Abstract
Lignin is an integral cell wall component of all vascular plants. Peroxidases are widely believed to catalyze the last enzymatic step in the biosynthesis of lignin, the dehydrogenation of the p-coumaryl alcohols. As the first stage in identifying lignin-specific peroxidase isoenzymes, the classical anionic peroxidases found in the xylem of poplar (Populus trichocarpa Trichobel) were purified and characterized. Five different poplar xylem peroxidases (PXP 1, PXP 2, PXP 3–4, PXP 5, and PXP 6) were isolated. All five peroxidases were strongly glycosylated (3.6% to 4.9% N-glucosamine), with apparent molecular masses between 46 and 54 kD and pI values between pH 3.1 and 3.8. Two of the five isolated peroxidases (PXP 3–4 and PXP 5) could oxidize the lignin monomer analog syringaldazine, an activity previously correlated with lignification in poplar. Because these isoenzymes were specifically or preferentially expressed in xylem, PXP 3–4 and PXP 5 are suggested to be involved in lignin polymerization.
Lignin is a polymeric constituent of the plant cell wall and, second to cellulose, is the most abundant organic compound in the biosphere. It is a complex aromatic polymer derived mainly from the polymerization of three different hydroxycinnamyl alcohols: p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol. Specific isoenzymes of cell wall-localized peroxidases are widely believed to be responsible for the final enzymatic step in lignification: the oxidative dehydrogenation of monolignols, which leads to free radical polymerization (Higuchi, 1985). Despite extensive studies, however, no well-defined link between a single peroxidase and the developmentally regulated lignification process has been established.
Classical secretory plant peroxidases (class III; EC 1.11.1.7; donor, hydrogen peroxide oxidoreductase) are heme-containing enzymes of approximately 300 amino acids. The majority are N-glycosylated and are believed to be localized in the cell wall or the vacuole (Welinder, 1992). Most peroxidases can oxidize a wide range of substrates at the expense of H2O2, albeit at somewhat different rates. Classical peroxidases have been implicated in several primary and secondary metabolic processes, including hormone catabolism (Kenten, 1955), pathogen defense (Moerschbacher, 1992), phenol oxidation (Lagrimini, 1991), cross-linking of cell wall-structural proteins and polysaccharides (Fry, 1986; Lamport, 1986), and particularly in lignin polymerization (Mäder 1992; McDougall, 1992; Baier et al., 1993).
The major reasons it has been difficult to assign a specific function to any particular peroxidase have been the very high redundancy found in peroxidase genes, the broad spectrum of substrates accepted by these enzymes, and the very similar immunological properties of different isoenzymes. It has now become evident that this complexity is even more pronounced than previously anticipated; the Arabidopsis genome was recently estimated to encode more than 40 different ER-targeted peroxidases grouped in families of high homology (Welinder et al., 1996). Additionally, because the down-regulation of specific peroxidases did not always lead to phenotypes, it was suggested that other isoenzymes were compensating for the reduced expression levels (Sherf et al., 1993). This phenomenon complicates the determination of physiological roles for peroxidases when using a molecular biology approach. Therefore, it seems necessary to choose methods that allow the study of these enzymes at different levels (e.g. transcription, translation, enzymatic properties, spatio-temporal expression), without interference from the gene redundancy.
To identify lignin-specific isoenzymes, we have focused on the anionic peroxidases from poplar (Populus spp.) xylem for the following reasons: between 15% and 36% of the dry weight of wood consists of lignin (Higuchi, 1985), so the lignification process is extensive. Poplar xylem can be easily separated from other tissues, resulting in a reduction of candidate peroxidases for this process. Poplar is amenable to genetic engineering and can be considered a model tree for molecular lignification studies (Boerjan et al., 1997). Also, in histochemical studies, anionic peroxidases with SYR-oxidizing activity have been shown to correlate strictly with lignifying cells in poplar (Populus × euramericana; Goldberg et al., 1983). It was shown that this SYR activity originates from fast-migrating anionic peroxidases (Imberty et al., 1985). Furthermore, the specific SYR and the sinapyl alcohol-oxidizing activities appear to be mediated by the same isoenzymes (Tsutsumi and Sakai, 1993, 1994; Tsutsumi et al., 1994). These researchers used differences in the kinetic constants for the oxidation of lignin monomers among poplar (Populus alba L.) callus isoenzymes to assign monomer specificity to different isoenzymes (Tsutsumi and Sakai, 1993, 1994; Tsutsumi et al., 1994). However, this kind of assignment has been questioned lately by the data from Takahama and Oniki (1996) and Takahama et al. (1996), who have demonstrated that peroxidase-generated radicals of hydroxycinnamic acids, coniferyl alcohol, and other cell wall components can function as mediators for the oxidation of sinapyl alcohol.
Here we present the purification and characterization of the five classical anionic peroxidases detected in poplar xylem. Two of the five isolated peroxidases were able to oxidize SYR, and because oxidation of SYR has been correlated with lignification in a wide range of woody species (Harkin and Obst, 1973), we suggest that these peroxidases represent lignifying isoenzymes.
MATERIALS AND METHODS
Unless stated otherwise, all chemicals, enzymes, and materials were purchased from Sigma.
Peroxidase Assays
Peroxidase activity was measured spectrophotometrically at 25°C by following the H2O2-dependent oxidation of DAB at 452 nm or of ABTS at 418 nm. The reaction mixture contained 20 mm sodium citrate, pH 5.5, 1% (v/v) protein sample, and 1 mm DAB and 0.03% (w/v) H2O2, or 0.04% (w/v) ABTS and 0.006% (w/v) H2O2.
For the SYR assay, peroxidase activity was measured spectrophotometrically at 25°C by determining the initial rate of the H2O2-dependent oxidation of SYR at 530 nm. The reaction mixture contained 5 mm Tris-HCl, pH 7.5, 1% (v/v) protein sample, 20 μm SYR, and 0.03% (w/v) H2O2. The method was adapted from that of Goldberg et al. (1983).
For peroxidase activity staining in polyacrylamide gels, protein samples (minimum of 2 fmol per isoenzyme) were dissolved in loading buffer without SDS and thiol-reducing agents and analyzed on 0.5-mm-thick SDS-polyacrylamide gels (10%) without prior boiling, according to the method of Laemmli (1970). Proteins were separated at 10 V cm−1. After separation, the gels were equilibrated for 30 min in 50 mm sodium citrate, pH 5.5, prior to incubation with 1 mm DAB and 0.03% (w/v) H2O2 in fresh sodium citrate buffer.
Plant material (poplar, Populus trichocarpa cv Trichobel) for initial analysis was harvested and stored at −70°C. Stems and roots were separated into bark and xylem. The tissues were ground in liquid nitrogen and extracted with 20 mm Tris adjusted to pH 7.5 with l-ascorbic acid. The debris were removed by centrifugation, and the extracts were analyzed by native PAGE as described above.
