Abstract
Systematic study of proteins requires the availability of thousands of proteins in functional format. However, traditional recombinant protein expression and purification methods have many drawbacks for such study at the proteome level. We have developed an innovative in situ protein expression and capture system, namely NAPPA (nucleic acid programmable protein array), where C-terminal tagged proteins are expressed using an in vitro expression system and efficiently captured/purified by antitag antibodies coprinted at each spot. The NAPPA technology presented in this chapter enable researchers to produce and display fresh proteins just in time in a multiplexed high-throughput fashion and utilize them for various downstream biochemical researches of interest. This platform could revolutionize the field of functional proteomics with it ability to produce thousands of spatially separated proteins in high density with narrow dynamic rand of protein concentrations, reproducibly and functionally.
1. Introduction
The advancement of proteomics has significantly accelerated the pace of biomedical research. To this end, there is great demand for high-throughput data-driven approaches to study proteins and their functions at the proteome level. Protein microarrays provide a key enabling technology where thousands of proteins are spotted in high spatial density on a microscopic glass slide and enable the assay of protein biochemical properties on a planar surface in a multiplexed fashion(MacBeath and Schreiber, 2000; Zhu et al., 2000). In contrast to DNA microarrays, which only measure the relative concentration of DNA molecules complementary to spotted “oligonucleotides” (Schena et al., 1995), protein microarrays can be employed not only to measure target protein concentrations in biological samples (antibody arrays; Haab et al., 2001) but also to study a broad range of biochemical properties for thousands of proteins in a high-throughput fashion (LaBaer and Ramachandran, 2005). These biochemical properties include protein functions and interactions with other molecules such as small-molecule drugs, DNA, or other proteins.
While holding tremendous potential to meet the pressing need to study thousands of proteins in parallel, high-density recombinant protein micro-arrays have not been widely deployed into the biomedical research community. The production of protein microarrays requires the expression and purification of proteins at a scale that traditional methodologies cannot deliver. Unlike DNA that can be produced en masse using Polymerase Chain Reaction (PCR) by one enzyme—DNA polymerase—or even synthesized chemically, proteins must be produced from cDNA by complex transcription and translation machinery. During and/or after translation, proteins also need suitable environment and machinery to promote and maintain their native functional conformations.
The production and purification of recombinant protein for biological studies typically includes cloning the gene into an expression vector, transforming it into an expression system, inducing the cells to produce protein, and isolating the protein through a laborious set of purification steps on affinity columns. Although commonly employed, and even automated in some circumstance, this approach has serious drawbacks. First, there is variable protein yield during production that can lead to >1000-fold differences in concentrations from one protein to another in preparation. Because most biochemical reactions are concentration dependent, this can lead to false negatives/positives if proteins of interest are under/overrepresented on the microarray. Second, prepared proteins require storage at −20 °C or even −80 °C to maintain functionality and still have limited shelf life. Third, the recombinant proteins are often expressed in Escherichia coli, yeast, or insect cell expression system that lacks the mammalian context sometimes essential for adequate production and proper protein folding. The manipulation during purification and storage of the recombinants may further limit proper protein folding. Lastly, the expression, purification, and spotting of tens of thousands recombinant proteins is a laborious task that elevates its cost, reduces its reproducibility, and limits its utilization.
To address the deficiencies of spotting purified proteins, we have developed a high-throughput just-in-time protein expression and in situ purification system, namely nucleic acid programmable protein array (NAPPA; Ramachandran et al., 2004). Several platforms have been developed to use cell-free systems to generate protein microarrays, such as protein in situ array (PISA) (He and Taussig, 2001) and DNA array to protein array (DAPA; He et al., 2008). However, to our knowledge, NAPPA is the only nonprotein printing platform that has advanced beyond proof-of-concept to produce arrays of >1000 unique proteins for biomedical studies at the proteome level. NAPPA will not only achieve protein expression and isolation at the proteome level but also deliver the end products in a microarray format for multiplexed high-throughput studies.
