Abstract
Overnight low-temperature exposure inhibits photosynthesis in chilling-sensitive species such as tomato (Lycopersicon esculentum) and cucumber by as much as 60%. In an earlier study we showed that one intriguing effect of low temperature on chilling-sensitive plants is to stall the endogenous rhythm controlling transcription of certain nuclear-encoded genes, causing the synthesis of the corresponding transcripts and proteins to be mistimed when the plant is rewarmed. Here we show that the circadian rhythm controlling the activity of sucrose phosphate synthase (SPS) and nitrate reductase (NR), key control points of carbon and nitrogen metabolism in plant cells, is delayed in tomato by chilling treatments. Using specific protein kinase and phosphatase inhibitors, we further demonstrate that the chilling-induced delay in the circadian control of SPS and NR activity is associated with the activity of critical protein phosphatases. The sensitivity of the pattern of SPS activity to specific inhibitors of transcription and translation indicates that there is a chilling-induced delay in SPS phosphorylation status that is caused by an effect of low temperature on the expression of a gene coding for a phosphoprotein phosphatase, perhaps the SPS phosphatase. In contrast, the chilling-induced delay in NR activity does not appear to arise from effects on NR phosphorylation status, but rather from direct effects on NR expression. It is likely that the mistiming in the regulation of SPS and NR, and perhaps other key metabolic enzymes under circadian regulation, underlies the chilling sensitivity of photosynthesis in these plant species.
Most warm-climate plant species are sensitive to brief exposures to low, nonfreezing temperatures. Low-temperature exposure in combination with high irradiance causes rapid, often very severe inhibition of photosynthesis in a broad range of plants, including maize (Baker et al., 1983), cucumber (Peeler and Naylor, 1988), and tomato (Lycopersicon esculentum) (Martin and Ort, 1985). Several elements that contribute to this inhibition have been identified and all may ultimately arise from the photosynthetic production of oxygen radicals (Wise, 1995). An inhibition of electron-transport capacity originating from damage to the reducing side of PSII is well documented (e.g. Powles et al., 1983; Kee et al., 1986; Percival et al., 1987) and, for moderately sensitive species such as maize, may be the major cause of impaired whole-plant photosynthesis after chilling. However, in the most severely chilling-sensitive species, such as domestic tomato, impaired reductive activation of the stromal bisphosphatases appears to be the dominating factor limiting carbon assimilation after chilling in the light (Sassenrath et al., 1990).
Low temperature at night can also cause severe reductions in CO2 fixation that persist on the day after the chill, even after optimal growth temperatures have been restored. Like the inhibition caused by chilling in the light, it is clear that the primary loss of activity is caused by direct impairment at the biochemical level, as opposed to interference with stomatal-mediated leaf gas exchange (Martin et al., 1981). However, in the case of dark chilling, it has not been possible to assign the cause to specific reactions of photosynthesis. Thus, although the inhibition of net photosynthesis by dark chilling in plants such as tomato and cucumber can be quite large, the underlying causes are subtle and are likely to involve disruption of the coordination among the component reactions of photosynthesis rather than direct inhibition of the reactions themselves. An intriguing effect of low temperature on the circadian regulation of transcription (Martino-Catt and Ort, 1992) indicates that the low-temperature-induced inhibition of photosynthesis in tomato may result from the loss of coordination in the expression of critical enzymes controlling photosynthetic metabolism.
In tomato, low-temperature treatment delays the progress of the circadian clock regulating the transcription of certain nuclear-encoded genes, including cab (chlorophyll a/b binding) and rca (Rubisco activase) (Martino-Catt and Ort, 1992). The clock stops for the duration of the chilling treatment and resumes upon rewarming, but the affected rhythms are then out of phase with the actual time of day. Notably, whereas cab and rca gene expression are circadian regulated in chilling-tolerant spinach, the rhythm is not affected in this fashion by low-temperature treatment (Ort et al., 1989). However, cab and rca are exceedingly abundant, stable proteins, and this transitory mistiming in their synthesis in chilling-sensitive plants does not lead to detectable changes in cellular protein level (Cooper and Ort, 1988). Therefore, this mistiming cannot be expected to cause the severe inhibition of net photosynthesis that is observed. Although the chilling-sensitive aspect of circadian regulation is unknown, it is an interesting possibility that low temperature could delay the timing of circadian-regulated activity of certain enzymes, as we observed for circadian-regulated gene transcription (Martino-Catt and Ort, 1992). Such delays in circadian activity rhythms would likely have a negative effect on photosynthetic performance if the affected enzymes were critical control points of cellular carbon or nitrogen metabolism.
SPS (EC 2.4.1.14) is a central enzyme in photosynthetic metabolism because it catalyzes a rate-limiting step in Suc biosynthesis. SPS is subject to multiple levels of regulation, including regulation by the allosteric effectors Glc-6-P and phosphate (Doehlert and Huber, 1984; McMichael et al., 1993) and regulation by protein phosphorylation (Huber et al., 1989). Our 1997 study showed that SPS activity exhibits diurnal and circadian rhythms in tomato, which are the result of corresponding oscillations in SPS protein phosphorylation state (Jones and Ort, 1997). To our knowledge, this was the first report of a circadian rhythm in SPS activity, although an endogenous ultradian rhythm with a period of about 12 h had been reported in soybean (Kerr et al., 1985). Our evidence indicates that the circadian rhythm in tomato SPS phosphorylation state is the result of circadian-regulated transcription of a protein phosphatase, possibly the one that dephosphorylates and thereby activates SPS. Even a transitory mistiming in transcription of this phosphatase gene as a result of low-temperature treatment might potentiate a change in the pattern of SPS activity. Because the capacity to use triose phosphate can limit photosynthesis (Herold, 1980; Sharkey, 1990), it would be anticipated that mistimed SPS activity as a result of low-temperature treatment could contribute significantly to the chilling-induced inhibition of photosynthesis.
NR (EC 1.6.6.1) activity exhibits a circadian rhythm in many plant species (Lillo, 1984; Deng et al., 1990; Cheng et al., 1991; Pilgrim et al., 1993). NR is a highly regulated cytosolic enzyme catalyzing the first and rate-limiting step in the nitrate assimilation pathway, reducing nitrate (NO3−) to nitrite (NO2−). The regulation of NR activity is complex and can involve modulation of the enzyme level through the regulation of synthesis and degradation (Solomonson and Barber, 1990; Hoff et al., 1994; Kaiser and Huber, 1997). However, immediately after a light-to-dark transition, rapid posttranslational modifications of the enzyme are thought to dominate the regulation of NR activity (Kaiser and Spill, 1991; MacKintosh, 1992; Kaiser and Huber, 1994). Inactivation of NR in the dark is initiated by phosphorylation of a specific seryl residue (Douglas et al., 1995; Bachmann et al., 1996; Su et al., 1996) followed by the Mg2+-dependent association of 14-3-3-type inhibitor proteins with phospho-NR (Spill and Kaiser, 1994; Bachmann et al., 1995; Glaab and Kaiser, 1995; MacKintosh et al., 1995; Lillo et al., 1997). The similarities between the regulation of SPS and NR in spinach (Huber et al., 1992a), especially the role of protein dephosphorylation in enzyme activation and similar diurnal activity dynamics, prompted us to investigate whether NR activity in tomato is regulated by a circadian rhythm and, if so, whether this rhythm is driven by changes in protein phosphorylation state, as we have shown for SPS (Jones and Ort, 1997). We further investigated the effect of low-temperature treatments on the timing of NR activity in tomato.