Protein Extraction and Extract Conditioning
Stems from 2-year-old poplars grown in a nursery were harvested in spring and stored at −70°C. The bark was peeled off and the xylem (275 g) was ground to a very fine powder by pressing it against a rotating sanding disc submerged in liquid nitrogen. The wood powder was extracted twice with 20 mm Tris adjusted to pH 7.5 with l-ascorbic acid (to minimize oxidation), and the extract was filtered twice through four layers of Miracloth (Calbiochem), once through Whatman 3MM paper, twice through a glass microfiber filter (GF/C, Whatman), and finally through a 0.45-μm Microsart module (SM 3021230601W, Sartorius, Göttingen, Germany) mounted in a Sartocon mini-cross-flow system (Sartorius). Subsequently, the protein extract was concentrated 5-fold using the mini-cross-flow system containing a 10-kD cutoff Ultrasart module (SM 3031453901E, Sartorius).
Protein Chromatography
The proteins were separated from low-molecular-mass substances by Sephadex G-25 (Pharmacia) gel-filtration chromatography on a 5- × 30-cm column (4 mL min−1 flow), equilibrated in 5 mm Tris-Cl, pH 7.5 (200 mL of extract run−1). Detection was carried out at 280 nm. The eluting proteins from each run were pooled and applied on a DEAE-Sepharose column (2.6 × 25 cm, Pharmacia), equilibrated in 5 mm Tris-Cl, pH 7.5. The column was washed with 5 bed volumes of equilibration buffer and the proteins were eluted with a short linear gradient (500 mL, 0–0.5 m NaCl in equilibration buffer) at 2 mL min−1. The peroxidase activity was identified in the eluting fractions by assaying with DAB, ABTS, and SYR. Peroxidase fractions were pooled, and ammonium sulfate was added to a final concentration of 1.7 m, before loading onto a Phenyl-Sepharose column (1.6 × 15 cm, Pharmacia), equilibrated in 20 mm Tris-Cl, pH 7.5, and 1.7 m ammonium sulfate. The column was washed with 5 bed volumes of equilibration buffer before the proteins were eluted with a 300-mL linear gradient from equilibration buffer to 5 mm Tris-Cl, pH 7.5 (1 mL min−1). Fractions containing peroxidase activity were pooled and applied to a concanavalin A column (1 × 5 cm, Pharmacia) equilibrated in 20 mm Tris-Cl, pH 7.5, 0.5 m ammonium sulfate. The column was washed with 5 bed volumes of 20 mm Tris-Cl, pH 7.5, 0.5 m NaCl; after a buffer change to 5 mm Tris-Cl, pH 7.5, the proteins were recovered with a 50-mL gradient of 0 to 100 mm Man in 5 mm Tris-Cl, pH 7.5 (0.5 mL min−1 flow). The fractions containing peroxidase activity were pooled. All steps in the purification were performed at room temperature. Protein samples were stored at 4°C.
Isoenzyme Separation
The protein sample resulting from the low-pressure purification scheme was applied to a Mono-Q anion-exchange column (Pharmacia) equilibrated in 10 mm Tris-Cl, pH 7.5. The column was washed with 10 bed volumes of equilibration buffer before the isoenzymes were eluted with a gradient of 0 to 0.5 m NaCl in equilibration buffer at 1 mL min−1. The column eluate was monitored at 280 nm for proteins and at 404 nm for peroxidases. The RZ values (ASORETmax/A280) were calculated from the chromatograms.
Protein Determination
Protein concentration was determined according to the method of Bradford (1976) using BSA as a standard. The molar concentration of the purified isoenzymes was estimated spectrophotometrically at the Soret maximum (404 nm), assuming an extinction coefficient of 100 mm−1 cm−1 (Welinder, 1992).
Peptide and Sequence Analysis
The purified isoenzymes were reduced and the Cys residues were blocked by a method adapted from that of Rüegg and Rudinger (1977). Essentially, protein samples were precipitated with 4 volumes of ethanol and dissolved in 100 μL of 100 mm Tris-Cl, pH 8.2, and 8 m urea; subsequently, 50 μL 1-propanol was added. Before the addition of 1 μL of tributylphosphine and incubation in the dark for 2 h, the samples were flushed with N2. After the proteins were reduced, 1 μL of 1 m iodoacetate was added. After another 5 min in the dark, 1 μL of 2-mercaptoethanol was added and the proteins were precipitated with 4 volumes of acetone. The precipitated proteins were then separated by SDS-PAGE (10%; Laemmli, 1970) and blotted onto PVDF membranes (Millipore). The excised proteins were digested in situ with trypsin and the generated peptides were subsequently separated by reversed-phase HPLC, as previously described (Bauw et al., 1987, 1989). Chemical fragmentation of the intact proteins was performed in 100 μL of 70% (v/v) formic acid containing 0.16 mg of cyanogen bromide for 24 h in the dark at room temperature. After cleavage, the reaction mixture was diluted with 10 volumes of water and lyophilized. The peptides were dissolved in SDS sample buffer, separated by SDS-PAGE (15%), transferred onto PVDF membranes, and subjected to protein-sequencing analysis as described by Bauw et al. (1989).
Amino Acid Analysis
Amino acid analysis was performed essentially as described by Barkholt and Jensen (1989). Protein samples (60 pmol) were hydrolyzed in 6 n HCl, 0.05% (v/v) phenol, 0.1% (w/v) 3,3′-dithiodipropanoic acid at 110°C, and the amino acids were then separated by ion-exchange HPLC. The number of replicas was dependent on the availability of the isoenzymes: for PXP 1, two for 6 h of hydrolysis, four for 20 h, and two for 72 h; for PXP 2, PXP 3, and PXP 4, two for 20 h; for PXP 5, two for 6 h, three for 20 h, and two for 72 h; and for PXP 6, one for 20 h.
Deglycosylation
The deglycosylation protocol was adapted from that of Edge et al. (1981) as follows. The protein sample (200 pmol of peroxidase) was lyophilized and dissolved in 200 μL of trifluoromethanesulfonic acid:anisole (2:1, v/v) in a glass tube with a Teflon-lined screw cap. The reaction mixture was flushed with N2 and left for 2 h at room temperature. Diethyl ether (400 μL) and 400 μL of 50% (v/v) aqueous pyridine were added, and the solution was extracted three times with diethyl ether. The pH of the aqueous phase was adjusted by the addition of 0.1% (v/v) TFA and applied to a C2 reversed-phase column (Reversed-Phase kit, Alltech, Deerfield, IL), previously equilibrated in 0.1% (v/v) TFA. The column was washed with 5 bed volumes of equilibration buffer and 5 bed volumes of 0.1% (v/v) TFA and 5% (v/v) acetonitrile, before the proteins were eluted with 0.1% (v/v) TFA and 70% (v/v) acetonitrile. The eluted peroxidase was lyophilized and analyzed by SDS-PAGE (10%; Laemmli, 1970).