In this method, full-length cDNAs corresponding to the proteins of interest are printed on the microarray substrate. The cDNAs are configured to append a common epitope tag to all of the proteins on the N- or C-termini so that they can be captured by a high-affinity capture reagent that recognizes the epitope tag and is immobilized along with the cDNA. To produce the protein at the time of assay, in vitro transcription and translation (IVTT)-coupled rabbit reticulocyte lysate is used. This approach offers the following advantages over traditional method:
Replaces preparing proteins with the more reliable and less expensive process of preparing DNA.
Avoids the need to express, purify, and store individual proteins.
Avoids concerns about protein shelf life because the proteins are made fresh at the time of assay.
Displays better than 95% of sequence-verified full-length genes, including membrane proteins.
Protein display levels are more consistent from protein to proteins; 93% of display levels are within twofold of the mean.
Assures protein integrity by using mammalian expression machinery to synthesize and fold proteins.
Easy to create custom arrays by simply rearranging plasmids.
Using this approach, ~20,000 different proteins have been expressed including human kinases, transcription factors, G-protein coupled receptors, and various druggable targets. Early studies demonstrated functional proteins by documenting 85% of the known protein interactions in the human DNA prereplication complex. More recently, we have demonstrated that kinases expressed on the array are active enzymes by measuring autophosphorylation activity that can be inhibited selectively by known kinase inhibitors (Festa and LaBaer, unpublished data). Since development, this technology has been effectively used for disease biomarker discovery and functional protein assays and successfully adopted by several other labs (Anderson et al., 2008, 2010; Ceroni et al., 2010; Montor et al., 2009; Ramirez et al., 2010; Wright et al., 2010).
In this chapter, we will detail the methods from plasmid DNA preparation to array production to protein display assessment. Rabbit reticulocyte lysates will be used as an example for the in vitro expression system. However, we have demonstrated that this protocol can be easily adapted to other expression systems, such as insect cell or human cell lysates. Our standard expression vector pANT7-cGST is freely available to the research community (Ramachandran et al., 2004). Genes of interest can be cloned into pANT7-cGST through recombinational cloning (Park and Labaer, 2006). Other versions of expression vectors are also available through DNASU (www.dnasu.asu.edu). Furthermore, ~ 10,000 human gene clones including ~ 500 human kinase genes together with tens of thousands genes from different pathogens that are already in NAPPA compatible vector are readily available from DNASU (Hu et al., 2007; Labaer et al., 2004; Murthy et al., 2007; Park et al., 2005; Rolfs et al., 2008).
2. Overview of NAPPA Chemistry
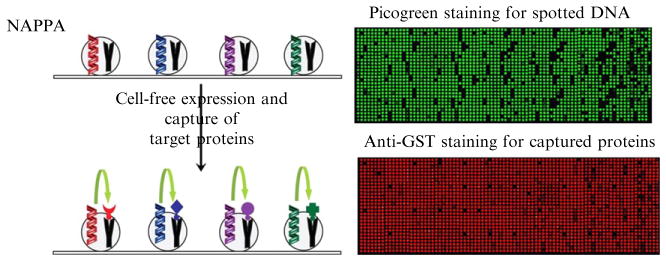
The innovation of NAPPA lies in the functional expression of proteins from immobilized plasmid DNA on a solid surface and efficient capture in situ by cospotted capture agents. Microscopic slides were treated with 3-aminopropyltriethoxysilane (APS) to attach a functional primary amine group to the surface. Plasmid DNAs and capture antibodies are immobilized on the slide surface with a homobifunctional primary amine cross-linker BS3 without compromise of integrity in terms of expression of cDNAs and binding of antibodies. The addition of Bovine Serum Albumin (BSA) in the printing mixture provides unexplained promoting effects on both effective immobilization and efficient expression (Ramachandran et al., 2008). Expressed proteins are tagged at the C-termini and captured by corresponding polyclonal antibody against the tag. The employment of the C-terminal tag ensures the capture of only full-length proteins (Fig. 9.1).
Figure 9.1.
On-array protein expression and capture. Left: Plasmid DNA is mixed with BSA, BS3 cross-linker, and the anti-GST capture antibody and arrayed on the aminosilane-coated glass slide. After blocking, cell-free expression mix is applied to the slide, and during a temperature-programmed incubation, the proteins are produced and bind to the capture antibody. Captured proteins can be detected by detecting the GST tag using a monoclonal anti-GST antibody, an HRP-labeled anti-mouse antibody, and Cy3-tyramide (TSA) HRP substrate. Right: Sample NAPPA images for PicoGreen staining of spotted DNA and anti-GST staining for captured proteins.
