl/well and each sample was run in duplicate.
Data analysis for ELISA assays
ELISA antibody titers were calculated as the reciprocal plasma dilution using serial 2-fold dilutions of plasma from 1:100 to 1:12,800 and are expressed as the reciprocal end-point titer. Geometric mean titers (GMT) were calculated from the reciprocal end-point titers. Data analyses and graphs were generated using Graphpad Prism version 5. Statistical comparisons were made using nonparametric tests and a two-sided p-value of <0.05 was considered significant.
Surface plasmon resonance
To confirm the findings of the ELISA studies, surface plasmon resonance (SPR) measurements were conducted with a Biacore T100 using CM5 or SA chips prepared as described below. Lysozyme was immobilized onto a CM5 chip using the amine coupling kit. Unbound free amines were quenched with ethanolamine. All solutions used during the immobilization steps had a flow rate of 10 μl/min and all experiments were performed at 25°C. The immobilization wizard packaged within the T100 control software was used to immobilize 500 nM lysozyme in 20 mM sodium phosphate, pH 7.4 (5 min contact time) in flow cell 1. Cyc V2 biotinylated peptides were prepared at 0.1–1 μM in Tris-buffered saline, pH 7.4, and allowed to flow for 2–10 min over the streptavidin-coated surface of the SA chip, which resulted in immobilization of approximately 895, 1,401, and 2,073 response units (RU), respectively, for the 16, 25, and 42 aa containing Cyc V2 peptides. Cyc V2 Scr MR (1900 RU) and Cyc V2 Scr Fl peptides (1,850 RU) were immobilized to the respective flow cells.
Plasma samples were heat inactivated (56°C, 45 min), centrifuged, and the supernatant filtered prior to use. The plasma was diluted 1:50 in Tris-buffered saline, pH 7.4. The diluted plasma samples were passed over the chip surface at 30 μl/min for 3 min followed by a 5-min dissociation period. At the end of the 5-min period, a 50 nM solution of affinity-purified gamma chain-specific sheep antihuman IgG antibody was passed over the peptide-coated Ig-bound surface for 2 min at a flow rate of 10 μl/min. After a 70-s dissociation period, the biotinylated peptides were regenerated using a 30-s pulse of 50 mM HCl, a 30-s pulse of 100 mM EDTA in 20 mM Tris, pH 7.4, another 30-s pulse of 50 mM HCl, followed by a 1 min injection of Tris-buffered saline, pH 7.4. Nonspecific binding was subtracted and data analysis was performed using the BIA evaluation 4.1 software. The reported RUs for the IgG-specific values are the difference between the average value of a 5-s window taken 60 s after the end of the anti-IgG injection and the average value of a 5-s window taken 10 s before the beginning of the anti-IgG injection. The data (RU) are presented as dot plots for individual plasma samples. A representative experiment of three independent experiments is shown.
Results
RV144 antibody responses to recombinant gp120 proteins
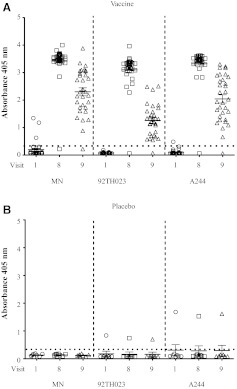
To investigate the antibody responses induced by the RV144 vaccine regimen, we performed ELISAs using the vaccine matched recombinant gp120 proteins from the three HIV-1 vaccine strains, MN, 92TH023, and A244. A244 and MN comprised the protein boost and 92TH023 was in the canarypox vector. Set Z plasma samples from visit 1 (prevaccination), visit 8 (two weeks), and visit 9 (28 weeks) post-last inoculation were tested against gp120 proteins (Fig. 2). The visit 1 responses were negative except for three vaccinees' samples where the responses were above the cutoff value for the assay (Fig. 2A). However, the visit 1 responses for these three samples were markedly lower than visit 8 and 9 samples and were considered nonspecific. Of the 32 vaccinees, 31 (97%) had robust antibody responses against all three recombinant proteins at visit 8 with reactivity against MN and A244 being the highest followed by 92TH023 (Fig. 2A). One of the eight placebo subjects had antibody responses to both 92TH023 and A244 proteins at all three visits but no responses to MN (Fig. 2B). Although the frequency of positive responses at visit 9 did not change 6 months after administration of the last vaccination, the magnitude of antibody responses against all three gp120 proteins declined with antibody levels to 92TH023 being the lowest followed by A244 and MN (Fig. 2A).
FIG. 2.
IgG antibody responses to recombinant HIV-1 Env proteins in RV144 vaccinees and placebo recipients. (A) Antibody responses to recombinant Env proteins gp120 MN, A244, and 92TH023 at visits 1, 8, and 9 determined by ELISA using plasma samples from set Z at a dilution of 1:100. Absorbance at 405 nm is shown on the y-axis. (B) Antibody responses to the same recombinant proteins and visits in placebo recipients. Dotted line indicates the cutoff for the assay in the absence of the capturing protein. Each sample was run in triplicate and data represent the average of two independent experiments. Bars represent the standard error of the mean.