Determination of Apparent Molecular Masses
The isoenzymes, together with molecular mass standards (Pharmacia), were analyzed by SDS-PAGE (10%; Laemmli, 1970) and visualized with Coomassie blue staining. Apparent molecular masses were determined according to the instructions in the calibration kit booklet (Pharmacia).
IEF
IEF gels for the low-pH range (pH 2.5–5) were cast using Pharmalytes according to the manufacturer's instructions (Pharmacia). The gel was mounted onto a water-cooled (15°C) 2117 Multiphor II electrophoresis unit (Pharmacia) and 0.5 pmol of each isoenzyme and 10 μL of pI standards (low range, Pharmacia) were focused for 3500 V h−1, 5 W, 1500 V, 25 mA. The gel was cut into parts, the peroxidases were visualized as for the DAB gel assay, and the pI standards were visualized with PhastGel Blue R (Pharmacia), following the manufacturer's instructions. The migration of all activity bands and standards was measured and the pIs were determined by interpolation.
RESULTS
Poplar Anionic Peroxidases
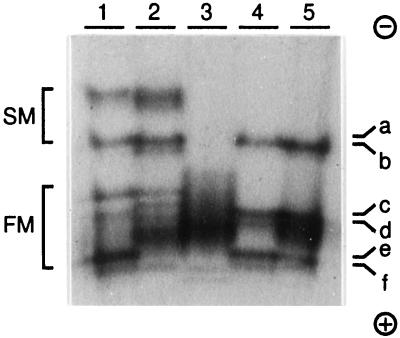
To identify putative lignin-specific peroxidases from poplar, proteins from different tissues of P. trichocarpa were extracted and the anionic peroxidases analyzed on activity gels (Fig. 1). The activity pattern obtained from the xylem of roots and stems was similar, with minor variation in the band intensities, suggesting that the same peroxidase isoenzymes were active in both tissues. In the bark of roots and stems, the slow-migrating activities were also similar, but the fast-migrating activities showed a different pattern. The leaf extract contained only poorly resolved, fast-migrating peroxidase activities. The xylem peroxidase activity bands c and d seemed to be specifically or preferentially expressed in the xylem, whereas the other activity bands from the xylem seemed to be present in other tissues as well.
Figure 1.
Anionic peroxidase activities from poplar (P. trichocarpa Trichobel). Extracts from root bark (1), stem bark (2), leaf (3), root xylem (4), and stem xylem (5) were analyzed on a native gel. Peroxidases were visualized with DAB and H2O2 as described in Methods. The letters a through f were chosen arbitrarily to represent the xylem gel activities. SM, Slow migrating; FM, fast migrating. The protein migration was from − to +.
Peroxidases are often classified according to their extractability as soluble, ionically bound, or covalently bound. To investigate whether other anionic peroxidases were present in the stem xylem that had escaped our extraction procedure, tissue that had been extracted four times with low-salt buffers was treated further with the cell wall-degrading enzyme mixture Driselase (Fluka) and reextracted with buffer containing 1 m NaCl. Peroxidase activity was released in this extract, but the activity pattern was identical to that of the low-salt extract (data not shown).
The activity ratio between the fast- and the slow-migrating bands varied within the season. In particular, the activity bands e and f varied strongly. In the material used for the purification described below, activity band f was not detected at all.
Isolation of Xylem Peroxidases
All anionic PXPs, detected by three different peroxidase substrates (ABTS, DAB, and SYR), were copurified in three chromatographic steps and subsequently separated and further purified on a high-resolution anion-exchange column, yielding six peaks of pure peroxidase isoenzymes.
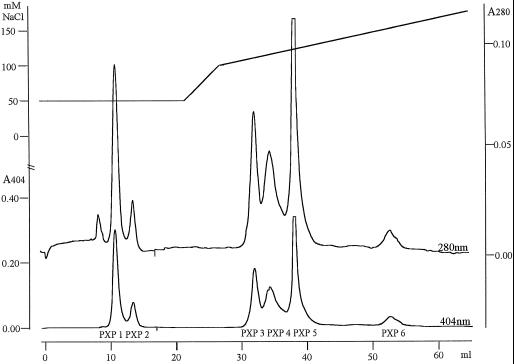
The purification scheme is presented in Table I. All fractions showing peroxidase activity by using DAB, ABTS, or SYR as substrates were pooled after each chromatographic step and applied to the next column. The activity of all isoenzymes was apparently fully preserved throughout the copurification procedure. This was judged visually from activity gels in which 1 × 10−5 of the pooled fraction from each chromatographic step was analyzed (data not shown). The three low-pressure chromatographic separation steps (anion-exchange, hydrophobic-interaction, and lectin-affinity chromatography) gave a 6-, 7-, and 4-fold purification, respectively, yielding 1.1 mg of protein from 275 g of xylem material, and a final peroxidase-to-protein enrichment of 232-fold (Table I). The peroxidase isoenzymes were separated and further purified by Mono-Q chromatography, yielding six peaks of pure peroxidase activity, designated PXP 1 to PXP 6. PXP 1 and PXP 2 eluted first during an isocratic step and PXP 3 to PXP 6 eluted during a linear gradient (Fig. 2).
Table I.
Copurification of anionic xylem peroxidases
| Separation | Volume | Protein | Peroxidase Enrichment |
|---|---|---|---|
| mL | mg | -fold | |
| Extract | 2900 | 255 | 1.0 |
| Ultra-filtrate | 600 | 171 | 1.5 |
| G25-permeate | 780 | 166 | 1.5 |
| DEAE-Sepharose | 180 | 29 | 8.8 |
| Phenyl-Sepharose | 150 | 4.2 | 61 |
| Concanavalin A | 32 | 1.1 | 232 |
Xylem (275 g) from 2-year-old poplar trees was ground and extracted.
Figure 2.
Mono-Q separation of the anionic peroxidase isoenzymes. The elution of the Mono-Q column was recorded at 404 nm (lower chromatogram, lower scale to the left) and at 280 nm (upper chromatogram, scale to the right). The upper line represents the NaCl concentration in the elution gradient (upper scale to the left). The horizontal scale represents the elution volume. The eluting peaks are designated PXP 1, PXP 2, PXP 3, PXP 4, PXP 5, and PXP 6.
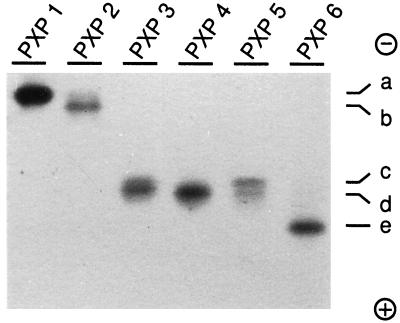
The recovery of peroxidase activity after the Mono-Q separation was typically between 60% and 70%. The purity of the peroxidase peaks was reflected directly by the chromatograms in which the specific peroxidase absorption (404 nm) corresponded perfectly to the protein absorption (280 nm), except for a small 280-nm absorption peak preceding the PXP 1 peak (Fig. 2). The RZ values ranged from 2.7 to 3.6 (Table II). The yield for each isoenzyme is indicated in Table II. To relate the purified isoenzymes with the gel activities in Figure 1, the separated peroxidases were analyzed on an activity gel (Fig. 3). A clear separation in two activity bands was seen for PXP 5. The two bands for PXP 5 were not due to tailing from PXP 4, because samples collected from different positions within the PXP 5 peak showed the same two bands (data not shown). Comparison of Figures 1 and 3 revealed that PXP 1, PXP 2, PXP 3, PXP 4, PXP 5, and PXP 6 represented activity bands a, b, c, d, c-d, and e, respectively.