We have shown that, whereas the circadian rhythm in SPS activity in tomato corresponds to oscillations in SPS phosphorylation state, the circadian rhythm in NR activity is primarily the result of oscillations in the amount of enzyme present in the leaves. We present data demonstrating that low temperature delays the circadian rhythms in both SPS and NR activity. Inhibitor treatments were used to investigate the mechanism of the low-temperature shift, and the results indicate that delayed activity involves low-temperature effects on the expression of critical protein phosphatase(s). We believe that the low-temperature inhibition of photosynthesis in chilling-sensitive plants is caused by low-temperature-induced mistiming of the normal diurnal activity pattern of key enzymes, thereby disrupting photosynthetic and cellular metabolism.
MATERIALS AND METHODS
Plant Growth Conditions
Tomato (Lycopersicon esculentum Mill. cv Floramerica) plants were grown from seed in growth chambers under a 14-h (26°C) light/10-h (21°C) dark cycle at 75% RH, as described by Jones and Ort (1997). Plants were fertilized twice weekly with a liquid formula (12-31-14, Plant Marvel Laboratories, Chicago, IL) supplemented with 10 mm KNO3. All samples were taken from young, fully expanded leaves of plants 21 to 28 d after planting.
Low-Temperature Treatments
Potted tomato plants were chilled at 4°C at 100% RH. During chilling the pots were enclosed in an insulated box fitted with a fan, which circulated warm air around the pots. This apparatus maintained the soil temperature at approximately 15°C and completely prevented the plants from wilting during treatment and rewarming.
SPS Assay
In vitro SPS activity was assayed colorimetrically by the anthrone method to monitor the formation of Suc (and Suc-6-P) from Fru-6-P and UDP-Glc (Huber et al., 1992b). Tomato extracts and assay conditions were as described by Jones and Ort (1997).
NR Assay
In vivo NR activity was assayed using the leaf disc method of Nicholas et al. (1976). Weighed leaf punches (approximately 200 mg) were vacuum infiltrated in 10 mL of incubation buffer (0.1 m potassium phosphate [pH 7.5], 0.05 m KNO3, 1% [v/v] propanol). Samples were incubated in a shaking water bath for 30 min at 30°C in the dark. After the incubation, 0.4 mL of the incubation buffer was diluted with water to 4 mL. Two milliliters of 1% sulfanilic acid in 1.5 m HCl was added to the diluted incubation buffer, followed by 2 mL of N-(1-naphthyl)ethylenediamine-HCl (200 mg/L). The samples were then incubated for at least 20 min at room temperature to allow full color development, after which the A540 was recorded.
The measurement of in vitro NR activity using crude tissue extracts followed a protocol that was modified from Kaiser and Brendle-Behnisch (1991). Excised leaf samples were immediately frozen in liquid nitrogen and stored at −80°C until use. The samples were ground under liquid nitrogen and then suspended in extraction buffer (1 g of tissue for each 2 mL of buffer) consisting of 50 mm Mops-NaOH (pH 7.5), 10 mm MgCl2, 1 mm EDTA, 5 mm DTT, and 0.1% Triton X-100. The homogenate was subsequently desalted on Sephadex G-25 spin columns that had been preequilibrated in 50 mm Mops-NaOH (pH 7.5) and 2 mm DTT. The desalted tissue extract (100 μL) was added to 900 μL of reaction solution containing 50 mm Mops-NaOH (pH 7.5), 10 mm KNO3, 0.1 mm NADH, and either 10 mm MgCl2 or 5 mm EDTA. The samples were incubated at 30°C for 3 min and the reaction stopped by the addition of 100 μL of 50 mm zinc acetate. NO2− was quantified by standard colorimetric reagent addition and the A540 was recorded (Lillo, 1983). The blanks were identical to the samples, but were quenched with zinc acetate before the addition of the leaf extract.
NR activity was calculated on a chlorophyll basis. The chlorophyll concentration was assayed from the crude homogenates after an 80% acetone extraction and calculated according to the method of Graan and Ort (1984).
Inhibitor Treatments
Attached tomato leaves were lightly abraded with 400-grit Duralum powder (Electro Minerals Corp., Niagara Falls, NY) and rinsed before application of cordycepin (200–500 μg/mL), cycloheximide (200 μg/mL), staurosporine (100 μm), or okadaic acid (10 μm). All inhibitors were dissolved in 0.5% to 1% Tween 20. Okadaic acid and staurosporine were applied in 100-μL droplets to the attached tomato leaves or the leaves were briefly submerged in the cordycepin or cycloheximide solutions. The incubation times were as described in the figure legends.
RNA Gel-Blot Hybridization
Total RNA (30 μg/lane) prepared from leaf samples was separated by electrophoresis on agarose-formaldehyde gels and transferred to nitrocellulose membranes. The membranes were hybridized overnight at 50°C with a NR probe prepared from a tobacco nia-2 cDNA insert (Vaucheret et al., 1989) isolated from an Escherichia coli plasmid and randomly labeled with dCTP32. The membranes were washed three times for 5 min at 50°C in a solution of 150 mm NaCl, 15 mm sodium citrate, and 0.1% SDS followed by three 20-min washes at 50°C in 30 mm NaCl, 3 mm sodium citrate, and 0.1% SDS. A corresponding ethidium bromide-stained gel was used to monitor the uniformity of total RNA loaded in each lane. The radioactivity bound to the membranes was quantified by 24 h of exposure on a phosphor screen before analysis with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
RESULTS
The Circadian and Diurnal Rhythms of SPS Activity in Tomato Are Delayed by Low-Temperature Treatment
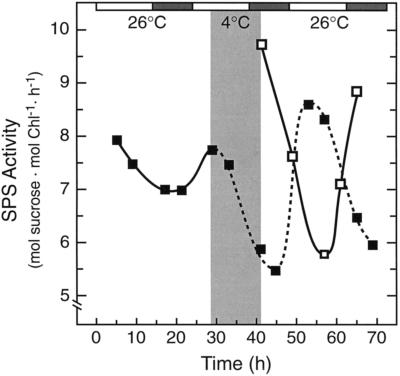
SPS activity oscillates with a circadian rhythm in tomato when plants are maintained under constant light (50 μmol quanta m−2 s−1) and temperature conditions (Jones and Ort, 1997). The rhythm of SPS activity was monitored after a 12-h low-temperature treatment (4°C, 50 μmol quanta m−2 s−1) depicted by the gray area in Figure 1. Samples were taken after rewarming, and the results demonstrate that the low-temperature treatment delayed the circadian rhythm in SPS activity by 12 h, indicating that the endogenous clock controlling SPS activity stopped for the duration of the chill and resumed upon rewarming.
