Abstract
Translocations are chromosomal rearrangements that are frequently associated with a variety of disease states and developmental disorders. We identified 2 families with brachydactyly type E (BDE) resulting from different translocations affecting chromosome 12p. Both translocations caused downregulation of the parathyroid hormone-like hormone (PTHLH) gene by disrupting the cis-regulatory landscape. Using chromosome conformation capturing, we identified a regulator on chromosome 12q that interacts in cis with PTHLH over a 24.4-megabase distance and in trans with the sex-determining region Y-box 9 (SOX9) gene on chromosome 17q. The element also harbored a long noncoding RNA (lncRNA). Silencing of the lncRNA, PTHLH, or SOX9 revealed a feedback mechanism involving an expression-dependent network in humans. In the BDE patients, the human lncRNA was upregulated by the disrupted chromosomal association. Moreover, the lncRNA occupancy at the PTHLH locus was reduced. Our results document what we believe to be a novel in cis– and in trans–acting DNA and lncRNA regulatory feedback element that is reciprocally regulated by coding genes. Furthermore, our findings provide a systematic and combinatorial view of how enhancers encoding lncRNAs may affect gene expression in normal development.
Introduction
Translocations are chromosomal rearrangements that can be inherited and are often associated with cancers and congenital disorders. One of the great difficulties in determining mechanistic causes is the fact that translocations can affect gene expression and regulatory elements and alter chromosomal architecture by changing the relative genomic position of genes and their regulators. Thus, structural genomic changes may effect the regulatory landscape. Such perturbations may be involved in the well-known observations of position effects (1, 2). We combined several methods to resolve some of the complex translocation-related position effects on gene regulation, based on 2 families with brachydactyly type E (BDE). BDE is characterized by a shortening of metacarpals and metatarsals (3). The morphogenesis gene PTHLH, encoding parathyroid hormone–like hormone on chromosome 12p11.2, has been linked to BDE with short stature (4, 5). PTHLH is involved in the regulation of cartilage differentiation and digit condensation (6). During digit condensation, PTHLH and other factors act in concert to balance an intricate signaling network, ultimately impinging on BMP-mediated and sex-determining region Y-box 9–directed (SOX9-directed) chondrogenesis (7). Dysregulated expressions of the haploinsufficient genes PTHLH or SOX9 can cause skeletal phenotypes (2, 4, 5, 8–10). In previous work with a BDE family with balanced translocation t(8;12)(q13;p11.2), we discovered a breakpoint-dependent PTHLH downregulation that elucidated new mechanisms resulting in a Mendelian phenotype (4). The cis regulation of gene expression is accompanied by distinct tissue- and cell type–specific histone modifications that enable or disable the binding of transcription factors to evolutionarily highly conserved DNA elements (11, 12). cis-regulatory elements (CREs) can be intra- or intergenic (13, 14). They are typically located up to 1 megabase (Mb) from their coregulated genes (8, 15–17), but may be farther away. Enhancers may be transcribed into lncRNAs that increase gene expression in cis (18). Furthermore, lncRNAs can themselves act as enhancers; their expression is then correlated with the expression of associated genes (19). The lncRNA transcripts have more than 200 nt, display high interspecies conservation, and are multiexonic (20). Finally, lncRNAs are essential for the development of complex organisms (21). The most accepted model for CRE communication involves direct interaction of CRE with a gene promoter through spatial proximity in chromatin loops (22).
We here investigated another family with autosomal-dominant BDE and a balanced t(4;12)(q13.2-13.3;p11.2) translocation. The translocation-mediated genomic disruption of a CRE caused PTHLH repression and led to BDE in both translocation families. We studied the native PTHLH CREs and determined how they were altered. We discovered what we believe to be a novel CRE that acts both in cis and in trans. Within this CRE exists a lncRNA that was pivotal to its function. lncRNAs are known to be important to developmental programs and tumor suppressor gene networks (23). We observed reciprocal gene and lncRNA regulation that provide a systematic and combinatorial view of how enhancers encoding lncRNAs may affect gene expression in normal development. Dysfunction of the CRE and its lncRNA caused a Mendelian disease.
Results
Molecular characterization of t(4;12)(q13.2-13.3;p11.2).
We analyzed a pedigree demonstrating autosomal-dominant inheritance. The roentgenograms were diagnostic of BDE (ref. 3, Figure 1A, and Supplemental Figure 1, A and B; supplemental material available online with this article; doi: 10.1172/JCI65508DS1). Affected individuals exhibited short stature (less than third percentile). A relative shortening of the extremities in comparison to the trunk was evident. The arm span/height ratio was reduced (Supplemental Table 1). High-resolution karyotyping identified the balanced translocation t(4;12)(q13.2-13.3;p11.2) (Figure 1A). BDE-causative HOXD13 and PTHLH mutations were excluded by sequencing (5, 24). Breakpoint allele-specific sequencing identified the derivative chromosome 4 [der(4)] breakpoint 145,460 bp downstream of PTHLH at position 28,147,744 bp (UCSC assembly hg18). The breakpoint was 45,433 bp downstream of the previously described der(8) breakpoint in t(8;12)(q13;p11.2) (4). Microdeletions and 7SL pseudogene insertions at the breakpoints did not eliminate or create transcription factor binding sites (TFBSs; Supplemental Figure 1, E and F). To focus on the chondrogenic function of PTHLH, we induced chondrogenic differentiation in fibroblasts of an affected patient and 2 nonaffected controls. Using quantitative RT-PCR, we observed downregulation of PTHLH (P = 0.04) and its downstream targets, ADAMTS-7 and ADAMTS-12 (Supplemental Figure 1, G and H). PTHLH was downregulated, and its insufficiency correlated with BDE in affected family members.
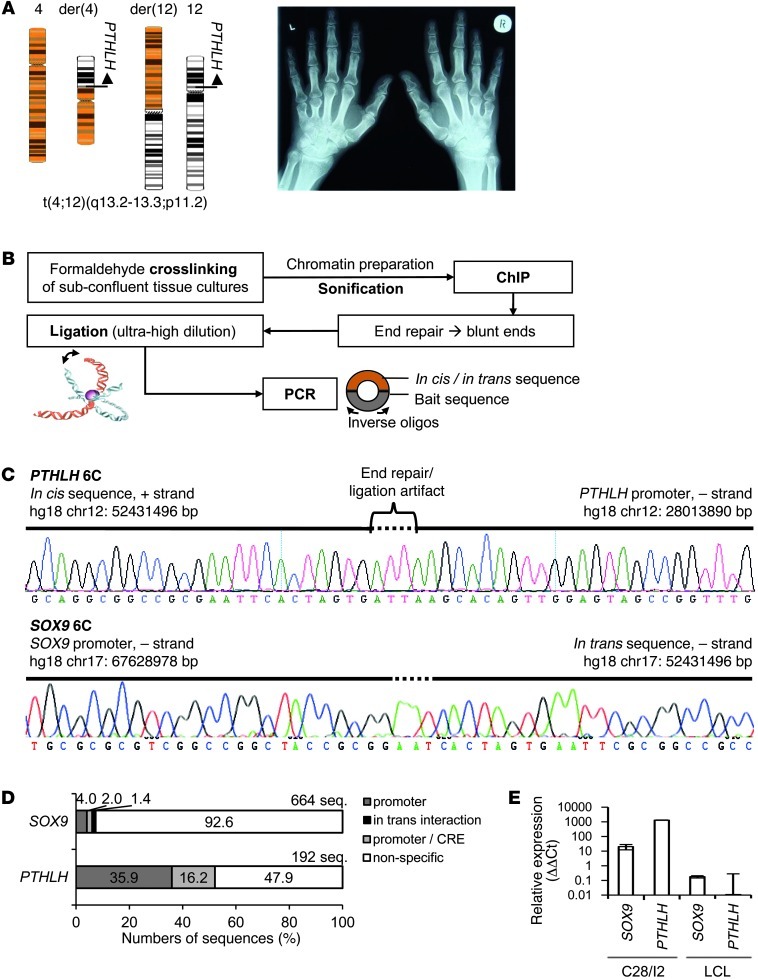
Figure 1. The translocation t(4;12)(q13.2-13.3;p11.2) and scheme of the 6C method.
(A) Translocation with der(4) and der(12). The hand roentgenogram, especially the shortened metacarpals of digits 4 and 5, are diagnostic of BDE. (B) 6C. Formaldehyde-crosslinked chromatin of C28/I2 chondrocytes was sonicated. After ChIP, the chromatin was blunt-end repaired and ligated at ultra-high dilution to favor intramolecular ligation events. Chromatin loops were PCR amplified with inverse oligos, and the amplicons were subcloned and sequenced. PTHLH and SOX9 promoters were the bait sequences. (C) Example of PTHLH 6C. Several bp were inserted due to the end repair of chromatin fragments. The interacting sequence was located at 52,431,496 bp on chromosome 12 (positions in bp; UCSC assembly hg18). In trans event in SOX9 6C. The interacting element on chromosome 12 was identified next to the bait sequence of the SOX9 promoter. (D) Restrictive classification of the 6C sequence reads. Either the promoter (bait) or the promoter interaction with any sequence was counted. (E) PTHLH and SOX9 mRNA expression in human C28/I2 and LCL.
