Abstract
We present a case of sexual transmission of HIV-1 predicted to have CXCR4-tropism during male-to-male sexual exposure. Phylogenetic analyses exclude cell-free virus in the seminal plasma of the source partner and possibly point to the seminal cells as the origin of the transmission event.
Introduction
Most HIV transmission events globally occur via mucosal exposure to HIV found in male genital secretions (Baeten et al., 2011), but it remains unclear whether cell-free, cell-associated virus, or both are the primary source for sexually transmitted HIV (Anderson et al., 2010; Butler et al., 2010). It is also unclear if the transmission bottleneck selects for CCR5 tropic (R5) viruses (Schuitemaker et al., 2011), yet such viruses predominate in the blood early after infection (Rieder et al., 2011; Zhu et al., 1993). Although recent studies provide some evidence of CXCR4 (X4) co-receptor use during acute infection (Chalmet et al., 2012; Eshleman et al., 2007; Wagner et al., 2012), reports of transmission of X4-tropic virus remains extremely rare. Recently, we described several cases where the virus transmitted during sexual exposure between men who have sex with men (MSM) originated from seminal plasma (Butler et al., 2010). Here, we present a case of sexual transmission of HIV that was genotypically predicted to be predominantly X4-tropic, and was not detected in the seminal plasma of the source partner.
Material and Methods
Paired blood and semen samples were collected from an epidemiologically and phylogenetically linked transmission pair, as described in (Butler et al., 2010). Single genome sequencing and ultra deep sequencing (UDS) of the HIV-1 env C2-V3 coding region (Genbank accession numbers: JQ087498-JQ087605) were performed on HIV RNA extracted from source and recipient blood plasma. Single genome amplification was performed using cloning on nucleic acid extracted from peripheral blood mononuclear cell (PBMC) (DNA), seminal plasma (RNA), and seminal cell (DNA) of the source partner, as previously described (Butler et al., 2010; Gianella et al., 2011). Phylogenetic maximum likelihood trees were generated using RAxML (Stamatakis et al., 2005) and clade support values were obtained using MrBayes (Ronquist and Huelsenbeck, 2003). Compartmentalization analyses were performed using Slatkin-Maddison and FST methods (Zarate et al., 2007). Cellular tropism was predicted from V3 loop sequences using Web PSSM (Jensen et al., 2006) and Geno2pheno (Daumer et al., 2011) with a false-positive rate set at 5.75% (Rieder et al., 2011).
Results and Discussion
Both subjects were men who reported sex with other men as their only HIV risk factor, and the recipient partner reported receptive and insertive unprotected anal and oral sex with his source partner. Seminal and blood samples were collected from both subjects 90 days after recipient’s estimated date of infection. At this time, the source partner was 22 years old, his HIV RNA viral load (VL) was 88,586 copies/ml of blood plasma, and his CD4 T cell count was 294 cells/ml. The recipient partner was 30 years old, his HIV RNA VL was 38,775 copies/ml of blood plasma, and CD4 T cell count was 370 cells/ml. Both subjects were infected with HIV-1 subtype B, as determined using SCUEAL (Kosakovsky Pond et al., 2009).
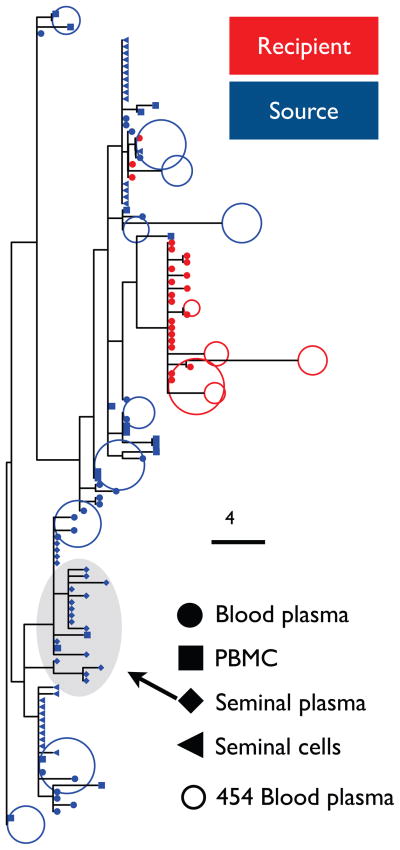
For the source partner, two statistical tests (i.e. Slatkin-Maddison and FST) provided clear evidence of viral compartmentalization (p<0.01) between sequences obtained from seminal plasma and seminal cells, as well as between seminal plasma and blood (both blood plasma and PBMC derived sequences). Subcompartmentalization between HIV-1 isolated from seminal plasma and from seminal cells has been described previously (Butler et al., 2010; Paranjpe et al., 2002) and is likely the consequence of different anatomic origin of the two component (seminal plasma is derived from the prostate and the seminal cells mostly originate from the rete testis and epididymis). Additionally, phylogenetic reconstruction strongly suggested that the viral population sampled from the recipient’s blood plasma originated from either the source partner’s seminal cells (HIV DNA) or blood rather than from the source’s seminal plasma (posterior support of reciprocal monophyly >95%) (figure 1).
