Abstract
Insects stimulate specific behaviors by the correct recognition of the chemicals in the external environment. Rhodojaponin III is a botanical grayanoid diterpenid oviposition deterrent isolated from Rhododendron molle. In this study we aimed to determine whether the CSPs involved in the recognition of Rhodojaponin III. A full-length cDNA encoding chemosensory protein was isolated from the antennae of Spodoptera litura Fabricius (CSPSlit, GenBank Accession No. DQ007458). The full-length cDNA of NlFoxA is 1789 bp and has an open reading frame (ORF) of 473 bp, encoding a protein of 126 amino acids, Northern blot analysis revealed that CSPSlit mRNA was mainly expressed in the antennae, legs, wings and female abdomens. A three-dimensional model of CSPSlit was constructed using homology modeling method, and its reliability was evaluated. The active site of CSPSlit was calculated using CDOCKER program indicated that the Tyr24, Ile45, Leu49, Thr64, Leu68, Trp79 and Leu82 were responsible ligand-binding active site on identifying Rhodojaponin III in the CSPSlit. The recombinant CSPSlit protein was expressed in Escherichia coli and purified using single-step Ni-NTA affinity chromatography. Fluorescence emission spectra revealed that the CSPSlit protein had significant affinity to rhodojaponin III. These results mean that CSPSlit is critical for insects identify the Rhodojaponin III.
Introduction
Insects can recognize a variety of plant compounds that stimulate specific behaviors, such as feeding and egg laying (oviposition) by chemoreceptive organs [1]–[4]. It is well known that some insects lay eggs on their host plants, and the oviposition behavior is induced by the recognition of the plant components with sensilla on these chemoreceptive organs [5]–[6], But the detailed mechanism of this identification is not clear. Chemosensory proteins (CSPs) are a class of small (10–15 kDa) soluble proteins containing 4 conserved cysteines which abundantly exist in the chemoreceptive organs and transmit chemical signals to nervous system [7]–[8]. The CSP was first in Drosophila melanogaster and confirmed that CSPs are capable of binding a range of aliphatic compounds, esters and other long chain compounds that are typical components of pheromonal blends [7], [9].
The first member of the CSP family was discovered more than a decade ago in Drosophila melanogaster and was called olfactory specific protein D (OS-D) due to its preferential expression in the antennae [9]. Later studies identified other members of this family in sensory appendages such as antennae, labial palps and legs in a variety of insects [10]–[11]. Several members of this class of protein have been described in insects of different orders, including Lepidoptera [11]–[19], Orthoptera [10], [20]–[22], Hymenoptera [7], [23]–[26], Diptera [27], Blattoidea [28]–[29], Phasmatodea [30]–[32], Hemiptera [33], etc. The function of CSPs as carrier proteins was strengthened by studies on the higher order structure of a CSP from Bombyx mori, which revealed a globular configuration of six alpha helices surrounding a hydrophobic binding pocket [34]. Recent studies confirmed that CSPs are capable of binding a range of aliphatic compounds, esters and other long chain compounds that are typical components of pheromonal blends [7], [14]–[15], [35].
The Spodoptera litura, is one major pest of agricultural crops in many Asia areas. It is a polyphagous pest and known about 150 host species [36]–[37]. The extensively use of synthesis pesticides has caused it to develop resistance against various chemicals. The residual pesticides have not only polluted the environment, but are also a threat to human life. And it is serious during the seedling stage, especially in upland rice and other crucifer and it is also regarded as a very good target for the applications of rhodojaponin III [38]. Rhodojaponin III, a grayanoid diterpene compound isolated from the flower of Rhododendron molle, has been reported to have high levels of oviposition deterrent, antifeedant, contact and/or stomach toxicity against more than 40 species of agricultural pests in laboratory bioassays and field trials [39]–[40]. However, the mechanism of rhodojaponin III as an oviposition deterrent is yet poorly understood.
The computer-aided structure-based study of molecular recognition is an important component of structure-based potential ligands screening [41]–[42]. The original DOCK algorithm addressed rigid body docking using a geometric matching algorithm to superimpose the ligand onto a negative image of the binding pocket [43]–[44]. A representative docking method is used to study these factors, namely, CDOCKER, a molecular dynamics (MD) simulated-annealing-based algorithm, which places a unique constraint on the development process [42].
The present study was designed to characterize and identify CSPSlit expression of the in Lepidoptera, S. litura, and the role of a grid representation of CSPSlit -rhodojaponin III interactions. We also intended to provide evidences to confirm the fundamental biological phenomena of CSPSlit and agricultural problems related to the S. litura.
Materials and Methods
2.1 Insects and Preparation of Tissues
The S. litura were reared on an artificial diet [45], at 25±1°C in a 14:10 light: dark photoperiod and 60–70% relative humidity. Adults were harvested within 3 days after emergence. Different tissues (antennae, de-antennated heads, forelegs, mesopedes, metapedes, thoraces, wings and abdomens) were dissected from newly emerged adults and immediately frozen in liquid nitrogen and stored at −80°C until used.