Detection of V2-specific antibody responses in RV144 by peptide microarray analysis
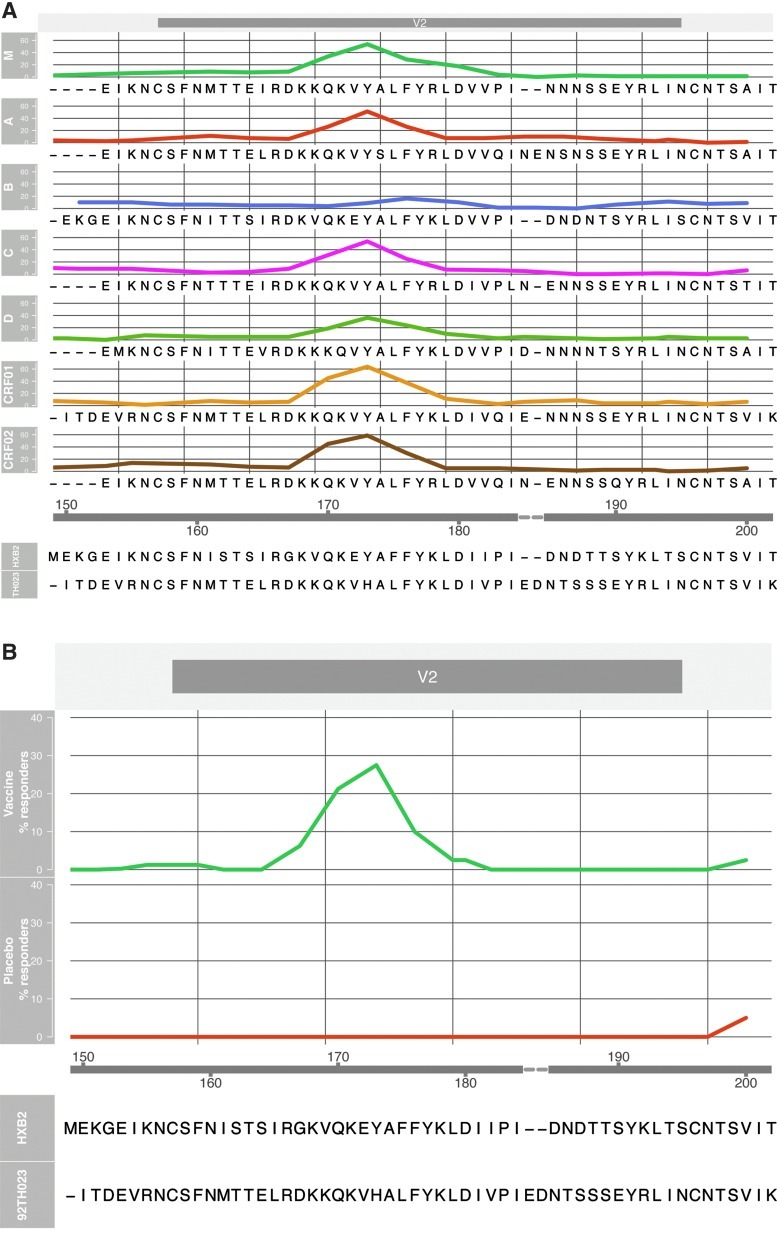
Binding antibodies in RV144 set A plasma samples were assessed by peptide microarray analysis using linear overlapping peptides covering the entire V2 loop of all major genetic subtypes and CRFs of gp120 (Fig. 3). Of the seven sets of HIV-1 gp120 peptides tested, six displayed antibody responses against V2. Notably, there was little or no reactivity with the V2 peptides of subtype B despite the fact that the vaccinees had received gp120 protein boosts derived from both subtypes B and CRF01_AE (Fig. 3A). Based on this analysis, the RV144 vaccine induced antibody responses that specifically targeted 15-mer peptides covering the V2 loop with 25% (20/80) of the vaccinated subjects having antibodies to this region (Fig. 3B).
FIG. 3.
Evaluation of antibody responses to overlapping peptides of V2 by peptide microarray analysis. Microarray peptide analysis was done using plasma from 80 HIV-1-uninfected vaccine recipients and 20 placebo recipients (set A). (A) Reactivity of plasma with peptides from seven HIV-1 Group M subtypes (M consensus, A, B, C, D, CRF01, and CRF02). (B) Overall reactivity of RV144 plasma samples to the V2 loop. Peptide positions are shown on the x-axis and percent responses are shown on the y-axis. The peptide positions are aligned with the amino acid sequence of HXB2 protein and the sequence of 92TH023 V2 loop is shown for comparison.
Antibody responses in RV144 target the mid-region of the V2 loop
Based on the antibody responses to linear peptides that span the V2 loop in peptide microarray analysis, we concluded that antibodies interacted with the mid-region of the V2 loop having the following aa sequence—KQKVHALFYKLDIVPI—and corresponding to HXB2 aa 169–184 (Fig. 1). The mid-region of the V2 loop contains highly conserved aa including the α4β7 integrin binding motif LDI/V23 and ALFY, but also a less conserved region (KQKVH). To investigate further the induction of V2 loop-specific antibodies in RV144, we synthesized Cyc V2 peptides based on the canarypox Env glycoprotein 92TH023 (see Materials and Methods, Table 2, and Fig. 1). 92TH023 and A244 V2 loops vary by two aa at positions 188 and 189 (HXB2 numbering) but the mid-region is identical. The Cyc V2 peptide contained 42 aa extending from aa 158 to 199 (corresponding to HIV-1 HXB2 aa 157–196). To analyze the antibody responses targeting the flanks and mid-region of the V2 loop, two additional Cyc peptides were used (Table 2).
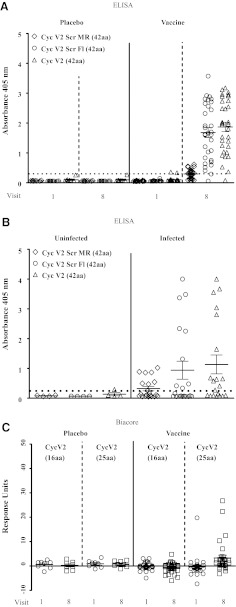
Plasma samples from visits 1 and 8 were tested against the Cyc V2 peptides by ELISA (Fig. 4). No antibody responses to Cyc V2 were detected in prevaccinated (visit 1) plasma. Of the 32 vaccinated subjects tested at visit 8, 31 had positive IgG antibody responses against Cyc V2 (97%) at visit 8 (Fig. 4A). Antibody responses to the Cyc V2 Scr Fl peptide were almost identical to those against the Cyc V2 peptide. Unlike Cyc V2 and Cyc V2 Scr Fl peptides, the IgG antibody responses against Cyc V2 Scr MR were significantly reduced with only 16/32 (50%) plasma samples being above background values (p<0.001) indicating that the majority of antibody responses against V2 targeted the mid-region of the loop (Fig. 4A). Identical responses were seen with Biacore for all three peptides (data not shown).
