SUMMARY
Extracellular signals in development, physiology, homeostasis and disease often act by regulating transcription. Herein we describe a general method and specific resources for determining where and when such signaling occurs in live animals and for systematically comparing the timing and extent of different signals in different cellular contexts. We used recombinase-mediated cassette exchange (RMCE) to test the effect of successively deleting conserved genomic regions of the ubiquitously active Rosa26 promoter and substituting the deleted regions for regulatory sequences that respond to diverse extracellular signals. We thereby created an allelic series of embryonic stem cells and mice, each containing a signal-responsive sentinel with different fluorescent reporters that respond with sensitivity and specificity to retinoic acids, bone morphogenic proteins, activin A, Wnts or Notch, and that can be adapted to any pathway that acts via DNA elements.
INTRODUCTION
Numerous types of extracellular signals exert their effect on cell function by regulating gene transcription. Transcriptional effects are mediated by cis-acting sequences at promoters and enhancers that play crucial roles in controlling development, modulating physiology in response to perturbations, and maintaining homeostatic control of ongoing cell processes. Monitoring, in a tissue, where and when extracellular signals exert their effects on gene regulatory sequences provides important insights into inductive processes. The cis-acting sequences that respond to particular cell signals (‘response elements’) normally occur amidst sequences for other types of regulatory sequences, e.g. for cell type specificity; hence, many eukaryotic enhancers and promoters are modular in composition (Yamamoto, 1985). Prior studies have isolated individual response elements from complex regulatory sequences and tested them as multimers (Schirm et al., 1987), so that the element would report the appearance of an individual signal when linked to a convenient marker. Using this approach, transgenic mice have been created that successfully report where and when cells in embryos and animals receive signals for retinoic acid (Rossant et al., 1991), Wnt (DasGupta and Fuchs, 1999; Maretto et al., 2003), Notch (Hansson et al., 2006; Mizutani et al., 2007) and bone morphogenic protein (BMP) signaling (Monteiro et al., 2008). However, different transgene integration sites are subject to position effects and thus, for a given cell type, the readout for transgenic signaling sentinels and the differences between one sentinel and another might vary by parameters unrelated to signaling. In addition, many prior studies employed reporters that function in fixed cells, and the unique nature of transgene integration makes it difficult to create isogenic lines employing variants of the original structure.
In the present study, we circumvented these problems by devising a system to conveniently insert multimerized signaling elements within deleted regions of the promoter for the ubiquitously expressed Rosa26 gene (Soriano, 1999). Our approach creates an allelic series of embryonic stem (ES) cell lines and mice that have different response element substitutions in a highly defined and consistent genomic context. To this end, we modified the Rosa26 locus in ES cells so that it contains heterologous Lox sites flanking the gene’s promoter and first exon, allowing directional targeting of plasmid DNA containing similarly oriented Lox sites flanking a desired signaling sentinel construct. Recombinase-mediated cassette exchange (RMCE) approaches (Long et al., 2004; Jones et al., 2005) have been used on the Rosa26 locus to express proteins that differ from the native Rosa26 promoter (Chen et al., 2011). We have employed RMCE to systematically modify the Rosa26 promoter itself with multimerized response elements for different developmental signaling pathways. To find optimally responsive arrangements, we inserted different signaling elements into a series of nested deletions of the endogenous Rosa26 promoter and discovered that different deletion points work best with different signaling reporters. We show that the system efficiently allows the production of an allelic series of genetic sentinels to monitor diverse types of signals in different cell types, including live and fixed ES cells and mouse embryos. The system has been used to monitor signaling for retinoic acid, Wnt, BMP, activin A and Notch pathways and can be adapted, in principle, for any signal in which transcriptional output occurs via a specific response element.
RESULTS
Rationale for Rosa26 promoter sequence deletion and replacement
Our approach was to partially delete the regulatory sequences of a ubiquitously expressed gene so that its high level of basal promoter activity would be impaired and replaced by the activity of a multimerized response element. The desired features of the system were: retention of the potential for ubiquitous expression upon induction, low basal promoter activity and rigorous dependence upon an exogenous cell signal for transcription. We hypothesized that for different signaling reporters, different extents of deletion of the endogenous promoter might be optimal. To this end, we selected the mouse Rosa26 locus, which exhibits apparently tissue-ubiquitous expression and into which reporter sequences have successfully been inserted downstream of the promoter (Zambrowicz et al., 1997; Soriano, 1999; Srinivas et al., 2001). Using the transcription start site as the 3′ boundary of the Rosa26 promoter (Zambrowicz et al., 1997), we compared the mouse, rat and human promoter sequences for similarity. Blocks of DNA sequence were >95% conserved to −3 kb between the mouse and rat Rosa26 promoters and >80% conserved between the mouse and human (Fig. 1A, vertical red blocks; see Methods for details). Within the proximal 3 kb of the start site, there was a marked increase in the density of transcription factor binding motifs, conserved from mouse to human, from −1.2 kb through the transcription start site (Fig. 1B, to right of vertical dotted line). More detailed analysis of the promoter-proximal sequences revealed that the first gap in mouse-human homology occurs at −228 bp and a conserved CCAAT motif at −56 (Fig. 1C, dashed lines and boxed, respectively). On the basis of these landmarks, we made four deletions of the mouse Rosa26 promoter, extending from within exon 1 to −60, to −228, to −1217 and to −3000 bp of the mRNA start site, with each deletion replaced by a particular multimerized response element and a TATA box (Fig. 1D). Promoter deletion landmarks were chosen to be conserved between mouse and human so that the same approach could be applied to the Rosa26 locus in human ES cells.
Fig. 1.
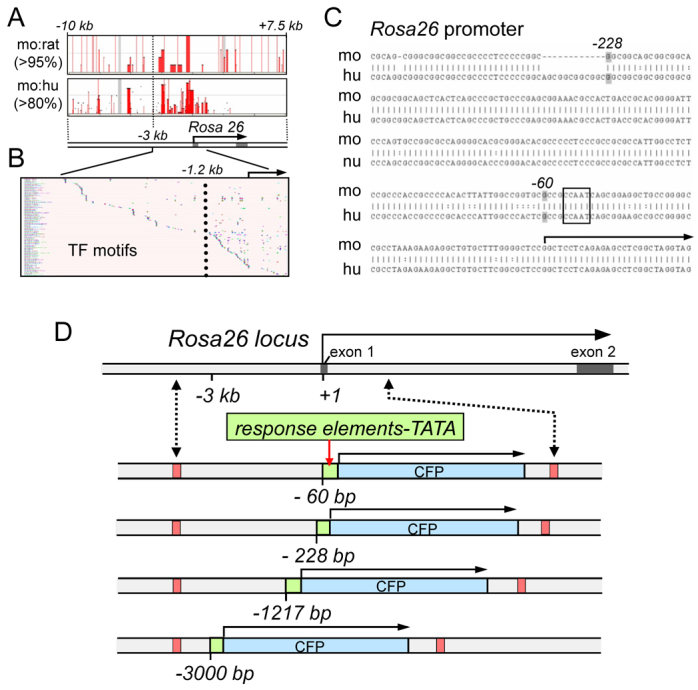
Rosa26 promoter analysis and design of deletions for signaling sentinel substitution. (A) Scheme depicts the Rosa26 locus; arrow, transcribed region. Boxes above depict sequence identities over 95% and 80% for mouse and rat (mo:ra) and mouse and human (mo:hu), respectively. Red bars within the boxes indicate conserved domains, largely extending to −3 kb of the start site of transcription. (B) VISTA plot of transcription factor motif distribution across the proximal 3 kb of the mouse Rosa26 promoter region. Vertical dotted line indicates a marked increase in motif density at −1.2 kb. (C) Comparison of the mouse and human Rosa26 promoter regions. A natural break occurs at −228 bp and a CCAAT box is evident downstream of −60 bp. (D) RMCE at the Rosa26 locus (upper scheme) results in substitutions that replace the regions from −4 kb to +1 kb with recombinant sequences where signaling response sentinels (green boxes) are inserted between deletion endpoints −60, −228, −1217 and −3000 and a TATA box fused to a coding sequence for CFP.
Engineering the Rosa26 locus to receive diverse signaling sentinels
We sought to create a system whereby it would be convenient to assay different deletions as well as to create different signaling sentinels integrated at the Rosa26 locus. Accordingly, we first used homologous recombination to construct a mouse ES line in which sequences between −4 kb and +1 kb relative to the Rosa26 start site were removed and replaced with lox71 and lox2272 sites flanking a positive and negative selection cassette (Fig. 2A). The resulting Loxed Cassette Acceptor allele (Rosa26LCA) serves as a generic recipient line for all signaling sentinel constructs via RMCE. With RMCE, the heterologous lox sites allow directional, targeted recombination, in the presence of cre recombinase, of plasmid DNA containing the lox66 and lox2272 sites flanking signaling sentinel constructs (Fig. 2B) (Long et al., 2004; Jones et al., 2005).
Fig. 2.