Figure 1.
Low-temperature treatment delays the circadian rhythm controlling SPS activity. Plants were transferred to constant low-light (50 μmol quanta m−2 s−1) and temperature (26°C) conditions. The light and dark bars above the figure represent day and subjective night periods, respectively. A low-temperature treatment (4°C), represented by the shaded area in the main body of the figure, was imposed for 12 h, between h = 29 and h = 41. After rewarming, samples were taken to determine the effect of low temperature on the circadian rhythm in SPS activity. Comparison of the reference circadian rhythm (▪) and the postchilling oscillation (□) demonstrates that the 12-h low-temperature treatment delayed the oscillation in SPS activity by 12 h. Each point represents the mean of two samples, and the experiment was repeated several times with consistent results. Chl, Chlorophyll.
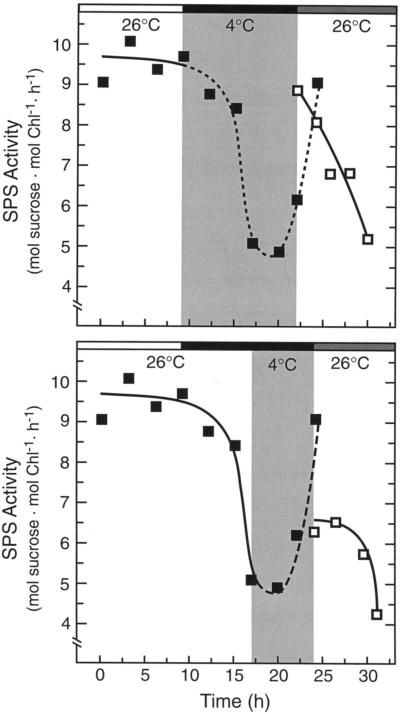
SPS activity also has a pronounced diurnal rhythm in tomato, with high activity during the day and lower activity at night (Jones and Ort, 1997). Figure 2 shows that the SPS diurnal rhythm was also delayed by low-temperature treatment. Dark chilling treatment initiated at midday (h = 9), when SPS activity was high, resulted in the maintenance of high activity upon rewarming, even though the plants were rewarmed at a time of day when SPS activity should be low (h = 22) (Fig. 2, top). Samples taken during the dark recovery after the chilling episode showed that, although SPS activity was initially high, the activity decreased to nighttime levels during the next 8 h (Fig. 2, top).
Figure 2.
The effect of dark chilling treatments initiated at different times in the diurnal cycle on SPS activity in tomato. The white bar above the figure represents the light cycle, the black bar represents the dark cycle, and the gray bar represents the continuation of darkness during the normal light cycle (i.e. subjective day). Top, The dark chilling treatment was initiated during the light cycle at h = 9 and plants were rewarmed at h = 22. Data points were normalized to the diurnal SPS activity profile at h = 24. Bottom, The dark chilling treatment (4°C) was initiated during the night at h = 17, plants were rewarmed (26°C) at h = 24, and samples were then taken in the dark after rewarming (□). Data were normalized to the diurnal activity profile at h = 22. In both experiments, the low-temperature treatment resulted in the maintenance of the prechill SPS activity, showing that low-temperature treatment delayed the normal rhythm in SPS activity. Chl, Chlorophyll.
Dark chilling treatment initiated during the night (h = 17), when SPS activity was low, resulted in low activity upon rewarming, even when plants were rewarmed at a time when SPS activity should be high (h = 24; Fig. 2, bottom). However, after rewarming, SPS activity remained low and did not continue the chill-delayed oscillation (Fig. 2, bottom). The reason that SPS activity did not recover as expected is not certain but is likely the result of the extended dark period. We have been unable to follow a circadian rhythm in SPS activity in constant darkness because, after an initial oscillation during the first subjective night, SPS activity decreased to low levels and remained there (data not shown).
Low-Temperature Treatment Delays SPS Activity by Delaying the Oscillation of the Protein Phosphorylation State
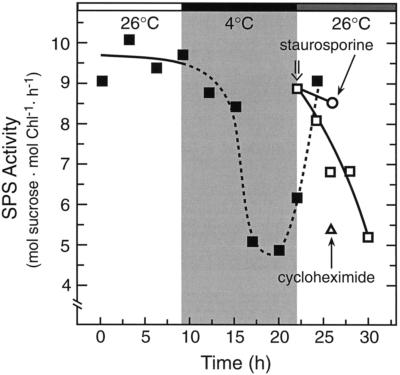
To determine if the inappropriately high SPS activity after the chilling treatment (Fig. 2, top) might be caused by maintenance of the prechilling phosphorylation state, we use the Ser/Thr kinase inhibitor staurosporine, which is an effective inhibitor of SPS deactivation in tomato (Jones and Ort, 1997). Staurosporine (100 μm) was applied (Fig. 3) to lightly abraded, attached tomato leaves immediately upon rewarming (h = 22) and samples were harvested 4 h later (h = 26). The staurosporine treatment prevented SPS deactivation after chilling, suggesting that the maintenance of high SPS activity was caused by maintenance of the prechilling dephosphorylated state of the enzyme. Cycloheximide (200 μg/mL), an effective inhibitor of cytoplasmic translation when applied under these conditions (Jones and Ort, 1997), did not prevent the postchilling deactivation of SPS. This result showed that, although SPS protein kinase activity is required for the postchilling deactivation of SPS, the kinase was already present and did not need to be synthesized de novo. Therefore, the delay in the circadian rhythm of SPS activity was the result of delayed changes in protein phosphorylation state, very possibly resulting from an effect of chilling on the circadian rhythm controlling the transcription of a protein phosphatase (perhaps SPS phosphatase) gene(s).
Figure 3.
Inhibitor studies show that SPS maintains the prechilling phosphorylation state after chilling. Light/dark cycles are represented by the shaded bars above the figure, as described in the legend to Fig. 2. A dark chill was imposed between h = 9 and h = 22, and immediately upon rewarming staurosporine (100 μm, ○) or cycloheximide (200 μg/mL, ▵) was applied to attached, abraded tomato leaves. The time of inhibitor application is indicated (⇓). After a 4-h dark incubation, samples were taken to determine SPS activity. The staurosporine treatment maintained SPS in the more active state, demonstrating that the prechilling, dephosphorylated form of SPS was maintained throughout the low-temperature treatment. Each point represents the mean of at least three samples, and the sd values were less than 15% of the mean values. Chl, Chlorophyll.