FIG. 4.
Antibody responses to cyclic V2 loop peptides by ELISA and Biacore. (A) Antibody responses at visits 1 and 8 to cyclic peptides Cyc V2, Cyc V2 Scr Fl, and Cyc V2 Scr MR by ELISA. (B) Antibody responses to Cyc V2, Cyc V2 Scr Fl, and Cyc V2 Scr MR by ELISA using 20 infected and four uninfected subjects. (C) Antibody responses to shorter cyclic V2 peptides by Biacore. In Biacore, plasma samples were used at a 1:50 dilution and the values are reported as response units. A representative experiment of three independent experiments is shown. ELISA and Biacore analyses were performed using the identical plasma samples (set Z). In ELISAs, plasma samples were used at a dilution of 1:100 and responses were considered positive if they exceeded 2.5 times the background (absence of capturing peptide) and are indicated by the dotted line. Each sample was tested in triplicate and the results represent the average of at least two independent experiments. Absorbance at 405 nm is shown on the y-axis.
To better characterize the antibody-binding epitope within the Cyc V2 peptide, two shorter Cyc V2 peptides, Cyc V2 (25aa) and Cyc V2 (16aa), were synthesized and tested by Biacore analysis (Table 2 and Fig. 4C). There was a single positive responder among the vaccinees at visit 1 to the Cyc V2 (25aa) peptide (Fig. 4C). Both low-frequency and weak binding were observed with the Cyc V2 (25aa) peptide (5/32, 16%; p<0.001) but no binding was observed with Cyc V2 (16aa) (Fig. 4C). No antibody responses were detected in plasma samples from visits 1 and 8 of placebo recipients to any of the Cyc V2 peptides (Fig. 4C).
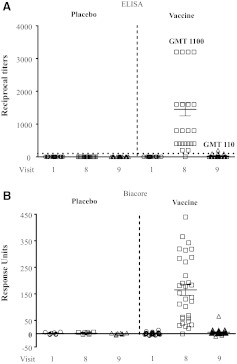
Durability of RV144 antibody responses to the V2 loop
To determine the durability of antibody responses against the V2 loop, the antibody responses at visits 8 and 9 were compared using the Cyc V2 peptide. The frequency of antibody responses at visit 8 was high 31/32 (97%), with a GMT of 1,100 and a range of 200–3,200 (Fig. 5A). The frequency and magnitude of antibody responses decreased significantly at visit 9 (6/32, 19%) compared to visit 8 (p<0.001) with a GMT of 110 and a range of 100–200 (p<0.001). Biacore analysis using the same peptide and plasma samples showed similar results with antibody responses to V2 diminishing by visit 9 (Fig. 5B). Reactivity was not detected in placebo recipients or vaccinees prior to immunization by either ELISA or Biacore assays (Fig. 5A and B).
FIG. 5.
Antibody responses to cyclic V2 loop decline over time. (A) Antibody responses to Cyc V2 peptide at visits 1, 8, and 9 by ELISA. Responses were considered positive if they exceeded 2.5 times the background (absence of capturing peptide) and are indicated by the dotted line. (B) Antibody responses to Cyc V2 peptide at visits 1, 8, and 9 by Biacore. Plasma samples were used at a 1:50 dilution and the values are reported as response units. A representative experiment of three independent experiments is shown. ELISA and Biacore analyses were done using plasma sample set Z.
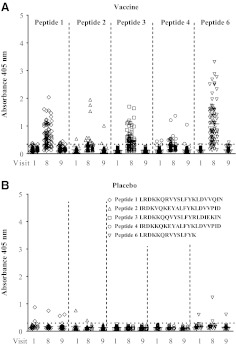
Antibody responses to the V2 loop in vaccinated subjects detected with linear peptides
Antibody responses to V2 were also examined using five linear biotinylated V2 loop-derived peptides that are heterologous in sequence to the RV144 immunogens (Table 2 and Fig. 6). In this ELISA, RV144 plasma samples from set C were used (80 vaccine and 20 placebo recipients). Antibody responses against all peptides were detected at visit 8 with peptide 6 having the highest frequency (93%) of responses followed by peptides 1 (84%), 3 (68%), 4 (39%), and 2 (38%), respectively (Fig. 6A). Poor responses against linear peptides 4, 6, and consensus B V2 peptides (microarray analysis) indicated that the presence of valine at position 172 is important for antibody binding. The responses to linear peptides declined to background levels at visit 9 and were similar to those observed with the V2 cyclic peptide using ELISA and Biacore (Fig. 5). Two samples from placebo recipients reacted with the linear peptides 1 and 6 and one sample reacted with peptide 2 (Fig. 6B). The sample reacting with peptides 1 and 6 (shorter version of peptide 1) was not the same as with peptide 2. These responses were considered nonspecific.
FIG. 6.
Antibody responses to linear V2 loop peptides decline over time. (A) Antibody responses to linear biotinylated peptides 1, 2, 3, 4, and 6 at visits 1, 8, and 9 by ELISA using plasma sample set C. Absorbance at 405 nm is shown on the y-axis. (B) ELISA using the same peptides and plasma of 20 placebos, set C. Plasma was used at a dilution of 1:100 and peptide aa sequences are shown in the right upper corner of (B). Samples were run in duplicate and are the average of two experiments.