Table II.
Yield and purity of the Mono-Q-separated isoenzymes
| Isoenzyme | Yielda | RZ (A404/A280) |
|---|---|---|
| pmol | ||
| PXP 1 | 2250 | 3.6 |
| PXP 2 | 650 | 3.3 |
| PXP 3 | 1800 | 2.8 |
| PXP 4 | 2500 | 2.7 |
| PXP 5 | 3350 | 3.2 |
| PXP 6 | 650 | 3.1 |
Yields were estimated assuming an extinction coefficient of 100 mm−1 cm−1 at 404 nm.
Figure 3.
Native gel analysis of the separated isoenzymes. The isoenzymes were separated in a native gel and visualized with DAB and H2O2 as in Figure 1. The numbers above the lanes correspond to the isoenzyme names given in Figure 2. The letters a through e represent the xylem gel activities, as described in the legend to Figure 1. The protein migration was from − to +.
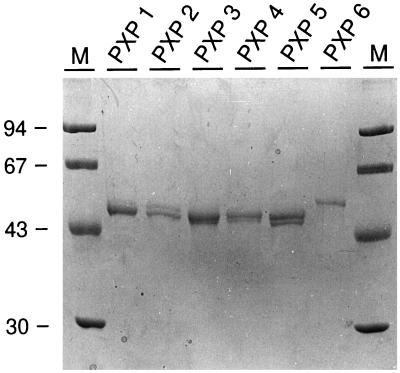
Figure 4.
Analysis of the purified isoenzymes on a denaturing gel. SDS-PAGE (10%) analysis of the purified isoenzymes (50 pmol), stained with Coomassie blue. Apparent molecular mass markers (M) are given with mass indications in kD. The numbers above the lanes correspond to the isoenzyme names given in Figure 2.
Figure 5.
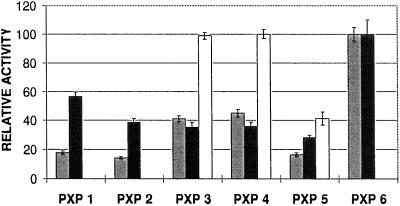
Analysis of the substrate specificity of the peroxidase isoenzymes. The rate of oxidation for the three substrates ABTS, DAB, and SYR was measured for the six purified isoenzymes as described in Methods. The activity of the most active isoenzyme for each substrate was set to 100. The measurements were repeated 10 times. The means ± sd values are indicated. Enzyme activity measurements were performed using 10 pmol of peroxidase isoenzyme in a 200-μL reaction volume. Gray bars, DAB; black bars, ABTS; white bars, SYR.
Figure 6.
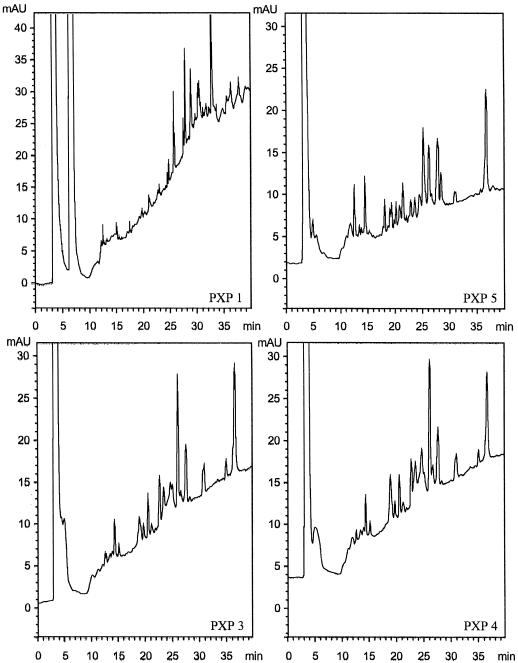
Analysis of the peroxidases by trypsin digestion. Comparison of the reversed-phase HPLC separation chromatograms of the trypsin-generated peptides from the four most abundant peroxidases. mAU, Milli-absorbance unit.
Protein Characterization
To determine the apparent molecular masses, the purified, denatured, and reduced isoenzymes were analyzed by SDS-PAGE. This revealed two polypeptide bands for each isoenzyme except PXP 6 (Fig. 4). The apparent molecular masses of the polypeptide bands are listed in Table III. The double-band pattern was also observed on gel analysis performed without a thiol-reducing agent in the sample buffer, excluding partial reduction of the disulfide bridges as an explanation for the doublets. The ratio between the two polypeptide bands did not change when the purification was performed at 4°C, indicating that degradation was not the cause. To further investigate the reason for these doublets in the SDS-PAGE experiment, two abundant isoenzymes (PXP 1 and PXP 5) were deglycosylated as described in Methods. After deglycosylation, the proteins migrated as a single polypeptide band, with apparent molecular masses of approximately 37 kD (data not shown), indicating that heterogeneity in the glycan part was the reason for the two polypeptide bands per isoenzyme.
Table III.
Molecular data of the Mono-Q-separated isoenzymes
| Isoenzyme | Apparent Molecular Mass | pI | Glycans |
|---|---|---|---|
| kD | pH | estimated no. | |
| PXP 1 | 50.3, 54.3 | 3.83 | 15 |
| PXP 2 | 49.1, 53.6 | 3.83 | 13 |
| PXP 3 | 46.9, 48.2 | 3.43, 3.53 | 11 |
| PXP 4 | 47.4, 48.5 | 3.43, 3.53 | 11 |
| PXP 5 | 46.5, 48.2 | 3.73 | 11 |
| PXP 6 | 53.4 | 3.13 | 13 |
The estimation of the glycan number was calculated from the amino acid hydrolysis assuming two residues of N-glucosamine per glycan, a 50% loss in the 20-h hydrolysis, and peroxidases of 300 amino acids.
The pI values for the purified isoenzymes ranged between pH 3.1 and 3.8 (Table III). Two bands with similar intensities were observed for PXP 3 and PXP 4.
All isoenzymes could oxidize ABTS and DAB, albeit to a different extent, whereas only PXP 3, PXP 4, and PXP 5 were able to oxidize SYR. PXP 3 and PXP 4 showed identical rates of oxidation for the three substrates tested (Fig. 5).