2.2 Cloning and Sequence Analysis of CSPSlit
Cloning and sequence analysis of NlFoxA Total RNA was isolated from four 2 nd day brachypterous female adults of N. lugens using the Trizol kit (Invitrogen, USA). Its integrity was detected using Agilent 2100 Bioanalyzer (USA). First-strand cDNA was synthesized with a first strand synthesis kit using reverse transcriptase X L (AMV) and an oligo dT 18 primer (TaKaRa, Japan). Two pairs of degenerate primers were designed based on the conserved amino acid sequences of chemosensory proteins from different Lepidoptera insects. The first-strand cDNA (1 µl) was used as a template for PCR using a general protocol. The reaction mixture contained 0.1 mM dNTP, 0.5 mM of each degenerate primer and 1.0 U of HiFi-Taq DNA polymerase (TransGen Biotech, Guangzhou, China) in a total volume of 25 µl. The first PCR was carried out with the following conditions: initial preheating for 5 min at 94°C, 35 cycles at 94°C for 30 s, 48°C for 30 s and 72°C for 1 min, and with a final extension at 72°C for 10 min using the primer pair ACC GAC MRS TAY GAC AGY GAG AC and TCY TTG AGT TCC TTC TCR TAC TT. The second PCR was performed using another degenerate pair, CAA CCG YCG CCT SWT GGT GCY TAT and TAC TTG GCC KTC AGC TSK TTC CA, with the before mentioned program. The amplified fragment was recovered in a 1% agarose gel and purified using the Gel Extraction Kit (Omega, USA). Purified DNA was ligated into the pMD18-T vector (TaKaRa, Japan), and recombinant clones were digested with EcoRI and PstI to screen the presence of inserted DNA. Positive clones were sequenced by Invitrogen Company (Shanghai, China). To obtain the full-length NlFoxA cDNA, we used a RACE Kit (CLONTECH, Japan). Specific primers for the 5′- and 3′- Rapid Amplification of cDNA Ends (RACE) were designed based on homologous PCR fragments. The specific primers CGA CTT CAA TTC CTT GGC ATC AGG TG and GTT TAG GAT CTC GTC CAG GTC GAT G were used for 5′ –RACE, while GTA CCC CAG CTC AGA AGG ATG GTA C and GCC AGA AAT GTG GAA CCA GCT CTG C were used for 3′ –RACE. Using the 5′- and 3′-RACE cDNAs as templates, PCR was performed using the 5NlFoxA1 primer and Universal Primer Mix (UPM, Clontech) by denaturing at 95°C for 30 s, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 2 min, and a final extension at 72°C for 10 min. Nested PCR was carried out with the first-round PCR product as a template and the Nested Universal Primer A (NUP, Clontech) and NlFoxA2 primer. The reaction conditions consisted of the followings: 6 min of initial preheating at 94°C, 30 cycles of 94°C for 30 s, 68°C for 30 s and 72°C for 40 s, and a final elongation at 72°C for 7 min. The RACE products were purified and sequenced as described above. Sequence homologous alignment and similarity searches were carried out by Blast biological software http://www.ncbi.nlm.nih.gov/blast. The signal peptide was analyzed by SignalP procedure.
2.3 Northern-blot Analysis
Total RNA was isolated as described above from the antennae, de-antennated heads, forelegs, mesopedes, metapedes, thoraces, wings and abdomens. Northern blot was carried out according to the method described by Sambrook [46]. Total RNA (20 µg/µl) was separated on 1.5% (W/V) denaturing formaldehyde agarose gels. The RNA was blotted onto NC membranes. The 405 bp fragment of CSPSlit was labeled with α-[32P]dCTP and used as a probe for hybridization at 68°C for 16 h. Final wash conditions for the RNA blots were 15 min at 68°C in 1×SSC, 0.2% (W/V) SDS, 15 min at 68°C in 0.5×SSC and 0.1% SDS. Washed membrane was dried at 80°C and exposed to X-ray film.
2.4 Expression and Refolding of CSPSlit
PCR was performed using the specific primers, 5′- CTG CCA TGG CAC ACC CAC ATG AG TCC-3′ and 5′- TTG AGT CTC GAG GTG TTT AAC GGA TTT G -3′ to obtain the open reading frame (ORF) of the CSPSlit. In order to facilitate sub-cloning of the ORF into the expression vector, the restriction sites of NcoI and XhoI were separately introduced into the forward and reverse primers. PCR products, purified by agarose gel electrophoresis, were digested with Nco I and Xho I enzymes and ligated into the Nco I/Xho I-digested pET-28a(+) (Novagen) to construct the expression vector pET-CSPSlit. The resultant plasmid was transformed into the competent E. coli BL21 (DE3). A single positive bacterial colony, which was confirmed by restriction enzyme digestion and sequencing, was inoculated into LB medium containing ampicillin (100 µg/ml) and grown overnight. The seed culture was diluted with 1∶100 of LB medium and grown at 37°C to the optical density of A600 = 0.4, and the cells were induced by addition of 0.5 mmol/L isopropyl-D-thiogalactoside (IPTG). After 6 h incubation at 28°C, cultures were harvested by centrifugation and lysed by the lysis solution (10 mM imidazole, 300 mM NaCl and 50 mM NaH2PO4). After sonication, the supernatants were recovered by centrifugation and subjected to the Ni2+-NTA column to purify the recombinant protein.
The recombinant protein was refold with the approach of Tsumoto et al with some modified [47]. Briefly, 10 ml recombinant protein was solubilized in denaturant buffer (6 M guanidine hydrochloride, 200 mM NaCl, 100 mM Tris-HCl, and 1 mM EDTA at pH 8.3). Denaturant was slowly removed by a series of overnight equilibrations with buffers of successive decreasing guanidine hydrochloride concentration. The guanidine hydrochloride concentration was reduced as follows: 6 M, 3 M, 2 M, 1 M, 0.5 M, 0 M. The 2 M guanidine hydrochloride equilibration buffer was supplemented with 400 mM L-arginine and 375 mM oxidized glutathione (GSSG) as folding additives. After the final overnight dialysis with buffer containing no denaturant, the sample was removed from the dialysis buffer and freeze-drying at −90°C, then stored at −80°C. The concentration of refolded CSPSlit was established with the Bradford method that a calculation based on the absorbance at 280 nm [48].
2.5 Molecular Modeling
The first step for modeling was to search for a number of related sequences to find the template structure which had a high sequence similarity with the target CSPSlit sequence on RCSB Protein Data Bank (PDB)(www.rcsb.org). The homology modeling of CSPSlit was performed using build homology model [49] protocol of Discovery Studio 2.0 (DS 2.0) (Accelrys Inc.). The best model was confirmed using the evaluation of PDF total energy, verify score and Ramachardran plot. After performing 1000 steps of steepest descend (SD) and 2000 steps of conjugate gradient (CG) minimization, a molecular dynamic (MD) simulation was carried out to examine the quality of the model structure, by checking its stability via performing 1000 ps simulations. An explicit solvent model TIP3P water was used [50], and the homology solvent model was constructed with a 20 Å water truncated octahedron from the center of CSPSlit mass.
2.6 Molecular Docking
The binding-site module is a suite of programs in DS 2.0 for identifying and characterizing protein binding sites and functional residues from protein. In this study, this program was used to search the protein binding site by locating cavities in CSPSlit structure, which can be used to guide the protein-ligand docking experiment. Molecular docking can fit molecules together in a favorable configuration to form a complex system. The structural information from the theoretically modeled complex may help us to clarify the binding mechanism between CSPSlit and rhodojaponin III. In order to gain insights into binding mode of CSPslit with rhodojaponin III, the advanced docking program CDOCKER [42] was used to perform the automated molecular docking. CDOCKER uses a CHARMm-based molecular dynamics scheme to dock ligands into a receptor binding site. Random ligand conformations were generated using high-temperature molecular dynamics. The conformations were translated into the binding site. Candidate poses were created using random rigid-body rotations followed by simulated annealing. A final energy minimization was used to refine the ligand poses. Finally, the docked complex of CSPslit with rhodojaponin III was selected according to the criteria of interacting energy combined with geometrical matching quality whereas the interaction energy between rhodojaponin III and key residues of CSPSlit was calculated. MD simulation was carried out to examine the binding quality of the complex structure, by checking its stability and performing 1000 ps simulations, as mentioned above.