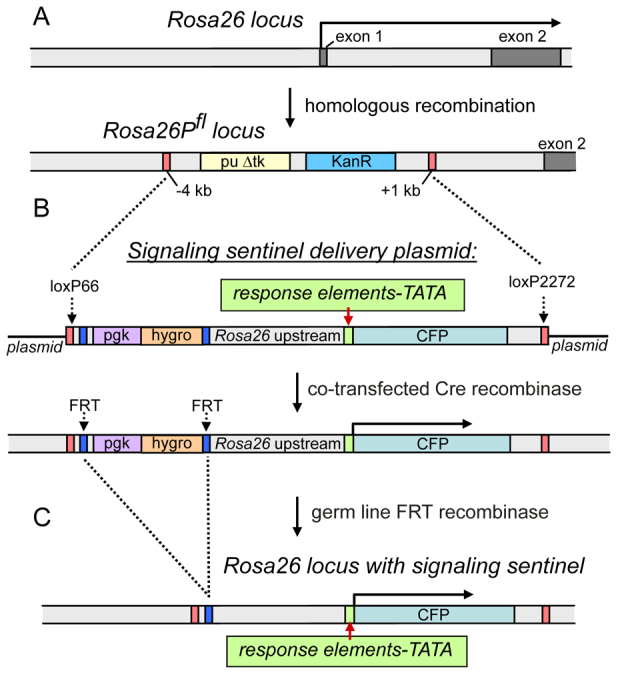
RMCE at the Rosa26 locus to generate signaling sentinel ES cells. (A) Upper scheme depicts the natural Rosa26 locus; lower scheme, Rosa26 with an acceptor cassette substitution generated by homologous recombination in ES cells. Orange boxes indicate heterologous lox target sites, allowing unidirectional insertion of donor cassette in step B. The puroR sequence allows positive selection for recombination. (B) Upper scheme indicates signaling sentinel delivery plasmid transfected into acceptor ES cells. When co-transfected with a plasmid expressing cre recombinase, the plasmid undergoes unidirectional insertion into the acceptor locus (lower scheme). The appropriately structured ES lines are confirmed by PCR and used to generate chimeric mice; the mice are bred for germ line transmission of the recombined locus. (C) When the resulting mouse strain is crossed to a line expressing FRT recombinase in its germ lineage, the FRT recombination sites (blue) allow the subsequent excision of the selectable markers, yielding a mouse with the appropriate signaling sentinel at the Rosa26 locus.
We constructed four signaling sentinel delivery plasmids with Rosa26 locus 5′ promoter sequences from −4 kb to −60, −228, −1217 and −3000 bp. Each promoter segment was followed by multimerized response elements inserted at a unique NheI site, a TATA box and a Cerulean fluorescent protein (CFP) reporter in the delivery plasmid (Fig. 1D, Fig. 2B). To select for properly recombined integrants, the signaling sentinel delivery plasmids contained a pgk-hygromycin gene flanked by FLP recombination (FRT) sites. After appropriate DNA analysis of stable ES cell transfectants to ascertain RMCE, the cells were used for in vitro studies as well as to create germ line chimeras. The latter were crossed with mice expressing FRT recombinase in the germ lineage (Jones et al., 2005) to generate progeny lacking the upstream selectable marker (Fig. 2C). In summary, we created a system whereby different multimerized elements were substituted for different deleted portions of the Rosa26 promoter, allowing us to investigate diverse constructs in an isogenic genomic background and thus normalize for position effects.
Test case for a signaling sentinel: retinoic acid response elements
In order to determine which Rosa26 promoter deletion(s) were optimal for signaling sensitivity, we tested retinoic acid response elements (RAREs). RAREs bind well-characterized heterodimers of the retinoic acid receptor (RAR) and retinoid X receptor (RXR) subunits (Rochette-Egly and Germain, 2009) that confer transcriptional induction; RAREs have been used successfully in transgenic sentinel reporter systems (Rossant et al., 1991) and thus we could compare our approach accordingly. For our studies, we used a 5× direct repeat of a DR5 (AGGTCAccaggAGGTCA) from pDR5-Luc (Stratagene), which is a consensus RARE direct repeat with a 5 bp spacer (Chambon, 1996), linked to a TATA box (TA); the latter providing a basal promoter element. As a control, we used the TATA box without the DR5 repeat. The DR5-TA sequence, fused to a CFP reporter and polyadenylation signal, was inserted at the −60, −228, −1217 and −3000 bp deletion points noted above (Fig. 1D). The TA-CFP sequence was inserted at the −228 and −60 bp deletion points.
In the mouse ES lines treated with 10−5 M all-trans retinoic acid (RA) for 24 hours, only the substitutions of DR5-TA into both the −60 and −228 deletions gave clear CFP-positive colonies (Fig. 3A; see 3B for magnified views). Weak background signals were observed only for the −60 sentinel, which could reflect sensitivity to endogenous RA signaling. CFP signals from the −1217 line were very faint and −3000 was negative, like the parent ES cells (Fig. 3A). Signals for only the −60/DR5-TA line were observed after a short 4.5-hour RA treatment (supplementary material Fig. S1A). In addition, the −228/TA-CFP line, lacking the DR5 elements, was negative (supplementary material Fig. S1B). Curiously, the -60/TA-CFP line, also lacking the DR5 elements, was highly active in both the presence and absence of RA (supplementary material Fig. S1B). Given the difference in activity of the −60/TA-CFP and −60/DR5-TA lines in the absence of RA, plus the fact that all other response elements inserted at −60 were basically inactive in the absence of agonist (see below), it indicates that insertion of DNA in the −60 construct essentially destroys promoter activity, in the absence of activator. On the basis of all of the above findings, we ceased further studies on the −1217 and −3000 constructs and the TA-CFP constructs and focused on lines with the −60/TA- and −228/TA-based sentinel insertions.
Fig. 3.
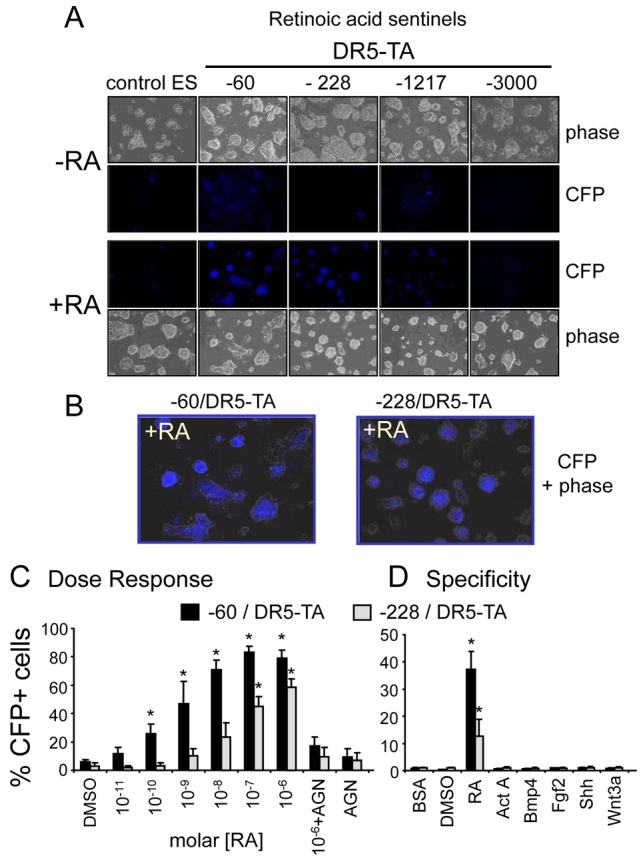
Dose-responsiveness and specificity of retinoic acid signaling sentinels. (A) ES lines containing the DR5 RA response elements substituted for deletions of the Rosa26 promoter to −60, −228, −1217 and −3000 bp, visualized by phase contrast microscopy and for CFP fluorescence, in the presence and absence of 10 μM all-trans RA for 24 hours. The −60 construct had evident background fluorescence and exhibited increased visual fluorescence with RA treatment. The −228 construct had no background fluorescence and lower induced fluorescence with RA. The −1217 and −3000 constructs exhibited little or no fluorescence under either condition. Separate TA-CFP constructs without DR5 elements, inserted at −60 and −228, exhibited no fluorescence (data not shown). An experiment conducted with a 4.5-hour induction time is shown in supplementary material Fig. S1. (B) Higher magnification views of induced panels in A. (C) Dose-response curves where the percentage of CFP fluorescence-positive cells above a threshold defined in supplementary material Fig. S2 are quantified by FACS over a range of RA concentrations, with and without 10−5 M antagonist (AGN193109). Individual FACS profiles for each experimental point are shown in supplementary material Fig. S2, indicating that the cell induction profile distributions appear similar at each concentration of RA. (D) We tested a range of different ligands: RA, 10−6 M; activin A (ActA), 100 ng/ml; BMP4, 10 ng/ml; Fgf2, 50 ng/ml; Shh, 15 nM; and Wnt3a, 100 ng/ml. The data show that the −60 construct is sensitive to lower concentrations of RA than the −228 construct (C), and that both constructs exhibit a high degree of specificity of response to RA versus the other pathway agonists tested (D). The extracts in D were assayed after freezing at length, explaining why the overall %CFP+ cells is lower than in C. One-tailed t-tests of drug-treated samples compared with DMSO controls; *P<0.05.