Molecular Analysis
The purified isoenzymes were subjected to protein hydrolysis. The subsequent amino acid analysis revealed that all isoenzymes were different except PXP 3 and PXP 4 (Table IV). The amino acid analysis also detected N-glucosamine and demonstrated that all isoenzymes were heavily glycosylated (Table IV). Assuming a 50% loss of N-glucosamine for a 20-h hydrolysis (K.G. Welinder, unpublished results), a protein size of 300 amino acids, and N-linked glycans containing two GlcNAc residues (Welinder, 1992), we can estimate the isoenzymes to have 11 to 15 glycan side chains attached (Table III). The hyperglycosylated state of these peroxidases explains the relatively high apparent molecular masses and could explain their high stability during the purification process.
Table IV.
Amino acid composition of the purified isoenzymes
| Amino Acid | PXP 1 | PXP 2 | PXP 3 | PXP 4 | PXP 5 | PXP 6 |
|---|---|---|---|---|---|---|
| mol % | ||||||
| Asn/Asp | 14.904 | 14.887 | 14.809 | 14.923 | 15.546 | 14.809 |
| Thr | 8.194 | 8.907 | 9.053 | 8.959 | 8.837 | 8.097 |
| Ser | 10.741 | 11.383 | 9.946 | 10.092 | 8.816 | 10.276 |
| Gln/Glu | 7.168 | 8.242 | 8.078 | 8.243 | 8.099 | 7.639 |
| Pro | 4.776 | 4.234 | 4.412 | 4.254 | 4.527 | 4.880 |
| Gly | 8.296 | 8.382 | 7.313 | 7.714 | 6.859 | 8.745 |
| Ala | 9.175 | 9.378 | 9.350 | 9.271 | 8.935 | 9.691 |
| Cys | 2.176 | 1.886 | 2.469 | 2.272 | 2.273 | 1.911 |
| Val | 5.176 | 4.695 | 5.204 | 5.146 | 6.167 | 4.465 |
| Met | 0.979 | 0.965 | 0.950 | 0.929 | 0.953 | 1.068 |
| Ile | 5.318 | 2.755 | 3.590 | 3.497 | 3.826 | 4.533 |
| Leu | 9.804 | 10.654 | 9.905 | 9.834 | 9.765 | 9.852 |
| Tyr | 1.295 | 1.372 | 1.381 | 1.379 | 1.242 | 1.428 |
| Phe | 4.737 | 5.434 | 5.764 | 5.733 | 6.122 | 4.719 |
| His | 1.055 | 1.211 | 1.126 | 1.139 | 1.027 | 1.165 |
| Lys | 1.644 | 1.693 | 2.174 | 2.178 | 2.017 | 2.174 |
| Arg | 4.562 | 3.923 | 4.477 | 4.437 | 4.988 | 4.548 |
| Trp | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sum | 100 | 100 | 100 | 100 | 100 | 100 |
| GlcN | 4.93 | 4.31 | 3.74 | 3.59 | 3.78 | 4.25 |
All isoenzymes were hydrolyzed and analyzed as described in Methods (60 pmol sample−1). N-Glucosamine (GlcN) content was derived from the 20-h hydrolysates. Corrections for degradation of Ser (10%) and Thr (5%) for the 20-h hydrolyses were performed.
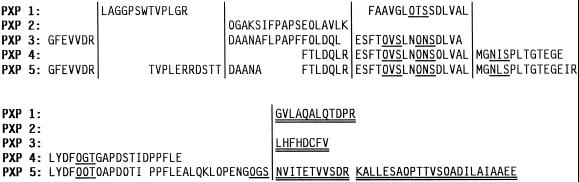
To obtain sequence information, the isolated isoenzymes were subjected to tryptic digestion, followed by peptide separation and sequencing. Before the samples were digested, it was necessary to perform an extensive reduction and modification of the disulfide bridges. The high level of glycosylation and the compact structure of plant peroxidases are probably the causes for this fragmentation problem. The HPLC chromatograms of the peptide separations showed only minor differences between PXP 3 and PXP 4, whereas the chromatograms for PXP 1 and PXP 5 were clearly unique (Fig. 6). For PXP 2 and PXP 6, no comparable chromatograms were obtained because of the limited amount of protein. Several tryptic peptides were selected for amino acid-sequencing analysis. These amino acid sequences, as well as the sequences obtained after cyanogen bromide fragmentation, are presented in Figure 7. The alignment of the amino acid sequences revealed that PXP 1 and PXP 2 were clearly distinct from PXP 3, PXP 4, and PXP 5, whereas the amino acid sequences of PXP 3, PXP 4, and PXP 5 showed that these three were very similar, with only one amino acid difference found in the 68 amino acids of the homologous peptides obtained (PXP 4, MGNISPLTGTEGE; PXP 5, MGNLSPLTGTEGEIR). PXP 1 was 68% identical to the group PXP 3, PXP 4, and PXP 5 in the homologous regions. The amino acid sequences also demonstrated the hyperglycosylated state of these proteins with 10 putative glycosylation sites 0XS/T (0 indicating that no amino acid was identified during that particular step in the sequencing) in the 22 peptides sequenced. In addition, two NXS/T sequences were identified, which apparently were either not glycosylated or only partially glycosylated (Fig. 7). All peptide sequences obtained showed homology to known peroxidases.
Figure 7.
Amino acid sequences of the trypsin- and cyanogen bromide-generated peptides. Underlined sequences are putative glycosylation sites (NXT/S or 0XT/S, where X represents any amino acid except P). If N was linked to a glycan, it was not detected during sequencing (indicated by 0). Sequences placed at the end (double underlined) showed no homology to other peptide sequences obtained.
DISCUSSION
It has often been suggested that anionic peroxidases are involved in the formation of lignin in poplar (Imberty et al., 1985; Tsutsumi et al., 1994) and in tobacco (Mäder et al., 1977; Lagrimini, 1992). To identify putative lignin-specific peroxidases we have purified and characterized the anionic peroxidases from the poplar stem xylem and identified SYR-specific isoenzymes. In this article we present the first example, to our knowledge, of an almost complete set of anionic peroxidases from a single tissue; the peroxidases were purified, separated, and characterized, and thus, we present a unique starting point to unravel the function of individual anionic peroxidases and their presumed involvement in lignification.
Six Anionic Peroxidase Fractions Purified from Poplar Xylem
A fairly standard procedure exploiting methods often used to purify plant peroxidases (ion-exchange, hydrophobic interaction, and lectin binding) allowed us to isolate six anionic peroxidase fractions from the xylem of poplar stems. We are confident that all of the major anionic peroxidases were detected and isolated by this purification, because all fractions after each chromatographic step were assayed with three different peroxidase substrates and no additional peroxidase activities were detected. In addition, extractions with high-salt buffers after digestion with cell wall-degrading enzymes did not reveal other anionic activities on activity gels. The minor gel activity (Fig. 1, f) was not detected in the material used for the present purification but was detected from purifications with material harvested at other periods of the year; it was demonstrated to coelute with PXP 6 from the Mono-Q column (data not shown). Whether band f corresponds to another gene product is currently unclear. Lignification is one of the major processes taking place during xylogenesis; thus, if anionic peroxidases are involved in this process, they should be among the peroxidases isolated here.