2.7 Fluorescence Assays
In order to investigate the interaction of rhodojaponin III and CSPSlit, fluorescence quenching method was used to evaluate their binding character according to Lartigue [35]. The fluorescence spectra were recorded on a F-4500 FL Fluorescence Spectrophotometer (HITACHI) at 23°C. The interaction of rhodojaponin III and CSPSlit was monitored by following the quenching of the intrinsic protein fluorescence (excitation at 295 nm and emission 300–550 nm, slit width of 5 nm were used for both excitation and emission). Spectra were recorded with 0.5 µM protein in 10 mM Tris buffer, pH 7.4 and under the same conditions in the presence of different concentrations of rhodojaponin III (0, 50, 100, 300, and 600 µM).
Results
3.1 cDNA Cloning and Sequence Analysis of CSPSlit
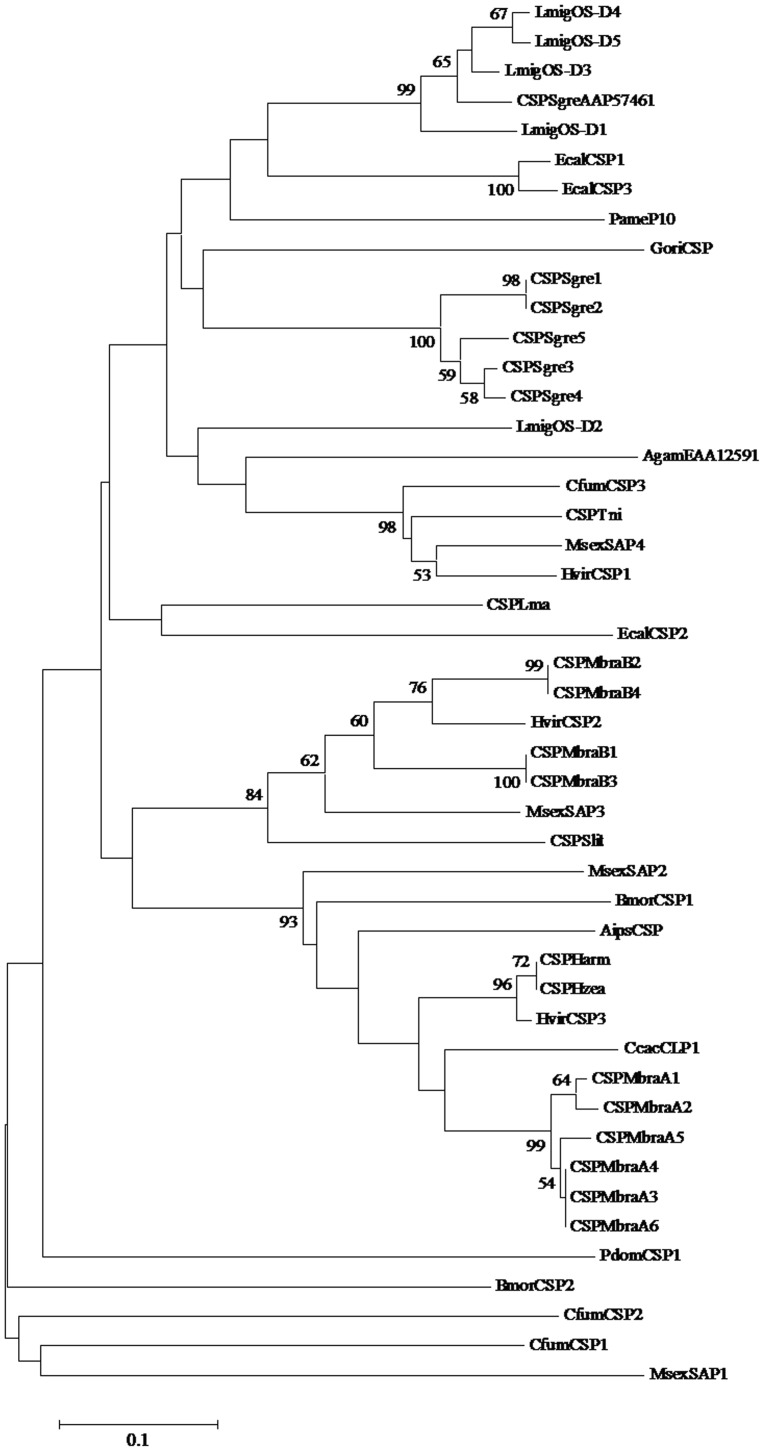
Two RACE fragments were amplified with four pairs of specific primers designed according to the nucleotide sequence of the fragment. By using rapid amplification of cDNA ends PCR (RACE-PCR), a full-length CSPSlit of 473 bp was obtained by overlapping the RACE fragments (GenBank Accession No: DQ007458). Sequence analysis showed that the full-length (ORF) of CSPSlit was 378 bp, encoding 126 amino acid residues, with a predicted MW of 14.67 kD. A 16-residue signal peptide in the CSPSlit was identified by SignalP, with a calculated molecular mass of a mature protein (110 amino acids) of 12.69 kD with an estimated pI of 6.66 by ExPASy [51] (Fig. 1). The phylogenetic tree was constructed based on the amino acid sequences CSP from S.litura and other insects (Fig. 2). The dendrogram showed that the CSPSlit had closer ancestry from the same order insects.
Figure 1. Nucleotide and deduced amino sequences of CSPSlit.
The predicted signal peptide is underlined. The asterisk marks the translation-termination codon.
Figure 2. Phylogenetic analysis of CSP amino acid sequences.
Bootstrap support values based on 1000 replicates are indicated.
3.2 Tissues-specificity Expession Analysis of CSPSlit
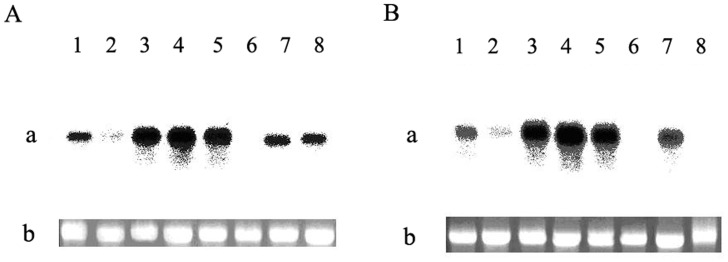
To determine whether CSPSlit is present in various tissues in the S. litura, we used northern blot to characterize the pattern of tissues-specificity expression of CSPSlit gene from different tissues (male & female antennae, de-antennated heads, forelegs, mesopedes, metapedes, thoraces, wings and abdomens). Total RNA of each sample was isolated and separated, an approximately of 500 bps α-[32P]dCTP labeled CSPSlit antisense RNA probe gave strong hybridization signals to the antennae, legs and wings, lower trace was detected from de-antennated heads and thoraces, and it was expessed in female abdomen but absent in male (Fig. 3).