The Circadian Pattern in NR Activity in Tomato Is Driven Primarily by Changes in NR Expression
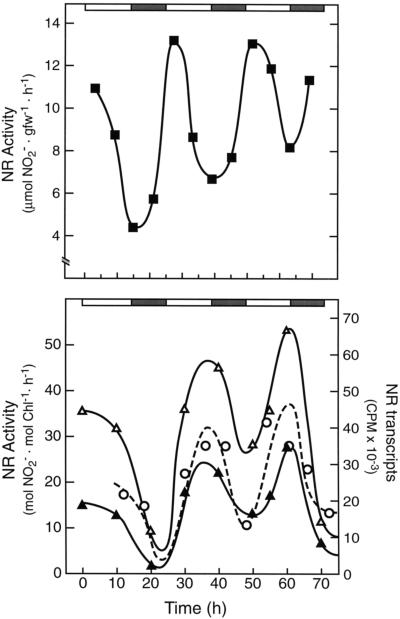
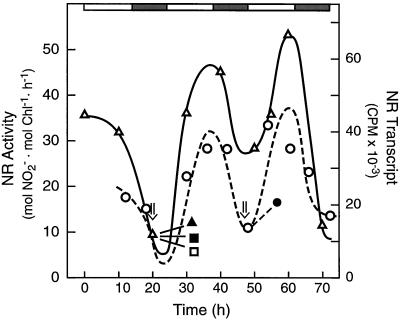
There was a robust circadian oscillation in NR activity in tomato under constant light (450–500 μmol quanta m−2 s−1) and temperature (26°C) whether assayed under in vivo (Fig. 4, top) or in vitro (Fig. 4, bottom) conditions. This circadian profile is very similar to that reported by Lillo (1984) for NR activity in barley.
Figure 4.
NR has a circadian pattern in transcript and protein levels, which is responsible for an endogenous rhythm in NR activity in tomato. The light and dark bars above the figure represent day and subjective night periods, respectively. Top, NR activity (▪) was assayed under in vivo conditions (see Methods) over a 3-d, constant light (450 μmol quanta m−2 s−1) and temperature (26°C) time course. The experiment was repeated twice, and the results shown are representative. gfw, Grams fresh weight. Bottom, NR activity was assayed under in vitro conditions in the absence (▵) and presence (▴) of Mg2+. The absence of Mg2+ prevents the Mg2+-dependent binding of 14-3-3-type inhibitor proteins to phospho-NR, thereby revealing changes in NR activity that are caused by changes in enzyme level (see text). Total leaf RNA was isolated from tomato leaves under constant-light circadian conditions and probed with a dCTP32-labeled tobacco nia-2 cDNA (○). The radioactivity was quantified on a phosphor imager. All experiments were repeated at least three times and representative results are shown. Chl, Chlorophyll.
Light activation of NR activity can involve both changes in NR protein level and changes in enzyme phosphorylation state (Solomonson and Barber, 1990; Huber et al., 1992a; Crawford and Arst, 1993; Pilgrim et al., 1993; Ramalho et al., 1995). We used the in vitro assay to resolve changes in activity associated with the NR phosphorylation status and subsequent Mg2+-dependent binding of 14-3-3-type inhibitor proteins to phospho-NR (Spill and Kaiser, 1994; Bachmann et al., 1995; Glaab and Kaiser, 1995; MacKintosh et al., 1995). Under continuous-light conditions, in the absence of Mg2+, NR activity was stimulated and still circadian (Fig. 4, bottom). The fact that the Mg2+-dependent inhibition of NR activity did not increase (Fig. 4, bottom) during the subjective night, when NR activity was lowest, is reasonably strong evidence that the circadian rhythm in NR activity is not primarily dependent on changes in the enzyme's phosphorylation state.
Kaiser and Huber (1997) showed that, when phosphorylated, full activation of spinach NR by the removal of Mg2+ can require as much as 30 min, which could obscure regulation of activity by changes in NR phosphorylation. For tomato leaf samples taken at the peak (32 h) or trough (24 h) of continuous-light circadian NR activity, there was no evidence of slow activation in the presence of EDTA (Fig. 5). Kaiser and Huber (1997) also included the NR activators AMP and Pi during the incubation with EDTA, but we observed no added effect of these activators in our experiments (data not shown). The decline in NR activity over the course of the incubation shown in Figure 5 was most likely caused by artificial inactivation of the enzyme and was considerably more pronounced in the absence of protease inhibitors or if the incubation was performed at room temperature (data not shown). An indication of slow activation of tomato NR upon removal of Mg2+ was observed in tomato leaves after 20 h of dark adaptation. The extent of the slow activation would be very pronounced if corrected for the time-dependent decline in activity seen in the continuous-light samples. It should be noted that the chlorophyll content on a fresh-weight basis was approximately 20% higher for tomato under continuous light than after 20 h in the dark, such that maximum NR activity on a fresh-weight basis was actually lower in dark-adapted leaves. This difference in slow activation of NR between light- and dark-adapted leaves is consistent with the report of Lillo et al. (1997). Thus, unlike SPS (Jones and Ort, 1997), the dynamics of NR phosphorylation do not play a dominant role in the circadian regulation of this enzyme.
Figure 5.
Continuous-light circadian samples do not show slow activation of NR in the presence of EDTA (i.e. in the absence of Mg2+). Leaf samples were taken at the peak (32 h, •) or trough (24 h, ○) of continuous-light circadian NR activity and after 20 h of dark adaptation (▵). The data plotted are the average of three separate experiments.
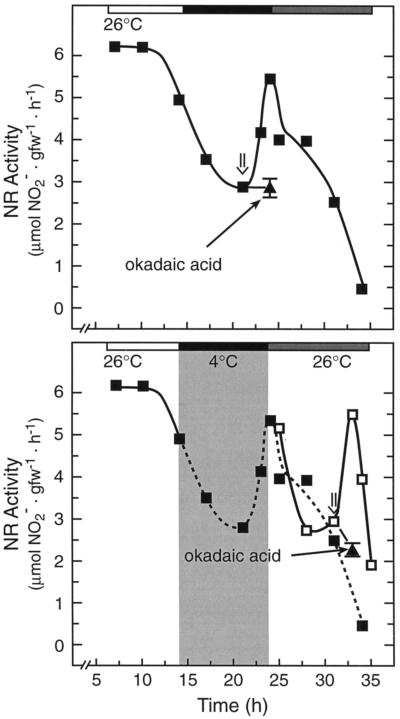
Figure 4 (bottom) further shows that the NR transcript level oscillated in synchrony with the phase of NR circadian activity. Whereas this may suggest circadian control of transcription, as we demonstrated previously in tomato for both cab and rca expression (Martino-Catt and Ort, 1992), there is evidence that circadian oscillations of NR mRNA in Arabidopsis (Pilgrim et al., 1993) and maize (Redinbaugh et al., 1996) are controlled primarily by posttranscriptional events. Figure 6 shows that the circadian increase in NR activity that would normally occur at 32 h was inhibited by pretreatment of the leaves with the transcription inhibitor cordycepin (300 μg/mL) and the translation inhibitor cycloheximide (200 μg/mL) whether activity was assayed in the presence (data not shown) or absence (Fig. 6) of Mg2+. As we observed for SPS (Jones and Ort, 1997), okadaic acid (10 μm), a potent inhibitor of types 1 and 2A protein phosphatase, also completely inhibited the circadian increase in NR activity whether Mg2+ was present or absent in the assay. Okadaic acid also significantly diminished the circadian increase in NR mRNA, as it has been observed to do for the NR mRNA increase after a dark-to-light transition in maize (Redinbaugh et al., 1996).