To rigorously assess the sensitivity of the RA sentinel lines, we treated them with different concentrations of RA for 24 hours, with or without the inverse agonist AGN193109 (Johnson et al., 1995), then performed FACS analysis for CFP expression on dissociated cells. The induction of CFP for the −60/DR5-TA line (Fig. 3C) began in the subnanomolar range (10−10 M RA) and was saturated by the submicromolar range (10−8 M RA); all within physiological signaling levels. The induction of CFP for the −228/DR5-TA line was about 100-fold less sensitive and was saturated at 10−7 M RA, which is within the pharmacological signaling range. Notably, FACS profiles of live cells showed that all concentrations of RA tested elicited remarkably uniform shifts in fluorescence of the original population, with nearly 100% of the cells responding (supplementary material Fig. S2).
In the presence of saturating amounts of RA (10−6 M), the antagonist AGN193109 suppressed most of the signal for the −60 and −228 sentinels in the FACS assay, demonstrating specificity (Fig. 3C; supplementary material Fig. S2). Despite its effectiveness in suppressing sentinel activity, we note that the antagonist alone had a slight agonist effect, in comparison with the DMSO vehicle control. Finally, we found that both the −60 and −228 RA sentinels were highly specific for RA and exhibited no response to activin A, BMP4, fibroblast growth factor 2 (Fgf2), sonic hedgehog (Shh) or Wnt3a (Fig. 3D). We conclude that the ES lines with two different DR5 signaling sentinels provide highly sensitive and specific reporting over the physiological and pharmacological ranges of RA signaling.
We generated stable DR5-TA mouse lines from the mouse ES cells and analyzed reporter expression in live embryos. The −60 and −228 RA sentinels gave similar CFP signals in E8.25 embryos (six somite pairs; 6S) along the foregut, midgut, hindgut and mid-ventral neural tube, with weak signals in the ventral heart and no signal in the developing head folds and open neural tube (Fig. 4A), as expected (Rossant et al., 1991). By E9.5 (25S), signals were in a similar distribution, except with signals now developing around the optic vesicle. Overall, similar expression patterns were seen with the original RARE-lacZ transgenics (Rossant et al., 1991). Anterior half-embryos (Wandzioch and Zaret, 2009) were cultured with 10−6 M RA for 24 hours, whereupon all embryos exhibited signals that extended more anteriorly (Fig. 4B, white arrows), as expected (Conlon and Rossant, 1992) and were overall stronger than the controls. To test the specificity of the RA signal in vivo, we compared the signals from −228/DR5-TA half-embryos that were cultured for 24 hours in RA antagonist, RA, BMP4, BMP4 inhibitor (noggin), TGFβ inhibitor (SB431542) or MAPK inhibitor (U0126). Whereas the antagonist suppressed the CFP signal and RA enhanced it anteriorly, none of the other reagents markedly changed the intensity or expression pattern of CFP (supplementary material Fig. S3B).
Fig. 4.
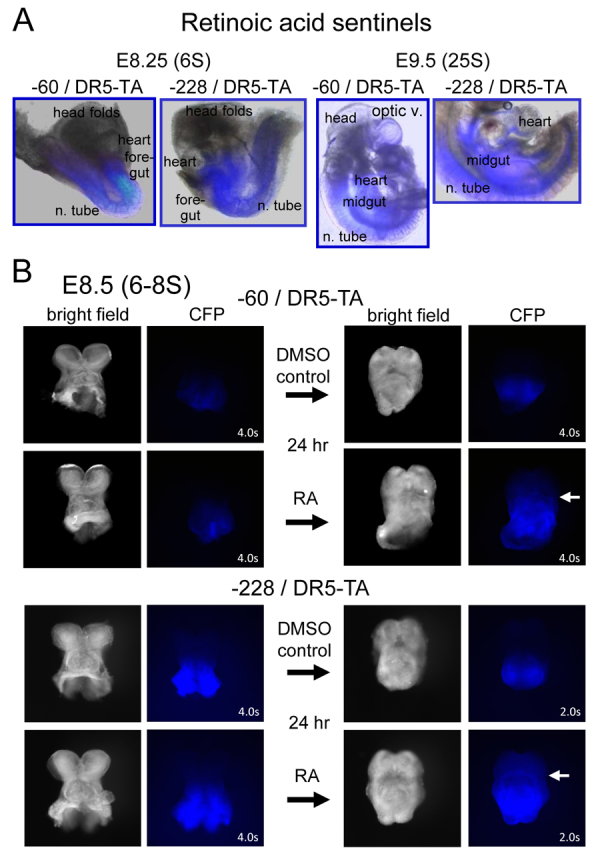
Retinoic acid sentinels in live embryos and responding to exogenous RA in half-embryo cultures. (A) Merged bright field and fluorescence images of live E8.25 and E9.5 embryos containing the retinoic response sentinel DR5-TA-CFP at the −60 and −228 sites of the endogenous Rosa26 promoter. Composite images were generated with Photoshop. Fluorescent domains are noted. (B) Half-embryo fragments from E8.5 embryos of the designated genotypes were cultured overnight with 1 μM RA or DMSO carrier (control). Images are shown before (left) and after (right) culture, revealing an anterior-ward extension of the fluorescent signal upon RA treatment for each construct (white arrows). In supplementary material Fig. S3, data are shown indicating that only RA extended the CFP expression domain anteriorly, and not the RA inhibitor AGN193109, BMP4, noggin, a TGFβ inhibitor or a MAPK inhibitor, demonstrating specificity.
Taking the data together, we conclude that the two different Rosa26 promoter deletions provide different ranges of responses to the multimers of the retinoic acid response elements in vitro and comparable ranges of responses in vivo; though further analysis might reveal subtle differences in the latter.
Signaling sentinels for closely related BMP and activin A pathways
We next sought to create signaling sentinels in −60 and −228 Rosa26 promoter deletions that would distinguish between closely related pathways. Given the important developmental roles of BMP and activin A signaling, which are mediated by heterodimeric receptors and SMAD transcription factors, and each of which has common and specific subunits for each pathway (Harrison et al., 2004), we generated signaling sentinels for each. Previous studies employed BMP sentinel transgenes that exhibited a degree of heterogeneity in signaling in different germline animals (Monteiro et al., 2004; Monteiro et al., 2008). We also used different fluorescent reporter constructs to ultimately allow the creation of heteroallelic reporters for different pathways within a single cell or mouse. For the BMP sentinels, we used a 4× multimer of the IBRE element from the Smad7 gene (Benchabane and Wrana, 2003) fused to TATA within each of the −60 and −228 CFP constructs. These were used to generate the −60/IBRE4-TA-CFP and −228/IBRE4-TA-CFP sentinel ES cell lines. For the activin A sentinels, we used an 8× multimer of the activin response (AR) element from the Xenopus laevis Mix.2 gene (Chen et al., 1997) fused to TATA within each of the −60 and −228 deletion backgrounds and an mCherry reporter (Shaner et al., 2004). These were used to generate −60/AR8-TA-mCherry and −228/AR8-TA-mCherry sentinel ES cell lines.
As seen in Fig. 5A, each of the signaling sentinel lines exhibited low or no background signals in control conditions. Fluorescent colonies were observed in the presence of the appropriate BMP and activin A agonists. Cellular fluorescence was not observed with respective antagonists (Fig. 5A) (Cuny et al., 2008). Qualitatively, both BMP and activin A reporters gave brighter signals in the −228 deletion background under the microscope. In quantitative FACS analyses, the dose-response curves for the −60 and −228 constructs were similar (Fig. 5B; supplementary material Fig. S4), with the −228 constructs exhibiting slightly better performance in certain experiments (Fig. 5C). All constructs yielded cells exhibiting high sensitivity to agonist, expressing CFP in response to 1–30 ng/ml BMP4 and expressing mCherry in response to 12-10 ng/ml activin A. The respective inhibitors suppressed agonist signals, as expected (Fig. 5B). We note that both the BMP and activin A sentinel ES cell lines exhibited weak responses to agonist by 24 hours and required 48 hours to maximally induce CFP or mCherry expression. Furthermore, whereas cells harboring the BMP sentinel at −60 or −228 exhibited a uniform fluorescence shift profile for the induced population, with virtually 100% of the cells responding, the cells harboring the activin A sentinel exhibited a different, much broader, profile in the induced population, exhibiting heterogeneity in response to this particular element; with a portion of the cells responding highly and a portion responding weakly or not at all (supplementary material Fig. S4).
Fig. 5.