Identification of putative CREs for PTHLH and SOX9 by the 6C technique.
Chromosome conformation capture (3C) techniques can identify gene-associated cis or trans regulators (25, 26). The 3C method was extended to include ChIP, circularization, and cloning (6C technique). We purposely used a non–genome-wide approach to investigate the cis-regulatory landscape of the candidate gene PTHLH. The ChIP was done on chromatin of the chondrocyte cell line C28/I2 with anti-RNA polymerase II (recognized phosphorylated S5) and anti-H3K4me1 antibodies (27). We searched for H3K4me1-enriched CREs that were linked by chromatin loops to the active PTHLH or SOX9 promoters, our bait sequences. SOX9, located on chromosome 17 and expressed in C28/I2, is the major transcription factor commanding chondrogenesis in the limb, known for long-range cis regulation (9, 28), and served as positive control. The intramolecular ultra-high dilution ligation resulted in circularized chromatin (29). Using inverse-orientated oligos within the baits, we PCR amplified any CREs in the circular chromatin, subcloned the nested PCR amplicons, and sequenced the plasmids (Figure 1B).
We restrictively classified the reads: the bait sequence had to be adjacent to a CRE. These specific sequences of approximately 50 nt of either the promoter or the promoter plus CRE were interspersed by several nt due to the blunt-end repair of chromatin ends (Figure 1C). Randomly distributed positions at all chromosomes without the baits were classified as nonspecific. The 2% interaction frequency of SOX9 reads identified 4 CREs on chromosome 17, whereas 16.2% showed 9 CREs for PTHLH on chromosome 12 (Figure 1D and Figure 2, A and B). The nonspecific sequences of 92.6% in SOX9 6C compared with 47.9% in PTHLH 6C could be explained by the lower SOX9 expression observed (Figure 1E). Surprisingly, we detected an interaction frequency of 1.4% in SOX9 6C for an in trans element on chromosome 12 (Figure 1D). The same element was also detected in PTHLH 6C (Figure 1C).
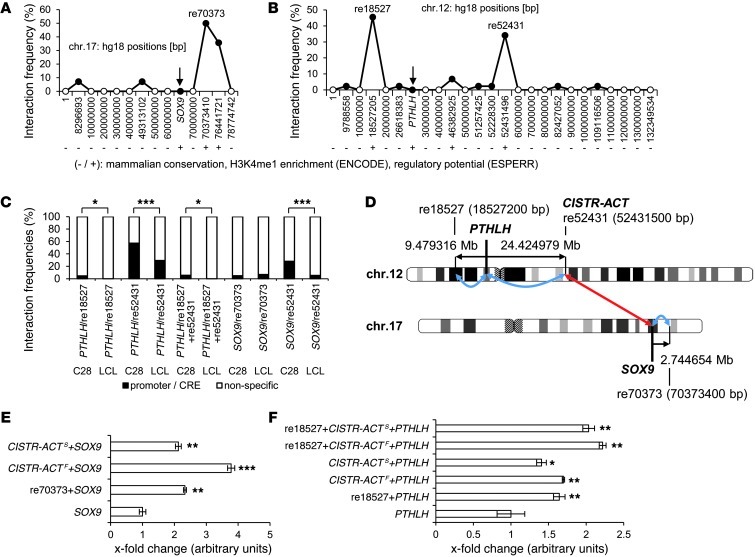
Figure 2. 6C results and validation.
(A and B) Interaction frequencies of SOX9 (A) and PTHLH (B) 6C data. In SOX9 6C, 4 CREs were detected; 9 were found in PTHLH 6C (black symbols). re70373, re18527, and re52431 were the most abundant, and further characterized by high mammalian conservation, H3K4me1 enrichment (ENCODE), and ESPERR regulatory potential (denoted by “+”) (positions in bp; UCSC assembly hg18). re52431 interacted with SOX9 in trans. (C) New 6C libraries validated prior 6C results; control was a LCL. A direct PCR approach amplified re70373, re18527, and re52431. In the LCL, the C28/I2 CRE interactions were significantly reduced; only for re70373 were interactions in C28/I2 and LCL detected equally. (D) Distances of cis- and trans-regulatory elements identified in PTHLH 6C and SOX9 6C. re18527 (18,527,200 bp) and re52431 (52,431,500 bp) interacted with PTHLH, re70373 (70,373,400 bp) interacted in cis and re52431 in trans with SOX9. re52431 was further named CISTR-ACT, due to its cis and trans interactions. (E) Luciferase reporter assays of re70373, CISTR-ACTF (full-length regulatory sequence), and CISTR-ACTS (containing the most conserved part) elements were placed in front of the SOX9 promoter; all 3 enhanced transcription. (F) re18527, CISTR-ACTF, and CISTR-ACTS, either alone or in combination, controlled the PTHLH promoter and enhanced luciferase transcription (n = 6). ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05.
We then used ChIP to quantify the H3K4me1 and H3K4me2 enrichment of the 6C-identified CREs by quantitative PCR (qPCR; Supplemental Figure 2 and ref. 30). To check for regulatory features of the putative 6C-identified CREs, we analyzed the interspecies PhastCons conservation, the ENCODE H3K4me1 enrichment, and the ESPERR regulatory potential in UCSC. We performed comparative analysis of TFBSs on homologous DNA sequences from different species (rVISTA 2.0; zPicture) and identified highly conserved TFBSs (Supplemental Table 2 and Supplemental Figures 3 and 4). SOX5, SOX9, PBX1, AP1, and HOXA motifs were among those detected. All are known to regulate limb bud development and chondrogenesis (reviewed in ref. 31). Based on significant H3K4me1 enrichment, the most abundantly found 6C interactions, and the in silico analysis, we selected 3 CREs for further experiments: re70373 for SOX9 and re18527 and re52431 for PTHLH (Figure 2, A and B). re52431 on chromosome 12q interacted with SOX9 in trans. Chromosomal aberrations (dup and del) in 3 patients with short stature and shortened phalanges in the DECIPHER database of chromosomal imbalances and phenotypes (patient nos. 248901, 248785, and 250426) included the re52431 region, which indicates that the region is important during chondrogenesis.
Validation of the 6C results.
New 6C libraries validated the reproducibility of the 6C technique. To determine bias, a human lymphoblastoid cell line (LCL), marginally expressing SOX9 and PTHLH, was the control (Figure 1E). In a direct PCR approach, oligos were aligned in each of the 3 selected CREs and within the PTHLH or SOX9 promoter. We reproduced the chondrogenic in cis and in trans interactions (Figure 2C). In the LCL, re18527 did not interact, and re52431 showed a significantly lesser interaction by χ2 test, with PTHLH or SOX9. re70373 interacted with SOX9 in both cell lines, 1.64 Mb more downstream than a previous reported CRE for SOX9 (9). The higher interaction frequencies in C28/I2 compared with those in LCL argued for the detection of chondrogenic-specific CREs. Due to the cis- and trans-chromosomal interactions of re52431, we termed this regulator CISTR-ACT.
The distances between the 6C-identified CREs and the associated gene promoter were far greater than previously reported (17). re70373 was 2.74 Mb downstream of SOX9. re18527 was 9.48 Mb upstream, and CISTR-ACT 24.43 Mb downstream, of PTHLH (Figure 2D). We constructed luciferase reporters with the specific SOX9 and PTHLH promoter sequences downstream of re70373, re18527, and CISTR-ACT to investigate their enhancing potential. Since the latter were found combined with PTHLH in 6C, we constructed composite plasmids: CISTR-ACTF represented all identified TFBSs, whereas CISTR-ACTS was from the most conserved part (Supplemental Figure 4). Significant enhancement of luciferase reporter activity for all CRE-PTHLH and CRE-SOX9 constructs was observed. re70373 combined with CISTR-ACTS and CISTR-ACTF increased SOX9 promoter activity 2.5- and 3.8-fold, respectively (Figure 2E). CISTR-ACTS and CISTR-ACTF activated the PTHLH promoter less effectively than they did SOX9. The combination of re18527 with either CISTR-ACTS or CISTR-ACTF showed an additive activation effect on the PTHLH promoter (Figure 2F).
Physically detectable in cis or in trans interactions.