Figure 6.
The circadian increase in NR activity (▵) is prevented by inhibitors of translation and transcription; the circadian increase in NR activity and transcript level is prevented by inhibitors of protein phosphatases. The light and dark bars above the figure represent day and subjective night periods, respectively. Tomato leaves were treated with okadaic acid (10 μm, ▴ and •), cordycepin (200 μg/mL, ▪), or cycloheximide (200 μg/mL, □) at the times indicated (⇓). NR activity was assayed under in vitro conditions. NR activity data are representative of at least four separate samples, and were normalized to the continuous-light circadian rhythm at h = 32. Total leaf RNA (○, •) was probed with a dCTP32-labeled tobacco nia-2 cDNA. NR transcript data are the average of two separate samples, and were normalized to the circadian rhythm at h = 55. Chl, Chlorophyll.
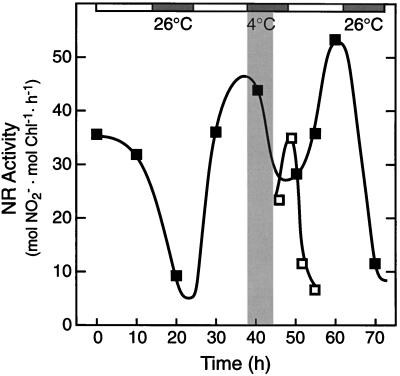
Low-Temperature Treatment Delays the Circadian Rhythm in NR Activity
The shaded area in Figure 7 depicts a 6-h low-temperature treatment imposed during continuous-light circadian conditions at a NR activity peak. Samples were taken after rewarming and assayed in the absence of Mg2+. The low-temperature treatment delayed the circadian rhythm in NR activity by approximately 6 h, indicating that the endogenous clock controlling NR activity stopped for the duration of the chill and resumed upon rewarming. Assays performed in the presence of Mg2+ showed an essentially identical response (at 50% lower NR activity) to the low-temperature treatment (data not shown).
Figure 7.
Low-temperature treatment delays the circadian rhythm controlling NR activity. The light and dark bars above the figure represent day and subjective night periods, respectively. A low-temperature treatment (4°C), represented by the shaded area in the main body of the figure, was imposed for 6 h, between h = 38 and h = 44. After rewarming, NR activity was assayed under in vitro conditions in the absence of Mg2+ (□). Comparison of the reference circadian rhythm (▪) and the postchilling oscillation (□) demonstrates that the 6-h low-temperature treatment delayed the oscillation in NR activity without Mg2+ (i.e. NR protein level). The experiment was repeated several times with consistent results. Chl, Chlorophyll.
Because chilling in conjunction with high light inhibits photosynthesis in tomato by a different and more rapidly developing mechanism than chilling in the dark (Sassenrath et al., 1990), we wanted to validate the result shown in Figure 7 under dark conditions. Using the in vivo assay, we observed a highly reproducible initial dark-circadian increase in NR activity at h = 24, subjective dawn, but the activity then decreased to very low levels (Fig. 8, bottom). Low-temperature treatment (4°C) was imposed during the diurnal dark cycle, h = 14 to 24, and the plants were rewarmed in the dark (26°C). NR activity was monitored after rewarming, and Figure 8 (bottom) reveals that the low-temperature pretreatment delayed the dark-circadian increase in NR activity just as it did in the light (Fig. 7). Comparing the control dark-circadian oscillation with the chill-delayed oscillation, it is clear that the circadian timing was offset by the 10-h duration of the chill.
Figure 8.
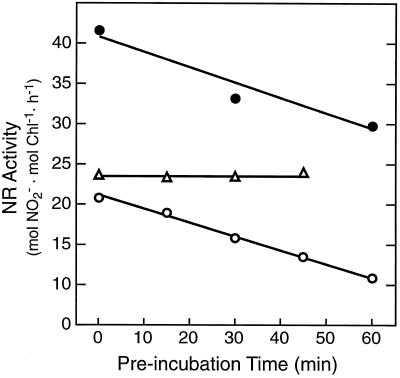
Low temperature delays the circadian control of the rise in in vivo NR activity that normally occurs before dawn. Top, For these experiments, plants grown under diurnal conditions (light/dark cycles are represented by the shaded bars above the figure, as described in the legend to Fig. 2) were given extended darkness in place of the normal dark-to-light transition at h = 24 (subjective day is represented by the gray bar). NR activity (▪) was assayed in the dark during the night and the following subjective day. Okadaic acid (10 μm, ▴) inhibited the circadian increase in NR activity. The experiment was repeated three times, and the profile shown is a representative result. Bottom, Plants were chilled at 4°C during the diurnal night cycle, between h = 14 and h = 24, and rewarmed in the dark. Samples were taken after rewarming (□) and demonstrate that the low-temperature treatment delayed the increase in NR activity by 10 h, indicating that the circadian clock stopped for the duration of the chill and resumed upon rewarming. During the dark recovery phase, okadaic acid (10 μm, ▴) was applied to attached tomato leaves (h = 32). Two hours later (h = 34), in vivo NR activity was assayed from the inhibitor-treated leaves and surfactant-treated controls. This experiment was repeated three times with consistent results. gfw, Grams fresh weight.
Inhibitor treatments indicated that the low-temperature delay in the NR circadian rhythm involved protein phosphatase activity (Fig. 8, bottom). After chilling treatment, okadaic acid (10 μm) was applied to attached leaves at h = 31, when NR activity was low. Samples were harvested 2 h later, when NR activity was expected to be high (h = 33). The okadaic acid treatment completely abolished the chill-delayed increase in NR activity and the normal diurnal increase in NR activity (Fig. 8, top). We obtained qualitatively indistinguishable results compared with those presented in Figure 8 using the in vitro NR assay in both the presence and the absence of Mg2+ (data not shown).
DISCUSSION
Several plant genes have circadian oscillations in transcription, including those for cab (Millar and Kay, 1991; Kellmann et al., 1993), Fd (Bringloe et al., 1995), catalase (Zhong et al., 1994), and rca (Martino-Catt and Ort, 1992). Frequently, however, there are not corresponding oscillations in enzyme activity because of large, stable protein pools. In fact, few proteins have been shown to have circadian oscillations in enzyme activity; those that have include NR (Lillo, 1984), SPS (Jones and Ort, 1997), and PEP carboxylase (Carter et al., 1991). Because circadian fluctuations are exhibited in many overt higher-plant processes, including leaf movement (Darwin, 1880; Satter and Galston, 1981), stomatal resistance (Martin and Meidner, 1971; Kerr et al., 1985; Holmes and Klein, 1986), and net photosynthesis (Chia-Looi and Cumming, 1972; Hennessey and Field, 1991), it seems unavoidable that many more enzymes must exhibit a circadian component to their regulation than the few so far identified. Circadian fluctuations in free cytosolic calcium (Johnson et al., 1995) may also play an important role in mediating the rhythms in these complex plant processes.