Figure 3. Northern blot analysis of RNA coding for CSPSlit in different tissues.
(a): The recovery and integrity of each RNA were assessed from the 18S rRNA pattern; (b): 1, antennas; 2, de-antennated heads; 3, foreleg; 4, mesopedes; 5, metapedes; 6, thoraces; 7, wings; 8, abdomens. (A): female; (B): male.
3.3 3D Modelling of CSPSlit Protein
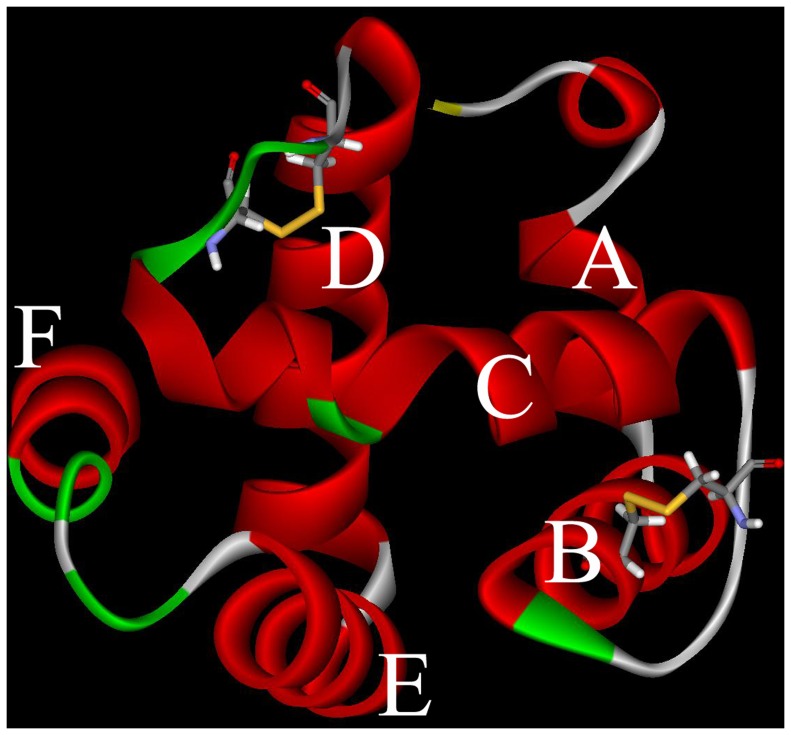
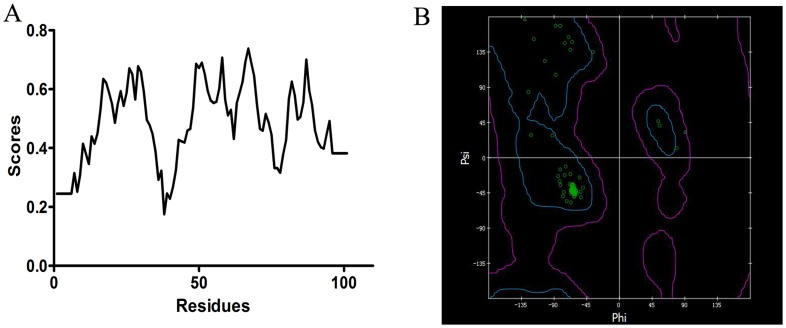
The sequence of CSPSlit was compared to all known proteins in PDB and the results showed that chemosensory protein A6 from Mamestra brassicae (CSPMbraA6) (PDB code 1N8V) had the sequence identity (52%) with CSPSlit, so CSPMbraA6 was chose as template to model the 3D structure of the CSPSlit. Following the homology modeling, the best model (Fig. 4) was chosen from 10 candidates, and its quality was further checked by Ramachardran plot and verify score (Fig. 5). Figure 6 shows the time series of potential energy and root-mean-square deviation (RMSD) of backbone for 700 ps MD simulation of CSPSlit structure. The potential energy of the model was stabilized at 200 ps production after 800 ps equilibration and the RMSD of backbone compared to the starting coordinate remained at 1.0 Å up and down fluctuations. These 2 properties converged at production, indicating that the model is stable and can be used for subsequent docking calculation.
Figure 4. 3D structure of the CSPSlit. Six α-helixes were marked A–F.
Two disulfide bridges are represented by yellow stick.
Figure 5. The evaluation of the CSPSlit 3D structure by verify score (A) and Ramachardran plot (B).
Square: proline residues; triangle: glycine residues; circle: all other residues. blue and purple: favorable regions; all else: unfavorable regions.
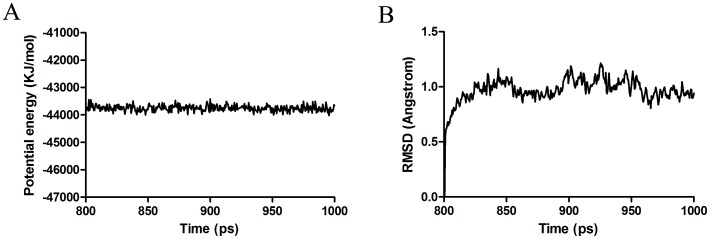
Figure 6. Potential energy (A) and root-mean-square deviation (A) with respect to simulation time for 1000 ps free MD simulation on the CSPSlit model.