Antibody responses to V2 in HIV-1-infected control subjects
V2-specific IgG antibody responses generated by the RV144 vaccine regimen were compared to those induced by HIV-1 infection. Antibodies to Cyc V2, Cyc V2 Scr MR, and Cyc V2 Scr Fl were measured using plasma samples from 20 HIV-1 CRF01_AE-infected individuals and four HIV-1-uninfected Thai subjects. Ten of the 20 infected subjects (50%) had antibodies to Cyc V2 (GMT: 400; range: 100–3,200) (Fig. 4B), which were significantly lower than the frequency and magnitude for the RV144 vaccine group (p<0.001). Similar antibody-binding responses in frequency and magnitude were measured with the Cyc V2 Scr Fl peptide suggesting that the antibody responses in HIV-1 infection also target the mid-region of V2 loop and are similar to those generated by the RV144 vaccine regimen (Figs. 4A and 5). The magnitude of antibody responses to the Cyc V2 Scr MR peptide was low, 30% (6/20, Fig. 4B), and antibody responses were not detected in HIV-1-uninfected controls (Fig. 4B).
Biacore analysis using plasma samples from 18 of the 20 HIV-1-infected subjects tested by ELISA gave similar results (data not shown).
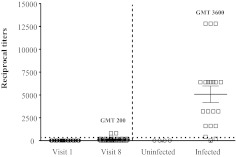
Antibody responses to the V3 loop in HIV-1-uninfected vaccinated subjects and HIV-1-infected subjects
Antibody responses to V2 were also compared to those generated against the V3 loop. Plasma from 32 vaccinated and eight placebo recipients at visits 1 and 8 were tested against Cyc V3 (Table 2 and Fig. 7). The frequency (18/32, 56%) and magnitude (GMT: 200; range: 100–800) of antibody responses against V3 were lower in vaccinated subjects (Fig. 7). In comparison, the Cyc V3 antibody responses in HIV-infected individuals were both higher in frequency (19/20, 95%) and magnitude (GMT: 3,600; range: 200–12,800) (Fig. 7). Antibody reactivity against the Cyc V3 peptide was not detected in uninfected control subjects or vaccine recipients at visit 1. Unlike HIV-1-uninfected RV144-vaccinated subjects, HIV-1-infected individuals had a higher frequency of antibody responses against 92TH023 V3 (95%) compared to V2 (50%; p=0.003). Antibody responses against MN Cyc V3 were not tested.
FIG. 7.
Antibody responses to cyclic V3 peptide in vaccinated uninfected subjects, HIV-1-infected and HIV-uninfected. Antibody responses to Cyc V3 peptide (strain 92TH023) using plasma samples from RV144 visits 1 and 8 (set Z) and from 20 HIV-1-infected and four HIV-1-uninfected subjects. Plasma samples were used at a starting dilution of 1:100. Cutoff values are indicated by the dotted line. Panels represent the average of at least two independent experiments.
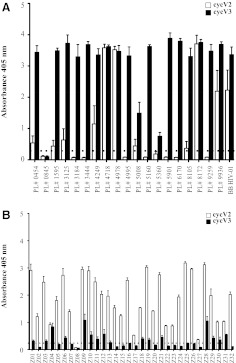
Comparison of V2 and V3 responses in HIV-1-infected and vaccinated subjects
The levels of Cyc V2 and V3 antibody responses were compared in the same HIV-1-infected individuals. Seven out of 20 (35%) subjects tested had Cyc V3 loop antibody responses with a GMT of 4,800 (range: 1,600–12,800) but undetectable levels of antibodies against the Cyc V2 loop (Fig. 8A). In 3/20 (15%) subjects, the V2 response was high (GMT: 3,200) but still below the V3 antibody responses (GMT: 5,100; range: 3,200–6,400). All HIV-1-infected subjects had higher responses to Cyc V3 than to Cyc V2 (Fig. 8A). Of the 20 infected subjects, one (5%) did not have an antibody response to either V2 or V3 loop peptides.
FIG. 8.
Comparison of antibody responses to V2 and V3 loops in RV144 vaccinees and HIV-1 naturally infected subjects. (A) Comparison of antibody responses to gp120 V2 and V3 loops from HIV-1-infected individuals. (B) Antibody responses to gp120 V2 and V3 loops in RV 144 vaccinated individuals. Plasma dilutions were at 1:100 and absorbance at 405 nm is shown on the y-axis. Cutoff values are indicated by the dotted line.
Of the RV144 vaccinated subjects, 31/32 (97%) had an antibody response to the Cyc V2 peptide, but only 18/32 (56%) had antibody responses to the Cyc V3 peptide (Fig. 8B). None of the 32 vaccinated subjects had a V3 loop antibody response that was higher than that of V2 (Fig. 8B). One of the vaccinated individuals did not have antibody responses to either the V2 or V3 loops.
Discussion
The estimated efficacy in the RV144 vaccine trial was 31.2% by 42 months in the modified intent to treat population,1 but data through months 12 and 18 showed estimated efficacy of 60% and 44%, respectively.54 Initial studies demonstrated that binding antibody to vaccine gp120 antigens was found in 95% of vaccine recipients 2 weeks post-final vaccination but waned substantially over the next 6 months. Using peptide microarray analysis, ELISA, and Biacore studies we demonstrated that the immunogens used in the RV144 trial induced antibodies to the V2 loop of gp120 from diverse HIV-1 subtypes. These responses were high in frequency and magnitude but also declined rapidly to nearly undetectable levels at 28 weeks post-last vaccination. Haynes et al.20 have shown that IgG against a conformational V1/V2 epitope from 2 weeks postvaccination was inversely correlated with infection rate in the RV144 trial. While diminution of the V2 antibody may have corresponded to waning vaccine efficacy in RV 144, a mechanistic link between the two observations is purely speculative at this point. Further research in the “time-dependent” correlates analysis will be required to pursue this hypothesis.