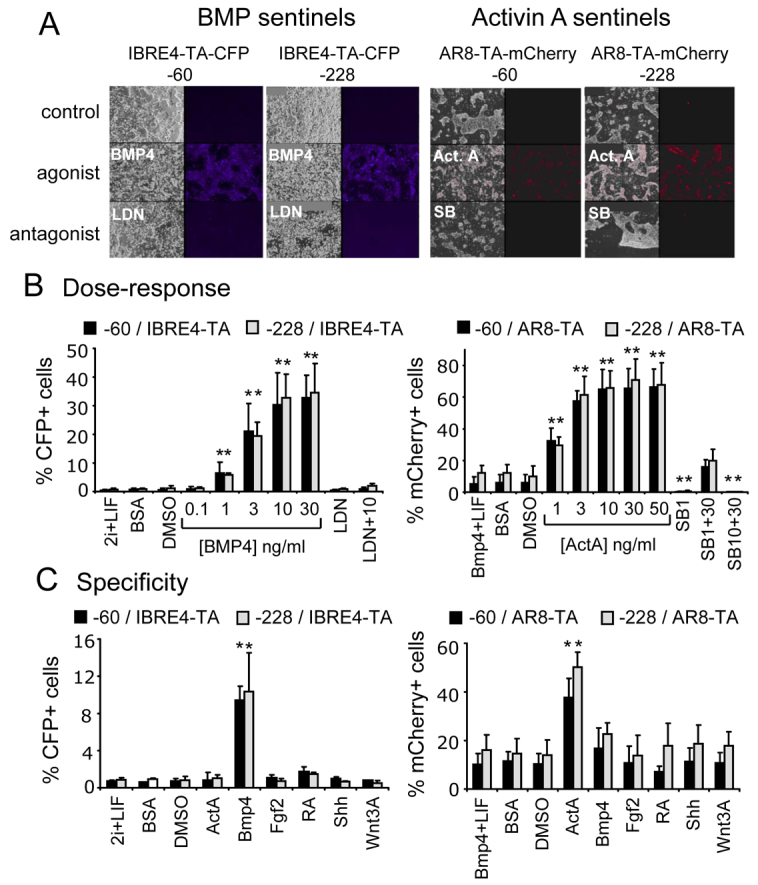
Dose-responsiveness and specificity of BMP and activin A signaling sentinels. (A) ES cell signaling sentinel lines with response elements for BMP (IBRE-4-CFP) or activin A (AR8-TA-mCherry) inserted at the −60 and −228 sites of the endogenous Rosa26 locus. All cells exhibited responses to the respective agonists and not their antagonists (100 nM LDN193189 and 10 μM SB431542, respectively). Assays were for 48 hours. (B) Dose-response tests of the BMP and activin A (ActA) sentinels substituted at −60 and −228, demonstrating high sensitivity of both constructs for each reporter type. Individual FACS profiles are shown in supplementary material Fig. S4. (C) Specificity tests demonstrate high specificity of responsiveness, particularly illustrating that the BMP reporter responds to BMP4 and not activin A, whereas the activin A reporter responds to activin A and not BMP (doses as for Fig. 3). One-tailed t-tests of drug-treated samples compared with DMSO controls; **P<0.01.
To assess the specificity of the BMP and activin A sentinel ES lines, we treated each of them with diverse signaling molecules, including BMP4, activin A, Fgf2, RA, Shh and Wnt3A. After 48 hours, we collected the cells and quantitated their mean fluorescence intensity. As seen in Fig. 5C, only the BMP sentinel lines were responsive to BMP and only the activin A sentinel lines were responsive to activin A. The background signal with the activin A sentinel lines (Fig. 5B,C) was eliminated by treating the cells with the TGFβ pathway inhibitor SB1 (Fig. 5B), consistent with endogenous nodal signaling occurring in mouse ES cultures (Ogawa et al., 2007). On the basis of these findings, we conclude that the BMP and activin A sentinels at the Rosa26 locus exhibit high sensitivity and selectivity, and that mCherry works as an alternative to CFP.
In vivo at E6.5, both the −60 and −228 BMP sentinels elicited clear extra-embryonic ectoderm and proximal epiblast fluorescence in live embryos, as expected (Ben-Haim et al., 2006) (Fig. 6A). At E8.5, the BMP sentinels revealed ventral-lateral fluorescence in the lateral plate mesoderm and in the foregut and heart (Fig. 6A), which are all sites of known BMP signaling (Winnier et al., 1995; Zhang and Bradley, 1996; Solloway and Robertson, 1999). Indeed, the live embryo fluorescence seen in the foregut and heart resembles that seen previously in those domains in fixed embryos for activated phosphorylated Smad1, Smad5 and Smad8 (Wandzioch and Zaret, 2009). At E9.5, the −60 BMP sentinel revealed clear zones of ventral signaling throughout the anterior-posterior axis, including the ventral somites, regions in the midgut, the heart, the branchial arches and into the rostral portion of the head (Fig. 6B). These live BMP sentinels exhibited fluorescence in more of these known regions of BMP signaling than a previous transgenic lacZ reporter for fixed cells (Monteiro et al., 2004), more comparable with that seen for a well-characterized GFP transgene reporter (Monteiro et al., 2008). The −60 and −228 activin A sentinels both exhibited fluorescence at E9.5 in the branchial arches, heart and midgut. The −60 sentinel was weak elsewhere whereas the −228 sentinel exhibited signal in the limb buds (Fig. 6B). We conclude that in vivo, the BMP and activin A sentinels reveal overlapping and distinct domains of signaling, and that the −228 sentinel is more useful for activin A.
Fig. 6.
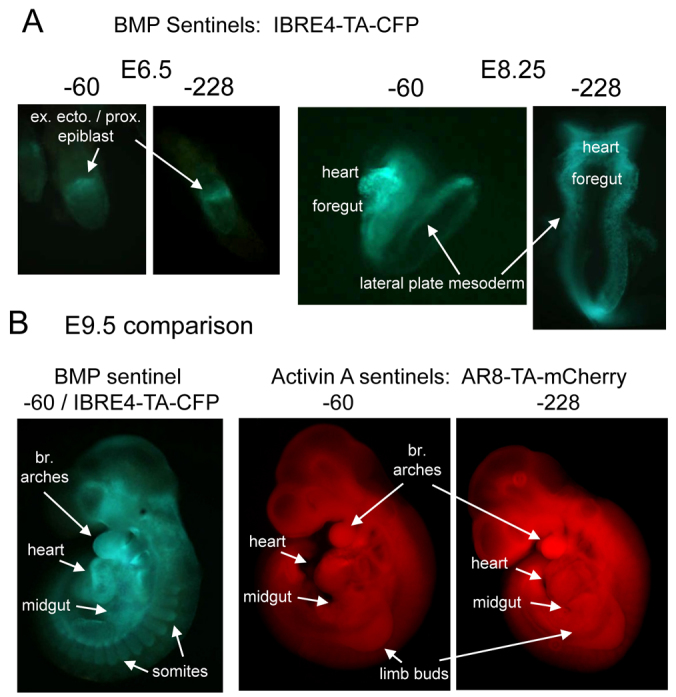
BMP and activin A sentinels in live embryos. (A) Live E6.5 and E8.25 embryos containing the BMP sentinel IBRE4-TA-CFP substituted at −60 and −228 of the endogenous Rosa26 promoter. Fluorescence is in embryo domains as shown. (B) Live E9.5 embryos containing the BMP or activin A sentinels at −60 (BMP) or −60 and −228 (activin A). The heart, midgut and branchial arches constitute overlapping expression domains, whereas BMP signaling is more sharply revealed in the somatic regions and into portions of the head. Activin A signaling is reported more diffusely in the limb buds and elsewhere.
Signaling sentinels for the Wnt pathway
To develop live signaling sentinels for the Wnt pathway, we employed the Wnt response elements from the SuperTOP-Flash construct, which uses a luciferase reporter assay with lysed cells (Veeman et al., 2003). The SuTOP element was placed in the context of the −60 and −228 deletions of the Rosa26 promoter, fused to CFP. Our preliminary studies with the −60/SuTOP construct have been described (Petersen et al., 2011). Comparing the −60 and −228 substitutions for SuTOP-TA-CFP gave markedly different results. In live ES cells, leukemia inhibitory factor (LIF) plus the 2i cocktail (Silva et al., 2008) containing the GSK3 inhibitor CHIR99021 activated the −60 construct well (as shown by microscopy) and less so for the −228 construct (Fig. 7A). LIF plus BMP4, or BSA alone, failed to activate either, as expected. Wnt3a weakly activated SuTOP only in the −60 but not in the −228 background. Presence of the Wnt signaling antagonist Dkk1 decreased the Wnt3a response (Fig. 7A). Notably, FACS profiles of the induced cells showed relatively uniform population profile shifts in the induced cells, with virtually 100% of the cells responding (supplementary material Fig. S5). The specificity for the Wnt response was very tight, with both construct backgrounds exhibiting induction only for Wnt agonist in the absence of antagonists and not exhibiting induction in response to activin A, BMP4, Fgf2, RA, Shh or N2B27 medium (Fig. 7B). Furthermore, the −60/SuTOP cells exhibited a uniform population shift in response to LIF plus 2i (supplementary material Fig. S5).
Fig. 7.