Figure 1. Phylogenetic analysis of source and recipient’s viral sequences.
Maximum likelihood phylogenies of source and recipient’s viral populations in the different compartments. Sequences from recipient are red, sequences from source are blue. Full dots show SGS sequences from blood plasma RNA, squares show clonal sequences from PBMC DNA, diamonds show clonal sequences from seminal plasma RNA, triangles show sequences from seminal cells DNA. Empty dots display a representative sample of UDS reads. Grey shadow shows the seminal plasma cluster, which is not the origin of the transmitted virus
Although the source of sexually transmitted virus might be expected to be from the genital compartment, it is possible that the observed HIV transmission occurred when the recipient partner was exposed to the blood of the source partner during sex, e.g. mucosal tears. Additionally, since the samples were collected 3 months after the estimated date of infection, we cannot role out that the virus sequenced in the seminal cell compartment from the source partner was present also in the seminal plasma at the time of transmission. It is possible that the virus integrated in the cellular compartment lives longer than the cell-free virus in seminal plasma.
Genotypic tropism analysis of clonal sequences predicted a mixed population of CXCR4 (X4) and R5 tropic viruses in blood plasma (14 X4; 9 R5), PBMC (14 X4; 5 R5) and seminal cells (16 X4; 10 R5) of the source partner, while 20 sequences within the source’s seminal plasma were exclusively R5 tropic. We recovered predominantly X4 virus from the recipient’s blood plasma (22 sequences were predicted as X4 and one was predicted as R5). Similar results were obtained from UDS data, with 49% predicted X4 tropism in source’s blood and 99.6% predicted X4 tropism in recipient’s blood plasma. As discussed above, the delay in collecting the specimens could have confounded our findings since the sampled virus has been replicating in the recipient for 90 days since transmission. However, it seems unlikely to observe a complete tropism switch to X4 in the recipient blood population within 3 months from the transmission event. Unfortunately, it was not possible to obtain phenotypic proof of tropism for these samples. However, several previous studies demonstrated a high concordance between genotypic and phenotypic co-receptor usage prediction especially for subtype B viruses and using 454 data (Daumer et al., 2011; Schuitemaker et al., 2011; Wagner et al., 2012).
This report provides evidence that (i) HIV sexually transmitted between MSM does not always originate from seminal plasma, (ii) seminal cells can present a compartmentalized viral populations in regard to seminal plasma, (iii) genotypically predicted X4 tropic virus can be transmitted during a sexual transmission.
Since transmitted virus represents the initial encounter of the immune system with the new infection, the understanding of its composition will be critical in the development and implementation of preventive measures.
Highlights.
We investigated the origin of sexually transmitted HIV-1 between MSM (cell-free versus cell-associated virus)
HIV sexually transmitted between MSM does not always originate from seminal plasma
Seminal cells can present a compartmentalized viral populations in regard to seminal plasma
X4 tropic virus may be transmitted during a sexual transmission.
Acknowledgments
We are grateful to all the participants in the San Diego Primary Infection Cohort, and to Caroline Ignacio, Parris Jordan and Gemma Caballero for excellent technical support.
Footnotes
Author Contributions
SG participated in the study design, performed laboratory experiments and data analyses, and wrote the first version of the manuscript; SRM participated in data analysis, and revised the manuscript; MVV performed laboratory experiments, SJL and DMS enrolled patients and revised the manuscript; SKP, MAS and JAY participated in the data analysis and revised the manuscript, DDR, SJL and DMS designed the study, participated in data analysis and revised the manuscript. All authors read and approved the final manuscript.
Competing Interests
SG does not have any commercial or other associations that might pose a conflict of interest. DDR has served as a consultant for Biota, Bristol-Myers Squibb, Chimerix, Gen-Probe, Gilead Sciences, J & J, Merck & Co, Monogram Biosciences, and Tobira Therapeutics and Vertex. DMS has received research support from ViiV Pharmaceuticals and has served as a consultant to Genprobe. SKP has served as a consultant to Genprobe. The remaining authors do not report any conflicts of interest.