Using scrambled cyclic and linear peptides of the V2 loop we determined that antibodies induced by RV144 target the mid-region of the loop, which includes highly conserved aa including the integrin-binding motif LDI/V but also less conserved regions. It is likely that RV144 antibodies have multiple binding epitopes within the mid-region and further studies are needed to characterize their binding specificities.55 Two monoclonal antibodies isolated from RV144, CH58, and CH59 have different but partly overlapping binding.56 It is likely that not all antibody responses targeting the V2 loop are protective and certain epitopes could be more protective than others in preventing HIV-1 acquisition. Furthermore, volunteers who develop antibodies to Cyc V2 may or may not develop antibodies to gp70, a scaffold glycosylated protein containing the V1/V2 loops used in the case control study.20 Scrambling the flanks of the V2 loop had minimal effect on the frequency or magnitude of binding in vaccinated and infected subjects indicating that the majority of the antibodies target epitopes within the mid-region. To further define the binding epitopes of these antibodies, Cyc V2 (16aa) and Cyc V2 (25aa) peptides were used. Of these two peptides, weak responses were observed only to Cyc V2 (25aa). Peptide Cyc V2 (25aa) includes all aa found in the V2 mid-region except KQKV at the N terminus of the peptide, suggesting that these four aa could be important for epitope recognition by V2 antibodies. Alternatively, conformational changes of shorter cyclized peptides may not bind to antibodies. It is possible that certain conformational antibodies induced by the recombinant proteins used in the vaccine regimen may not bind to peptides. Furthermore, antibodies targeting glycosylated epitopes within the V1/V2 loops were not investigated in this study.
Recently, a proposed structure of scaffolded V1/V2 loops complexed with the broadly neutralizing monoclonal antibody PG9 has been reported.47 V1/V2 loops form an intertwined four-stranded β sheet domain composed of four antiparallel β-strands designated A, B, C, and D. PG9 interacts with N-linked glycans on the V1/V2 loop but also interacts with aa within strand C, which is part of the V2 loop region. Strand C is part of the mid-region of the V2 loop and interacts strongly with antibodies induced by the RV144 vaccine regimen. In addition to its interaction with PG9, strand C contributes to the formation of a nonglycosylated hydrophobic core and participates with the other three strands to form a single topological entity.47 The consequence of interaction of this topological entity with antibodies is currently unknown.
Comparing antibody responses and sequence alignments of peptides used in the study, we conclude that valine at position 172 (HXB2 numbering) could be important in epitope formation and reactivity. Valine at this position is characteristic of CRF01_AE V2 loops and differs from the glutamate found most commonly at this position in subtype B strains. Linear and cyclic peptides lacking valine at position 172 were poorly reactive or nonreactive with the samples tested.
The generation of antibody responses against the mid-region of the V2 loop was also confirmed by linear biotinylated peptides. Of the five peptides tested, peptide 6 had the highest response followed by peptides 1 and 3. Peptide 6 is a shorter version of peptide 1 and lacks the LDI motif. The high antibody response to this peptide suggests that the core epitope recognized by antibodies to linear peptides does not include LDI; however, this does not imply that either the LDI is not targeted by antibodies to conformational epitopes in V2 or is not sterically blocked by V2 antibodies to the core epitope. In general, linear peptides had weaker responses than the 92TH023 Cyc V2 peptide possibly due to the lack of high homology with the aa sequences of the V2 loop used in the vaccine and to conformational differences as cyclic peptides may assume a more complex conformational folding.57–61
Using microarray analysis, RV144 vaccinee plasmas were reactive with six of the seven sets of overlapping V2 peptides from consensus gp160 proteins from group M and subtypes A, C, D, CRF01, and CRF02; only those from subtype B were negative. The frequency of antibody responses to the V2 loop by microarray peptide analysis was much lower than that observed for cyclic V2 peptides using ELISAs and Biacore. The difference in binding could be attributed to aa sequence variability but also to conformation, adsorption, or peptide length.
In contrast to the V2-specific IgG antibody responses induced by the RV144 vaccine regimen, the Cyc V3 loop responses in RV144 were of lower magnitude (GMT of 200) and frequency (56%), thus indicating that RV144 did not induce vigorous antibody responses to the clade E V3 loop region. However, responses to the cyclic V3 loop in HIV-1-infected individuals were higher in magnitude (GMT of 3,600) and frequency (95%). Infected subjects had robust antibody responses to the Cyc V3, but 35% of those having strong antibody responses to V3 lacked antibodies to the Cyc V2. There was an 18-fold higher response to the Cyc V3 peptide in HIV-1-infected individuals compared to the RV144 vaccinees. However, V2 loop-specific antibodies in infected individuals were much less frequent (50%) and of lower magnitude than those induced by the RV144 vaccine regimen. Whether this observation of high V2 and low V3 responses in vaccinees compared to HIV-1-infected subjects is related to the vaccine antigen is unknown. Vaccine-induced antibody responses have many similarities to antibodies found in infected individuals but generally are not neutralizing in cell assays.62
Heterosexual transmission is usually a single infectious event and productive infection reflects the expansion of the founder virus.36,63,64 The V2 loop has a number of features that might involve initial events in infection. It has a highly conserved length among early or transmitted/founder isolates and length and glycosylation of the V2 loop are commonly associated with escape from neutralizing antibodies.30,35,65 Furthermore, V2 contains a putative α4β7 integrin-binding motif of uncertain significance.39 Although V2 has been implicated in early virus transmission the precise mechanisms of attachment and cell-to-cell dissemination are not well defined.23,24,36,65–67 Rolland et al. have reported that sieve mutations in HIV-1 breakthrough infections in RV144 contain mutations at positions 169 and 181 in V2.50 T cell epitope mapping studies using PBMCs from HIV-1-uninfected RV144 vaccine recipients and the interferon-gamma ELISpot assay also showed preferential targeting of the V2 loop and that the responding cells were CD4+.49 Taken together with the potential role of antibody to V2 as a correlate of protection,17,20 these findings suggest that the RV144 vaccine regimen may have induced protection by targeting the V1/V2 loops.
The generation of humoral immune responses by ALVAC-HIV and AIDSVAX B/E to the mid-region of the V2 loop raises hypotheses to test in regard to correlates of protection against HIV-1 infection. The V2 loop (like the V3 loop) contains many conserved features, and several anti-V2 monoclonal antibodies show broad cross-reactivity41,48,68 (S. Zolla-Pazner, unpublished), suggesting that antibodies can target conserved elements in the V2 loop and that this region of Env might be pivotal for the induction of protective antibodies by successful vaccines. Since replacement of various aa in the V2 loop can alter the structure and function of the Env rendering HIV-1 less infectious or more sensitive to neutralization,33,48,69 the choice of Env and/or V2 sequences used in rational vaccine design should be carefully considered.