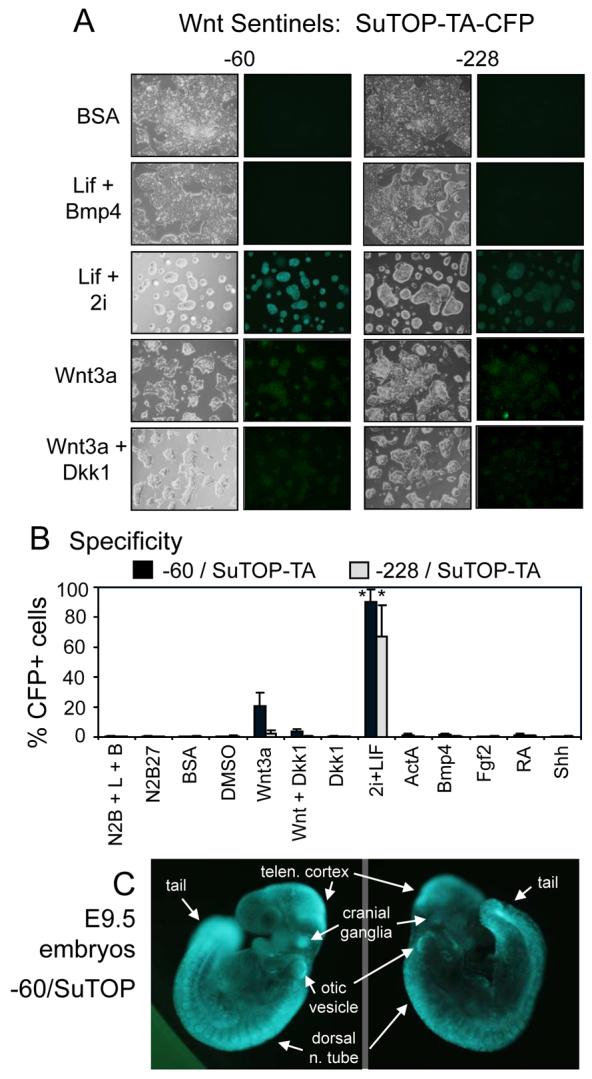
Specificity of Wnt signaling sentinels and activity in live embryos. (A) Cell signaling sentinel lines with response elements for Wnt (suTOP-TA-CFP) inserted at the −60 and −228 sites of the endogenous Rosa26 locus. All cells exhibited responses to medium containing LIF plus 2i, which contains 3 μM CHIR99021, a GSK3β antagonist that promotes Wnt signaling, and not to medium containing LIF plus BMP4, with the −60 construct exhibiting a stronger fluorescence signal. The cells harboring the −60 construct responded more weakly to Wnt3a added to the medium, whereas the −228 cells exhibited minimal induction. The presence of the Dkk1 inhibitor of Wnt signaling suppressed the Wnt3a-dependent response in the −60 cells. (B) Dose-responsive tests of the −60 and −228 reporters, as quantified from FACS profiles shown in supplementary material Fig. S5. The data show that the Wnt reporters do not respond to N2B27 medium, N2B27 medium with 1500 U/ml LIF (L) and 10 ng/ml BMP4 (B), 320 ng/ml Dkk1, 100 ng/ml activin A (ActA), 10 ng/ml BMP4, 50 ng/ml Fgf2, 10−6 M RA or 15 mM Shh. (C) Different live E9.5 embryos harboring the −60/SuTOP-TA-CGP construct, exhibiting fluorescence in designated areas. One-tailed t-tests of drug-treated samples compared with DMSO controls; *P<0.05.
Given the success with the −60 construct, we used the ES line to generate embryos with −60/SuTOP-TA. At E9.5, the live embryos consistently exhibited fluorescence in the dorsal neural tube, somites, tail, optic vesicles, cranial ganglia and midbrain-hindbrain boundary, among other sites (Fig. 7C). This is similar to results seen with the BAT-Gal construct using β-galactosidase staining of fixed embryos (Maretto et al., 2003). Taken together, these studies demonstrate once again the specificity of the response elements in the Rosa26 promoter deletion background and the utility of having two different promoter deletions to test, giving optimal signal sensitivity in vitro and live imaging in vivo.
Signaling sentinel for the Notch pathway
To explore the development of a sentinel for the Notch pathway and the utility of an additional fluorescent marker, we inserted a 12× binding site repeat for the CSL transcription factor, downstream of Notch signaling (Hansson et al., 2006), into the −60 and −228 deletion constructs with the TATA element and mOrange in ES cells. To test the resulting −60/12×CSL-TA-mOrange and −228/12×CSL-TA-mOrange sentinels, we transfected the cells with an empty IRES-GFP vector (control); the vector encoding a constitutively active Notch intracellular domain (NICD) and then with the IRES-GFP vector; the vector encoding NEXT, a membrane-bound form that is not yet cleaved to generate the NICD (Mumm et al., 2000); or the vector with NEXT and a γ-secretase inhibitor (Comp XVIII) to block NEXT cleavage and, thereby, Notch signaling. Transfected cells were sorted and quantitated for GFP fluorescence as well as mOrange (Fig. 8; supplementary material Fig. S6). In the subset of GFP-positive cells that were transfected with the Notch signaling effectors and/or controls, we found that the NICD induced mOrange in about 25% of the cells, the NEXT fragment more weakly so and the γ-secretase inhibitor blocked such signaling for both the −60 and −228 constructs. Though further work is needed to determine optimal conditions and in vivo expression, these initial data indicate that it should be possible to develop a live animal Notch reporter using the Rosa26 deletion-based signaling sentinel system.
Fig. 8.
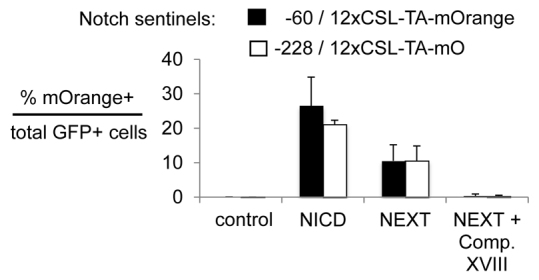
Developing live cell sentinel for Notch signaling. Cell signaling sentinel lines with 12×CSL-TA-mOrange response elements for Notch were inserted at the −60 and −228 sites of the endogenous Rosa26 locus. The ES sentinel lines were transfected with an empty IRES-GFP vector; the vector encoding a constitutively active NICD, followed by the IRES-GFP; the vector encoding the membrane-bound NEXT; and the vector with NEXT in the presence of the γ-secretase inhibitor Compound XVIII (1 μM). Transfected cells were quantified by GFP fluorescence, which was then used to normalize for mOrange fluorescence (see supplementary material Fig. S6). The NICD induced mOrange significantly, the NEXT fragment more weakly so, and the γ-secretase inhibitor blocked such signaling for both the −60 and −228 constructs.
DISCUSSION
We present a system by which diverse transcriptional reporter elements, downstream of different signal transduction pathways, elicit cellular fluorescence in an agonist-dependent and agonist-selective fashion in live cells and embryos. By employing the constitutively active Rosa26 gene and analyzing its upstream promoter sequences, we made successive deletions with the intention of preserving the inherent ‘openness’ of the locus and its constitutive activity, while diminishing core promoter function sufficiently to provide a low background of transcription in the absence of signal pathway activity. The −228 deletion endpoint satisfied these criteria, whereas the −60 deletion retained high constitutive activity. Fortunately, we discovered that insertion of all signaling elements studied here at −60 resulted in a marked diminution of basal activity. Not unexpectedly, the different −228 and −60 constructs provided slightly different results in terms of basal and induced activity with different regulatory element insertions. We suggest that further use of this model with new signaling sentinels would be best served by trying both the −60 and −228 deletions in ES cells. The RMCE method makes it relatively simple to swap a desired signaling sentinel into the Rosa26 locus and the exchanged clones can be screened readily for proper insertion and signal responsiveness.
As noted in the Introduction and Results, signaling reporters have been generated for many of the pathways studied here. However, to date they have utilized conventional transgene technology, where transgenes are inserted at random locations throughout the genome. The random locations result in position effects, such that different transgenic lines with the same construct do not yield identical signaling readouts and might also exhibit variegated expression. Moreover, many of the existing reporters have employed lacZ or luciferase and thus are used on fixed or lysed cells. By utilizing fluorescent reporters integrated into the same locus, for different signaling sentinels, we eliminate position effects and allow systematic modification of response elements in a constant genomic location. By demonstrating that the system works with Cerulean, mCherry and mOrange reporters, it paves the way for crossing distinct signaling reporter lines of mice to create heteroallelic animals. This would allow different signaling pathways to be assessed simultaneously in live animals. We chose fluorescent reporters with their native half-life determinants, rather than high-turnover derivatives, because we felt that the latter would be less sensitive overall and we wished for maximum signal to allow visualization of the earliest times of induction and in intact embryos (e.g. Fig. 4A; Fig. 6A–C; Fig. 7C).
Along these lines, we were able to successfully create sentinels that distinguish BMP and activin A signaling, despite the fact that their pathways possess components in common. Whereas the BMP sentinels exhibited uniform fluorescence shift profiles for the induced population, the activin A sentinel exhibited a much broader profile in the induced population. Further analysis of the AR8 element will be required to determine its unusual profile of activity. However, because the function of the AR8 element was assessed here in an allelic fashion with diverse other constructs that gave more uniform cellular response shifts, we can attribute the difference to unexpected factors in the composition of the AR8 element, i.e. additional factors that bind. Our approach allows for subsequent dissection of regulatory elements and optimization, in a systematic fashion, by performing RMCE with variant AR8 elements at the originally targeted locus.
