Abstract
Background
Clostridium difficile is the main cause of antibiotic associated diarrhea. In the past decade, the number of C. difficile patients has increased dramatically, coinciding with the emergence of two PCR ribotypes 027 and 078. PCR ribotype 078 is also frequently found during C. difficile outbreaks in pigfarms. Previously, the genome of the PCR ribotype 078 strain M120, a human isolate, was described to contain a unique insert of 100 kilobases.
Results
Analysis of this insert revealed over 90 open reading frames, encoding proteins originating from transposons, phages and plasmids. The insert was shown to be a transposon (Tn6164), as evidenced by the presence of an excised and circularised molecule, containing the ligated 5’and 3’ends of the insert. Transfer of the element could not be shown through filter-mating experiments. Whole genome sequencing of PCR ribotype 078 strain 31618, isolated from a diarrheic piglet, showed that Tn6164 was not present in this strain. To test the prevalence of Tn6164, a collection of 231 Clostridium difficile PCR ribotype 078 isolates from human (n = 173) and porcine (n = 58) origin was tested for the presence of this element by PCR. The transposon was present in 9 human, tetracycline resistant isolates, originating from various countries in Europe, and none of the pig strains. Nine other strains, also tetracycline resistant human isolates, contained half of the transposon, suggesting multiple insertion steps yielding the full Tn6164. Other PCR ribotypes (n = 66) were all negative for the presence of the transposon. Multi locus variable tandem repeat analysis revealed genetic relatedness among transposon containing isolates. Although the element contained several potential antibiotic resistance genes, it did not yield a readily distinguishable phenotype.
Conclusions
Tn6164 is a newly described transposon, occurring sporadically in C. difficile PCR ribotype 078 strains. Although no transfer of the element could be shown, we hypothesize that the element could serve as a reservoir of antibiotic resistance genes for other bacteria. Further research is needed to investigate the transfer capabilities of the element and to substantiate the possible role of Tn6164 as a source of antibiotic resistance genes for other gut pathogens.
Keywords: Clostridium difficile, Transposable element, Phage, Antimicrobial resistance, Virulence
Background
Over the past decade, Clostridium difficile has emerged as an important gut pathogen, causing hospital- and community-acquired diarrhea. The number of patients and the severity of disease have increased dramatically, due to the emergence of two hypervirulent PCR ribotype, 027 [1] and 078 [2,3]. Traditionally, PCR ribotype 027 has been linked to nosocomial outbreaks. In contrast, PCR ribotype 078 has been detected frequently in farming animals, especially pigs [2,4], and is found more during community acquired infection. The increase in C. difficile infections (CDI) of humans has boosted interest in C. difficile biology, diagnostics and pathogenesis.
In the past few years, multiple genome sequences of several PCR ribotypes have been determined [5-8]. The analyses of the genomes, aided by comparative genomics of DNA-DNA microarrays [9,10] has shown that the genomes of C. difficile are highly variable with inserts of mobile DNA from phage, plasmid or transposon origin. These mobile DNA elements are actively moving within C. difficile genomes and are frequently passed on to neighboring bacteria, harboring mosaic genomes [7,11]. It is unclear what role the mobile elements play in the virulence of C. difficile. Some virulence linked genes, for example the holin-like tcdE, have a phage origin [12]. In fact, it has been suggested that the whole pathogenicity locus (PaLoc), encoding the major C. difficile virulence factors TcdA and TcdB, is of phage origin [13,14]. Recently, phages have been shown to upregulate toxin production in C. difficile, thereby increasing the virulence [15]. C. difficile transposons have been shown to contain antibiotic resistance genes [5,7,16,17], and therefore acquiring such an element could increase the virulence and/or colonization potential of a particular strain.
Mobile elements play an important role in the diversification of bacterial genomes. One important group of mobile genetic elements is the Tn916 family of conjugative transposons (also known as integrative and conjugative elements [ICEs]) [18]. These conjugative transposons usually code for tetracycline resistance and are found primarily in the Firmicutes. Numerous transposons have been described to be present in C. difficile genomes [5,7,11,17,19]. Several elements closely related to Tn916 are present in diverse C. difficile strains, including Tn5397 which confers tetracycline resistance [20,21]. Other transposons have been described to confer resistance to chloramphenicol and erythromycin [5].