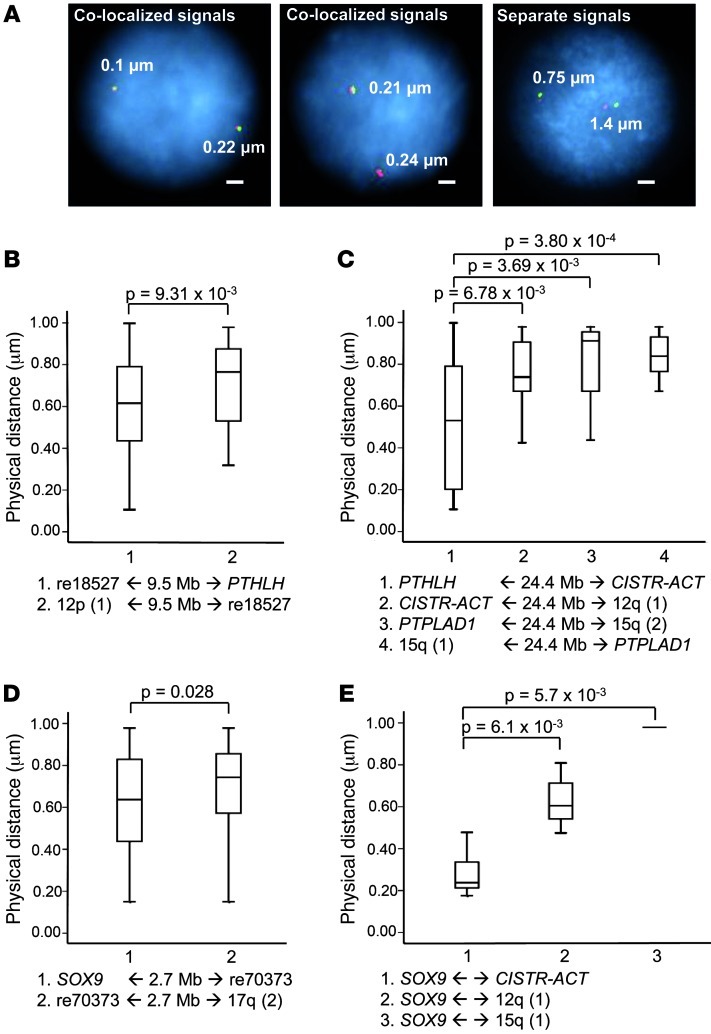
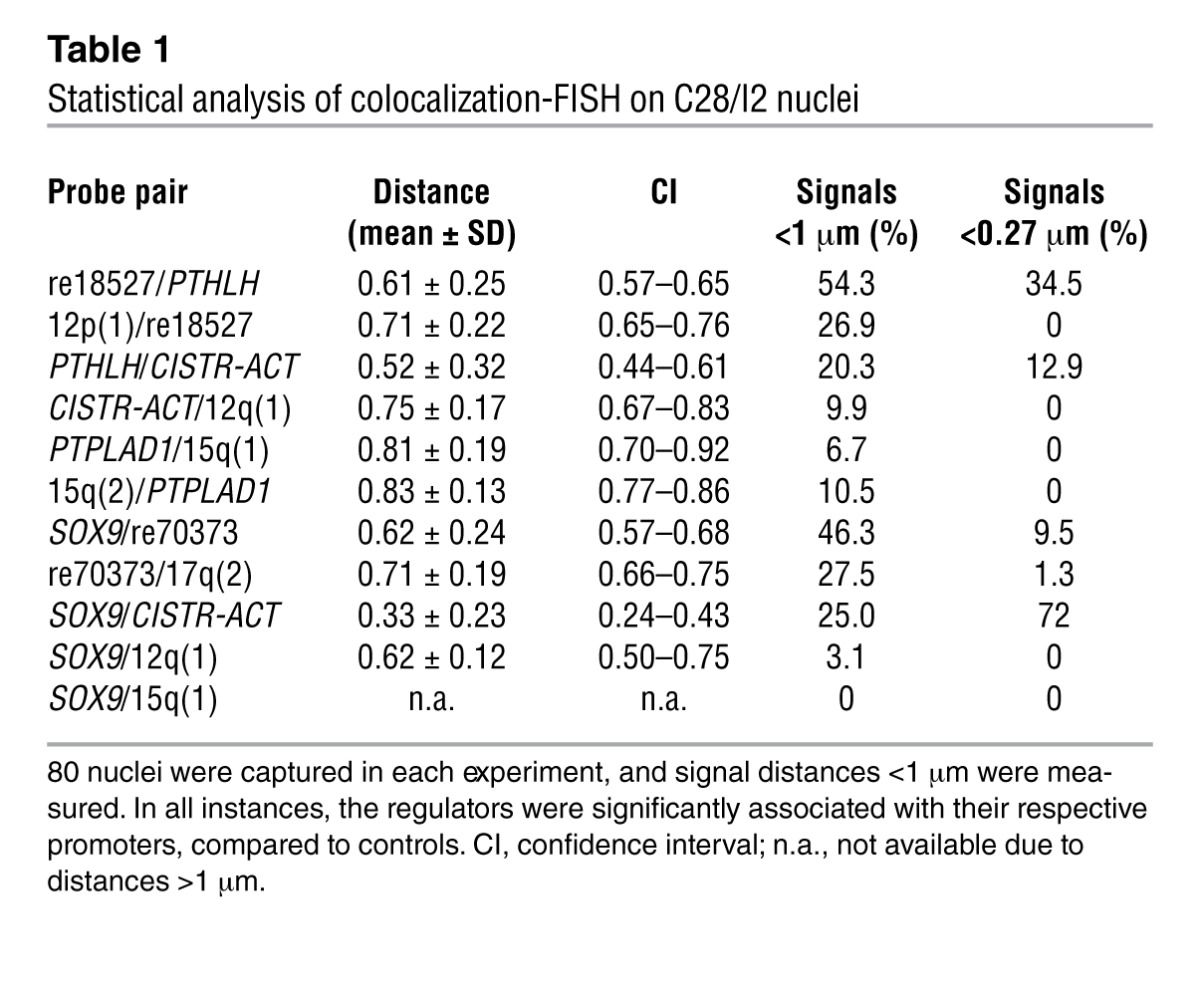
To address spatial chromatin loops that might form between the selected CREs and the promoters as a prerequisite of gene regulation, we performed colocalization-FISH on C28/I2 nuclei (8). We captured 80 nuclei in each experiment and analyzed signal distances less than 1 μm. Bona fide colocalization signals merged below 0.27 μm (Mann-Whitney rank-sum test; Figure 3A). For negative controls, we chose intervening probes between the gene and the CREs and equidistant loci from the CREs in the other direction on chromosome 12 or 17 (Supplemental Figure 5).
Figure 3. Colocalization-FISH.
(A) Examples of colocalized vs. separate signal pairs in C28/I2 nuclei (distances in μm; scale bars: 2 μm). (B) Accumulated colocalized signals were found in all experiments. Shown are genomic distances between probes (in Mb); Mann-Whitney test was used to determine significance. PTHLH was in physical proximity to re18527 compared with the re18527 negative control, with a 9.5-Mb equidistant upstream probe 12p(1). (C) CISTR-ACT significantly showed interaction with PTHLH compared with the 3 controls. PTPLAD1 was not expressed in C28/I2. (D) re70373 was marginally in spatial proximity to SOX9. (E) Significant in trans actions of CISTR-ACT with SOX9. In the control of SOX9 with a chromosome 15q(1) probe, only 2 signal pairs had distances less than 1 μm.
The measured distances between re18527 and PTHLH were significantly shorter than the negative controls (P = 9.31 × 10–3; Figure 3B and Supplemental Figure 5A). For investigation of the cis distance of 24.43 Mb between PTHLH and CISTR-ACT, probe pairs with accordant genomic distances on chromosome 15 around the protein tyrosine phosphatase–like A domain containing 1 (PTPLAD1) locus were used as further controls. PTPLAD1 was not expressed in C28/I2. Compared with all controls, CISTR-ACT was in spatial proximity to PTHLH (Figure 3C and Supplemental Figure 5A). The colocalization of the re70373 element and SOX9 was barely significant; the intervening probe detected closer proximity (P = 0.028; Figure 3D and Supplemental Figure 5B). The SOX9–CISTR-ACT in trans contact was compared with SOX9–chromosome 12q and SOX9–chromosome 15q probe pairs (Figure 3E). Moreover, 2 probe pairs of SOX9 with positions on the exterior nucleus–located chromosome 1 showed no spatial distance less than 1 μm. In all instances, CISTR-ACT had significant contact with SOX9. Putative chromosome 12 with 17 translocations were excluded in metaphase FISH (Supplemental Figure 5C). The statistical analysis showed undetermined signal distances, but specific cis or trans interactions of CISTR-ACT with PTHLH or SOX9 happened in a fraction of cells. The frequency distributions of colocalized or very close signals were significantly higher in the experimental measurements than in the negative controls (Table 1). Overall, the in cis genomic distances between promoter and CRE were inversely proportional to the interaction frequencies in cells. Consistent with the 6C results, the FISH data excluded the possibility that incidental intrachromosomal folding or interchromosomal territorial contacts occurred.
Table 1.
Statistical analysis of colocalization-FISH on C28/I2 nuclei
CISTR-ACT expresses the lncRNA DA125942.