The purity of the isolated peroxidases was confirmed directly by the Mono-Q chromatography: the protein and the peroxidase peaks (280 nm and 404 nm) had identical elution times and peak shapes, indicating that the peaks contained exclusively peroxidases. The high RZ values (between 2.7 and 3.6; Table II) are also indicative of relatively pure peroxidase fractions. Furthermore, the purity was supported by SDS-PAGE, with which all isoenzymes appeared as one or two polypeptide bands (Fig. 4). Finally, all amino acid sequences obtained match very well with known peroxidase sequences. When analyzed on activity gels, all isoenzymes behaved homogeneously, except for PXP 5 for which two activity bands were observed (Fig. 3). The separation of one gene product into different apparent molecular masses and pIs has previously been shown for peroxidases and was suggested to arise from heterogeneity in posttranslational modifications (Lagrimini et al., 1987, 1990). The doublets seen by SDS-PAGE (Fig. 4) are probably caused by heterogeneity in the glycans, because they disappeared after deglycosylation for PXP 1 and PXP 5.
Five Closely Related Peroxidases
We have shown that all of the isoenzymes characterized here were closely related at the biochemical level. This was to be expected from their similar behavior on low-pressure chromatographic columns, on which all of the isoenzymes eluted as closely overlapping peaks: they had pI values between pH 3.1 and pH 3.8 (Table II), they had relatively high apparent molecular masses between 46.9 and 54.3 kD (Table II), and the amino acid compositions were similar (Table III). The peptide sequences obtained and the amino acid analysis strongly indicated that PXP 3 and PXP 4 are identical and that PXP 5 is closely related to these two isoenzymes. This conclusion was supported by the fact that both PXP 3 and PXP 4 generated very similar chromatograms of the tryptic peptides (Fig. 6) and, within the experimental error, had identical apparent molecular masses, pI values (Table II), and enzymatic properties (Fig. 5). PXP 3 and PXP 4 differed only in their migration in native gels and in the elution time from the Mono-Q column. These small differences were probably due to heterogeneity in the glycan part or other posttranslational modifications. Therefore, we conclude that the peroxidase fractions PXP 3 and PXP 4 are derived from one peroxidase gene. For convenience, we designated this peroxidase PXP 3–4.
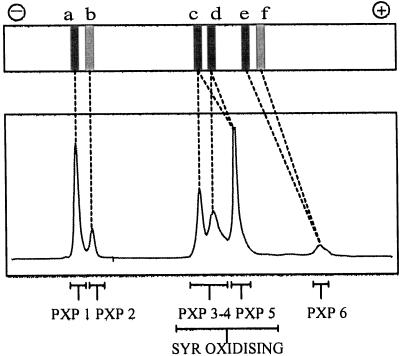
By comparing the activity pattern of xylem extracts (Fig. 1), the chromatogram from the Mono-Q separation (Fig. 2), and the native PAGE gel (Fig. 3), we concluded that PXP 1, PXP 2, PXP 3–4, PXP 5, and PXP 6 correspond to gel activities a, b, c-d, c-d, and e, respectively (Fig. 8). The kind of heterogeneity seen within these peroxidase fractions, i.e. polypeptide bands with different apparent molecular masses (Fig. 4), the two pI values for PXP 3–4 (Table III), and the double-activity band of PXP 5 in native gels (Fig. 3), is rather common for plant peroxidases and is probably caused by heterogeneity in the glycan part. We therefore consider PXP 1, PXP 2, PXP 3–4, PXP 5, and PXP 6 to be single gene products.
Figure 8.
Correlation of peroxidase isoenzymes, gel activities, and SYR oxidation. The top panel represents a stem xylem lane from Figure 1. The bottom panel represents the 404-nm chromatogram from Figure 2. The dashed lines indicate the relation between gel activities and the Mono-Q-separated peroxidase isoenzymes. PXP 1 to PXP 6 represent the gene products determined from the molecular characterization. The SYR-oxidizing isoenzymes are indicated.
PXP 3–4 and PXP 5 Are Homologous to Lignifying Peroxidases
Among the tobacco peroxidases, the fast-migrating, low-salt-extractable anionic isoenzymes (GI) are believed to be responsible for the polymerization of lignin (Mäder et al., 1975, 1977). This function was suggested based on their cell wall localization (Mäder et al., 1975), their high expression levels in stems (Lagrimini et al., 1987), and their high rate of oxidation of the lignin monomer coniferyl alcohol (Mäder et al., 1977). The role of the major fast-migrating anionic isoenzyme from tobacco has been investigated with sense and antisense technologies and seems to be responsible for lignification and/or phenol oxidation in relation to pathogen defense and physical damage but not for the developmentally regulated lignification process (Lagrimini et al., 1997). The peroxidase-dependent oxidation of SYR has been shown to be specific for lignifying cells in tobacco (Pang et al., 1989), but until now no correlation of this activity with a specific group of tobacco isoenzymes has been reported. In poplar xylem, fast-migrating anionic peroxidases (AII) are believed to polymerize lignin as in tobacco. Goldberg et al. (1981, 1983) showed a strict histochemical correlation between peroxidase-dependent SYR oxidase activity and the lignification process in P. × euramericana. SYR oxidase activity was observed in the primary cell walls at the onset of secondary cell wall deposition and in the secondary cell walls. This activity decreased as the xylem matured and no staining was seen in mature tissues. Additionally, it was reported that the SYR-oxidizing activity increased slightly during the growing season in young xylem.
Our data and the plant material used here, i.e. xylem from 2-year-old field-grown trees from P. trichocarpa, are comparable with the previous data obtained from P. × euramericana (2-year-old branches; Goldberg et al., 1983). Extraction of the anionic peroxidases could in both cases be performed with low-salt buffers, and subsequent reextraction after digestion with cell wall-degrading enzymes released activities with identical native gel mobilities, as for the low-salt extracts (Imberty et al., 1985). The fact that the activities released after treatment with cell wall-degrading enzymes are identical to the soluble activities was supported by Goldberg et al. (1983), who observed identical Km values for the SYR-oxidizing activities in both the cytoplasmic (soluble) and the cell wall (bound) fractions. Finally, the SYR-oxidizing activities correspond in both cases to the fast-migrating anionic peroxidases and were shown to be very stable (Goldberg et al., 1983; Imberty et al., 1985).
A comparison of the results obtained in P. trichocarpa with those from P. × euramericana suggests that PXP 3–4 and PXP 5 from P. trichocarpa (Fig. 1, c-d) are homologous to the fast-migrating peroxidase group AII from P. × euramericana and that PXP 1 and PXP 2 (Fig. 1, a-b) are homologous to the slow-migrating anionic group (AI; Imberty et al., 1985). The activity corresponding to PXP 6 (activity e and f) was not described for P. × euramericana, possibly because of the low abundance of these isoenzymes. A functional correlation between AII from P. × euramericana and GI from tobacco has previously been suggested (Imberty et al., 1985) and was based mainly on their similar electrophoretic behaviors.