Recently, the first full length genome of a PCR ribotype 078 strain was published [5]. This M120 strain has been isolated from an Irish diarrheic patient. It was shown that PCR ribotype 078 is highly divergent from PCR ribotype 027, 001, 017 and 012. In addition, this PCR ribotype 078 strain was described to contain a unique 100 kb insert that showed 80% similarity to sequences of Thermoanaerobacter species and Streptococcus pneumoniae[5]. In this paper we show that the 100 kb insert is a mobile element that is only sporadically present in PCR ribotype 078 strains. Furthermore, we show that the 100 kb consists of at least two independent mobile elements that were fused during evolution.
Results
Previously, an insert, unique for C. difficile, was described in the genome of strain M120, a PCR ribotype 078 strain, isolated from an Irish diarrheic patient [5]. We analyzed the open reading frames (ORFs) present in the insert to investigate their nature and origin (see Figure 1 and Table 1).
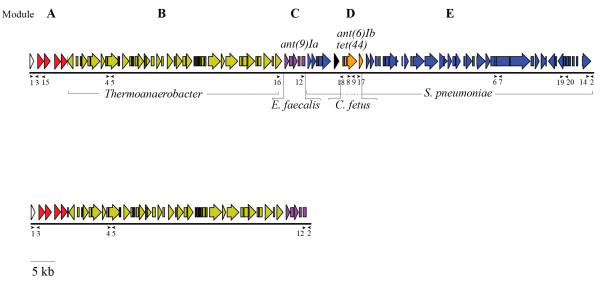
Figure 1.
Schematic view of full Tn 6164 (top panel) and half the element (bottom panel) and its open reading frames, flanked by C. difficile regions. Various parts of the insert are colored according to their homology. White, C. difficile; Red, Module A; Yellow, Module B; Purple, Module C; Orange, Module D; Blue, Module E; black, unknown. Location of the oligonucleotides used for the data in Table 2 is indicated by arrowheads
Table 1.
Open reading frames encoded by Tn 6164
| Gene | Position on Tn 6164 | Module | Sequence identity to | Annotation | Gene | Position on Tn 6164 | Module | Sequence identity to | Annotation |
|---|---|---|---|---|---|---|---|---|---|
|
Orf1 |
650-1930 |
A |
- |
putative modification methylase |
Orf25 |
26793-27122 |
B |
- |
conserved hypothetical protein |
|
Orf2 |
1915-3186 |
A |
- |
putative modification methylase |
Orf26 |
27189-28451 |
B |
Thermoanaerobacter sp. |
HK97 family phage portal protein |
|
Orf3 |
3252-3962 |
A |
- |
hypothetical protein |
Orf27 |
28448-29128 |
B |
Thermoanaerobacter sp. |
Peptidase S14, ClpP |
|
Orf4 |
3952-5031 |
A |
- |
ATPase associated with various cellular activities |
Orf28 |
29140-30339 |
B |
Thermoanaerobacter sp. |
HK97 family phage major capsid protein |
|
Orf5 |
5047-6312 |
A |
- |
LlaJI restriction endonuclease |
Orf29 |
30585-30899 |
B |
Thermoanaerobacter sp. |
uncharacterized phage protein |
|
Orf6 |
C 7557-6361 |
A |
- |
Protein with unknown function, contains a C-terminal CGNR Zinc finger motif |
Orf30 |
30903-31238 |
B |
Thermoanaerobacter sp. |
phage head-tail adaptor, putative |
|
Orf7 |
8000-8494 |
B |
Thermoanaerobacter sp. |
ECF RNA polymerase sigma-24 factor |
Orf31 |
31252-31662 |
B |
Thermoanaerobacter sp. |
HK97 family phage protein |
|
Orf8 |
8809-9126 |
B |
Thermoanaerobacter sp. |
rRNA biogenesis protein rrp5, putative |
Orf32 |
31659-32012 |
B |
Thermoanaerobacter sp. |
Protein of unknown function (DUF806); |
|
Orf9 |
9123-10250 |
B |
Thermoanaerobacter sp. |
Phage associated protein |
Orf33 |
32016-32618 |
B |
Thermoanaerobacter sp. |
DUF3647 Phage protein (HHPred) |
|
Orf10 |
10256-10816 |
B |
Thermoanaerobacter sp. |
phage-associated protein |
Orf34 |
33330-35786 |
B |
Thermoanaerobacter sp. |
Phage tape measure protein |
|
Orf11 |