3.4 Molecular Interaction Analysis between Rhodojaponin III and CSPSlit
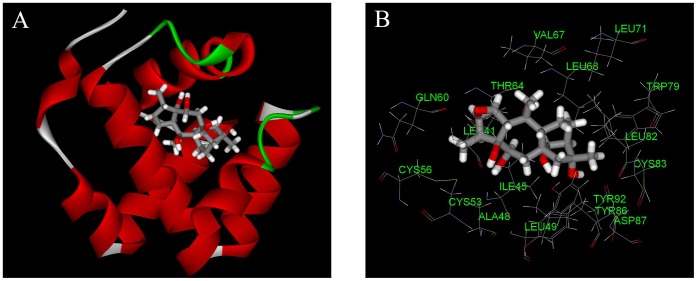
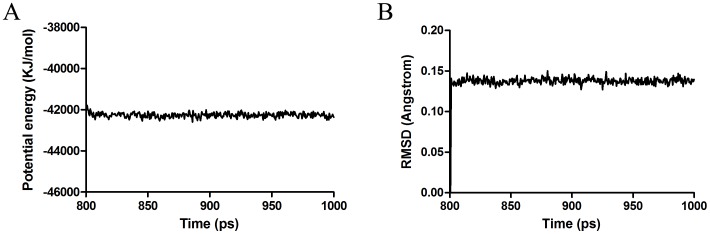
Considering the sequence conservation of CSPSlit with CSPMbraA6, the binding site was confirmed for its hydrophobicity. These binding poses were evaluated by score ligand pose program and ranked by Consensus score program. Finally, the optimal 3D binding conformations of complexes were selected and shown in Figure 7. The interaction energies between key residues and the ligand are listed in the Table 1. All of these residues are located in the cavity formed by six helices. Several residues including Ile45, Leu49, Thr64, Leu68 and Leu82 identified by current docking simulations have been shown to play an important role in the binding of CSPSlit and rhodojaponin III. Figure 8 shows the time series of potential energy and RMSD of backbone for 1000 ps MD simulation of CSPSlit structure. The potential energy of the complex was stabilized at 200 ps production after 800 ps equilibration, and the RMSD of backbone compared to the starting coordinate remained at 0.14 Å up and down fluctuations. These two properties converged at production, indicating that the complex is stable.
Figure 7. The complex (A) and detailed binding mode (B) of CSPSlit with rhodojaponin III.
The residues within 6 Å from ligand are shown.
Table 1. VdW Energy (EvdW) and Electrostatic Energy (Eele) between rhodojaponin III and CSPSlit.
| Residue | EvdW (kcal/mol) | Eele (kcal/mol) |
| Leu41 | −0.830793 | 0.017039 |
| Ile45 | −3.700930 | −0.346153 |
| Ala48 | −1.080190 | 0.739731 |
| Leu49 | −2.234000 | −0.098858 |
| Cys53 | −0.358571 | 0.154808 |
| Cys56 | −0.318546 | −0.240461 |
| Gln60 | −1.109890 | 0.331317 |
| Thr64 | −2.165880 | −0.800064 |
| Val67 | −2.477020 | 0.549604 |
| Leu68 | −2.773280 | −0.523853 |
| Leu71 | −0.885637 | 0.179657 |
| Trp79 | 0.174320 | 0.187195 |
| Leu82 | −2.684090 | 0.219521 |
| Cys83 | −1.524360 | 0.297607 |
| Tyr86 | −1.640490 | 0.447274 |
| Asp87 | −0.697000 | −0.607238 |
| Tyr92 | −0.572799 | 0.349116 |
Figure 8. Potential energy (A) and root-mean-square deviation (B) with respect to simulation time for 1000 ps molecular dynamics simulation on the CSPSlit- rhodojaponin III complex model.
3.5 Binding Assay of Rhodojaponin III and CSPSlit
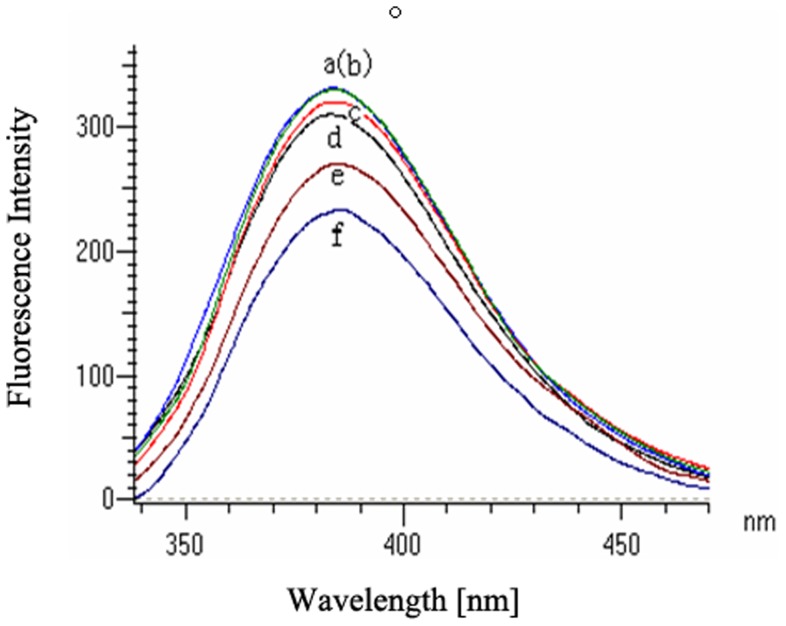
Purified CSPSlit protein solution contained 5.0 mg/ml was used to analyse the binding property of CSPSlit with rhodojaponin III. When excited at 295 nm, the fluorescence emission spectra showed maximally relative fluorescence intensity at 383.6 nm for CSPSlit. Following, with the increasing concentration of rhodojaponin III, CSPSlit peak underwent a blue shift, but no peak intensity increaseing was observed. When the final concentration of rhodojaponin III was 600 µM and 300 µM, the fluorescence intensity decreased to 29.43% and 18.47%, respectively (Fig. 9). These results showed that CSPSlit could be intensely bound with rhodojaponin III.
Figure 9. Fluorescence spectra obtained titrating a 0.5 µM solution of CSPSlit with increasing amounts of 50 µM rhodojaponin III in methanol to final concentrations of 0, 50, 100, 150, 300 and 600 µM (a–f).
The intensity of this peak was used to measure the amount of rhodojaponin III bound to the protein.
Discussion
The olfactory system of insects is essential for Lepidoptera as well as in other insect orders to initiate behavioral responses, such as searching for food sources, mating, oviposition and feeding [52]–[53]. Chemosensory proteins (CSPs), known as another class of soluble protein, share no sequence homology with either PBPs or general OBPs of many insects [11]–[12], [18]. The CSPs are smaller proteins which contain four cysteines instead of six with conserved interval spacing involved in two disulfide bonds [35], [54]. In the present study, a cDNA sequence encoding the CSP of S. litura was cloned. The CSPSlit has 4 typical conservative cysteines in the sequence (Fig.1). It is consistent with previous report. The CSPSlit was expressed in antennae, legs, wings and female abdomens (Fig.3), these results is similar with the research in other insect [9], [21]. In M. brassicae, the CSPs has a abundant expression in proboscis [15], [30], but in this study, there is no hybridization signal was detected in the de-antennated head (Fig. 3). These results indicated that the CSPs could not only recognize the odours but tastes. The result of Northern blot in this study revealed that the CSPSlit was found not only in antennas but also in legs, deantennated heads, thoraces, wings and abdomens. Especially in legs, its specifically high expression suggested that CSPSlit was perhaps functionally associated with contact chemoreception. CSPSlit with preferentially expression in female abdomen but absent in male suggested that CSPSlit might involve in female-specific chemical senses during mating or oviposition.