The data obtained for PXP 3–4 and PXP 5 and the parallel observations in both poplar and tobacco (fast-migrating anionic enzymes, SYR-oxidizing, low-salt extractable, and stable enzymes) suggest that these isoenzymes are functionally and/or genetically homologous to the peroxidase isoenzymes believed to be involved in lignification in poplar and tobacco. This conclusion is further supported by the xylem-specific expression of PXP 3–4 and PXP 5 (Fig. 1, c and d).
Conclusion and Perspectives
Because peptide sequence information has been obtained from individual peroxidase isoenzymes, it is now possible to identify and isolate the cDNAs and genes corresponding to the SYR-oxidizing isoenzymes and to study their involvement in the lignification process. Two partial cDNAs and four genes encoding peroxidases have already been cloned from Populus kitakamiensis (Kawai et al., 1993; Osakabe et al., 1994, 1995), and experiments to overexpress the anionic peroxidase prxA1 have been initiated (Kajita et al., 1994). However, no information is available concerning the catalytic activities of the corresponding isoenzymes and none of the clones corresponds fully to the SYR-oxidizing isoenzymes that were identified here. To obtain further insight into the role of the SYR-oxidizing isoenzymes, it will be necessary to prove their colocalization with lignifying cells. For this purpose, immunolocalization using peptide tag-fused peroxidases or isoenzyme-specific antibodies can be considered. cDNAs and genes for a number of the purified peroxidases have been obtained for this purpose (J.H. Christensen, unpublished data). Finally, our data provide the possibility for a molecular study of individual peroxidases within an isoenzyme group. Previous studies of peroxidases in planta have been limited to the study of isoenzyme groups or single isolated isoenzymes. It has often been suggested that isoenzymes within groups (classified following gel mobility [pI] and extractability) should have similar functions, but, to our knowledge, this has never been demonstrated.
ACKNOWLEDGMENTS
The authors thank Martine De Cock, Rebecca Verbanck, Christiane Germonprez, Karel Spruyt, Antje Rohde, Jens Østergaard, and Mark Davey for helping with the figures and the manuscript, and Geert Persiau, Arne Jensen, and Yvonne Berger for technical assistance.
Abbreviations:
- ABTS
2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid)
- DAB
3,3′-diaminobenzidine
- PXP
poplar xylem peroxidase
- RZ
Reinheitszahl
- SYR
syringaldazine
- TFA
trifluoroacetic acid
Footnotes
This research was supported by funds from the Danish Agricultural and Veterinary Research Council, the European Commission (AIR2-CT93-1661), and Novo Nordisk (Denmark).
LITERATURE CITED
- Baier M, Goldberg R, Catesson AM, Francesch C, Rolando C. Seasonal changes of isoperoxidases from poplar bark tissues. Phytochemistry. 1993;32:789–793. [Google Scholar]
- Barkholt V, Jensen AL. Amino acid analysis: determination of cysteine plus half-cystine in proteins after hydrochloric acid hydrolysis with a disulfide compound as additive. Anal Biochem. 1989;177:318–322. doi: 10.1016/0003-2697(89)90059-6. [DOI] [PubMed] [Google Scholar]
- Bauw G, De Loose M, Inzé D, Van Montagu M, Vandekerckhove J. Alterations in the phenotype of plant cells studied by NH2-terminal amino acid-sequence analysis of proteins electroblotted from two-dimensional gel-separated total extracts. Proc Natl Acad Sci USA. 1987;84:4806–4810. doi: 10.1073/pnas.84.14.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauw G, Van Damme J, Puype M, Vandekerckhove J, Gesser B, Ratz GP, Lauridsen JB, Celis JE. Protein-electroblotting and microsequencing strategies in generating protein databases from two-dimensional gels. Proc Natl Acad Sci USA. 1989;86:7701–7705. doi: 10.1073/pnas.86.20.7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Baucher M, Chabbert B, Petit-Conil M, Leplé J-C, Pilate G, Cornu D, Monties B, Inzé D, Van Doorsselaere J, and others (1997) Genetic modification of lignin biosynthesis in quaking aspen (Populus tremuloides) and poplar (Populus tremula × Populus alba). In NB Klopfenstein, YW Chun, M-S Kim, MR Ahuja, eds, Micropropagation, Genetic Engineering, and Molecular Biology of Populus (General Technical Report RM-GTR-297). Rocky Mountain Forest and Range Experiment Station, Fort Collins, CO, pp 193–205
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Edge ASB, Faltynek CR, Hof L, Reichert LE, Jr, Weber P. Deglycosylation of glycoproteins by trifluoromethanesulfonic acid. Anal Biochem. 1981;118:131–137. doi: 10.1016/0003-2697(81)90168-8. [DOI] [PubMed] [Google Scholar]
- Fry SC. Cross-linking of matrix polymers in the growing cell walls of angiosperms. Annu Rev Plant Physiol. 1986;37:165–186. [Google Scholar]
- Goldberg R, Catesson A-M, Czaninski Y (1981) Histochemical and biochemical characteristics of peroxidases involved in lignification processes in poplar. In DG Robinson, H Quader, eds, Cell Walls '81. Wissenschaftliche Verlagsgesellschaft, Stuttgart, Germany, pp 251–260
- Goldberg R, Catesson A-M, Czaninski Y. Some properties of syringaldazine oxidase, a peroxidase specifically involved in the lignification processes. Z Pflanzenphysiol. 1983;110:267–279. [Google Scholar]
- Harkin JM, Obst JR. Lignification in trees: indication of exclusive peroxidase participation. Science. 1973;180:296–298. doi: 10.1126/science.180.4083.296. [DOI] [PubMed] [Google Scholar]
- Higuchi T. Biosynthesis of lignin. In: Higuchi T, editor. Biosynthesis and Biodegradation of Wood Components. Orlando, FL: Academic Press; 1985. pp. 141–160. [Google Scholar]
- Imberty A, Goldberg R, Catesson A-M. Isolation and characterization of Populus isoperoxidases involved in the last step of lignin formation. Planta. 1985;164:221–226. doi: 10.1007/BF00396085. [DOI] [PubMed] [Google Scholar]
- Kajita S, Osakabe K, Katayama Y, Kawai S, Matsumoto Y, Hata K, Morohoshi N. Agrobacterium-mediated transformation of poplar using a disarmed binary vector and the overexpression of a specific member of a family of poplar peroxidase genes in transgenic poplar cell. Plant Sci. 1994;103:231–239. [Google Scholar]
- Kawai S, Matsumoto Y, Kajita S, Yamada K, Katayama Y, Morohoshi N. Nucleotide sequence for the genomic DNA encoding an anionic peroxidase gene from a hybrid poplar, Populus kitakamiensis. Biosci Biotechnol Biochem. 1993;57:131–133. doi: 10.1271/bbb.57.131. [DOI] [PubMed] [Google Scholar]