Interestingly, previous studies of transfected or injected reporter constructs found that Mix.2-based AR elements required co-transfected or injected Forkhead activin signal transducer (Fast-1) in order to exhibit TGFβ- or activin-based induction (Weisberg et al., 1998; Yeo et al., 1999; Osada et al., 2000). By contrast, our AR8 reporter did not need additional Fast-1 for highly specific activin A responsiveness. We attribute the difference to the monoallelic nature of our reporter and its apparent ability to function sufficiently with endogenous Fast-1 (Sirard et al., 2000) or with Fast-2 (Nagarajan et al., 1999), in comparison with transfected or injected constructs that involve many DNA copies per cell. This emphasizes the effectiveness of our targeted integration approach, in contrast to transfected or transgenic reporters that typically involve multiple gene copies per cell. Additionally, the ability of SB431542 to inhibit the activin response shows that the response is dependent on receptor activation.
We note that by having the same signaling reporter in ES cells and mice, it is now possible to directly compare inductive signaling events seen in particular lineages and stages in development with signaling events occurring during in vitro stem cell differentiation. Similarly, signaling that occurs during homeostasis and regenerative responses in vivo should be informative for the maintenance and further manipulation of cell types generated in vitro.
We based our Rosa26 promoter deletion endpoints on landmarks that are conserved between mouse and human. Thus, it should be straightforward to apply the same promoter deletion-substitution approach to the Rosa26 locus in human ES cells and thereby provide a sensitive means by which cell programming can be monitored and guided in living human ES cells. The high signaling sensitivity of most of the sentinels we created and their high signaling specificity suggests that the −60 and −228 deletion sites for the Rosa26 locus should be useful for the assessment of diverse types of signal response elements and pathways in vivo and in vitro.
METHODS
Rosa26 promoter analysis and signaling sentinel delivery plasmids
The Rosa26 locus in mouse, rat and human were initially scanned for sequence similarities 30 kb upstream of the transcription start site, using Blastz alignment tool in PipMaker (zpicture.dcode.org). Adjusting the minimum mouse:rat homology to 95% and the mouse:human homology to 80% revealed a common conserved sequence to −3 kb, shown by the red vertical bars in Fig. 1A. Subsequent RVISTA analysis of the proximal 3 kb region revealed a qualitative increase in transcription factor binding motifs within −1.2 kb of the transcription start site (Fig. 1B). Visual inspection of the mouse and human sequences of the transcription start to −1.2 kb revealed the first break in sequence identity after −228 and a CCAAT box just before −60 (Fig. 1C). Hence, we made deletion endpoints of the Rosa26 promoter to −3000, −1217, −228 and −60 bp with respect to the transcription start site. PCR was used to clone Rosa26 upstream promoter fragments with a BstB1 site added at −4000 to NheI sites added at −3000, −1217, −228 or −60 bp.
The resulting BstB1-Nhe1 fragments from Rosa26 were ligated with the Nhe1-SacII fragments with DR5-TA-CFP or TA-CFP (see below) to the 6.1 kb BstB1-SacII partial digest fragment from pRosa26.Ex1.fH, which encodes an FRT site, pgk, hygR, an FRT site, a loxP66 site, Escherichia coli replication and selection sequences, a loxP2272 site and sequences from the SacII site in exon 1 of Rosa26 to 1 kb within the gene’s intron (Jones et al., 2005). The resulting signaling sentinel delivery plasmids (see Fig. 2B) were ascertained by DNA sequencing and used for RMCE in mouse ES cells in which the resident Rosa26 locus had been modified by gene targeting to introduce both a lox66 and lox2272 sites, as shown in Fig. 2A, and as confirmed by PCR analysis and Southern blot analysis (Jones et al., 2005; Chen et al., 2011). Correct insertion of the signaling sentinels into the Rosa26 locus was ascertained by PCR and restores the Rosa26 locus from +81 to the downstream insertion of the loxP2272 site at +1 kb. A resulting modified ROSA26LCA locus is depicted in Fig. 1D and Fig. 2C. The unique NheI site upstream of the TATA box −60/TA-CFP and −228/TA-CFP signaling sentinel delivery plasmids allows other response elements to be inserted readily.
DR5-TA-CFP
We first constructed a plasmid containing a minimal TATA box (from pTA-Luc; Clontech) and CFP (gift from David Piston, Vanderbilt University, Nashville, TN) (Rizzo et al., 2004), with the SV40 poly(A) site (from pTA-Luc; Clontech). The CFP coding region was amplified by PCR (forward primer, 5′-CACCATGGTGAGCAAGGGCGAGGAG-3′; reverse primer, 5′-GGCCGGCCTACTTGTACAGCTCGTCC-3′), digested with NcoI and FseI, and ligated into the vector fragment of pTA-luc to generate pTA-Cerulean. Similarly, the mOrange coding region (Shaner et al., 2004) (gift from Roger Tsien, University of California, San Diego, CA) was amplified by PCR (forward primer, 5′-CCATGGTGAGCAAGGGCGAGGAG-3′; reverse primer, 5′-GGCCGGCCTTACTTGTACAGCTCGTC-3′), digested with NcoI and FseI, and ligated into the vector fragment of pTA-luc to generate pTA-mOrange.
The RA-responsive element from pDR5-luc (Stratagene), containing five repeats of the RA-binding sequence 5′-AGGTCA-3′ separated by five nonspecific bases, was inserted into the pTA-CFP between unique SacI and XhoI restriction sites and used for PCR to create the original signaling sentinel delivery plasmids described above.
R26 −60 and −228 IBRE4 TA-Cerulean
Four repeats of a 130 bp intronic BMP response element (IBRE) from the first intron of the Smad7 gene (Benchabane and Wrana, 2003) was used to generate the BMP responsive sentinel. One repeat was amplified from mouse genomic DNA and was TOPO-cloned and PCR-amplified to yield IBRE fragments flanked by SacI-MluI, MluI-XmaI, XmaI-XhoI and XhoI-BglII, respectively, to allow for sequential insertion of the fragments into the pTA-based vectors to generate the pIBRE4-TA-Cerulean. The entire IBRE4-TA-Cerulean cassette was then excised with EcoICRI and FseI inserted into MscI-FseI-digested pR26-60 TA-CFP and pR26-228 TA-CFP RMCE shuttle vectors to generate the pR26-60 IBRE4-TA-CFP and pR26-228 IBRE4-TA-CFP, respectively.
R26 −60 and −228 AR8-TA-mCherry
A 50-bp activin response element (ARE) from the Xenopus laevis Mix.2 promoter (Huang et al., 1995) containing two FoxH1 binding sites (Chen et al., 1996) was used to generate a nodal-activin reporter. We first generated a pAR8-luc reporter plasmid by amplification using forward (5′-GAGCTCTATCT-GCTGCCCTAAAATGTGTATTCCATGGAAATGTCTGCCCT-TCTCTCCCTAGTATTGCTGCCCTAAAATGTGTATTCCATG-GAAATGTCTGCCCTTCTCTC-CCTAGTATCT-3′) and reverse (5′-CTCGAGGGAGAGAAGGGCAGACATTTCCATGGAAT-ACACATTTTAGGGCAGCAGATACTAGGAGAGAAGGGCA-GACATTTCCATGGAATACACATTTTAGGGCGCAATACTA-GGGAGA-3′) oligonucleotides and PFU-polymerase (Stratagene). The amplicons were inserted in the pCR-Blunt2-TOPO (Invitrogen). Several clones were sequenced (MWG-Biotech) and a clone with eight copies of the ARE was identified and excised with SacI and XhoI (New England Biolabs), gel purified and annealed with SacI-XhoI-digested pTA-Luc (Clontech) to generate pAR8-luc. The luciferase gene of pAR8-luc was then replaced with mCherry (Shaner et al., 2004). The mCherry fragment was PCR generated using an mCherry plasmid (gift from Roger Tsien) as template and the primers: forward 5′-CCATGGTGAGCAAGGGCGAGGAG-3′ and reverse 5′-GGCCGGCCTTACTTGTACAGCTCGTC-3′ followed by digestion with NcoI and FseI and ligation into NcoI-FseI-digested pAR8-luc to generate pAR8-mCherry. The entire AR8-mCherry cassette was then excised with EcoICRI and FseI and inserted into MscI-FseI-digested pR26-60 TA-Cerulean and pR26-228 TA-Cerulean RMCE shuttle vectors to generate the pR26-60 AR8-TA-mCherry and pR26-228 AR8-TA-mCherry, respectively (Chen et al., 1996).
R26 −60 and −228 SuTOP-TA-CFP
The pSuper8×TOPFlash reporter plasmid (Veeman et al., 2003) was a gift from Randall T. Moon (University of Washington, Seattle, WA). We exchanged the luciferase reporter in the pSuper8×TOPFlash with CFP by digesting with NcoI and FseI and isolating the vector fragment. CFP was amplified by PCR (forward primer, 5′-CACCATGGTGAGCAAGGGCGAGGAG-3′; reverse primer, 5′-GGCCGGCCTACTTGTACAGCT-CGTCC-3′), digested with NcoI and FseI and ligated into the vector fragment of pSuper8×TOPFlash to generate pSuper8×TOP-TA-CFP. The entire Super8×TOP-TA-CFP cassette was then excised with EcoICRI and FseI inserted into MscI-FseI-digested pR26-60 TA-CFP and pR26-228 TA-CFP RMCE shuttle vectors to generate the pR26-60 SuTOP-TA-CFP and pR26-228 SuTOP-TA-CFP, respectively.