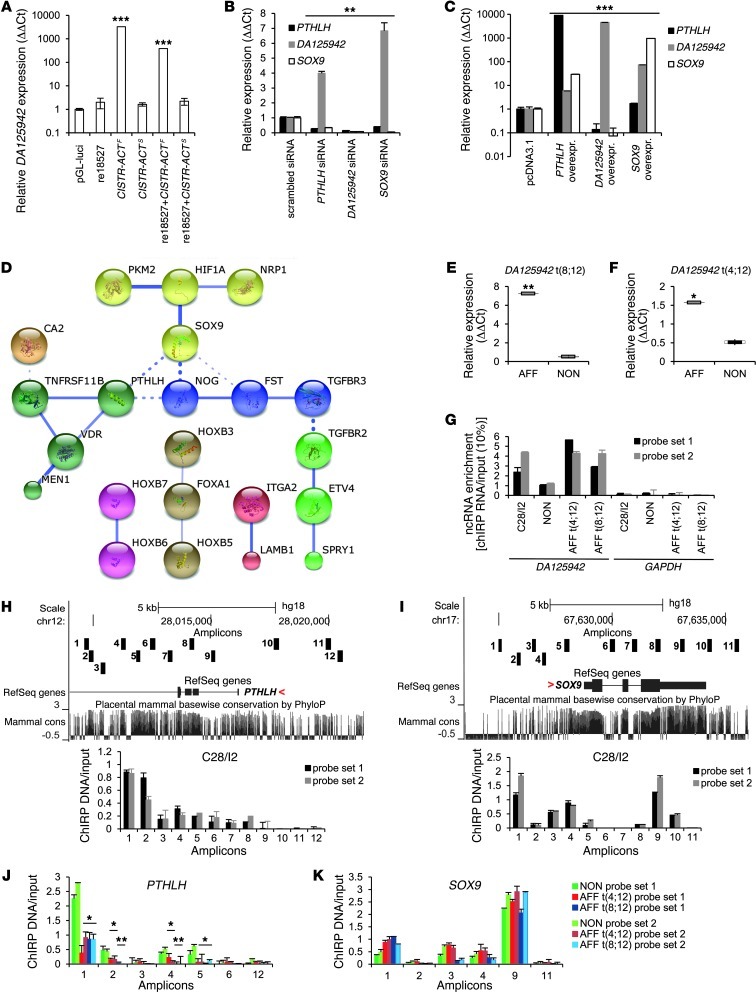
Interestingly, we found an expressed sequence tag, DA125942, with 2 exons overlapping with CISTR-ACT on 12q in the UCSC Genome Browser. DA125942 represented a nonpolyadenylated lncRNA of 581 bp (Supplemental Figure 4). Rapid amplification of cDNA ends (RACE) experiments confirmed the full-length transcript in C28/I2 cells. To determine whether DA125942 transcription was independent of an external promoter and regulator, we constructed a vector lacking regulatory sequences (pGL-luci) and added re18527, CISTR-ACTS, or CISTR-ACTF alone or in combination. Using quantitative RT-PCR, we observed highly upregulated DA125942 in C28/I2 cells transfected with CISTR-ACTF (Figure 4A). This result was suggestive of 2 concepts: first, CISTR-ACTF harbors an internal promoter driving the transcription; and second, mature DA125942 results from splicing. Kim and colleagues reported similar observations (18). In disrupted CISTR-ACTS plasmids, DA125942 was not expressed, and re18527 negatively influenced DA125942 expression (Figure 4A).
Figure 4. DA125942 transcription and dependent regulation of SOX9, DA125942, and PTHLH.
(A) CISTR-ACTS and CISTR-ACTF, with or without re18527, were cloned in a promoter and regulator lacking plasmid (pGl2-luci). In CISTR-ACTF–transfected C28/I2 cells, DA125942 was highly expressed. re18527 blocked the transcription; CISTR-ACTS produced no transcription. (B) SOX9 expression was knocked down relative to the scrambled siRNA control. Depleted PTHLH or DA125942 downregulated SOX9. siRNA-mediated depletion of PTHLH and SOX9 upregulated DA125942, and PTHLH expression was significantly reduced by PTHLH, DA125942, and SOX9 siRNA (n = 6). (C) Overexpression of PTHLH, SOX9, and DA125942 confirmed the expression-dependent network (n = 4). (D) STRING network. Expression array analysis revealed 22 organ morphogenesis genes (GO:0009887) showing interactions. Line thickness is indicative of physical or functional interaction confidence. Mesenchymal and prechondrogenic genes were differentially regulated. (E and F) Fibroblasts of affected BDE patients (AFF) and 3 nonaffected subjects (NON) were chondrogenically induced. DA125942 was upregulated in patients of both BDE families (n = 3). (G) RNA-ChIRP on C28/I2, nonaffected subject, and BDE patient chromatin. DA125942 was retrieved in contrast to GAPDH. (H and I) ChIRP on eluted C28/I2 DNA detected DA125942 binding at the PTHLH and SOX9 loci. Each amplicon was mapped by the UCSC custom tracks (numbered black boxes). (J and K) In t(4;12) and t(8;12), reduced lncRNA occupancy at the PTHLH locus was observed; binding at SOX9 was not affected (n = 2). ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05.
lncRNAs can enhance and specifically influence gene regulation in cis over several Mb (19). We proposed that DA125942 could conduct in cis regulation of PTHLH expression and simultaneously could also be involved in the in trans regulation of SOX9. To test this issue, we used siRNA knockdown in C28/I2 cells to deplete DA125942, PTHLH, and SOX9. Depletion of DA125942 resulted in downregulation of PTHLH and SOX9. Surprisingly, knocking down PTHLH and SOX9 significantly increased DA125942 expression by 4- and 6.8-fold, respectively, suggestive of reverse action of these gene products on lncRNA transcription. Silencing of SOX9 led to significantly reduced PTHLH expression, and vice versa (Figure 4B). These results implied an interactive influence of DA125942, PTHLH, and SOX9 on transcript levels. Overexpression of PTHLH or SOX9 led to upregulation of DA125942, SOX9, or PTHLH. DA125942 overexpression repressed the expression of the coding genes (Figure 4C). These data confirmed the expression dependency of the network.
The HOXC cluster is localized 187 kb downstream of DA125942, and several lncRNAs are encoded by, and interact with, HOX loci (32, 33). We tested whether DA125942 regulated HOXC or other genes either in cis or in trans by 3′ expression profiling. We overexpressed and depleted DA125942 in C28/I2 cells. The combined ANOVA resulted in 1,842 differentially expressed probes (5% false discovery rate [FDR]; Supplemental Table 4). To distinguish up- and downregulation patterns, k-means clustering was performed (Supplemental Figure 6A). Chromatin-remodeling genes, i.e., protein arginine methyltransferase 1 (PRMT1), high mobility group nucleosomal binding domain 2 (HMGN2), polycomb group ring-finger 6 (PCGF6), and enhancer of zeste homolog 2 (EZH2), and several transcription factors were differentially expressed in clusters 1, 3, 4, and 5 (Supplemental Figure 6B). In cluster 2, 33 genes had annotations linking them to organ morphogenesis (GO:0009887; P = 7.51 × 10–7); 11 genes as singletons were excluded in STRING network (Figure 4D). In addition to PTHLH and SOX9, we found essential morphogenesis genes for early limb patterning and chondrogenesis. Hypoxia inducible factor 1 (HIF1A) regulates SOX9 (34). Noggin (NOG) and follistatin (FST) regulate outgrowth and chondrogenesis in the chicken limb bud (35, 36). ETS variant 4 (ETV4) is involved in limb outgrowth and in the anterior-posterior patterning of the autopode (37). The HOXB genes HOXB3, HOXB5–HOXB7, and HOXB8 (singleton; data not shown) and the HOXC genes code for transcription factors involved in diverse morphogenetic processes, including limb formation (38, 39). None of the HOXC genes was differentially expressed. Osteoprotegerin (TNFRSF11B) and the vitamin D receptor (VDR) maintain mineral bone density in osteoblasts (40). The HOXB cluster, ETV4, NOG, and SOX9 were all located on chromosome 17, indicative of several DA125942-mediated gene regulations.
DA125942 in BDE patients and its binding at the PTHLH and SOX9 loci.