Acknowledgments
The authors thank SCHARP for assisting with the microarray analysis, James Swetnam for providing the biotinylated subtype B linear peptides, Kelly Soderberg for coordination of sample shipment and project management, Rob O'Connell for constructive reading of the manuscript, Robert Parks, Ryan Meyerhoff, and Krissey Lloyd for ELISA technical assistance, and Ellen Turk for performing the peptide microarray analysis. These studies were supported in part by an Interagency Agreement Y1-AI-2642-12 between U.S. Army Medical Research and Materiel Command (USAMRMC) and the National Institutes of Allergy and Infectious Diseases. In addition, this work was supported by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine and the U.S. Department of Defense. This research was funded, in part, by the U.S. National Institute of Allergy and Infectious Diseases. Peptide microarray work was funded by the Bill and Melinda Gates Foundation's Collaboration for AIDS Vaccine Discovery (OPP38744) and the NIH Vaccine Research Center. S.Z.P. and T.J.C. receive funding from NIH Grants HL59725 (S.Z.P.) and AI084119 (T.J.C.) and the Department of Veterans Affairs. Supported in part by grants from the Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery to Dr. Haynes.
MOPH-TAVEG Collaboration: MOPH: Dr. Supachai Rerks-Ngarm, Dr. Prayura Kunasol, Dr. Nakorn Premsri, Dr. Chawetsan Namwat, Prof. Prasert Thongcharoen Mahidol, Prof. Punnee Pitisuttithum, Dr. Jaranit Kaewkungwal. RTA: MG Sorachai Nitayaphan, MG (ret) Chirapa Eamsila. MHRP: Dr. Nelson L. Michael, Dr. Jerome H. Kim, Dr. Robert C O'Connell, Dr. Merlin L. Robb, Dr. Robert Paris, Charla Andrews, Dr. Jeffrey Currier.
AFRIMS: Dr. Viseth Ngauy, Dr. Mark S. de Souza, Dr. Nicos Karasavvas, Ms. Rapee Trichavaroj, Ms. Susan T. Mason, Ms. Bessara Nuntapinit, Ms. Nampueng Churikanont, Dr. Michael Benenson, Ms. Patricia Morgan.
Abstract presentations: AIDS Vaccine 2011: Global Collaboration and Coordination to Advance HIV Vaccine Research, September 12–15, 2011, Bangkok, Thailand.
Disclaimer: The views expressed in this article are those of the authors and do not reflect the official policy of the Department of the Army, Department of Defense, the Department of Veterans Affairs, or the U.S. Government.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Rerks-Ngarm S. Pitisuttithum P. Nitayaphan S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361(23):2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 2.Kim JH. Rerks-Ngarm S. Excler JL. Michael NL. HIV vaccines: Lessons learned and the way forward. Curr Opin HIV AIDS. 2010;5(5):428–434. doi: 10.1097/COH.0b013e32833d17ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes BF. Liao HX. Tomaras GD. Is developing an HIV-1 vaccine possible? Curr Opin HIV AIDS. 2010;5(5):362–367. doi: 10.1097/COH.0b013e32833d2e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nitayaphan S. Pitisuttithum P. Karnasuta C, et al. Safety and immunogenicity of an HIV subtype B and E prime-boost vaccine combination in HIV-negative Thai adults. J Infect Dis. 2004;190(4):702–706. doi: 10.1086/422258. [DOI] [PubMed] [Google Scholar]
- 5.Karnasuta C. Paris RM. Cox JH, et al. Antibody-dependent cell-mediated cytotoxic responses in participants enrolled in a phase I/II ALVAC-HIV/AIDSVAX B/E prime-boost HIV-1 vaccine trial in Thailand. Vaccine. 2005;23(19):2522–2529. doi: 10.1016/j.vaccine.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 6.Cao H. Kaleebu P. Hom D, et al. Immunogenicity of a recombinant human immunodeficiency virus (HIV)-canarypox vaccine in HIV-seronegative Ugandan volunteers: Results of the HIV Network for Prevention Trials 007 Vaccine Study. J Infect Dis. 2003;187(6):887–895. doi: 10.1086/368020. [DOI] [PubMed] [Google Scholar]
- 7.Gupta K. Hudgens M. Corey L, et al. Safety and immunogenicity of a high-titered canarypox vaccine in combination with rgp120 in a diverse population of HIV-1-uninfected adults: AIDS Vaccine Evaluation Group Protocol 022A. J Acquir Immune Defic Syndr. 2002;29(3):254–261. doi: 10.1097/00126334-200203010-00005. [DOI] [PubMed] [Google Scholar]
- 8.Evans TG. Keefer MC. Weinhold KJ, et al. A canarypox vaccine expressing multiple human immunodeficiency virus type 1 genes given alone or with rgp120 elicits broad and durable CD8+ cytotoxic T lymphocyte responses in seronegative volunteers. J Infect Dis. 1999;180(2):290–298. doi: 10.1086/314895. [DOI] [PubMed] [Google Scholar]
- 9.Belshe RB. Gorse GJ. Mulligan MJ, et al. Induction of immune responses to HIV-1 by canarypox virus (ALVAC) HIV-1 and gp120 SF-2 recombinant vaccines in uninfected volunteers. NIAID AIDS Vaccine Evaluation Group. AIDS. 1998;12(18):2407–2415. doi: 10.1097/00002030-199818000-00009. [DOI] [PubMed] [Google Scholar]
- 10.Pitisuttithum P. Gilbert P. Gurwith M, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194(12):1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 11.Flynn NM. Forthal DN. Harro CD, et al. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 2005;191(5):654–665. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert PB. Peterson ML. Follmann D, et al. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis. 2005;191(5):666–677. doi: 10.1086/428405. [DOI] [PubMed] [Google Scholar]
- 13.Hessell AJ. Poignard P. Hunter M, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15(8):951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibata R. Igarashi T. Haigwood N, et al. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;5(2):204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 15.Baba TW. Liska V. Hofmann-Lehmann R, et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6(2):200–206. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 16.Mascola JR. Stiegler G. VanCott TC, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6(2):207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 17.Barouch DH. Liu J. Li H, et al. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012;482:89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barnett SW. Burke B. Sun Y, et al. Antibody-mediated protection against mucosal simian-human immunodeficiency virus challenge of macaques immunized with alphavirus replicon particles and boosted with trimeric envelope glycoprotein in MF59 adjuvant. J Virol. 2010;84(12):5975–5985. doi: 10.1128/JVI.02533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert JS. Mofenson LM. Fletcher CV, et al. Safety and pharmacokinetics of hyperimmune anti-human immunodeficiency virus (HIV) immunoglobulin administered to HIV-infected pregnant women and their newborns. Pediatric AIDS Clinical Trials Group Protocol 185 Pharmacokinetic Study Group. J Infect Dis. 1997;175(2):283–291. doi: 10.1093/infdis/175.2.283. [DOI] [PubMed] [Google Scholar]