We demonstrated circadian regulation of SPS (Jones and Ort, 1997) and NR activity (Fig. 4) in tomato. The circadian pattern of NR activity under continuous-light conditions was driven primarily by changes in the protein level, which mimicked oscillations in the NR transcript level. That the percentage of inhibition by Mg2+ did not change in phase with the rhythm showed that, unlike those of SPS, the dynamics of NR phosphorylation play no significant role in the circadian regulation of this enzyme.
Although circadian control of SPS and NR activity responded similarly to inhibitors that affect the phosphorylation status of proteins and to inhibitors of translation and transcription, it is likely that they did so for quite different reasons. In contrast to that in NR, the circadian rhythm in SPS activity occurs in the absence of any corresponding changes in SPS protein level, and corresponds to circadian oscillations in the SPS phosphorylation state, which in turn appears to be driven by a circadian pattern of SPS-phosphatase gene transcription (Jones and Ort, 1997). Whereas the transcription inhibitor cordycepin and the translation inhibitor cycloheximide blocked the circadian increase in NR activity (Fig. 6), this was very likely caused by direct effects of these inhibitors on NR expression. Okadaic acid treatment inhibited the circadian increase in NR activity even though the increase was clearly not caused by the dephosphorylation of NR. Okadaic acid also inhibited the circadian increase in NR mRNA accumulation (Fig. 6). It has been proposed that separate protein phosphatases are involved in regulating NR translation and transcript accumulation (Redinbaugh et al., 1996). Thus, the transcriptional and/or translational components to the circadian regulation of NR expression may be at multiple levels.
The circadian rhythm in SPS and NR activity in tomato was delayed by exposure to low temperature (Figs. 1 and 7) in a manner analogous to the chilling-induced delay in the circadian rhythm of cab and rca transcription that we described earlier (Martino-Catt and Ort, 1992). Perhaps more physiologically relevant, the diurnal rhythm in SPS (Fig. 2) and NR (Fig. 8) activity was also delayed by low-temperature treatments, so a cool night will result in mistiming of activity the next day.
Impaired photosynthesis as a result of mistimed SPS activity can be reconciled with current understanding of the regulation of photosynthetic carbon metabolism. Insufficient SPS activity would be expected to limit the rate of photosynthesis (Herold, 1980; Sharkey, 1990; Micallef et al., 1995) by causing an accumulation of triose phosphate in the chloroplast at the expense of free phosphate (Sharkey, 1990). It is proposed that the limited availability of free phosphate in the chloroplast would reduce the ATP-generating capacity of the chloroplast ATP synthase, thereby ultimately leading to the inhibition of photosynthesis (Sharkey, 1990).
Genetically transformed plants with modified levels of enzymes in Suc biosynthesis have been examined for altered photosynthesis. Overexpressed maize SPS results in substantial increases in SPS activity, as well as an increased capacity for Suc synthesis in transformed tomato (Worrell et al., 1991; Galtier et al., 1993; Micallef et al., 1995). Under standard atmospheric CO2 conditions there was no statistically significant increase in photosynthesis; however, when grown under doubled atmospheric CO2, the maize SPS-transformed lines supported a 20% greater rate of photosynthesis (Galtier et al., 1993; Micallef et al., 1995). Increasing atmospheric CO2 concentrations increases the likelihood that plants will experience feedback inhibition because this condition allows photosynthesis to outpace end-product (triose phosphate) utilization. The higher photosynthetic rates in the SPS-overexpressing plants indicates that increased Suc synthesis can alleviate some of this feedback inhibition. Similar results are seen in Flaveria linearis transformants with altered levels of cytosolic Fru-1,6-bisphosphatase (Micallef et al., 1996), another key regulatory enzyme in Suc biosynthesis. Photosynthesis at atmospheric levels of CO2 is not significantly affected in these mutants (either over- or underexpressing cytosolic Fru-1,6-bisphosphatase) compared with controls; however, at elevated CO2 levels the predicted effects based on feedback inhibition are seen. The lines overexpressing cytosolic Fru-1,6-bisphosphatase showed increased levels of photosynthesis, whereas those underexpressing this enzyme were unable to take full advantage of the elevated CO2.
Although the mistiming of SPS alone might be expected to lead to the inhibition of photosynthesis in tomato, our results suggest that the underlying basis for the inhibition of photosynthesis in chilling-sensitive plants is the cumulative result of the mistiming of numerous circadian-controlled enzyme activities. Here we show that the circadian rhythm in both SPS and NR activity was arrested in tomato by chilling and that an effect of low temperature on critical protein phosphatases was involved in each case. For both enzymes, it appears that it is a protein phosphatase that is under circadian regulation, and that this circadian rhythm is delayed by low-temperature treatment. These results imply that other, as-yet-unidentified circadian-regulated enzyme activities, particularly those regulated by protein phosphorylation, may be affected by low-temperature treatment as well.
Because the circadian rhythm in SPS activity seems to be controlled by the transcription of SPS phosphatase, and the SPS activity rhythm is delayed by low-temperature treatment, it is a sensible hypothesis that the mechanism for the low-temperature delay in SPS activity is a delay in the circadian rhythm in SPS-phosphatase transcription. However, the fact that NR behaves identically to chilling exposure even though phosphorylation of NR does not contribute to the circadian regulation of activity indicates that low temperature may act at an earlier, common step in the regulation pathway of these enzymes. Because okadaic acid-sensitive protein phosphatase activity is critical to the circadian regulation of both of these enzymes, it would be a plausible chill-sensitive common link that could have far-reaching effects on the photosynthetic metabolism in chill-sensitive plants.
ACKNOWLEDGMENTS
We thank Erin Grant and Sonja Kemmis for their able assistance in conducting a portion of the SPS and NR assays, and Jim Harper's laboratory for assistance with the in vivo NR assays.
Abbreviations:
- NR
nitrate reductase
- SPS
Suc phosphate synthase
Footnotes
This work was supported in part by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (grant no. 91-37100-6620 to D.R.O.) and by an Integrative Photosynthesis Research Training Grant from the Department of Energy (no. DEFGO2-92ER20095), funded under the program for Collaborative Research in Plant Biology.