Financial Disclosure
This work was supported by grants from the US National Institutes of Health AI69432, AI043638, MH62512, MH083552, AI077304, AI36214, AI047745, AI74621, GM093939 and AI080353, the James Pendleton Trust, and the Swiss National Science Foundation grant PASMP3_136983.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson DJ, Politch JA, Nadolski AM, Blaskewicz CD, Pudney J, Mayer KH. Targeting Trojan Horse leukocytes for HIV prevention. AIDS. 2010;24:163–187. doi: 10.1097/QAD.0b013e32833424c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeten JM, Kahle E, Lingappa JR, Coombs RW, Delany-Moretlwe S, Nakku-Joloba E, Mugo NR, Wald A, Corey L, Donnell D, Campbell MS, Mullins JI, Celum C. Genital HIV-1 RNA predicts risk of heterosexual HIV-1 transmission. Sci Transl Med. 2011;3:77ra29. doi: 10.1126/scitranslmed.3001888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler DM, Delport W, Kosakovsky Pond SL, Lakdawala MK, Cheng PM, Little SJ, Richman DD, Smith DM. The origins of sexually transmitted HIV among men who have sex with men. Sci Transl Med. 2010;2:18re11. doi: 10.1126/scitranslmed.3000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmet K, Dauwe K, Foquet L, Baatz F, Seguin-Devaux C, Van Der Gucht B, Vogelaers D, Vandekerckhove L, Plum J, Verhofstede C. Presence of CXCR4-using HIV-1 in patients with recently diagnosed infection: correlates and evidence for transmission. The Journal of infectious diseases. 2012;205:174–184. doi: 10.1093/infdis/jir714. [DOI] [PubMed] [Google Scholar]
- Daumer M, Kaiser R, Klein R, Lengauer T, Thiele B, Thielen A. Genotypic tropism testing by massively parallel sequencing: qualitative and quantitative analysis. BMC medical informatics and decision making. 2011;11:30. doi: 10.1186/1472-6947-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshleman SH, Husnik M, Hudelson S, Donnell D, Huang Y, Huang W, Hart S, Jackson B, Coates T, Chesney M, Koblin B. Antiretroviral drug resistance, HIV-1 tropism, and HIV-1 subtype among men who have sex with men with recent HIV-1 infection. AIDS. 2007;21:1165–1174. doi: 10.1097/QAD.0b013e32810fd72e. [DOI] [PubMed] [Google Scholar]
- Gianella S, Delport W, Pacold ME, Young JA, Choi JY, Little SJ, Richman DD, Pond SLK, Smith DM. Detection of Minority Resistance during Early HIV-1 Infection: Natural Variation and Spurious Detection Rather than Transmission and Evolution of Multiple Viral Variant. J Virol. 2011;85:8359–8367. doi: 10.1128/JVI.02582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MA, Coetzer M, van‘t Wout AB, Morris L, Mullins JI. A reliable phenotype predictor for human immunodeficiency virus type 1 subtype C based on envelope V3 sequences. Journal of virology. 2006;80:4698–4704. doi: 10.1128/JVI.80.10.4698-4704.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Stawiski E, Chappey C, Poon AF, Hughes G, Fearnhill E, Gravenor MB, Leigh Brown AJ, Frost SD. An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol. 2009;5:e1000581. doi: 10.1371/journal.pcbi.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjpe S, Craigo J, Patterson B, Ding M, Barroso P, Harrison L, Montelaro R, Gupta P. Subcompartmentalization of HIV-1 Quasispecies between Seminal Cells and Seminal Plasma Indicates Their Origin in Distinct Genital Tissues. AIDS Res Hum Retroviruses. 2002;18:1271–1280. doi: 10.1089/088922202320886316. [DOI] [PubMed] [Google Scholar]
- Rieder P, Joos B, Scherrer A, Kuster H, Braun D, Grube C, Niederöst B, Leemann C, Gianella S, Metzner K, Böni J, Weber R, Günthard H. Characterization of Human Immunodeficiency Virus Type 1 (HIV-1) Diversity and Tropism in 145 Patients With Primary HIV-1 Infection. Clin Infect Dis. 2011 doi: 10.1093/cid/cir725. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schuitemaker H, van‘t Wout AB, Lusso P. Clinical significance of HIV-1 coreceptor usage. J Transl Med. 2011;9(Suppl 1):S5. doi: 10.1186/1479-5876-9-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- Wagner G, Gianella S, Pacold M, Kosakovsky Pond SL, Vigil E, Little SJ, Morris SR, Richman DD, Smith DM. HIV-1 CXCR4 Co-receptor Tropism in Primary and Dual Infection Estimated with Ultra-deep Pyrosequencing. 19th Conference of Retroviruses and Opportunistic Infections.2012. [Google Scholar]
- Zarate S, Pond SL, Shapshak P, Frost SD. Comparative study of methods for detecting sequence compartmentalization in human immunodeficiency virus type 1. J Virol. 2007;81:6643–6651. doi: 10.1128/JVI.02268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Mo H, Wang N, Nam DS, Cao Y, Koup RA, Ho DD. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]