Gene targeting and RMCE
The derivation of ROSA26LCA mouse ES cells has been previously described (Chen et al., 2011). All RMCE experiments were performed using clone 5B9 and a positive-negative selection strategy as previously described (Long et al., 2004). Briefly, 5.6×106 mouse ES cells containing the ROSA26LCA allele were co-electroporated with 40 μg of an exchange plasmid and 40 μg of pBS185, a cre-expression plasmid (Sauer and Henderson, 1989). Positive selection was achieved using 200 μg/ml hygromycin (Invitrogen, Carlsbad, CA) and negative selection using 8 μM gancyclovir (Sigma, St Louis, MO). Surviving clones were screened for correct exchange using a DNA PCR strategy in order to validate cre-mediated recombination at both the lox66 and lox 2272 sites, as previously described (Chen et al., 2011).
Assays on signaling sentinel ES lines
ES cells were kept undifferentiated by culture on gelatin coated tissue-culture dishes (Nunc) in KO-DMEM supplemented with N2B27, 0.1 mM nonessential amino acids, 2 mM L-glutamine, penicillin/streptomycin (all from Invitrogen), 0.1 mM 2-mercaptoethanol (Sigma-Aldrich), 1500 U/ml leukemia inhibitory factor (LIF; Chemicon) and 10 ng/ml BMP4 (R&D Systems), essentially as described (Ying et al., 2003). The cells were split using trypsin (0.05% trypsin-EDTA; Invitrogen).
To assay induction of reporters, the cells were cultured in gelatin-coated 6-, 12- or 24-well plates (Nunc) in KO-DMEM supplemented with N2B27, 0.1 mM nonessential amino acids, 2 mM L-glutamine, penicillin/streptomycin (all from Invitrogen), 0.1 mM 2-mercaptoethanol (Sigma-Aldrich) together with the indicated factors for 48 hours, with daily change of medium, before being analyzed on a FACS-Aria flow cytometer (BD Biosciences). The following lasers and filters were used to detect fluorescent protein expression: Cerulean, 405 nm laser and 485/30 emission filter; mCherry, 530 nm laser with 600 LP filter and a 610/20 emission filter; mOrange, 530 nm laser with 560/20 emission filter; and GFP, 488 nm laser with 495 LP filter and a 525/50 emission filter. Gating was set so that less than 1% of the (unstimulated) negative control population fell within the positive gate. At least three independent experiments were performed for each condition, and data are presented as % positive cells ± s.d. The percentage of positive cells was calculated as (counts in positive gate/total counts) × 100%. For two-color analysis (12×CSL sentinel), the percentage of mOrange-positive cells were calculated relative to the total number of GFP-positive cells as (counts in Q2/counts in Q2+Q4) × 100%.
The following growth factors and inhibitors were used: activin A (1–50 ng/ml), BMP4 (0.1–30 ng/ml), Dkk1 (320 ng/ml), Shh (15 nM) and Wnt3a (100 ng/ml); all from R&D Systems. Fgf2 (50 ng/ml) was from Invitrogen, LIF (1500 U/ml) from Chemicon, retinoic acid (10−6 M unless otherwise indicated) from Sigma and Compound XVIII (γ-secretase inhibitor; 1 μM) from Calbiochem. PD0325901 (1 μM), CHIR99021 (3 μM) and SB431542 (1, 10 μM) were all from Axon Medchem. The antagonists AGN193109 (10−5 M) and LDN193189 (100 nM), were both synthesized by Novo Nordisk.
Mouse derivations and husbandry
Mouse ES cells containing the mutated Rosa26 promoter regions were microinjected into blastocysts at 3.5 days post-coitum (dpc) obtained from natural matings of C57BL/6J mice. The microinjected embryos were transferred to the uterine horns of pseudo-pregnant ICR females (2.5 dpc; Taconic) using standard methods. The resulting chimeric male offspring were bred with C57BL/6J females to achieve germline transmission. Then, the FRT-flanked pgk-hygromycin resistance cassette was removed by interbreeding with ACTB:FLPe mice (kindly provided by Susan M. Dymecki, Harvard Medical School, Boston, MA). The entire series of mutant Rosa26 sentinel alleles was established on an inbred C57BL/6J background. All mice were maintained in a specific pathogen-free state with a 12-hour light-dark cycle. Experimental protocols were approved by the Vanderbilt Institutional Animal Care and Use Committee and the institutional and Novo Nordisk animal welfare guidelines.
RESOURCE IMPACT.
Background
Understanding where and when inductive signals impinge on cells is crucial for determining the basis for developmental processes, homeostatic function, physiological responses and the course of many diseases. However, there is a lack of systematic ways to monitor signals that elicit transcriptional changes in cells and that enable different types of signals to be assessed quantitatively, temporally and in parallel. Previous studies have isolated individual genetic response elements from complex regulatory sequences and tested them as multimers, and showed that certain elements report the appearance of an individual signal when linked to a convenient marker. Using this approach, transgenic cells and mice have been created that report where and when cells in embryos and animals receive signals for a few known pathways. However, different transgene integration sites are subject to position effects, so that a given cell type’s readout for transgenic signaling sentinels can vary by parameters unrelated to signaling. Likewise, one sentinel cannot reliably be compared with another. In addition, many previous studies employed reporters that function in fixed cells, and the unique nature of transgene integration makes it difficult to create isogenic lines employing variants of the original structure.
Results
To overcome these issues, the authors devised a system to conveniently insert multimerized signaling elements into deleted regions of the promoter for the ubiquitously expressed Rosa26 gene. They thereby created an allelic series of embryonic stem cell lines and mice that have different response element substitutions in a highly defined and consistent genomic context. They show that this ‘signaling sentinel system’ allows the efficient production of genetic sentinels to monitor diverse types of signals in different cell types, including in live and fixed embryonic stem cells and mouse embryos.
Implications and future directions
As shown here, the signaling sentinel system can be used to monitor responses to the retinoic acid, Wnt, BMP, activin A and Notch pathways, and can be adapted, in principle, for any other signal in which transcriptional output occurs via a specific response element. In addition, the design is such that an identical reporter system could be generated in human embryonic stem cells. It is anticipated that this approach will allow systematic analyses of inductive, homeostatic and pathologic signaling inputs, and thereby allow more precise definition of mechanisms underlying biological processes and phenotypes than is possible with existing approaches.
Assays on signaling sentinel mice
Fully ES-cell-derived embryos were harvested at E6.5, E8.25 and E9.5. Epifluorescence in chimeric embryos was visualized on an Olympus IX71 microscope using the following objectives: 2× PlanAPo, 4× UPlanFLN, 10× CPlanFLN and 20× LCAchN (all from Olympus) and filters as described above, or for CFP on a Nikon microscope with a Nikon C-FL CFP cube (#96341) using a 433 nm excitation wavelength (420–440 filter) and a 470 nm U-MNBV2 emissions filter. For half-embryo cultures, embryos were harvested at E8.25 (2-6S) and processed as described previously (Wandzioch and Zaret, 2009). Briefly, after removal of the yolk sac, dissected embryos were cut at the level of first somite pair and the anterior half cultured at 37°C for 24 hours in DMEM containing 10% calf serum (not heat inactivated; Hyclone). The −60/DR5-TA or −228/DR5-TA half-embryos were cultured for 24 hours with DMSO solvent alone (control), 10−6 M RA in DMSO, RA antagonist, BMP4, BMP4 inhibitor (recombinant noggin/Fc chimera; R&D Systems), BMP inhibitor LDN193189 (Cuny et al., 2008), TGFβ inhibitor (SB431542; Sigma) and MAPK inhibitor (U0126; Cell Signaling Technologies) with doses as given in the figure legends.
Resources
The −60/TA-CFP and −228/TA-CFP signaling sentinel delivery plasmids for inserting new response elements, and the −60/DR5-CFP and −228/DR5-CFP ES cells and mice, are available from K.S.Z. The ROSA26LCA mouse ES cells and plasmids used for RMCE to derive the signaling sentinel plasmids are available from M.A.M. The BMP, activin A, Wnt and Notch sentinel ES cells and mice described herein are available from P.S. In time, mice for signaling pathways will be available from the Mutant Mouse Regional Resource Centers (MMRRC).
Acknowledgments
We thank Søren Lindskog, Ragna Jørgensen and Kathy D. Shelton for expert technical assistance and Cathy Mendelsohn for comments.
Footnotes
COMPETING INTERESTS
The authors declare that they do not have any competing or financial interests.
AUTHOR CONTRIBUTIONS
The work was conceived, designed and written into manuscript form by K.S.Z., M.A.M., P.S., R.J.M. and A.G.-B., and was performed by P.S., C.G., T.K., L.A.P., R.L., N.M., E.W., A.H., A.D., C.B., N.E., D.R.P. and J.M.L.P.
FUNDING
The work was supported by a National Institutes of Health Collaborative Bridging Project grant from the Beta Cell Biology Consortium [grant numbers U01 DK072473 to M.A.M., U19 DK072495 to P.A.S. and U01 DK072503 to K.S.Z.].