LITERATURE CITED
- Bachmann M, McMichael RW, Huber JL, Kaiser WM, Huber SC. Partial purification and characterization of a calcium-dependent protein kinase and an inhibitor protein required for inactivation of spinach leaf nitrate reductase. Plant Physiol. 1995;108:1083–1092. doi: 10.1104/pp.108.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann M, Shiraishi N, Campbell WH, Yoo BC, Harmon AC, Huber SC. Identification of Ser-543 as the major regulatory phosphorylation site in spinach leaf nitrate reductase. Plant Cell. 1996;8:127–131. doi: 10.1105/tpc.8.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NR, East TM, Long SP. Chilling damage to photosynthesis in young Zea mays. II. Photochemical function of thylakoids in vivo. J Exp Bot. 1983;34:189–197. [Google Scholar]
- Bringloe DH, Dyer TA, Gray JC. Developmental, circadian and light regulation of wheat ferredoxin gene expression. Plant Mol Biol. 1995;27:293–306. doi: 10.1007/BF00020184. [DOI] [PubMed] [Google Scholar]
- Carter PJ, Nimmo HG, Fewson CA, Wilkins MB. Circadian rhythms in the activity of a plant protein kinase. EMBO J. 1991;10:2063–2068. doi: 10.1002/j.1460-2075.1991.tb07737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-L, Acedo GN, Dewdney J, Goodman HM, Conking MA. Differential expression of the two Arabidopsis nitrate reductase genes. Plant Physiol. 1991;96:275–279. doi: 10.1104/pp.96.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia-Looi A-S, Cumming BG. Circadian rhythms of dark respiration, flowering, net photosynthesis, chlorophyll content and dry weight changes in Chenopodium rubrum. Can J Bot. 1972;50:2219–2226. [Google Scholar]
- Cooper P, Ort DR. Changes in protein synthesis induced in tomato by chilling. Plant Physiol. 1988;88:454–461. doi: 10.1104/pp.88.2.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM, Arst HNJ. The molecular genetics of nitrate assimilation in fungi and plants. Annu Rev Genet. 1993;27:115–146. doi: 10.1146/annurev.ge.27.120193.000555. [DOI] [PubMed] [Google Scholar]
- Darwin C (1880) On the Power of Movement in Plants. John Murray, London
- Deng M-D, Moureaux T, Leydecker M-T, Caboche M. Nitrate-reductase expression is under the control of a circadian rhythm and is light inducible in Nicotiana tabacum leaves. Planta. 1990;180:257–261. doi: 10.1007/BF00194005. [DOI] [PubMed] [Google Scholar]
- Doehlert DC, Huber SC. Phosphate inhibition of spinach leaf sucrose-phosphate synthase as affected by glucose-6-phosphate and phosphoglucose isomerase. Plant Physiol. 1984;76:250–253. doi: 10.1104/pp.76.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Morrice N, MacKintosh C. Identification of a regulatory phosphorylation site in the hinge I region of nitrate reductase from spinach (Spinacia oleracea) leaves. FEBS Lett. 1995;377:113–117. doi: 10.1016/0014-5793(95)01300-8. [DOI] [PubMed] [Google Scholar]
- Galtier N, Foyer CH, Huber J, Voelker TA, Huber SC. Effects of elevated sucrose-phosphate synthase activity on photosynthesis, assimilate partitioning, and growth in tomato (Lycopersicon esculentum var UC82B) Plant Physiol. 1993;101:535–543. doi: 10.1104/pp.101.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaab J, Kaiser WM. Inactivation of nitrate reductase involves NR-protein phosphorylation and subsequent ‘binding’ of an inhibitor protein. Planta. 1995;195:514–518. [Google Scholar]
- Graan T, Ort DR. Quantitation of the rapid electron donors to P700, the functional plastoquinone pool, and the ratio of the photosystems in spinach chloroplasts. J Biol Chem. 1984;259:14003–14010. [PubMed] [Google Scholar]
- Hennessey TL, Field CB. Circadian rhythms in photosynthesis. Oscillations in carbon assimilation and stomatal conductance under constant conditions. Plant Physiol. 1991;96:831–836. doi: 10.1104/pp.96.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold A. Regulation of photosynthesis by sink activity: the missing link. New Phytol. 1980;86:131–144. [Google Scholar]
- Hoff T, Truon HM, Caboche M. The use of mutants and transgenic plants to study nitrate assimilation. Plant Cell Environ. 1994;17:489–506. [Google Scholar]
- Holmes MG, Klein WH. Photocontrol of dark circadian rhythms in stomata of Phaseolus vulgaris L. Plant Physiol. 1986;82:28–33. doi: 10.1104/pp.82.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JLA, Huber SC, Nielsen TH. Protein phosphorylation as a mechanism for regulation of spinach leaf sucrose-phosphate synthase activity. Arch Biochem Biophys. 1989;270:681–690. doi: 10.1016/0003-9861(89)90551-1. [DOI] [PubMed] [Google Scholar]
- Huber SC, Huber JL, Campbell WH, Redinbaugh MG. Comparative studies of the light modulation of nitrate reductase and sucrose-phosphate synthase activities in spinach leaves. Plant Physiol. 1992a;100:706–712. doi: 10.1104/pp.100.2.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC, Huber JL, Campbell WH, Redinbaugh MG. Apparent dependence of the light activation of nitrate reductase and sucrose-phosphate synthase activities in spinach leaves on protein synthesis. Plant Cell Physiol. 1992b;33:639–646. doi: 10.1104/pp.100.2.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH, Knight MR, Kondo T, Masson P, Sedbrook J, Haley A, Trewavas A. Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science. 1995;269:1863–1865. doi: 10.1126/science.7569925. [DOI] [PubMed] [Google Scholar]
- Jones TL, Ort DR. Circadian regulation of sucrose phosphate synthase activity in tomato by protein phosphatase activity. Plant Physiol. 1997;113:1167–1175. doi: 10.1104/pp.113.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WM, Brendle-Behnisch E. Rapid modulation of spinach leaf nitrate reductase activity by photosynthesis. Plant Physiol. 1991;96:363–367. doi: 10.1104/pp.96.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WM, Huber SC. Posttranslational regulation of nitrate reductase in higher plants. Plant Physiol. 1994;106:817–821. doi: 10.1104/pp.106.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WM, Huber SC. Correlation between apparent activation state of nitrate reductase (NR), NR hysteresis and degradation of NR protein. J Exp Bot. 1997;48:1367–1374. [Google Scholar]
- Kaiser WM, Spill D. Rapid modulation of spinach leaf nitrate reductase by photosynthesis. In vitro modulation by ATP + AMP. Plant Physiol. 1991;96:368–375. doi: 10.1104/pp.96.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee SC, Martin B, Ort DR. The effects of chilling in the dark and in the light on photosynthesis of tomato: electron transfer reactions. Photosynth Res. 1986;8:41–51. doi: 10.1007/BF00028475. [DOI] [PubMed] [Google Scholar]
- Kellmann J-W, Merforth N, Wiese M, Pichersky E, Piechulla B. Concerted circadian oscillation in transcript levels of nineteen Lha/b (cab) genes in Lycopersicon esculentum (tomato) Mol Gen Genet. 1993;237:439–448. doi: 10.1007/BF00279449. [DOI] [PubMed] [Google Scholar]