- Kenten RH. The oxidation of indolyl-3-acetic acid by waxpod bean root sap and peroxidase systems. Biochem J. 1955;59:110–121. doi: 10.1042/bj0590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lagrimini LM. Wound-induced deposition of polyphenols in transgenic plants overexpressing peroxidase. Plant Physiol. 1991;96:577–583. doi: 10.1104/pp.96.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini LM (1992) Plant peroxidases: under- and over-expression in transgenic plants and physiological consequences. In C Penel, T Gaspar, H Greppin, eds, Plant Peroxidases 1980–1990, Topics and Detailed Literature on Molecular, Biochemical, and Physiological Aspects. Université de Genève, Geneva, Switzerland, pp 59–69
- Lagrimini LM, Bradford S, Rothstein S. Peroxidase-induced wilting in transgenic tobacco plants. Plant Cell. 1990;2:7–18. doi: 10.1105/tpc.2.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini LM, Burkhart W, Moyer M, Rothstein S. Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: molecular analysis and tissue-specific expression. Proc Natl Acad Sci USA. 1987;84:7542–7546. doi: 10.1073/pnas.84.21.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini LM, Gingas V, Finger F, Rothstein S, Liu T-TY. Characterization of antisense transformed plants deficient in the tobacco anionic peroxidase. Plant Physiol. 1997;114:1187–1196. doi: 10.1104/pp.114.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport DTA (1986) Roles for peroxidases in cell wall genesis. In H Greppin, C Penel, T Gaspar, eds, Molecular and Physiological Aspects of Plant Peroxidases. Université de Genève, Geneva, Switzerland, pp 199–208
- Mäder M (1992) Compartmentation of peroxidase isoenzymes in plant cells. In C Penel, T Gaspar, H Greppin, eds, Plant Peroxidases 1980–1990, Topics and Detailed Literature on Molecular, Biochemical, and Physiological Aspects. Université de Genève, Geneva, Switzerland, pp 37–46
- Mäder M, Meyer Y, Bopp M. Lokalisation der Peroxidase-Isoenzyme in Protoplasten und Zellwänden von Nicotiana tabacum L. Planta. 1975;122:259–268. doi: 10.1007/BF00385274. [DOI] [PubMed] [Google Scholar]
- Mäder M, Nessel A, Bopp M. Über die physiologische Bedeutung der Peroxidase-Isoenzymgruppen des Tabaks anhand einiger biochemischer Eigenschaften. II. pH-Optima, Michaelis-Konstanten, maximale Oxidationsraten. Z Pflanzenphysiol. 1977;82:247–260. [Google Scholar]
- Mäder M, Ungemach J, Schloss P. The role of peroxidase isoenzyme groups of Nicotiana tabacum in hydrogen peroxide formation. Planta. 1980;147:467–470. doi: 10.1007/BF00380189. [DOI] [PubMed] [Google Scholar]
- McDougall GJ (1992) Plant peroxidases and cell differentiation. In C Penel, T Gaspar, H Greppin, eds, Plant Peroxidases 1980–1990, Topics and Detailed Literature on Molecular, Biochemical, and Physiological Aspects. Université de Genève, Geneva, Switzerland, pp 101–115
- Moerschbacher BM (1992) Plant peroxidases: involvement in response to pathogens, In C Penel, T Gaspar, H Greppin, eds, Plant Peroxidases 1980–1990, Topics and Detailed Literature on Molecular, Biochemical, and Physiological Aspects. Université de Genève, Geneva, Switzerland, pp 91–99
- Osakabe K, Koyama H, Kawai S, Katayama Y, Morohoshi N. Molecular cloning and the nucleotide sequences of two novel cDNAs that encode anionic peroxidases of Populus kitakamiensis. Plant Sci. 1994;103:167–175. [Google Scholar]
- Osakabe K, Koyama H, Kawai S, Katayama Y, Morohoshi N. Molecular cloning of two tandemly arranged peroxidase genes from Populus kitakamiensis and their differential regulation in the stem. Plant Mol Biol. 1995;28:677–689. doi: 10.1007/BF00021193. [DOI] [PubMed] [Google Scholar]
- Pang A, Catesson A-M, Francesch C, Rolando C, Goldberg R. On substrate specificity of peroxidases involved in the lignification process. J Plant Physiol. 1989;135:325–329. [Google Scholar]
- Rüegg UT, Rudinger J. Reductive cleavage of cystine disulfides with tributylphosphine. Methods Enzymol. 1977;47:111–122. doi: 10.1016/0076-6879(77)47012-5. [DOI] [PubMed] [Google Scholar]
- Sherf BA, Bajar AM, Kolattukudy PE. Abolition of an inducible highly anionic peroxidase activity in transgenic tomato. Plant Physiol. 1993;101:201–208. doi: 10.1104/pp.101.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahama U, Oniki T (1996) Enhancement of peroxidase-dependant oxidation of sinapyl alcohol by esters of 4-coumaric and ferulic acids. In C Obinger, U Burner, R Ebermann, C Penel, H Greppin, eds, Plant Peroxidases: Biochemistry and Physiology. Université de Genève, Geneva, Switzerland, pp 118–123
- Takahama U, Oniki T, Shimokawa H. A possible mechanism for the oxidation of sinapyl alcohol by peroxidase-dependent reactions in the apoplast: enhancement of the oxidation by hydroxycinnamic acids and components of the apoplast. Plant Cell Physiol. 1996;37:499–504. [Google Scholar]
- Tsutsumi Y, Nishida T, Sakai K. Lignin biosynthesis in woody angiosperm tissues. III. Isolation of substrate-specific peroxidases related to the dehydrogenative polymerization of sinapyl and coniferyl alcohols from Populus callus cultures. Mokuzai Gakkaishi. 1994;40:1348–1354. [Google Scholar]
- Tsutsumi Y, Sakai K. Lignin biosynthesis in woody angiosperm tissues I. Lignification and peroxidase activity stimulated in water-stressed Populus callus cultures. Mokuzai Gakkaishi. 1993;39:214–220. [Google Scholar]
- Tsutsumi Y, Sakai K. Lignin biosynthesis in woody angiosperm tissues. II. Peroxidase related to syringyl and guaiacyl lignin biosynthesis in Populus callus cultures. Mokuzai Gakkaishi. 1994;40:744–750. [Google Scholar]
- Welinder KG (1992) Plant peroxidases: structure, function rela-tionships. In C Penel, T Gaspar, H Greppin, eds, Plant Peroxidases 1980–1990, Topics and Detailed Literature on Molecular, Biochemical and Physiological Aspects. Université de Genève, Geneva, Switzerland, pp 1–24
- Welinder KG, Jespersen HM, Kjærsgård IVH, Østergaard L, Abelskov AK, Hansen LN, Rasmussen SK (1996) What can we learn from Arabidopsis peroxidases? In C Obinger, U Burner, R Ebermann, C Penel, H Greppin, eds, Plant Peroxidases: Biochemistry and Physiology. Université de Genève, Geneva, Switzerland, pp 173–178