SUPPLEMENTARY MATERIAL
Supplementary material for this article is available at http://dmm.biologists.org/lookup/suppl/doi:10.1242/dmm.009696/-/DC1
REFERENCES
- Ben-Haim N., Lu C., Guzman-Ayala M., Pescatore L., Mesnard D., Bischofberger M., Naef F., Robertson E. J., Constam D. B. (2006). The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev. Cell 11, 313–323 [DOI] [PubMed] [Google Scholar]
- Benchabane H., Wrana J. L. (2003). GATA- and Smad1-dependent enhancers in the Smad7 gene differentially interpret bone morphogenetic protein concentrations. Mol. Cell. Biol. 23, 6646–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P. (1996). A decade of molecular biology of retinoic acid receptors. FASEB J. 10, 940–954 [PubMed] [Google Scholar]
- Chen S. X., Osipovich A. B., Ustione A., Potter L. A., Hipkens S., Gangula R., Yuan W., Piston D. W., Magnuson M. A. (2011). Quantification of factors influencing fluorescent protein expression using RMCE to generate an allelic series in the ROSA26 locus in mice. Dis. Model. Mech. 4, 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Rubock M. J., Whitman M. (1996). A transcriptional partner for MAD proteins in TGF-beta signalling. Nature 383, 691–696 [DOI] [PubMed] [Google Scholar]
- Chen X., Weisberg E., Fridmacher V., Watanabe M., Naco G., Whitman M. (1997). Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature 389, 85–89 [DOI] [PubMed] [Google Scholar]
- Conlon R. A., Rossant J. (1992). Exogenous retinoic acid rapidly induces anterior ectopic expression of murine Hox-2 genes in vivo. Development 116, 357–368 [DOI] [PubMed] [Google Scholar]
- Cuny G. D., Yu P. B., Laha J. K., Xing X., Liu J. F., Lai C. S., Deng D. Y., Sachidanandan C., Bloch K. D., Peterson R. T. (2008). Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg. Med. Chem. Lett. 18, 4388–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta R., Fuchs E. (1999). Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 126, 4557–4568 [DOI] [PubMed] [Google Scholar]
- Hansson E. M., Teixeira A. I., Gustafsson M. V., Dohda T., Chapman G., Meletis K., Muhr J., Lendahl U. (2006). Recording Notch signaling in real time. Dev. Neurosci. 28, 118–127 [DOI] [PubMed] [Google Scholar]
- Harrison C. A., Wiater E., Gray P. C., Greenwald J., Choe S., Vale W. (2004). Modulation of activin and BMP signaling. Mol. Cell. Endocrinol. 225, 19–24 [DOI] [PubMed] [Google Scholar]
- Huang H. C., Murtaugh L. C., Vize P. D., Whitman M. (1995). Identification of a potential regulator of early transcriptional responses to mesoderm inducers in the frog embryo. EMBO J. 14, 5965–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. T., Klein E. S., Gillett S. J., Wang L., Song T. K., Pino M. E., Chandraratna R. A. (1995). Synthesis and characterization of a highly potent and effective antagonist of retinoic acid receptors. J. Med. Chem. 38, 4764–4767 [DOI] [PubMed] [Google Scholar]
- Jones J. R., Shelton K. D., Magnuson M. A. (2005). Strategies for the use of site-specific recombinases in genome engineering. Methods Mol. Med. 103, 245–257 [DOI] [PubMed] [Google Scholar]
- Long Q., Shelton K. D., Lindner J., Jones J. R., Magnuson M. A. (2004). Efficient DNA cassette exchange in mouse embryonic stem cells by staggered positive-negative selection. Genesis 39, 256–262 [DOI] [PubMed] [Google Scholar]
- Maretto S., Cordenonsi M., Dupont S., Braghetta P., Broccoli V., Hassan A. B., Volpin D., Bressan G. M., Piccolo S. (2003). Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc. Natl. Acad. Sci. USA 100, 3299–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K., Yoon K., Dang L., Tokunaga A., Gaiano N. (2007). Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 449, 351–355 [DOI] [PubMed] [Google Scholar]
- Monteiro R. M., de Sousa Lopes S. M., Korchynskyi O., ten Dijke P., Mummery C. L. (2004). Spatio-temporal activation of Smad1 and Smad5 in vivo: monitoring transcriptional activity of Smad proteins. J. Cell Sci. 117, 4653–4663 [DOI] [PubMed] [Google Scholar]
- Monteiro R. M., de Sousa Lopes S. M., Bialecka M., de Boer S., Zwijsen A., Mummery C. L. (2008). Real time monitoring of BMP Smads transcriptional activity during mouse development. Genesis 46, 335–346 [DOI] [PubMed] [Google Scholar]
- Mumm J. S., Schroeter E. H., Saxena M. T., Griesemer A., Tian X., Pan D. J., Ray W. J., Kopan R. (2000). A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197–206 [DOI] [PubMed] [Google Scholar]
- Nagarajan R. P., Liu J., Chen Y. (1999). Smad3 inhibits transforming growth factor-beta and activin signaling by competing with Smad4 for FAST-2 binding. J. Biol. Chem. 274, 31229–31235 [DOI] [PubMed] [Google Scholar]
- Ogawa K., Saito A., Matsui H., Suzuki H., Ohtsuka S., Shimosato D., Morishita Y., Watabe T., Niwa H., Miyazono K. (2007). Activin-Nodal signaling is involved in propagation of mouse embryonic stem cells. J. Cell Sci. 120, 55–65 [DOI] [PubMed] [Google Scholar]
- Osada S. I., Saijoh Y., Frisch A., Yeo C. Y., Adachi H., Watanabe M., Whitman M., Hamada H., Wright C. V. (2000). Activin/nodal responsiveness and asymmetric expression of a Xenopus nodal-related gene converge on a FAST-regulated module in intron 1. Development 127, 2503–2514 [DOI] [PubMed] [Google Scholar]
- Petersen D. R., Gustavsen C., Lindskog S. R., Magnuson M. A., Zaret K. S., Serup P. (2012). Engineering artificial signaling centers to polarize embryoid body differentiation. Stem Cells Dev. 21, 647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo M. A., Springer G. H., Granada B., Piston D. W. (2004). An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22, 445–449 [DOI] [PubMed] [Google Scholar]
- Rochette-Egly C., Germain P. (2009). Dynamic and combinatorial control of gene expression by nuclear retinoic acid receptors (RARs). Nucl. Recept. Signal. 7, e005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J., Zirngibl R., Cado D., Shago M., Giguère V. (1991). Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 5, 1333–1344 [DOI] [PubMed] [Google Scholar]
- Sauer B., Henderson N. (1989). Cre-stimulated recombination at loxP-containing DNA sequences placed into the mammalian genome. Nucleic Acids Res. 17, 147–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm S., Jiricny J., Schaffner W. (1987). The SV40 enhancer can be dissected into multiple segments, each with a different cell type specificity. Genes Dev. 1, 65–74 [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T. W., Smith A. (2008). Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 6, e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirard C., Kim S., Mirtsos C., Tadich P., Hoodless P. A., Itié A., Maxson R., Wrana J. L., Mak T. W. (2000). Targeted disruption in murine cells reveals variable requirement for Smad4 in transforming growth factor beta-related signaling. J. Biol. Chem. 275, 2063–2070 [DOI] [PubMed] [Google Scholar]
- Solloway M. J., Robertson E. J. (1999). Early embryonic lethality in Bmp5;Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development 126, 1753–1768 [DOI] [PubMed] [Google Scholar]
- Soriano P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 [DOI] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C. S., William C. M., Tanabe Y., Jessell T. M., Costantini F. (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman M. T., Slusarski D. C., Kaykas A., Louie S. H., Moon R. T. (2003). Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 13, 680–685 [DOI] [PubMed] [Google Scholar]
- Wandzioch E., Zaret K. S. (2009). Dynamic signaling network for the specification of embryonic pancreas and liver progenitors. Science 324, 1707–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg E., Winnier G. E., Chen X., Farnsworth C. L., Hogan B. L., Whitman M. (1998). A mouse homologue of FAST-1 transduces TGF beta superfamily signals and is expressed during early embryogenesis. Mech. Dev. 79, 17–27 [DOI] [PubMed] [Google Scholar]
- Winnier G., Blessing M., Labosky P. A., Hogan B. L. M. (1995). Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 9, 2105–2116 [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R. (1985). Steroid receptor regulated transcription of specific genes and gene networks. Annu. Rev. Genet. 19, 209–252 [DOI] [PubMed] [Google Scholar]
- Yeo C. Y., Chen X., Whitman M. (1999). The role of FAST-1 and Smads in transcriptional regulation by activin during early Xenopus embryogenesis. J. Biol. Chem. 274, 26584–26590 [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Nichols J., Chambers I., Smith A. (2003). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292 [DOI] [PubMed] [Google Scholar]
- Zambrowicz B. P., Imamoto A., Fiering S., Herzenberg L. A., Kerr W. G., Soriano P. (1997). Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. USA 94, 3789–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Bradley A. (1996). Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development 122, 2977–2986 [DOI] [PubMed] [Google Scholar]