- Kerr PS, Rufty TW, Jr, Huber SC. Endogenous rhythms in photosynthesis, sucrose phosphate synthase activity, and stomatal resistance in leaves of soybean (Glycine max [L.] Merr) Plant Physiol. 1985;77:275–280. doi: 10.1104/pp.77.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillo C. Studies of diurnal variations of nitrate reductase activity in barley leaves using various assay methods. Physiol Plant. 1983;57:357–362. [Google Scholar]
- Lillo C. Circadian rhythmicity of nitrate reductase activity in barley leaves. Physiol Plant. 1984;61:219–223. [Google Scholar]
- Lillo C, Kazazaic S, Ruoff P, Meyer C. Characterization of nitrate reductase from light- and dark-exposed leaves. Plant Physiol. 1997;114:1377–1383. doi: 10.1104/pp.114.4.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKintosh C. Regulation of spinach-leaf nitrate reductase by reversible phosphorylation. Biochim Biophys Acta. 1992;1137:121–126. doi: 10.1016/0167-4889(92)90109-o. [DOI] [PubMed] [Google Scholar]
- MacKintosh C, Douglas P, Lillo C. Identification of a protein that inhibits the phosphorylated nitrate reductase form of nitrate reductase from spinach (Spinacia oleracea) leaves. Plant Physiol. 1995;107:451–457. doi: 10.1104/pp.107.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B, Ort DR. The recovery of photosynthesis subsequent to chilling exposure. Photosynth Res. 1985;6:121–132. doi: 10.1007/BF00032787. [DOI] [PubMed] [Google Scholar]
- Martin B, Ort DR, Boyer JS. Impairment of photosynthesis by chilling temperatures in tomato. Plant Physiol. 1981;68:329–334. doi: 10.1104/pp.68.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ES, Meidner H. Endogenous stomatal movements in Tradescantia virginiana. New Phytol. 1971;70:923–928. [Google Scholar]
- Martino-Catt S, Ort DR. Low temperature interrupts circadian regulation of transcriptional activity in chilling-sensitive plants. Proc Natl Acad Sci USA. 1992;89:3731–3735. doi: 10.1073/pnas.89.9.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael RW, Jr, Klein RR, Salvucci ME, Huber SC. Identification of the major regulatory phosphorylation site in sucrose-phosphate synthase. Arch Biochem Biophys. 1993;307:248–252. doi: 10.1006/abbi.1993.1586. [DOI] [PubMed] [Google Scholar]
- Micallef BJ, Haskins KA, Vanderveer PJ, Roh K-S, Shewmaker CK, Sharkey TD. Altered photosynthesis, flowering, and fruiting in transgenic tomato plants that have an increased capacity for sucrose synthesis. Planta. 1995;196:327–334. [Google Scholar]
- Micallef BJ, Vanderveer PJ, Sharkey TD. Responses to elevated CO2 of Flaveria linearis plants having a reduced activity of cytosolic fructose-1,6-bisphosphatase. Plant Cell Environ. 1996;19:10–16. [Google Scholar]
- Millar AJ, Kay SA. Circadian control of cab gene transcription and mRNA accumulation in Arabidopsis. Plant Cell. 1991;3:541–550. doi: 10.1105/tpc.3.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas JC, Harper JE, Hageman RH. Nitrate reductase activity in soybeans (Glycine max [L.] Merr.). I. Effects of light and temperature. Plant Physiol. 1976;58:731–735. doi: 10.1104/pp.58.6.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort DR, Martino S, Wise RR, Kent J, Cooper P. Changes in protein synthesis induced by chilling and their influence on the chilling sensitivity of photosynthesis. Plant Physiol Biochem. 1989;27:785–793. [Google Scholar]
- Peeler TC, Naylor AW. A comparison of the effects of chilling on thylakoid electron transfer in pea (Pisum sativum L.) and cucumber (Cucumis sativus L.) Plant Physiol. 1988;86:147–151. doi: 10.1104/pp.86.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival MP, Bradbury M, Hayden DB, Baker NR. Modification of the photochemical apparatus in maize by photoinhibitory stress at low temperature. In: Biggins J, editor. Progress in Photosynthesis Research, Vol 4. Dordrecht, The Netherlands: Martinus Nijhoff; 1987. pp. 47–50. [Google Scholar]
- Pilgrim ML, Caspar T, Quail PH, McClung CR. Circadian and light-regulated expression of nitrate reductase in Arabidopsis. Plant Mol Biol. 1993;23:349–364. doi: 10.1007/BF00029010. [DOI] [PubMed] [Google Scholar]
- Powles SB, Berry JA, Björkman O. Interaction between light and chilling temperature on inhibition of photosynthesis in chilling sensitive plants. Plant Cell Environ. 1983;6:117–123. [Google Scholar]
- Ramalho CB, Hastings JW, Colepicolo P. Circadian oscillation of nitrate reductase activity in Gonyaulax polyedra is due to changes in cellular protein levels. Plant Physiol. 1995;107:225–231. doi: 10.1104/pp.107.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redinbaugh MG, Huber SC, Huber JL, Hendrix KW, Campbell WH. Nitrate reductase expression in maize leaves (Zea mays) during dark-light transitions: complex effects of protein phosphatase inhibitors on enzyme activity, protein synthesis and transcript levels. Physiol Plant. 1996;98:67–76. [Google Scholar]
- Sassenrath GF, Ort DR, Portis AR., Jr Impaired reductive activation of stromal bisphosphatases in tomato leaves following low-temperature exposure at high light. Arch Biochem Biophys. 1990;282:302–308. doi: 10.1016/0003-9861(90)90121-e. [DOI] [PubMed] [Google Scholar]
- Satter RL, Galston AW. Mechanisms of control of leaf movements. Annu Rev Plant Physiol. 1981;32:83–110. [Google Scholar]
- Sharkey TD. Feedback limitation of photosynthesis and the physiological role of ribulose bisphosphate carboxylase carbamylation. Bot Mag Tokoyo Special Issue. 1990;2:87–105. [Google Scholar]
- Solomonson LP, Barber MJ. Assimilatory nitrate reductase: functional properties and regulation. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:225–253. [Google Scholar]
- Spill D, Kaiser WM. Partial purification of two proteins (100 kDa and 67 kDa) cooperating in the ATP-dependent inactivation of spinach leaf nitrate reductase. Planta. 1994;192:183–188. [Google Scholar]
- Su W, Huber SC, Crawford NM. Identification in vitro of a post-translational regulatory site in the hinge I of Arabidopsis nitrate reductase. Plant Cell. 1996;8:519–527. doi: 10.1105/tpc.8.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vincentz M, Kronenberger J, Caboche M, Rouzé P. Molecular cloning and characterisation of the two homologous genes coding for nitrate reductase in tobacco. Mol Gen Genet. 1989;216:10–15. doi: 10.1007/BF00332224. [DOI] [PubMed] [Google Scholar]
- Wise RR. Chilling-enhanced photooxidation: the production, action and study of reactive oxygen species produced during chilling in the light. Photosynth Res. 1995;45:79–97. doi: 10.1007/BF00032579. [DOI] [PubMed] [Google Scholar]
- Worrell AC, Bruneau JM, Summerfelt K, Boersig M, Voelker T. Expression of maize sucrose phosphate synthase in tomato alters carbohydrate partitioning. Plant Cell. 1991;3:1121. doi: 10.1105/tpc.3.10.1121. 1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong HH, Young JC, Pease EA, Hangarter RP, McClung CR. Interactions between light and the circadian clock in the regulation of CAT2 expression in Arabidopsis. Plant Physiol. 1994;104:889–898. doi: 10.1104/pp.104.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]