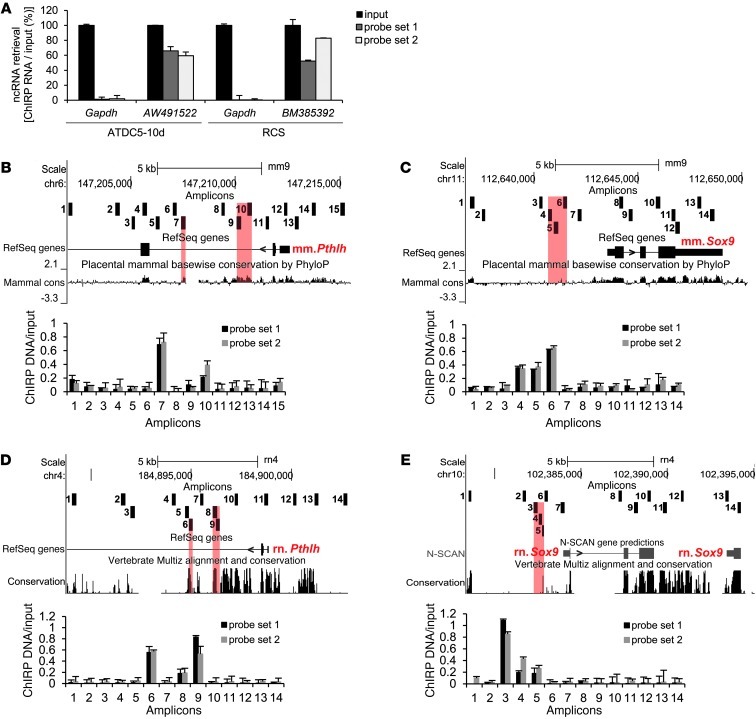
We investigated whether the 2 independent translocations on 12p influenced, next to the PTHLH downregulation, the distant DA125942 and SOX9 expressions in BDE patients. Significant DA125942 upregulation in chondrogenically differentiated fibroblasts of BDE patients with t(8;12) or t(4;12) was found (Figure 4, E and F). SOX9 was marginally, but not significantly, downregulated (P = 0.059; Supplemental Figure 7A). Next, we addressed whether DA125942 occupied the PTHLH and SOX9 loci and whether the interaction was altered in the translocations t(8;12) and t(4;12). We used the recently described chromatin isolation by RNA purification (ChIRP) method with split probe sets 1 and 2, tiling DA125942 (∼80% tiled) on chromatin of C28/I2 and chondrogenically differentiated BDE fibroblasts (41). For qPCR, we eluted RNA to measure lncRNA enrichment and DNA to detect DA125942 occupancy at the PTHLH or SOX9 loci; approximately 12 kb of each gene was analyzed. A probe set against lacZ, the GAPDH and CISTR-ACT loci, and a healthy proband served as controls; nonprecipitated RNA or chromatin was the input control. DA125942 was 1- to 5.6-fold enriched compared with the input. As expected, in chromatin from BDE patients, more lncRNA was retrieved; GAPDH and lacZ were not detected (Figure 4G). Since the lncRNA occupancy at genomic loci is not well understood, we analyzed low and highly conserved, intronic, exonic, and promoter sequences especially in regions of ENCODE-annotated TFBSs. We detected DA125942 binding in the eluted C28/I2 DNA at the PTHLH and SOX9 loci (Figure 4, H and I). In qPCR of the restricted chromatin from BDE patients, we analyzed the highest and lowest enriched positions that we had found in C28/I2 cells. Compared with the healthy proband, binding of DA125942 at the PTHLH locus was significantly reduced in the BDE patients’ chromatin (Figure 4J). Binding at the SOX9 locus was not significantly altered (Figure 4K); the GAPDH or DA125942 loci were not detected (Supplemental Figure 7, B and C).
CISTR-ACT is an evolutionarily conserved mechanism.
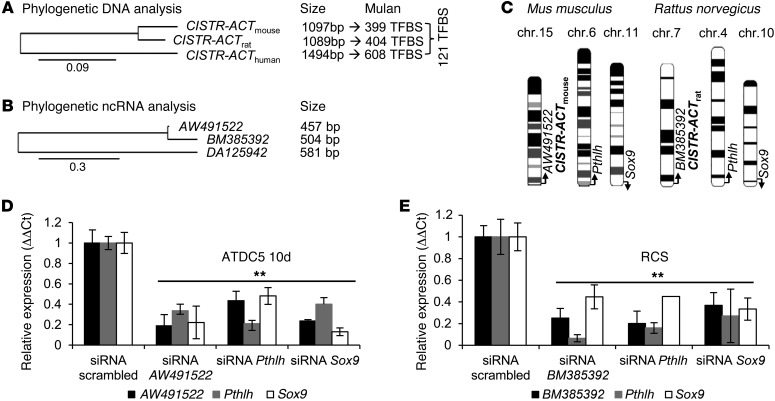
The homologous regions to the human CISTR-ACT in mouse and in rat were detected on the basis of the DNA sequence, including 1-exon lncRNA transcripts in both species (mouse, AW491522; rat, BM385392). By phylogenetic and comparative analysis of TFBSs by Mulan (42, 43), we found that 121 multiconserved TFBSs overlapped in humans, mice, and rats. The lncRNA sequences were evolutionarily more divergent (Figure 5, A and B). In contrast to the human genome, the lncRNAs AW491522 and BM385392 were not located on the chromosome encoding Pthlh (Figure 5C), raising the possibility of solely in trans–acting regulation. The approved mouse ATDC5 cells and rat chondrocytes (RCSs) were used to study chondrogenesis (44, 45). ATDC5 cells were chondrogenically differentiated for 10 days to induce major chondrogenic markers (Supplemental Figure 8, A and B). We again used siRNA transfections to knock down Pthlh, Sox9, and each lncRNA to investigate whether there is an expression-associated mechanism corresponding with that found in humans. AW491522 or BM385392 knockdown led to Pthlh and Sox9 downregulation. In contrast to the observations in human C28/I2 cells, Pthlh or Sox9 depletion caused lncRNA downregulation in ATDC5 cells and RCSs (Figure 5, D and E), indicative of an evolutionarily more complex gene regulation in humans. By ChIRP experiments with 2 probe sets targeting the homologous regions of the rodent lncRNAs (∼80% tiled), we retrieved approximately 60% (Figure 6A). In qPCRs on eluted DNA, homologous regions at the rodent Pthlh or Sox9 loci were occupied by either AW491522 or BM385392 (Figure 6, B–E). The regions corresponded to the human regions occupied by DA125942; enrichment of the lncRNA loci and Gapdh was not detected (Supplemental Figure 8, C–F). Further investigation is required to determine whether the exposed CISTR-ACT locations on the outer chromosomal q arms support the trans regulations. Overall, these data indicate that the lncRNAs act in a contact-based manner and that the network is conserved among the mammals studied herein.
Figure 5. CISTR-ACT in rodents.
(A and B) Phylogenetic analysis of the CISTR-ACT DNA and lncRNA sequences. 121 conserved TFBSs were bioinformatically predicted. The differences of the scales indicate that the lncRNAs are evolutionarily more divergent than the conserved DNA sequences. (C) Genomic arrangement of murine and rat Pthlh, Sox9, and CISTR-ACT–encoded lncRNAs AW491522 and BM385392, respectively, suggested an in trans network. (D) siRNA transfections of mouse ATDC5 (10 days differentiated) chondrocytes. Depletion of the mouse lncRNA AW491522 caused Pthlh or Sox9 downregulation. Knockdown of either Pthlh or Sox9 depleted the lncRNA (n = 4). (E) siRNA assays in RCSs. An analogous expression-dependent network was observed compared with the ATDC5 results (n = 5).**P ≤ 0.01.
Figure 6. ChIRP on ATDC5 and RCS chromatin.
(A) AW491522 and BM385392 retrieval. Mouse or rat Gapdh was not precipitated. (B and C) ChIRP on eluted DNA detected AW491522 binding at the murine Pthlh (B) and Sox9 (C) loci. Each amplicon was mapped by the UCSC custom tracks (numbered black boxes). (D and E) BM385392 binding at the rat Pthlh (D) and Sox9 (E) loci. The lncRNA’s occupancy was observed at homologous regions in mouse and rat (red shading). Annotated mammalian conservation is shown; genomic positions correspond to mouse assembly mm9 and rat assembly rn4 (n = 2).
CISTR-ACT shows mesenchymal and prechondrogenic involvement in transgenic embryos.
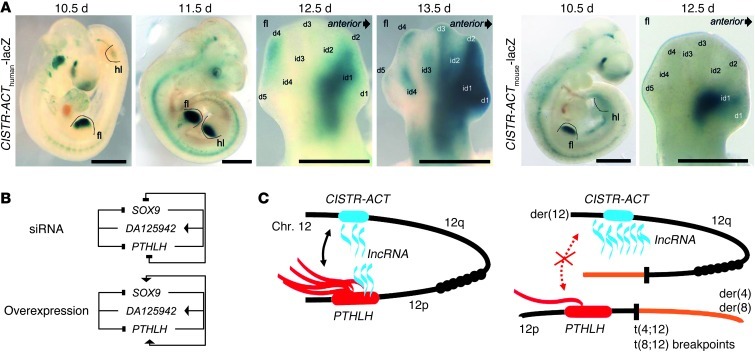
To investigate whether the human and mouse CISTR-ACTs act as classical enhancers in the developing limb buds, we generated transgenic embryos using pronuclei microinjection. The CISTR-ACTs were cloned in front of a lacZ reporter. All stages analyzed showed consistent lacZ expression patterns during limb development. Human transgene-driven lacZ expression was detected in the distal limb bud mesenchyme at E10.5 and E11.5, with a recognizable shift toward the anterior part (Figure 7A). At E12.5, lacZ was expressed overlapping with condensations of digits 1, 2 and 4 and in the first and fourth interdigital mesenchyme (Figure 7A). At E13.5, the strongest expression was maintained overlapping with the condensations of digits 1 and 2 and the first interdigit. Weaker staining was visible overlapping the condensations of digit 3 and the distal tip of digit 4 as well as in interdigits 2, 3, and 4. Thus, lacZ expression overlapped with domains known to be involved in skeletal patterning, as well as those undergoing and/or regulating chondrogenesis (46). Mouse transgene-driven lacZ expression analyzed at E10.5 and E12.5 showed a strong overlap with the staining of the human counterparts in both cases. At E12.5, staining from the mouse transgene showed restriction to the first interdigit and regions overlapping condensations of digits 1 and 2. The domains, in part, correlated with previous documentation of the early limb patterning and chondrogenesis genes identified in our expression profiling, including Hoxb8, Etv4, Pthlh, and Sox9 (37, 47–49).