Homology modeling is based on the assumption that the proteins with similar sequences might have analogous 3D structures. Thus, selection of a suitable template is the first step. The structural characterization of CSPMbraA6 by NMR for the first time; The crystal structure of CSPMbraA6 in complex with one of these compounds, 12-bromo-dodecanol, reveals extensive conformational changes on binding, resulting in the formation of a large cavity filled by three ligand molecules [8]. However, in the absence of an experimentally determined crystal structure, it is generally recognized that homology modeling of proteins is currently the most accurate method for 3D structure prediction [55]–[58]. To study the binding mechanism of rhodojaponin III-CSPSlit complex in depth, the best way is to acquire the 3D crystal structure of it. In PDB, CSPMbraA6 was chosen as the template for constructing the 3D model of CSPSlit for it has 52% amino acid sequence identity with CSPSlit. To assess the reasonable of the 3D model of CSPSlit, Ramachandran plot, verify score and molecular dynamic were used [59]–[60] and showed positive results (Fig. 5, 6). The results of quality assessment suggest that the model of CSPSlit structure is of reasonable quality compared to the crystal structure of the CSPMbraA6 complex [8].
The 3D model of CSPSlit showed that CSPSlit had typical structure of CSPs,six α-helices has been described in 3D structure of CSPSlit, and four of six cysteine residues are conserved in sequence alignments and formed 2 disulphide bridges (C1–C3 and C4–C5), which enforce the organization of the helices [7]–[8], [34]–[35]. All helices are amphiphilic, with the hydrophobic sides formed mostly by leucines and isoleucines. In 3D model of CSPSlit, helices A and B as well as helices D and E form two V shaped structures as a ‘binding pocket’. Helix C is perpendicular to these two planes and positioned in between the four ends of the two V-shaped structures to close one end of this pocket. The final helix (F) is located packed against the external face of the D-E helices and does not take part in the core assembly. This cavity is delimited by the hydrophobic sides of these helices, and therefore well-suited to constitute a binding site for hydrophobic ligands [7]–[8], [15], [20], [34].
The interaction energies between the rhodojaponin III and the important residues of CSPSlit are listed in Table 1. Although the interaction energies we obtained in the current docking study do not reproduce the real binding free energies, the relative values should be meaningful [59], [61]. Our docking results indicated that Ile 45 could have strong hydrophobic interactions with rhodojaponin III. The residue in this position has been shown to affect substrate binding or specificity in CSPSlit. Analysis of interactions between rhodojaponin III and CSPSlit reveals that vdW energy has a larger contribution to ligand binding than the electrostatic energy. This is in line with the fact that the binding pocket of CSPSlit is mainly composed of hydrophobic residues [7]–[8], [15], [34]. The Tyr 24 side chain might be rotated towards the protein surface like a door to open or close the entrance of the cavity. Although the Tyr 24 is not involved in the key residues we predicted in the binding of rhodojaponin III and CSPSlit, it might be play the role as mentioned above. The interaction of Tyr 24 and rhodojaponin III might occur before the ligand coming into the cavity. When this happened, the Tyr 24 is rotated and exposed the cavity to rhodojaponin III. Another interaction occurs after rhodojaponin III was imbedded in the cavity and the door will closed. Rhodojaponin III is fully imbedded and fit well in the cavity. It positions with the hydrophilic group turned to the outside are also in agreement with the hydrophobic character of the channel [8], [35], [34]. The hydrogen atoms of the rhodojaponin III are closer to the Trp 79 indole rings than to Trp 6. This orientation accounts well for the fluorescence of Trp 79 being mostly quenched. Our docking results were in good agreement with the experimental data. The more details in the binding modes of CSPSlit and its substrates need further investigations through combination of molecular, genetics and chemical methods.
Acknowledgments
We thank Dr. Zhao Fangming for technical assistance.
Funding Statement
The authors gratefully acknowledge the grants from the National Natural Science Foundation (31071713) and Foundation for Young Teacher in Higher School, Fok Ying Tong Education Foundation (111027), People’s Republic of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Awmack CS, Leather SR (2002) Host plant quality and fecundity in herbivorous insects. Annu Rev Entomol 47: 817–844. [DOI] [PubMed] [Google Scholar]
- 2. Hallem EA, Dahanukar A, Carlson JR (2006) Insect odor and taste receptors. Annu Rev Entomol 51: 113–135. [DOI] [PubMed] [Google Scholar]
- 3. Simmonds MS (2001) Importance of flavonoids in insect–plant interactions: feeding and oviposition. Phytochemistry 56: 245–252. [DOI] [PubMed] [Google Scholar]
- 4. Tsuchihara K, Ueno K, Yamanaka A, Isono K, Endo K, et al. (2000) A putative binding protein for lipophilic substances related to butterfly oviposition. FEBS Lett 478: 299–303. [DOI] [PubMed] [Google Scholar]
- 5. Nishida R, Fukami H (1989) Oviposition stimulants of an Aristolochiaceae-feeding swallowtail butterfly, Atrophaneura alcinous . J Chem Ecol 15: 2565–2575. [DOI] [PubMed] [Google Scholar]
- 6. Renwick J, Chew F (1994) Oviposition behavior in lepidoptera. Annu Rev Entomol 39: 377–400. [Google Scholar]
- 7. Briand L, Swasdipan N, Nespoulous C, Bezirard V, Blon F, et al. (2002) Characterization of a chemosensory protein (ASP3c) from honeybee (Apis mellifera L.) as a brood pheromone carrier. Eur J Biochem 269: 4586–4596. [DOI] [PubMed] [Google Scholar]