NAPPA chemistry is robust, reproducible, and versatile. Our standard NAPPA system uses an expression vector that encodes the protein of interest with a C-terminal glutathione-S-transferase (GST) fusion protein under the transcriptional control of the T7 promoter. Cospotted polyclonal anti-GST antibody is used to capture the expressed target proteins. Captured proteins can be confirmed by incubating the slides with an antibody that recognizes a different epitope on the tag than the antibody used for capture. Alternative tags such as HA and FLAG, alternative promoters such as SP6, and alternative expression systems such as wheat germ and insect cell lysates have also been demonstrated compatibility with our NAPPA system to accommodate different research needs and clone availability. Moreover, the schemes outlined here can be also altered by the user to accommodate different immobilization chemistries for the plasmid DNA and/or target proteins. Printed slides are stable when stored in a dry environment at room temperature (RT).
3. Array Production
Required materials
Automated liquid handling system
Filtered compressed air
Tabletop centrifuge capable of processing multiwell plates
3.1. Preparation of slides
Glass slides
3-APS (Pierce 80370)
Stainless steel 30-slide Wheaton rack (VWR 25461–014), handle removed
Glass staining dish (Wheaton 900201)
Lock & Lock 1.5 cup boxes (Heritage Mint Ltd., ZHPL810)
Prepare 300 ml of aminosilane coating solution (2% 3-APS in acetone) in a Wheaton glass staining dish.
Put glass slides in a 30-slide Wheaton metal rack.
Immerse slides in the aminosilane coating solution for 15 min at RT on a shaker.
Rinse with acetone in a Wheaton glass staining dish by dipping the slides in rack for five times.
Rinse with deionized water.
-
Dry with filtered compressed air.
It is important to dry slides quickly and evenly across slide surfaces to prevent water stain that might cause high background during assay.
Store at RT in metal rack in Lock & Lock boxes with desiccant packs until use.
3.2. Preparation of nucleobond anion exchange resin plate for DNA preparation
Nucleobond resin (Machery-Nagel custom order)
Solution N2 (Equilibration buffer): 100 mM Tris, 15% EtOH, 900 mM KCl, 0.15% Triton X-100, add phosphoric acid until pH is 6.3
96-well deep-well block (Marsh AB-0661)
800 μl glass fiber MBPP 25 μm filter plate (Whatman 13503–040)
-
In a 1-l glass bottle, add the nucleobond resin to 300 ml, and then add solution N2 to 900 ml.
This step should be done in a hood.
Mix the resin with N2 solution until it is homogeneous.
Pour the resin solution out into a Wheaton glass staining dish.
Place an 800 μl Whatman MBPP filter plate on top of a 96 deep well block (Marsh).
-
Using wide-bore P1000 tips and a multichannel pipette, transfer 450 μl resin slurry to into each well of the Whatman MBPP filter plate.
Gently mix the resin solution with the multichannel pipette laterally several times before each transfer to avoid sedimentation and ensure consistent transfer.
-
Spin the filter plate with the deep well block (Marsh) at the bottom for 131×g for 5 min.
The resin is ready to use at step 14 in Section 3.3.
3.3. Preparation of plasmid DNA
Terrific Broth media
Luria–Bertani media
Ampicillin stock: 100 mg/ml in H2O. Store at −20 °C
Agar
Omni plate (NUNC 242811)
96-pin device (Boekel 140500)
96-well deep-well block (Marsh AB-0661)
Gas permeable plate seal (VWR 47749-924)
Multitron shaker (Appropriate Technical Resources, Inc.)