Figure 7. Transgenic embryos expressing the human and mouse CISTR-ACT–lacZ constructs.
(A) E10.5, E11.5, E12.5, and E13.5 embryos showed consistent lacZ expression patterns during limb bud development. fl, fore limb; hl, hind limb; d, digit; id, interdigit. Scale bars: 1 mm. (B) Results of siRNA and overexpression experiments elucidated a reciprocal expression-associated network. T-bars denote downregulation of expression; arrows denote upregulating effects. (C) Scheme of the cis-regulatory network between PTHLH on chromosome 12p and the CISTR-ACT locus with its lncRNA DA125942 on 12q. Translocations altered the chromosome 12 territory (right), leading to dysregulation of PTHLH and the lncRNA. Orange denotes translocated chromosomal arms.
These results showed that the translocation-mediated disruption of the PTHLH–CISTR-ACT chromosomal association affected the chromosomes’ feedback control and caused PTHLH and DA125942 dysregulation. The findings in the BDE patients were analogous to those from the in vitro and in vivo studies described herein and expand our knowledge about remote-controlled gene and lncRNA regulation in altered chromatin architecture (Figure 7, B and C).
Discussion
Our results elucidate how the translocation-mediated disruption of the PTHLH–CISTR-ACT association caused disease and shed light on the complexity of a molecular pathogenic mechanism that we believe to be novel. These basic chromatin-related epigenetic findings could be of great translational value, especially in other diseases with chromosomal rearrangements. We demonstrated consistent results in patient material and in both in vitro and in vivo models. We documented that chromatin loops involving conserved DNA elements and lncRNA existed for intra- and inter-chromosomal communication, resulting in the regulation of gene and lncRNA expression. We elucidated CISTR-ACT to be a chondrogenic regulator, encoding lncRNA.
The human CISTR-ACT pinpointed SOX9 in trans on chromosome 17. We observed spatial nuclear proximity of the cis elements with distances of 9.48 and 24.43 Mb to PTHLH with colocalization-FISH. Proximal CRE promoter signals occurred. These temporary interactions happened in a limited number of cells, which indicates that adequate target gene expression and maintenance of tissue specificity were effectively fulfilled. In studies of a human ectopic β-globin locus control region (LCR), similar observations were made. Rare ultra-long range regulations and endogenous trans β-globin activation happened only in “jackpot” cells with enriched interchromosomal interaction (50). Amano and colleagues studied murine limbs and observed that sonic hedgehog (Shh) cis regulation was only active in some fractions within the area of Shh expression, but not in others. These findings confirm that chromatin looping is also tissue and developmental stage specific (51). Based on our observations using FISH and 6C, we suggest that tissue-specific fine regulation mediated by chromatin loops occurs infrequently and unpredictably.
We characterized the function of the CISTR-ACT–encoded lncRNAs in siRNA and overexpression experiments. We recapitulated the expression-associated network in 3 species. lncRNA silencing led to PTHLH or SOX9 repressions in humans and rodents, whereas their altered expression coincided with lncRNA dysregulation. Some lncRNAs have enhancer-like functions and are associated with gene expression (19). In contrast to this prior report, we here present evidence for a regulatory feedback expression network. Coding genes could regulate lncRNAs, and vice versa (Figure 7B). lncRNAs may be necessary to establish transcription by recruiting secondary mediators, as previously shown for the epigenetic state regulation of rRNA genes (52). In ChIRP experiments, we documented that the lncRNAs bound at the PTHLH and SOX9 loci. This finding indicates that the lncRNAs act directly on the coding genes’ loci. The translocation-altered chromatin architecture seemed to suppress the looping of CISTR-ACT necessary to find and adequately regulate PTHLH by its lncRNA or vice versa. PTHLH has a nonclassical nuclear localization signal and can bind RNA in the nucleus (53, 54).
Wang and colleagues observed that the long intergenic ncRNA (lincRNA) HOTTIP activates several HOXA genes by proximity and bound protein mediators, thereby maintaining active chromatin. Ectopic HOTTIP did not regulate HOXA genes in their study and generated a dominant-negative effect (33). Supported by their findings, we believe that regular chromosomal contacts bring the CISTR-ACT DNA locus in proximity of its target genes to bring down its lncRNA as an intermediate for targeting the regulated coding genes. Hence, the translocation-mediated altered chromosomal architecture disrupted the chromatin looping and the ability of CISTR-ACT’s lncRNA to find PTHLH. Moreover, DA125942 overexpression led to PTHLH and SOX9 repression. The lncRNA’s overexpression may lead to the binding of proteins that can generate a dominant-negative effect, as the repressed PTHLH leading to autosomal-dominant BDE.
lincRNAs interact with chromatin-remodeling complexes in a well-documented manner for HOTAIR at the HOXC gene cluster and XIST for the X chromosome inactivation (32, 55, 56). Chromatin-remodeling genes and ncRNAs determine tissue-specific epigenetic states and gene expression (57). In studies of HOTAIR or XIST ncRNA, others have proposed that lncRNAs mediate genetic traffic to establish specific epigenetic marks, resulting in guided gene expression (58). The differentially expressed genes suggest that DA125942 has several functions. The preparation of transcriptionally active euchromatin through the lncRNA might go hand in hand with specific morphogene regulation, mediated by the lncRNA and by DNA- or RNA-bound factors. The underlying highly conserved CISTR-ACT DNA sequences, with the predicted chondrogenic TFBSs, showed tissue-specific regulation in the transgenic embryos. The CISTR-ACT–driven lacZ expression domains partially overlapped with the expression patterns of Hoxb8, Etv4, Pthlh, and Sox9. These morphogenesis genes were also differentially expressed in expression profiling. Moreover, the domains anatomically covered the most apparent human BDE regions.
Translocation-mediated chromosome 12 disruption led to the dysregulation of PTHLH and DA125942. The zinc-finger protein CTCF is the best-described regulator mediating either intra- or interchromosomal genome organization (59, 60). Aberrant genomic architecture results in disorganization and failed recruitment of secondary mediators to genomic locations. Thus, the tissue-specific feedback expression of PTHLH and DA125942 does not occur. We suggest that SOX9 and PTHLH share common sites of transcription and belong to a chondrogenic transcription factory (61, 62). As far as we know from studying chromosomal aberration syndromes and intact chromosomes, certain chromosomal territories must be maintained to ensure proper gene regulation (63, 64). In a 3D map of human fibroblasts, the chromosome 12 and 17 territories are adjacent, although unambiguous deterministic chromosomal neighborhoods are not observed (65). We hypothesize that the trans-mediated regulation of SOX9 can occur because the major der(12)s carrying CISTR-ACT remain in their chromosomal territory. CISTR-ACT is still able to communicate with SOX9 on 17q, although the PTHLH and lncRNA dysregulations marginally affect SOX9 expression. PTHLH on 12p appears to be translocated to the chromosomal territories of chromosomes 4 and 8, respectively. Access to the CISTR-ACT is prevented by the translocation (Figure 7C).
Taken together, these observations broaden our awareness of regulatory distances across the centromere, further elucidate the incidence of trans-regulatory DNA-DNA and RNA-lncRNA communication, and suggest that molecular dysfunctions that result in clinical phenotypes are highly complex. We provided genetic evidence that chromosomal communicators work synergistically at both the DNA and RNA levels in a combinatorial and responsive fashion. The reciprocal fine regulation of gene and lncRNA expression is a systems biological process, accounting for effective development or tissue perpetuation. We believe that nucleus-based networks probably serve to maintain temporary, consecutive, and tissue-specific expression of certain gene products. The recruitment mechanism supporting the locus-specific contact must be investigated further. We argue that biophysical and biochemical processes work together stochastically and dynamically. Chromosomes own their territories; although mitotic and meiotic conformational changes occur, they are not randomly organized. Nuclear organization is evolutionarily conserved and directed (64, 66). Brownian motion, directed biochemical mechanisms, and effects that can be described using chaos theory could regulate the diverse interactions (67). We propose that determination meets variability.
Methods
Patients.
After obtaining written informed consent, we studied 2 affected (III:1 and III:4) and 2 nonaffected (III:3 and IV:1) individuals. Skin fibroblasts from 1 affected individual (III:4) were obtained.
Cell culture.
Fibroblasts were cultured in MEM 199, C28/I2 and ATDC5 cells were cultured in DMEM/F12 at a 1:1 ratio (68), and RCSs were cultured in DMEM; all were supplemented with 10% FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin. The chondrogenic differentiation of fibroblasts was done as previously reported (4); ITS-X (Life Technologies) induced ATDC5 chondrogenic differentiation.