10813-12747 |
B |
Thermoanaerobacter sp. |
DNA-directed DNA polymerase |
Orf35 |
35800-36573 |
B |
Thermoanaerobacter sp. |
phage putative tail component |
|
Orf12 |
12795-13625 |
B |
Thermoanaerobacter sp. |
Prophage antirepressor |
Orf36 |
36692-39100 |
B |
Thermoanaerobacter sp. |
phage minor structural protein |
|
Orf13 |
13629-14048 |
B |
Thermoanaerobacter sp. |
DUF 4406 (HHPred) |
Orf37 |
39320-39901 |
B |
Thermoanaerobacter sp. |
Putative Sipho Phage tail protein (HHPred) |
|
Orf14 |
14045-16390 |
B |
Thermoanaerobacter sp. |
virulence-associated E protein |
Orf38 |
39928-42369 |
B |
Thermoanaerobacter sp. |
glycosyl hydrolase-like protein |
|
Orf15 |
16910-18259 |
B |
Thermoanaerobacter sp. |
SNF2-related protein |
Orf39 |
42430-42855 |
B |
Thermoanaerobacter sp. |
toxin secretion/phage lysis holin |
|
Orf16 |
18264-18722 |
B |
Thermoanaerobacter sp. |
phage-associated protein |
Orf40 |
42855-43556 |
B |
Thermoanaerobacter sp. |
N-acetylmuramoyl-L-alanine amidase |
|
Orf17 |
18842-19201 |
B |
Thermoanaerobacter sp. |
HNH endonuclease |
Orf41 |
43975-45540 |
B |
Thermoanaerobacter sp. |
phage integrase family site-specific recombinase/resolvase |
|
Orf18 |
19314-19865 |
B |
Thermoanaerobacter sp. |
Phage terminase, small subunit |
Orf42 |
45541-45954 |
B |
Thermoanaerobacter sp. |
recombinase/integrase |
|
Orf19 |
19883-21058 |
B |
Thermoanaerobacter sp. |
S-adenosylmethionine synthetase |
Orf43 |
46222-47529 |
B |
Thermoanaerobacter sp. |
phage integrase family site-specific recombinase |
|
Orf20 |
21039-22283 |
B |
Thermoanaerobacter sp. |
DNA methylase N-4/N-6 domain-containing protein |
Orf44 |
47987-48856 |
C |
E. faecalis pEF418 |
Nucleotidyl transferase |
|
Orf21 |
22384-23076 |
B |
Thermoanaerobacter sp. |
hypothetical/virulence-related protein |
Orf45 |
48837-49571 |
C |
E. faecalis pEF418 |
methyltransferase |
|
Orf22 |
23445-24344 |
B |
Thermoanaerobacter sp. |
Putative amidoligase enzyme |
Orf46 |
49604-50467 |
C |
E. faecalis pEF418 |
putative aminoglycoside 6-adenylyltansferase |
|
Orf23 |
24382-24843 |
B |
Thermoanaerobacter sp. |
AIG2/GGCT-like protein |
Orf47 |
50511-51038 |
C |
E. faecalis pEF418 |
putative adenine phosphoribosyltransferase |
|
Orf24 |
25462-26685 |
B |
Thermoanaerobacter sp. |
phage terminase |
Orf48 |
51251-51979 |
C |
E. faecalis pEF418 |
putative spectinomycin/streptomycin adenyltransferase |
|
Orf49 |
52403-53176 |
E |
S. pneumoniae |
phage protein/replication initiator |
Orf71 |
77648-79216 |
E |
S. pneumoniae |
putative surface protein |
|
Orf50 |
53176-54000 |
E |
S. pneumoniae |
DNA replication protein |
Orf72 |
79231-80088 |
E |
S. pneumoniae |
putative bacteriocin |
|
Orf51 |
53993-54478 |
E |
S. pneumoniae |
DUF 3801 |
Orf73 |
80162-80773 |
E |
S. pneumoniae |
Predicted transcriptional regulator |
|
Orf52 |
54475-55209 |
E |
S. pneumoniae |
phage antirepressor protein |
Orf74 |
80766-81749 |
E |
S. pneumoniae |
Protein with unknown function |
|
Orf53 |
55202-56890 |
E |
S. pneumoniae |
TraG/TraD family protein |
Orf75 |
82268-82621 |
E |
S. pneumoniae |
transcriptional regulator, ArsR family |
|
Orf54 |
57454-58486 |
E |
- |
DUF 318 Predicted Permease (HHPred) |
Orf76 |
82696-83940 |
E |
S. pneumoniae |
major facilitator superfamily MFS_1 |
|
Orf55 |
59048-59398 |
D |
C. fetus |
glyoxalase family protein |