- 8. Campanacci V, Lartigue A, Hallberg BM, Jones TA, Giudici-Orticoni MT, et al. (2003) Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding. Proc Natl Acad Sci U S A 100: 5069–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mckenna MP, Hekmat-Scafe DS, Gaines P, Carlson JR (1994) Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system. J Biol Chem 269: 16340–16347. [PubMed] [Google Scholar]
- 10. Angeli S, Ceron F, Scaloni A, Monti M, Monteforti G, et al. (1999) Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria . Eur J Biochem 262: 745–754. [DOI] [PubMed] [Google Scholar]
- 11. Maleszka R, Stange G (1997) Molecular cloning, by a novel approach, of a cDNA encoding a putative olfactory protein in the labial palps of the moth Cactoblastis cactorum . Gene 202: 39–43. [DOI] [PubMed] [Google Scholar]
- 12. Bohbot J, Sobrio F, Lucas P, Nagnan-Le Meillour P (1998) Functional characterization of a new class of odorant-binding proteins in the moth Mamestra brassicae . Biochem Biophys Res Commun 253: 489–494. [DOI] [PubMed] [Google Scholar]
- 13. Gong DP, Zhang HJ, Zhao P, Lin Y, Xia QY, et al. (2007) Identification and expression pattern of the chemosensory protein gene family in the silkworm, Bombyx mori . Insect Biochem Mol Biol 37: 266–277. [DOI] [PubMed] [Google Scholar]
- 14. Jacquin-Joly E, Vogt RG, Francois MC, Nagnan-Le Meillour P (2001) Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae . Chem Senses 26: 833–844. [DOI] [PubMed] [Google Scholar]
- 15. Nagnan-Le Meillour P, Cain AH, Jacquin-Joly E, Francois MC, Ramachandran S, et al. (2000) Chemosensory proteins from the proboscis of Mamestra brassicae . Chem Senses 25: 541–553. [DOI] [PubMed] [Google Scholar]
- 16. Olivier V, Monsempes C, Francois MC, Poivet E, Jacquin-Joly E (2011) Candidate chemosensory ionotropic receptors in a Lepidoptera. Insect Mol Biol 20(2): 189–199. [DOI] [PubMed] [Google Scholar]
- 17. Ozakia K, Utoguchia A, Yamadaa A, Yoshikawaa H (2008) Identification and genomic structure of chemosensory proteins (CSP) and odorant binding proteins (OBP) genes expressed in foreleg tarsi of the swallowtail butterfly Papilio xuthus . Insect Biochem Mol Biol 38(11): 969–976. [DOI] [PubMed] [Google Scholar]
- 18. Picimbon JF, Dietrich K, Angeli S, Scaloni A, Krieger J, et al. (2000) Purification and molecular cloning of chemosensory proteins from Bombyx mori . Arch Insect Biochem Physiol 44: 120–129. [DOI] [PubMed] [Google Scholar]
- 19. Picimbon JF, Dietrich K, Krieger J, Breer H (2001) Identity and expression pattern of chemosensory proteins in Heliothis virescens (Lepidoptera, Noctuidae). Insect Biochem MolBiol 31: 1173–1181. [DOI] [PubMed] [Google Scholar]
- 20. Ban L, Scaloni A, Brandazza A, Angeli S, Zhang L, et al. (2003) Chemosensory proteins of Locusta migratoria . Insect Mol Biol 12: 125–134. [DOI] [PubMed] [Google Scholar]
- 21. Picimbon JF, Dietrich K, Breer H, Krieger J (2000) Chemosensory proteins of Locusta migratoria (Orthoptera: Acrididae). Insect Biochem Mol Biol 30: 233–241. [DOI] [PubMed] [Google Scholar]
- 22. Zhou SH, Zhang J, Zhang SG, Zhang L (2008) Expression of chemosensory proteins in hairs on wings of Locusta migratoria (Orthoptera: Acrididae). J Appl Entomol 132: 439–450. [Google Scholar]
- 23. Danty E, Arnold G, Huet JC, Huet D, Masson C, et al. (1998) Separation, characterization and sexual heterogeneity of multiple putative odorant-binding proteins in the honeybee Apis mellifera L. (Hymenoptera: Apidea). Chem Senses 23: 83–91. [DOI] [PubMed] [Google Scholar]
- 24. Gonzalez D, Zhao Q, McMahan C, Velasquez D, Haskins WE, et al. (2009) The major antennal chemosensory protein of red imported fire ant workers. Insect Mol Biol 18(3): 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ishida Y, Chiang V, Leal WS (2002) Protein that makes sense in the Argentine ant. Naturwissenschaften 89: 505–507. [DOI] [PubMed] [Google Scholar]
- 26. Maleszka J, Foret S, Saint R, Maleszka R (2007) RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev Genes Evol 217(3): 189–196. [DOI] [PubMed] [Google Scholar]
- 27. Hu LM, Shen JM, Hu MY, Rizwan-UI-Haq M, Hao WN (2010) Screening of T7 phage displayed Bactrocera dorsalis (Hendel) antenna cDNA library against chemosensory protein. Arch Insect Biochem 75(3): 174–186. [DOI] [PubMed] [Google Scholar]
- 28.Kitabayashi AN, Arai T, Kubo T, Natori S (1998) Molecular cloning of cDNA for p10, a novel protein that increases in regenerating legs of Periplaneta americana (American cockroach). Insect Biochemistry and Molecular Biology, 28, 785–790. [DOI] [PubMed]
- 29. Picimbon JF, Leal WS (1999) Olfactory soluble proteins of cockroaches. Insect Biochem Mol Biol 29: 973–978. [Google Scholar]
- 30. Mameli M, Tuccini A, Mazza M, Petacchi R, Pelosi P (1996) Soluble proteins in chemosensory organs of phasmids. Insect Biochem Mol Biol 26: 875–882. [DOI] [PubMed] [Google Scholar]
- 31. Marchese S, Angeli S, Andolfo A, Scaloni A, Brandazza A, et al. (2000) Soluble proteins from chemosensory organs of Eurycantha calcarata (Insecta, Phasmatodea). Insect Biochem Mol Biol 30: 1091–1098. [DOI] [PubMed] [Google Scholar]
- 32. Tuccini A, Maida R, Rovero P, Mazza M, Pelosi P (1996) Putative odorant-binding protein in antennae and legs of Carausius morosus (Insecta, Phasmatodea). Insect Biochem Mol Biol 26: 19–24. [DOI] [PubMed] [Google Scholar]
- 33.Jacobs SP, Liggins AP, Zhou JJ, Pickett JA, Jin X, et al.. (2005) OS-D-like genes and their expression in aphids (Hemiptera: Aphididae). Insect Molecular Biology, 14, 423–432. [DOI] [PubMed]
- 34. Jansen S, Chmelik J, Zidek L, Padrta P, Novak P, et al. (2007) Structure of Bombyx mori chemosensory protein 1 in solution. Arch Insect Biochem 66(3): 135–145. [DOI] [PubMed] [Google Scholar]
- 35. Lartigue A, Campanacci V, Roussel A, Larsson AM, Jones TA, et al. (2002) X-ray structure and ligand binding study of a moth chemosensory protein. J Biol Chem 277: 32094–32098. [DOI] [PubMed] [Google Scholar]
- 36.Feakin SD (1973) Pest control in groundnuts, Centre for Overseas Pest Research, Foreign and Commonwealth Office Overseas Development Administration, London.