Thermomixer (Eppendorf)
Matrix WellMate (ThermoFisher)
Aluminum plate seal (CIC FS-100)
Solution 1 (Resuspension buffer): 50 mM Tris (pH 8.0), 10 mM EDTA, and 0.1 mg/ml RNAse. Store at 4 °C
Solution 2 (Lysis buffer): 0.2 N NaOH with 1% SDS
Solution 3 (Neutralization buffer): 3 M Potassium Acetate (KOAc), add glacial acetic acid until pH is 5.1. Store at 4 °C
Solution N2 (Equilibration buffer): 100 mM Tris, 15% EtOH, 900 mM KCl, 0.15% Triton X-100, add phosphoric acid until pH is 6.3
Solution N3 (Wash buffer): 100 mM Tris, 15% EtOH, 1.15 M KCl, add phosphoric acid until pH is 6.3
Solution N5 (Elution buffer): 100 mM Tris, 15% EtOH, 1 M KCl, add phosphoric acid until pH is 8.5
800 μl 96-well block (Abgene AB-0859)
350 μl 96-well plate (Greiner 651201)
Note: Liquid transferring between plates can be done either by a multi-channel pipette or a robotic liquid handling system.
Take out the 96-well glycerol stock plate from −80 °C freezer.
Thaw at RT and pulse spin at 233 × g for 3 min to spin down media from the wall and the aluminum foil cover to the bottom of wells.
Open carefully to avoid cross contamination.
Spot 3 μl from the glycerol stock onto a prewarmed LB/Agar omni plate. Overnight incubation at 37 °C.
Sterilize a 96-pin device in 80% ethanol, then flame. Let it cool down before use.
-
Inoculate the culture block from the omni-agar plate to a 96-well deep plate (Marsh) with 1.5 ml TB supplied with 100 μg/ml ampicillin using the 96-pin device.
Sterilize the pin device between each inoculation.
Approximately six inoculates can be done from on a single agar plate.
-
Cover the 96-deep-well plate (Marsh) with a gas permeable seal and put it on an ATR multitron shaker for 24 h at 37 °C, 800 rpm.
Dilute the culture by 1:10 to check OD600 periodically.
Ideal OD600 should be 0.16–0.25 at the end of incubation.
Spin the 96-well deep plate (Marsh) for 20 min at 3724 × g on a tabletop centrifuge.
Decant supernatant and blot the plate upside down briefly on paper towels.
Add 200 μl of solution 1 to each well using wellmate and resuspend by putting on a thermomixer for 1 min at 2000 rpm.
-
Add 200 μl of solution 2 to each well using wellmate, seal with an aluminum plate seal, and mix by inverting the plate for five times.
Solution 2 has to be made fresh.
-
Five minutes later, add 200 μl of solution 3, seal with an aluminum plate seal, and mix by inverting the plate for five times.
Wipe the top of plate with a piece of clean Kimwipes before applying the aluminum foil plate seal may prevent leak and cross contamination among wells.
Spin the 96-well deep plate (Marsh) for 20 min at 3724 × g.
-
Transfer 600 μl supernatant lysates to the nucleobond resin plate using a robotic liquid handling system.
150 μl each for four times.
If encountering the block problem for lysate transfer using a robotic liquid handling system, use toothpick to remove the pellet and spin again.
Spin the plate for 5 min at 21×g using a tabletop centrifuge with slow start.
-
Add 400 μl washing solution N3 to each well using a wellmate, place the plate on the top of a vacuum manifold, and drain by vacuum.
Repeat the washing step four times.
Spin for 5 min at 131×g at RT.
Place the resin plate onto a clean 800 μl collection plate (Abgene). Add 300 μl elution buffer N5 to each well.
-
Spin the stacked plates for 5 min at 21×g with a slow start followed by 1 min at 233×g with a fast start.
Eluted DNA can be quantified using Nanodrop or Hoechst Dye. >30 ng/μl is desirable before precipitation.
Eluted DNA can be stored −20 °C before precipitation.
To precipitate DNA, add 40 μl of 3 M NaOAc, and then add 240 μl of isopropanol to each well.
Seal the plate with aluminum plate seal and mix by inverting the plate for three times.
Put the plate at −80 °C for 15 min.
Warm the plate up at RT for 10 min before spinning at 3724×g for 30 min at 20 °C.
Decant the supernatant.
Add 300 μl of 80% ethanol to each well.
Seal with aluminum plate seal and shake at 1200 rpm for 30 min on a Thermomixer.