Metaphase FISH and colocalization-FISH.
Metaphase FISH analysis was performed using standard protocols. BACs and fosmids were ordered at RZPD ( www.imagenes-bio.de). 2D colocalization-FISH was performed as previously reported (69). The Olympus-BX61 microscope with U-TV1X2 and PlanApo 60×/1.4 oil lens in combination with the ASI CCD-camera 1300QDS and FISHView software (version 6.0.0.18) were used for signal detection and measurements. Measurements were made independently by 2 different investigators.
6C and ChIRP.
See Supplemental Methods for the detailed 6C protocol. ChIRP was done according to a previously published protocol (41); see Supplemental Table 5 for probes.
Reporter assay, siRNA knockdown, and expression plasmids.
5 × 104 cells were seeded 12–24 hours prior to transfection. Equal molarities in transiently transfected C28/I2 cells compensated for the diverse plasmid sizes. For reporter assays, 75 fmol each of the CRE promoter luciferase constructs were transfected with Fugene HD according to the manufacturer’s recommendations (Roche). The SOX9 promoter sequence was on chromosome 17: 67,626,313–67,629,147 bp (UCSC assembly hg18). The vector backbone was pGL2-basic (Promega); 12.5 ng pRL-TK (Promega) was added to control transfection efficiency. Lysates were analyzed using the Dual-Glo Assay (Promega). 100 nM siRNA were transfected with Dharmafect (Dharmacon) for 48 hours in C28/I2 and ATDC5 cells, 120 nM siRNA were used for RCS transfections. Scrambled, GAPDH, SOX9, and PTHLH siRNAs were validated (Ambion). Different custom-designed siRNAs for the lncRNAs were obtained. For visual simplicity, results using only 1 siRNA are shown.
DA125942 siRNAs were as follows: siRNA-1 sense, GCACAUGACCACAUGGAAATT; siRNA-1 antisense, UUUCCAUGUGGUCAUGUGCGA; siRNA-2 sense, GUUUCUCUCAGCAAAUUCATT; siRNA-2 antisense, UGAAUUUGCUGAGAGAAACTG; siRNA-3 sense, CAAGGACACUGAAAAAGCUTT; siRNA-3 antisense, AGCUUUUUCAGUGUCCUUGAG. AW491522 siRNAs were as follows: siRNA-1 sense, AGGCCCUGAUGCUAGUGAATT; siRNA-1 antisense, UUCACUAGCAUCAGGGCCUTG; siRNA-2 sense, GCACGCAUCUCUUCCACCATT; siRNA-2 antisense, UGGUAAGAGAUGCGUGCGG. BM385392 siRNAs were as follows: siRNA-1 sense, GAGAUCUGGUCAGAGCAAATT; siRNA-1 antisense, UUUGCUCUGACCAGAUCUCTA; siRNA-2 sense, CAAUACUUCUGGACUCUCUTT; siRNA-2 antisense, AGAGAGUCCAGAAGUAUUGCA. Negative control was scrambled siRNA. SOX9, DA125942, and PTHLH were significantly downregulated (>75%). The siRNA for GAPDH was a reference for transfection efficiency and downregulation. Overexpression transfections were done as previously described by Fugene X-tremeGENE HP (Roche). PTHLH (SC307833) and SOX9 (SC321884) expression plasmids were obtained from OriGene.
Bioinformatics and 3′ expression array analysis.
Genomic sequences were analyzed by NCBI BLAST, UCSC BLAT, repeat masking at GIRI Institute, and in the UCSC Genome Browser (March 2006; NCBI36/hg18) (70, 71). rVISTA version 2.0 and Transfac Professional version 10.2 were used to determine mammalian conserved TFBSs (72, 73). Functional experiments were done at least 3 times. Number of experiments (n) are shown in figure legends. Significance was determined by Student’s t test. Signal detections in colocalization-FISH were statistically analyzed by Mann-Whitney rank-sum test; data represent mean ± SD. Expression profiling was done on ht12 arrays (Illumina) in biological triplicates, followed by ANOVA, multiple testing (FDR 5%), cluster, GProfiler enrichment, and STRING analysis. The microarray dataset was deposited in GEO (accession no. GSE39896; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39896).
On the BeadStudio 1.0.2.20706 Platform array, data were quantile normalized at the probe level without background correction, and consistently low-expressed transcripts (minimum detection P > 0.05) were rejected. Data were log2 transformed after an offset addition (16) and analyzed on significant expression differences among the 4 groups (siRNA-depleted and overexpressed DA125942 C28/I2 RNA, controls with empty expression vector, and scrambled siRNA) in triplicate using ANOVA followed by a FDR (74) multiple testing correction. Probes that underwent 5% FDR were k means clustered with average Euclidean distance function and k = 5, according to a local minimum in the Davies-Bouldin k estimation (75). Clusters 1, 2, and 4 were selected according to those outstanding expression statuses in DA125942 silencing and further investigated by functional enrichment and interaction. Enrichment analysis was performed using G:Profiler (76), with a simulation-based analytical threshold for significance estimation. Physical and functional interactions between proteins were determined using the String platform (77) and a medium confidence score of 0.4. Resulting singletons were rejected before network visualization.
Transgenic mice.
Human (chromosome 12; 52,430,503–52,431,992 bp; assembly hg18) and mouse (chromosome 15; 102,576,412–102,577,508 bp; assembly mm9) CISTR-ACT were cloned in the Sfi-Hsp68LacZ reporter construct (78). Gel-extracted linearized DNA was injected in fertilized mouse oocytes according to standard procedures. At each time point, 25–42 embryos were analyzed by whole-mount lacZ staining.
Statistics.
Data are presented as mean ± SD unless otherwise indicated. Differences between experimental groups were analyzed by 2-tailed Student’s t test unless otherwise specified. A P value less than 0.05 was considered statistically significant.
Study approval.
The local ethics committee (Charité-Universitätsmedizin Berlin, Ethikausschuss 1; EA3/009/07) approved the study. All subjects provided written informed consent. The local regulatory animal authority (Landesamt für Gesundheit und Soziales Berlin — LAGeSo; 0143/07) approved the pronuclear injections and oviduct transfers in mice.
Supplementary Material
Acknowledgments
We thank all family members for their cooperation. We thank I. Hollfinger, E. Bartels-Klein, Y. Wefeld-Neuenfeld, A. Mühl, A. Käbner-Frensel, and E.M. Gerlach for technical assistance. The Deutsche Forschungsgemeinschaft (DFG; grants BA1773/5-1, MA5028/1-2, and RU1508/1-1) supported the study. P.G. Maass was supported by a Max-Delbrück-Center and a DFG scholarship. This study made use of data generated by the DECIPHER Consortium. A full list of centers that contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. The Wellcome Trust provided funding.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(11):3990–4002. doi:10.1172/JCI65508.
See the related Commentary beginning on page 3837.