- 37. Ranga Rao GV, Wightman JA, Ranga Rao DV (1993) World review of the natural enemies and diseases of Spodoptera litura(F.) (Lepidoptera: Noctuidae). Insect Science and its Application 14: 273–284. [Google Scholar]
- 38. Li XD, Chen WK, Hu MY (1995) Studies on the effects and mechanisms of azadirachtin and rhodojaponin-III on Spodoptera litura . Journal of South China Agricultural University 16: 80–85. [Google Scholar]
- 39. Zhong GH, Hu MY, Wei XY, Weng QF, Xie JJ, et al. (2005) Grayanane diterpenoids from the flowers of Rhododendron molle with cytotoxic activity against a Spodoptera frugiperda cell line. J Nat Prod 68: 924–926. [DOI] [PubMed] [Google Scholar]
- 40. Zhong GH, Liu JX, Weng QF, Hu MY, Luo JJ (2006) Laboratory and field evaluations of rhodojaponin III against the imported cabbage worm Pieris rapae (L.) (Lepidoptera: Pieridae). Pest Manag Sci 62: 976–981. [DOI] [PubMed] [Google Scholar]
- 41. Ewing TJ, Makino S, Skillman AG, Kuntz ID (2001) DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des 15: 411–428. [DOI] [PubMed] [Google Scholar]
- 42. Wu G, Robertson DH, Brooks CL, Vieth M (2003) Detailed analysis of grid-based molecular docking: A case study of CDOCKER-A CHARMm-based MD docking algorithm. J Comput Chem 24: 1549–1562. [DOI] [PubMed] [Google Scholar]
- 43. Desjarlais RL, Sheridan RP, Seibel GL, Dixon JS, Dkuntz I (1988) Using shape complementarity as an initial screen in designing ligands for a receptor binding site of known three-dimensional structure. J Med Chem 31: 722–729. [DOI] [PubMed] [Google Scholar]
- 44. Meng EC, Shoichet BK, Kuntz ID (1992) Automated docking with grid-based energy evaluation. J Comput Chem 13: 505–524. [Google Scholar]
- 45. Chen QJ, Li GH, Pang Y (2000) A simple artificial diet for mass rearing of some noctuid species. Entomological Knowledge 37: 325–327. [Google Scholar]
- 46.Sambrook KJ, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual 2nd ed, Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
- 47. Tsumoto K, Shinoki K, Kondo H, Uchikawa M, Juji T, et al. (1998) Highly efficient recovery of functional single-chain Fv fragments from inclusion bodies overexpressed in Escherichia coli by controlled introduction of oxidizing reagent–application to a human single-chain Fv fragment. J Immunol Methods 219: 119–129. [DOI] [PubMed] [Google Scholar]
- 48. Kruger NJ (1994) The Bradford method for protein quantitation. Methods Mol Biol 32: 9–15. [DOI] [PubMed] [Google Scholar]
- 49. Baqi Y, Hausmann R, Rosefort C, Rettinger J, Schmalzing G, et al. (2011) Discovery of potent competitive antagonists and positive modulators of the P2X2 receptor. J Med Chem 54(3): 817–830. [DOI] [PubMed] [Google Scholar]
- 50. Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water, J Chem Phys. 79: 926–935. [Google Scholar]
- 51. Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, et al. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 3l: 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pelosi P, Maida R (1995) Odorant-binding proteins in insects. Comp Biochem Physiol B Biochem Mol Biol 111: 503–514. [DOI] [PubMed] [Google Scholar]
- 53. Vogt RG, Prestwich GD, Lerner MR (1991) Odorant-binding-protein subfamilies associate with distinct classes of olfactory receptor neurons in insects. J Neurobiol 22: 74–84. [DOI] [PubMed] [Google Scholar]
- 54. Picone D, Crescenzi O, Angeli S, Marchese S, Brandazza A, et al. (2001) Bacterial expression and conformational analysis of a chemosensory protein from Schistocerca gregaria . Eur J Biochem 268: 4794–4801. [DOI] [PubMed] [Google Scholar]
- 55. Monteiro A (2008) Alternative models for the evolution of eyespots and of serial homology on lepidopteran wings. Bioessays 30(4): 358–366. [DOI] [PubMed] [Google Scholar]
- 56. Jones RT, Bakker SE, Stone D, Shuttleworth SN, Boundy S, et al. (2010) Homology modelling of Drosophila cytochrome P450 enzymes associated with insecticide resistance. Pest Manag Sci 66(10): 1106–1115. [DOI] [PubMed] [Google Scholar]
- 57. Tohidi-Esfahani D, Lawrence MC, Graham LD, Hannan GN, Simpson AM, et al. (2011) Isoforms of the heteropteran Nezara viridula ecdysone receptor: protein characterisation, RH5992 insecticide binding and homology modeling. Pest Manag Sci 67(11): 1457–1467. [DOI] [PubMed] [Google Scholar]
- 58. Zotti MJ, Christiaens O, Rouge P, Grutzmacher AD, Zimmer PD, et al. (2012) Sequencing and structural homology modeling of the ecdysone receptor in two chrysopids used in biological control of pest insects. Ecotoxicology 21(3): 906–918. [DOI] [PubMed] [Google Scholar]
- 59. Li WH, Tang Y, Liu H, Cheng JG, Zhu WL, et al. (2008) Probing ligand binding modes of human cytochrome p450 2j2 by homology modeling, molecular dynamics simulation, and flexible molecular docking. Proteins 71(2): 938–949. [DOI] [PubMed] [Google Scholar]
- 60. Kumara R, Kumarb S, Sangwanc S, Yadava IS, Yadavd R (2011) Protein modeling and active site binding mode interactions of myrosinase–sinigrin in Brassica juncea–An in silico approach. J Mol Graph Model 29(5): 740–746. [DOI] [PubMed] [Google Scholar]
- 61. Zhang XN, Hu YW, Yuan ZH (2008) Computational analyses of JAK1 kinase domain: Subtle changes in the catalytic cleft influence inhibitor specificity. Biochem Bioph Res Co 370: 72–76. [DOI] [PubMed] [Google Scholar]