-
Decant the supernatant.
Spin for 5 min at 233×g 20 °C to bring all pellets down to the bottom of well.
-
Air-dry the plate.
Precipitated DNA can now be stored at −20 °C before use.
3.4. Production of arrays
Contact arrayer with humidity control
Mater mix: 3.67 mg/ml BSA (Sigma), 1.25 mg/ml BS3 (bis[sulfosuccinimidyl] suberate, Pierce 21580), 50 μg/ml polyclonal anti-GST antibody (Amersham Biosciences 27457701)
384-well plate for arraying (Genetix x7020)
Purified GST protein (Sigma G5663)
Whole mouse IgG (Pierce 31202)
-
Using P100 multichannel pipette to transfer 25 μl of master mix to each well of 96-well DNA plate prepared as in Section 3.2.
Pre-equilibrate the DNA plate to RT if taking out from −20 °C.
Various control samples such as master mix, purified GST protein, and mouse IgG can be included quality assurance and data analysis purposes.
Seal with an aluminum plate seal and shake for 45 min at 1000 rpm on a thermomixer.
Spin at 524×g for 1 min at 20 °C.
Transfer 23 μl resuspending DNA in master mix to a 384-well plate (Genetix x7020) using a robotic liquid handling system.
Spin the 384-well plate for 1 min at 524×g at 20 °C to remove bubbles.
Array, using humidity control at 60%.
Put barcode labels at the end of the nonspotting side of printed slides.
Store printed slides in a Lock & Lock box with desiccant packs at RT until use.
4. Detection of Protein Expression
4.1. Detection of spotted DNA
SuperBlock blocking solution in TBS (Pierce 37535)
Lifterslips (VWR 100499–632)
PicoGreen (Molecular Probes P11495) stock solution: to the 100 μl/vial that comes, add 200 μl TE buffer. Before use do a 1:600 dilution in SuperBlock
4-well dish (NUNC, 73521–424)
Block a slide with SuperBlock for 1 h at RT in a 4-well dish. Use ~4 ml per slide. Rock gently.
-
Before applying the PicoGreen reagent, tap the slides gently and wipe the nonspotting side on clean paper towels to remove excess water from the slides, but do not let dry.
If te slides are too wet, it will dilute the PicoGreen reagent.
For a single slide: apply 500 μl PicoGreen mix, and apply a lifterslip slowly to avoid trapping bubbles.
Incubate for 10 min at RT.
Rinse with deionized water for three times.
Dry with filtered compressed air- or spin-dry at 131×g for 3 min at 20°C.
-
Scan in microarray scanner at proper settings for Cy3.
Should avoid saturated pixels for quantitative analysis.
4.2. Expression of proteins
EchoTherm™ IN35 Bench Top Chilling/Heating Incubators with Fully Programmable Controls (Torrey Pines Scientific)
HybriWell gaskets (Grace Bio-Labs, 44904)
TNT® Quick Coupled Transcription/Translation Systems (Rabbit Reticulocyte Lysate) (Promega TM045)
SuperBlock blocking solution in TBS (Pierce 37535)
Milk blocking solution: 5% Milk in PBS with 0.2% Tween 20
4-well dish (NUNC, 73521–424)
Microarray scanner
Block slides for ~1 h at RT with SuperBlock in a 4-well dish. Use ~4 ml per slide. Rock gently.
Quickly rinse with deionized water.
Dry with filtered compressed air or spin at 131×g for 3 min at 20 °C.
-
While blocking, take out Promega T7 Coupled Rabbit Reticulocyte Lysates from −80 °C and thaw on ice.
Good expression of the NAPPA arrays depends on high quality rabbit reticulocyte lysates, which may vary from lot to lot. It is recommended to run one set of experiments with well-tested comparable lots.
-
Prepare IVTT mix. 160 μl is needed for 1 slide.
128 μl rabbit reticulocyte lysates
28.8 μl DEPC water
3.2 μl 1 mM Met
Apply a HybriWell gasket to each slide. Use the wooden stick to rub the areas where the adhesive is to make sure it is well stuck all around.