- 20.Haynes BF. Gilbert PB. McElrath MJ, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366(14):1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montefiori DC. Karnasuta C. Huang Y, et al. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J Infect Dis. 2012;206(3):431–441. doi: 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen B. Vogan EM. Gong H, et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433(7028):834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 23.Arthos J. Cicala C. Martinelli E, et al. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9(3):301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 24.Cicala C. Martinelli E. McNally JP, et al. The integrin alpha4beta7 forms a complex with cell-surface CD4 and defines a T-cell subset that is highly susceptible to infection by HIV-1. Proc Natl Acad Sci USA. 2009;106(49):20877–20882. doi: 10.1073/pnas.0911796106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker LM. Simek MD. Priddy F, et al. A limited number of antibody specificities mediate broad, potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 2010;6(8) doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doores KJ. Burton DR. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J Virol. 2010;84(20):10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu G. Liu J. Taylor KA. Roux KH. Structural comparison of HIV-1 envelope spikes with and without the V1/V2 loop. J Virol. 2011;85(6):2741–2750. doi: 10.1128/JVI.01612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spurrier B. Sampson JM. Totrov M, et al. Structural analysis of human and macaque mAbs 2909 and 2.5B: Implications for the configuration of the quaternary neutralizing epitope of HIV-1 gp120. Structure. 2011;19(5):691–699. doi: 10.1016/j.str.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch RM. Shen T. Gnanakaran S. Derdeyn CA. Appreciating HIV type 1 diversity: Subtype differences in Env. AIDS Res Hum Retroviruses. 2009;25(3):237–248. doi: 10.1089/aid.2008.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sagar M. Wu X. Lee S. Overbaugh J. Human immunodeficiency virus type 1 V1–V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J Virol. 2006;80(19):9586–9598. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore PL. Ranchobe N. Lambson BE, et al. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 2009;5(9):e1000598. doi: 10.1371/journal.ppat.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Rourke SM. Schweighardt B. Phung P, et al. Mutation at a single position in the V2 domain of the HIV-1 envelope protein confers neutralization sensitivity to a highly neutralization-resistant virus. J Virol. 2010;84(21):11200–11209. doi: 10.1128/JVI.00790-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinter A. Honnen WJ. He Y, et al. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol. 2004;78(10):5205–5215. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sagar M. HIV-1 transmission biology: Selection and characteristics of infecting viruses. J Infect Dis. 2010;202(Suppl 2):S289–296. doi: 10.1086/655656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chohan B. Lang D. Sagar M, et al. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1–V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol. 2005;79(10):6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derdeyn CA. Decker JM. Bibollet-Ruche F, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303(5666):2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 37.Wagner N. Lohler J. Kunkel EJ, et al. Critical role for beta7 integrins in formation of the gut-associated lymphoid tissue. Nature. 1996;382(6589):366–370. doi: 10.1038/382366a0. [DOI] [PubMed] [Google Scholar]
- 38.von Andrian UH. Mackay CR. T-cell function and migration. Two sides of the same coin. N Engl J Med. 2000;343(14):1020–1034. doi: 10.1056/NEJM200010053431407. [DOI] [PubMed] [Google Scholar]
- 39.Parrish NF. Wilen CB. Banks LB, et al. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin alpha4beta7. PLoS Pathog. 2012;8(5):e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker LM. Phogat SK. Chan-Hui PY, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zolla-Pazner S. Cardozo T. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat Rev Immunol. 2010;10(7):527–535. doi: 10.1038/nri2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Changela A. Wu X. Yang Y, et al. Crystal structure of human antibody 2909 reveals conserved features of quaternary structure-specific antibodies that potently neutralize HIV-1. J Virol. 2011;85(6):2524–2535. doi: 10.1128/JVI.02335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore PL. Gray ES. Sheward D, et al. Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop. J Virol. 2011;85(7):3128–3141. doi: 10.1128/JVI.02658-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorny MK. Stamatatos L. Volsky B, et al. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J Virol. 2005;79(8):5232–5237. doi: 10.1128/JVI.79.8.5232-5237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Honnen WJ. Krachmarov C. Kayman SC, et al. Type-specific epitopes targeted by monoclonal antibodies with exceptionally potent neutralizing activities for selected strains of human immunodeficiency virus type 1 map to a common region of the V2 domain of gp120 and differ only at single positions from the clade B consensus sequence. J Virol. 