References
- 1.Wijchers PJ, de Laat W. Genome organization influences partner selection for chromosomal rearrangements. Trends Genet. 2011;27(2):63–71. doi: 10.1016/j.tig.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76(1):8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell J. The Treasury Of Human Inheritance. 1951. Brachydactyly and symphalangism. In: Penrose LS, ed. pp. 1–31. Cambridge, United Kingdom: University of Cambridge Press; [Google Scholar]
- 4.Maass PG, et al. A cis-regulatory site downregulates PTHLH in translocation t(8;12)(q13;p11.2) and leads to Brachydactyly Type E. Hum Mol Genet. 2010;19(5):848–860. doi: 10.1093/hmg/ddp553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klopocki E, et al. Deletion and point mutations of PTHLH cause brachydactyly type E. Am J Hum Genet. 2010;86(3):434–439. doi: 10.1016/j.ajhg.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273(5275):613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 7.Stricker S, Mundlos S. Mechanisms of digit formation: Human malformation syndromes tell the story. Dev Dyn. 2011;240(5):990–1004. doi: 10.1002/dvdy.22565. [DOI] [PubMed] [Google Scholar]
- 8.Velagaleti GV, et al. Position effects due to chromosome breakpoints that map approximately 900 Kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. Am J Hum Genet. 2005;76(4):652–662. doi: 10.1086/429252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bien-Willner GA, Stankiewicz P, Lupski JR. SOX9cre1, a cis-acting regulatory element located 1.1 Mb upstream of SOX9, mediates its enhancement through the SHH pathway. Hum Mol Genet. 2007;16(10):1143–1156. doi: 10.1093/hmg/ddm061. [DOI] [PubMed] [Google Scholar]
- 10.Wagner T, et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79(6):1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 11.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20(17):R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbasi AA, et al. Human GLI3 intragenic conserved non-coding sequences are tissue-specific enhancers. PLoS One. 2007;2(4):e366. doi: 10.1371/journal.pone.0000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loots GG, et al. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288(5463):136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 15.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12(14):1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 16.Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132(4):797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- 17.Vavouri T, McEwen GK, Woolfe A, Gilks WR, Elgar G. Defining a genomic radius for long-range enhancer action: duplicated conserved non-coding elements hold the key. Trends Genet. 2006;22(1):5–10. doi: 10.1016/j.tig.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28(5):503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144(3):327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin L, Chang HY. Uncovering the role of genomic “dark matter” in human disease. J Clin Invest. 2012;122(5):1589–1595. doi: 10.1172/JCI60020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson D, et al. Missense mutations in the homeodomain of HOXD13 are associated with brachydactyly types D and E. Am J Hum Genet. 2003;72(4):984–997. doi: 10.1086/374721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 26.van Steensel B, Dekker J. Genomics tools for unraveling chromosome architecture. Nat Biotechnol. 2010;28(10):1089–1095. doi: 10.1038/nbt.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 28.Ng LJ, et al. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol. 1997;183(1):108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- 29.Gondor A, Rougier C, Ohlsson R. High-resolution circular chromosome conformation capture assay. Nat Protoc. 2008;3(2):303–313. doi: 10.1038/nprot.2007.540. [DOI] [PubMed] [Google Scholar]
- 30.Pekowska A, Benoukraf T, Ferrier P, Spicuglia S. A unique H3K4me2 profile marks tissue-specific gene regulation. Genome Res. 2010;20(11):1493–1502. doi: 10.1101/gr.109389.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldring MB, Tsuchimochi K, Ijiri K. The control of chondrogenesis. J Cell Biochem. 2006;97(1):33–44. doi: 10.1002/jcb.20652. [DOI] [PubMed] [Google Scholar]
- 32.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amarilio R, Viukov SV, Sharir A, Eshkar-Oren I, Johnson RS, Zelzer E. HIF1alpha regulation of Sox9 is necessary to maintain differentiation of hypoxic prechondrogenic cells during early skeletogenesis. Development. 2007;134(21):3917–3928. doi: 10.1242/dev.008441. [DOI] [PubMed] [Google Scholar]
- 35.Merino R, Ganan Y, Macias D, Economides AN, Sampath KT, Hurle JM. Morphogenesis of digits in the avian limb is controlled by FGFs, TGFbetas, and noggin through BMP signaling. Dev Biol. 1998;200(1):35–45. doi: 10.1006/dbio.1998.8946. [DOI] [PubMed] [Google Scholar]
- 36.Merino R, Rodriguez-Leon J, Macias D, Ganan Y, Economides AN, Hurle JM. The BMP antagonist Gremlin regulates outgrowth, chondrogenesis and programmed cell death in the developing limb. Development. 1999;126(23):5515–5522. doi: 10.1242/dev.126.23.5515. [DOI] [PubMed] [Google Scholar]
- 37.Mao J, McGlinn E, Huang P, Tabin CJ, McMahon AP. Fgf-dependent Etv4/5 activity is required for posterior restriction of Sonic Hedgehog and promoting outgrowth of the vertebrate limb. Dev Cell. 2009;16(4):600–606. doi: 10.1016/j.devcel.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson CE, et al. Analysis of Hox gene expression in the chick limb bud. Development. 1996;122(5):1449–1466. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
- 39.Lu HC, Revelli JP, Goering L, Thaller C, Eichele G. Retinoid signaling is required for the establishment of a ZPA and for the expression of Hoxb-8, a mediator of ZPA formation. Development. 1997;124(9):1643–1651. doi: 10.1242/dev.124.9.1643. [DOI] [PubMed] [Google Scholar]
- 40.Mencej-Bedrac S, et al. The combinations of polymorphisms in vitamin D receptor, osteoprotegerin and tumour necrosis factor superfamily member 11 genes are associated with bone mineral density. J Mol Endocrinol. 2009;42(3):239–247. doi: 10.1677/JME-08-0108. [DOI] [PubMed] [Google Scholar]
- 41.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dereeper A, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ovcharenko I, et al. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 2005;15(1):184–194. doi: 10.1101/gr.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mukhopadhyay K, Lefebvre V, Zhou G, Garofalo S, Kimura JH, de Crombrugghe B. Use of a new rat chondrosarcoma cell line to delineate a 119-base pair chondrocyte-specific enhancer element and to define active promoter segments in the mouse pro-alpha 1(II) collagen gene. J Biol Chem. 1995;270(46):27711–27719. doi: 10.1074/jbc.270.46.27711. [DOI] [PubMed] [Google Scholar]
- 45.Shukunami C, Ishizeki K, Atsumi T, Ohta Y, Suzuki F, Hiraki Y. Cellular hypertrophy and calcification of embryonal carcinoma-derived chondrogenic cell line ATDC5 in vitro. J Bone Miner Res. 1997;12(8):1174–1188. doi: 10.1359/jbmr.1997.12.8.1174. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki T, Hasso SM, Fallon JF. Unique SMAD1/5/8 activity at the phalanx-forming region determines digit identity. Proc Natl Acad Sci U S A. 2008;105(11):4185–4190. doi: 10.1073/pnas.0707899105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Charite J, de Graaff W, Shen S, Deschamps J. Ectopic expression of Hoxb-8 causes duplication of the ZPA in the forelimb and homeotic transformation of axial structures. Cell. 1994;78(4):589–601. doi: 10.1016/0092-8674(94)90524-X. [DOI] [PubMed] [Google Scholar]
- 48.Niedermaier M, et al. An inversion involving the mouse Shh locus results in brachydactyly through dysregulation of Shh expression. J Clin Invest. 2005;115(4):900–909. doi: 10.1172/JCI200523675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wright E, et al. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat Genet. 1995;9(1):15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]
- 50.Noordermeer D, et al. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat Cell Biol. 2011;13(8):944–951. doi: 10.1038/ncb2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell. 2009;16(1):47–57. doi: 10.1016/j.devcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 52.Bierhoff H, Schmitz K, Maass F, Ye J, Grummt I. Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harb Symp Quant Biol. 2010;75:357–364. doi: 10.1101/sqb.2010.75.060. [DOI] [PubMed] [Google Scholar]
- 53.Cingolani G, Bednenko J, Gillespie MT, Gerace L. Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol Cell. 2002;10(6):1345–1353. doi: 10.1016/S1097-2765(02)00727-X. [DOI] [PubMed] [Google Scholar]
- 54.Aarts MM, Rix A, Guo J, Bringhurst R, Henderson JE. The nucleolar targeting signal (NTS) of parathyroid hormone related protein mediates endocytosis and nucleolar translocation. J Bone Miner Res. 1999;14(9):1493–1503. doi: 10.1359/jbmr.1999.14.9.1493. [DOI] [PubMed] [Google Scholar]
- 55.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rajasekhar VK, Begemann M. Concise review: roles of polycomb group proteins in development and disease: a stem cell perspective. Stem Cells. 2007;25(10):2498–2510. doi: 10.1634/stemcells.2006-0608. [DOI] [PubMed] [Google Scholar]
- 58.Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr Opin Genet Dev. 2010;20(2):142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128(6):1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137(7):1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Osborne CS, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36(10):1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 62.Xu M, Cook PR. Similar active genes cluster in specialized transcription factories. J Cell Biol. 2008;181(4):615–623. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Osborne CS, et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5(8):e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2(3):a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bolzer A, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3(5):e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lawrence JB, Clemson CM. Gene associations: true romance or chance meeting in a nuclear neighborhood? J Cell Biol. 2008;182(6):1035–1038. doi: 10.1083/jcb.200808121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pollak E, Talkner P. Reaction rate theory: what it was, where is it today, and where is it going? Chaos. 2005;15(2):26116. doi: 10.1063/1.1858782. [DOI] [PubMed] [Google Scholar]
- 68.Finger F, Schorle C, Zien A, Gebhard P, Goldring MB, Aigner T. Molecular phenotyping of human chondrocyte cell lines T/C-28a2, T/C-28a4, and C-28/I2. Arthritis Rheum. 2003;48(12):3395–3403. doi: 10.1002/art.11341. [DOI] [PubMed] [Google Scholar]
- 69.Klink B, Schlingelhof B, Klink M, Stout-Weider K, Patt S, Schrock E. Glioblastomas with oligodendroglial component - common origin of the different histological parts and genetic subclassification. Cell Oncol. 2011;34(3):261–275. doi: 10.1007/s13402-011-0034-8. [DOI] [PubMed] [Google Scholar]
- 70.King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15(8):1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loots GG, Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32(Web Server issue):W217–W221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ovcharenko I, Loots GG, Hardison RC, Miller W, Stubbs L. zPicture: dynamic alignment and visualization tool for analyzing conservation profiles. Genome Res. 2004;14(3):472–477. doi: 10.1101/gr.2129504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125(1–2):279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 75.Davies DL, Bouldin DW. A cluster separation measure. IEEE Trans Pattern Anal Mach Intell. 1979;1(2):224–227. doi: 10.1109/TPAMI.1979.4766909. [DOI] [PubMed] [Google Scholar]
- 76.Reimand J, Arak T, Vilo J. g:Profiler -- a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res. 2011;39(Web server issue):W307–W315. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mortlock DP, Guenther C, Kingsley DM. A general approach for identifying distant regulatory elements applied to the Gdf6 gene. Genome Res. 2003;13(9):2069–2081. doi: 10.1101/gr.1306003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.