Apply IVTT mix through one of the sample application ports on the Hybriwell gasket. Pipette the mix in slowly. Gently massage the HybriWell with both hands to get the IVTT mix to spread out and cover all of the area of the array. Get rid of any bubbles in the center by tapping with the wooden stick. Apply the small round port seals to both ports.
Incubate for 1.5 h at 30 °C for protein expression, followed by 30 min at 15 °C for the query protein to bind to the immobilized protein.
Remove the HybriWell; rinse with milk blocking buffer five times in a pipette box.
Slides are ready for downstreaming assays and should be rinsed with an assay compatible buffer instead of milk blocking buffer.
Block with milk in PBST0.2 at RT for 1 h.
4.3. Detection of captured proteins
Primary AB solution: mouse anti-GST (Cell Signaling 2624) 1:200 in milk blocking solution.
Secondary AB solution: HRP-conjugated anti-mouse (JacksonImmunoResearch 515-035-062, 1 mg/ml) 1:400 in milk blocking solution.
Tyramide signal amplification (TSA) stock solution: use TSA reagent (PerkinElmer SAT704B001EA). Prepare per kit directions. Keep this solution at 4 °C.
PBST0.2: PBS with 0.2% Tween 20
Milk blocking solution: 5% milk in PBS with 0.2% Tween 20.
Lifterslips (VWR 100499–632).
Incubate with 3 ml primary antibody mouse anti-GST solution in a 4-well dish for 1 h at RT with gentle rocking.
Rinse with milk blocking solution for three times.
Wash with milk blocking solution for 3 × 5 min.
Incubate with 3 ml secondary antibody HRP-labeled goat anti-mouse IgG solution in a 4-well dish for 1 h at RT with gentle rocking.
Rinse with PBST0.2 for three times.
-
Wash with PBST0.2 for 3 × 5 min.
While washing, prepare the TSA reagent and keep in dark.
Rinse with deionized water for three times.
-
Before applying the TSA reagent, tap the slides gently on clean paper towels to excess water from the slides, but do not let dry.
. If the slides are too wet, it will dilute the TSA reagent.
-
Apply the TSA reagent on the slide and gently cover with a lifter slip.
Avoid trapping bubbles under the lifter slip.
Incubate for 10 min at RT.
-
Rinse in deionized water; dry with filtered compressed air- or spin-dry at 131×g for 3 min at 20 °C.
Do not let slides air-dry slowly as the water marks may form on array.
-
Scan in a microarray scanner at proper settings for Cy3.
Avoid saturated pixels for quantitative analysis.
Acknowledgments
The research relevant to this chapter was supported by grants from National Cancer Institute (1-R21/R33-CA099191-01), the Early Detection Research Network (5U01CA117374-02), the Juvenile Diabetes Research Foundation (JDRF 17-2007-1045), and a contract from National Institute of Allergy and Infectious Diseases (HHS2662004000053). The authors would like to thank Eliseo Mendoza Garcia for providing sample NAPPA images.