2007;81(3):1424–1432. doi: 10.1128/JVI.02054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson JE. Franco K. Elliott DH, et al. Quaternary epitope specificities of anti-HIV-1 neutralizing antibodies generated in rhesus macaques infected by the simian/human immunodeficiency virus SHIVSF162P4. J Virol. 2010;84(7):3443–3453. doi: 10.1128/JVI.02617-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McLellan JS. Pancera M. Carrico C, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gorny MK. Moore JP. Conley AJ, et al. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J Virol. 1994;68(12):8312–8320. doi: 10.1128/jvi.68.12.8312-8320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Souza MS. Ratto-Kim S. Chuenarom W, et al. The Thai phase III trial (RV144) vaccine regimen induces T cell responses that preferentially target epitopes within the V2 region of HIV-1 envelope. J Immunol. 2012;188(10):5166–5176. doi: 10.4049/jimmunol.1102756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rolland M. Larsen B. Edlefsen P. Sequence Analysis of HIV-1 Breakthrough Infections in the RV144 Trial. Presented at AIDS Vaccine 2011; Bangkok, Thailand. Sep 12–15;; 2011. Abstract. [Google Scholar]
- 51.Gaschen B. Taylor J. Yusim K, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296(5577):2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 52.Sandberg M. Eriksson L. Jonsson J, et al. New chemical descriptors relevant for the design of biologically active peptides. A multivariate characterization of 87 amino acids. J Med Chem. 1998;41(14):2481–2491. doi: 10.1021/jm9700575. [DOI] [PubMed] [Google Scholar]
- 53.Droit A. Cheung C. Gottardo R. rMAT—an R/Bioconductor package for analyzing ChIP-chip experiments. Bioinformatics. 2010;26(5):678–679. doi: 10.1093/bioinformatics/btq023. [DOI] [PubMed] [Google Scholar]
- 54.Robb ML. Rerks-Ngarm S. Nitayaphan S, et al. Risk behaviour and time as covariates for efficacy of the HIV vaccine regimen ALVAC-HIV (vCP1521) and AIDSVAX B/E: A post-hoc analysis of the Thai phase 3 efficacy trial RV 144. Lancet Infect Dis. 2012;12(7):531–537. doi: 10.1016/S1473-3099(12)70088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haynes BF. Case Control Study of the RV144 Trial for Immune Correlates: The Analysis Way Forward. Presented at AIDS Vaccine 2011; Bangkok, Thailand. Sep 12–15;; 2011. [Google Scholar]
- 56.Liao H-X. Bonsignori M. Alam SM, et al. HIV-1 Envelope Antibodies Induced by ALVAC-AIDSVAX B/E Vaccine Target a Site of Vaccine Immune Pressure Within the C β Strand of gp120 V1V2. Presented at Keystone Symposia 2012; Keystone, Colorado. Mar 21–26;; 2012. Abstract. [Google Scholar]
- 57.Belhadj Jrad B. Bahraoui E. Antigenicity of linear and cyclic peptides mimicking the disulfide loops in HIV-2 envelope glycoprotein: Synthesis, reoxidation and purification. J Pept Res. 1998;51(5):370–385. doi: 10.1111/j.1399-3011.1998.tb01228.x. [DOI] [PubMed] [Google Scholar]
- 58.Misumi S. Endo M. Mukai R, et al. A novel cyclic peptide immunization strategy for preventing HIV-1/AIDS infection and progression. J Biol Chem. 2003;278(34):32335–32343. doi: 10.1074/jbc.M301209200. [DOI] [PubMed] [Google Scholar]
- 59.Bogdanowich-Knipp SJ. Jois DS. Siahaan TJ. The effect of conformation on the solution stability of linear vs. cyclic RGD peptides. J Pept Res. 1999;53(5):523–529. doi: 10.1034/j.1399-3011.1999.00055.x. [DOI] [PubMed] [Google Scholar]
- 60.Mezo G. Majer Z. Vass E, et al. Conformational study of linear and cyclic peptides corresponding to the 276–284 epitope region of HSV gD-1. Biophys Chem. 2003;103(1):51–65. doi: 10.1016/s0301-4622(02)00232-6. [DOI] [PubMed] [Google Scholar]
- 61.Hoogerhout P. Donders EM. van Gaans-van den Brink JA, et al. Conjugates of synthetic cyclic peptides elicit bactericidal antibodies against a conformational epitope on a class 1 outer membrane protein of Neisseria meningitidis. Infect Immun. 1995;63(9):3473–3478. doi: 10.1128/iai.63.9.3473-3478.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beddows S. Lister S. Cheingsong R, et al. Comparison of the antibody repertoire generated in healthy volunteers following immunization with a monomeric recombinant gp120 construct derived from a CCR5/CXCR4-using human immunodeficiency virus type 1 isolate with sera from naturally infected individuals. J Virol. 1999;73(2):1740–1745. doi: 10.1128/jvi.73.2.1740-1745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abrahams MR. Anderson JA. Giorgi EE, et al. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J Virol. 2009;83(8):3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sagar M. Laeyendecker O. Lee S, et al. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J Infect Dis. 2009;199(4):580–589. doi: 10.1086/596557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rong R. Bibollet-Ruche F. Mulenga J, et al. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J Virol. 2007;81(3):1350–1359. doi: 10.1128/JVI.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nawaz F. Cicala C. Van Ryk D, et al. The genotype of early-transmitting HIV gp120s promotes alphabeta-reactivity, revealing alphabetaCD4+ T cells as key targets in mucosal transmission. PLoS Pathog. 2011;7(2):e1001301. doi: 10.1371/journal.ppat.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cicala C. Arthos J. Fauci AS. HIV-1 envelope, integrins and co-receptor use in mucosal transmission of HIV. J Transl Med. 2010;9(Suppl 1):S2. doi: 10.1186/1479-5876-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Israel ZR. Gorny MK. Palmer C, et al. Prevalence of a V2 epitope in clade B primary isolates and its recognition by sera from HIV-1-infected individuals. AIDS. 1997;11(1):128–130. [PubMed] [Google Scholar]
- 69.Kowalski M. Potz J. Basiripour L, et al. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237(4820):1351–1355. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]