References
- Anderson KS, Ramachandran N, Wong J, Raphael JV, Hainsworth E, Demirkan G, Cramer D, Aronzon D, Hodi FS, Harris L, Logvinenko T, LaBaer J. Application of protein microarrays for multiplexed detection of antibodies to tumor antigens in breast cancer. J Proteome Res. 2008;7:1490–1499. doi: 10.1021/pr700804c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KS, Wong J, Vitonis A, Crum CP, Sluss PM, Labaer J, Cramer D. p53 autoantibodies as potential detection and prognostic biomarkers in serous ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:859–868. doi: 10.1158/1055-9965.EPI-09-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceroni A, Sibani S, Baiker A, Pothineni VR, Bailer SM, LaBaer J, Haas J, Campbell CJ. Systematic analysis of the IgG antibody immune response against varicella zoster virus (VZV) using a self-assembled protein microarray. Mol Biosyst. 2010;6:1604–1610. doi: 10.1039/c003798b. [DOI] [PubMed] [Google Scholar]
- Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2001;2:RESEARCH0004. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Taussig MJ. Single step generation of protein arrays from DNA by cell-free expression and in situ immobilisation (PISA method) Nucleic Acids Res. 2001;29:e73. doi: 10.1093/nar/29.15.e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Stoevesandt O, Palmer EA, Khan F, Ericsson O, Taussig MJ. Printing protein arrays from DNA arrays. Nat Methods. 2008;5:175–177. doi: 10.1038/nmeth.1178. [DOI] [PubMed] [Google Scholar]
- Hu Y, Rolfs A, Bhullar B, Murthy TV, Zhu C, Berger MF, Camargo AA, Kelley F, McCarron S, Jepson D, Richardson A, Raphael J, et al. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 2007;17:536–543. doi: 10.1101/gr.6037607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBaer J, Ramachandran N. Protein microarrays as tools for functional proteomics. Curr Opin Chem Biol. 2005;9:14–19. doi: 10.1016/j.cbpa.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Labaer J, Qiu Q, Anumanthan A, Mar W, Zuo D, Murthy TV, Taycher H, Halleck A, Hainsworth E, Lory S, Brizuela L. The Pseudomonas aeruginosa PA01 gene collection. Genome Res. 2004;14:2190–2200. doi: 10.1101/gr.2482804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- Montor WR, Huang J, Hu Y, Hainsworth E, Lynch S, Kronish JW, Ordonez CL, Logvinenko T, Lory S, LaBaer J. Genome-wide study of Pseudomonas aeruginosa outer membrane protein immunogenicity using self-assembling protein microarrays. Infect Immun. 2009;77:4877–4886. doi: 10.1128/IAI.00698-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy T, Rolfs A, Hu Y, Shi Z, Raphael J, Moreira D, Kelley F, McCarron S, Jepson D, Taycher E, Zuo D, Mohr SE, et al. A full-genomic sequence-verified protein-coding gene collection for Francisella tularensis. PLoS One. 2007;2:e577. doi: 10.1371/journal.pone.0000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Labaer J. Recombinational cloning. Curr Protoc Mol Biol. 2006;74:3.20.1–3.20.22. doi: 10.1002/0471142727.mb0320s74. [DOI] [PubMed] [Google Scholar]
- Park J, Hu Y, Murthy TV, Vannberg F, Shen B, Rolfs A, Hutti JE, Cantley LC, Labaer J, Harlow E, Brizuela L. Building a human kinase gene repository: Bioinformatics, molecular cloning, and functional validation. Proc Natl Acad Sci USA. 2005;102:8114–8119. doi: 10.1073/pnas.0503141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran N, Hainsworth E, Bhullar B, Eisenstein S, Rosen B, Lau AY, Walter JC, LaBaer J. Self-assembling protein microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- Ramachandran N, Raphael JV, Hainsworth E, Demirkan G, Fuentes MG, Rolfs A, Hu Y, LaBaer J. Next-generation high-density self-assembling functional protein arrays. Nat Methods. 2008;5:535–538. doi: 10.1038/nmeth.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez AB, Loch CM, Zhang Y, Liu Y, Wang X, Wayner EA, Sargent JE, Sibani S, Hainsworth E, Mendoza EA, Eugene R, Labaer J, et al. Use of a single-chain antibody library for ovarian cancer biomarker discovery. Mol Cell Proteomics. 2010;9:1449–1460. doi: 10.1074/mcp.M900496-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfs A, Hu Y, Ebert L, Hoffmann D, Zuo D, Ramachandran N, Raphael J, Kelley F, McCarron S, Jepson DA, Shen B, Baqui MM, et al. A biomedically enriched collection of 7000 human ORF clones. PLoS One. 2008;3:e1528. doi: 10.1371/journal.pone.0001528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Wright C, Sibani S, Trudgian D, Fischer R, Kessler B, Labaer J, Bowness P. Detection of multiple autoantibodies in patients with ankylosing spondylitis using nucleic acid programmable protein arrays. Mol Cell Proteomics. 2010 doi: 10.1074/mcp. M900384-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Klemic JF, Chang S, Bertone P, Casamayor A, Klemic KG, Smith D, Gerstein M, Reed MA, Snyder M. Analysis of yeast protein kinases using protein chips. Nat Genet. 2000;26:283–289. doi: 10.1038/81576. [DOI] [PubMed] [Google Scholar]