Orf77 |
83927-84403 |
E |
S. pneumoniae |
toxin-antitoxin system, toxin component, GNAT domain protein |
|
Orf56 |
59411-59938 |
D |
C. fetus |
transcriptional regulator |
Orf78 |
84758-86491 |
E |
S. pneumoniae |
DNA topoisomerase III |
|
Orf57 |
59988-61910 |
D |
C. fetus |
tetracycline resistance protein |
Orf79 |
86484-87449 |
E |
S. pneumoniae |
possible DNA (cytosine-5-)-methyltransferase |
|
Orf58 |
62225-63082 |
D |
C. fetus |
aminoglycoside 6-adenylyltransferase (AAD(6) |
Orf80 |
87436-95079 |
E |
S. pneumoniae |
superfamily II DNA and RNA helicase |
|
Orf59 |
63575-64348 |
E |
S. pneumoniae |
replication initiator/phage |
Orf81 |
95123-95779 |
E |
S. pneumoniae |
putative single-stranded DNA binding protein |
|
Orf60 |
64345-65172 |
E |
S. pneumoniae |
replicative DNA helicase |
Orf82 |
95939-96841 |
E |
S. pneumoniae |
transcriptional regulator, XRE family |
|
Orf61 |
65314-65814 |
E |
S. pneumoniae |
TnpX site-specific recombinase family protein |
Orf83 |
97071-98282 |
E |
S. pneumoniae |
transporter, major facilitator family/multidrug resistance protein 2 |
|
Orf62 |
65938-66399 |
E |
S. pneumoniae |
flavodoxin |
Orf84 |
C 99739-98462 |
E |
S. pneumoniae |
relaxase/type IV secretory pathway protein VirD2 |
|
Orf63 |
66817-67302 |
E |
S. pneumoniae |
putative conjugative transposon protein |
Orf85 |
C 101169-99795 |
E |
S. pneumoniae |
conjugal transfer relaxosome component TraJ |
|
Orf64 |
67299-68033 |
E |
S. pneumoniae |
phage antirepressor protein |
Orf86 |
C 101403-100321 |
E |
S. pneumoniae |
toxin-antitoxin system, toxin component, Fic family |
|
Orf65 |
68026-69816 |
E |
S. pneumoniae |
TraG/TraD family protein/putative conjugal transfer protein |
Orf87 |
C 101878-101396 |
E |
S. pneumoniae |
putative membrane protein |
|
Orf66 |
70395-70706 |
E |
S. pneumoniae |
putative single-strand binding protein |
Orf88 |
C 102435-101887 |
E |
S. pneumoniae |
putative toxin-antitoxin system, toxin component |
|
Orf67 |
70934-71797 |
E |
S. pneumoniae |
conjugative transposon membrane protein |
Orf89 |
C 102845-102444 |
E |
S. pneumoniae |
regulator/toxin-antitoxin system, antitoxin component |
|
Orf68 |
72099-72509 |
E |
S. pneumoniae |
conjugative transposon membrane protein |
Orf90 |
103034-103555 |
E |
S. pneumoniae |
conserved hypothetical protein |
|
Orf69 |
72580-74823 |
E |
S. pneumoniae |
type IV conjugative transfer system protein |
Orf91 |
103825-104235 |
E |
S. pneumoniae |
sigma-70, region 4 |
| Orf70 | 74831-77410 | E | S. pneumoniae | conjugative transposon cell wall hydrolase/NlpC/P60 family | Orf92 | 104966-106712 | E | S. pneumoniae | site-specific recombinase, resolvase family |
The 100 kb insert has a modular composition
Bioinformatic analysis revealed that the insert has a modular composition. The 3’ end of the insert (module E) is homologous to Tn1806 of S. pneumoniae which confers erythromycin resistance. Although this element has not been shown to transfer via conjugation, transfer via transformation was shown [22]. In C. difficile strain M120 this element appears to be the backbone into which several other elements have been inserted (see Figure 1 top panel). The first 7.3 kb on the 5’ end of the insert (module A) has only moderate homology (60–70% maximum sequence identity) to known sequences. Interestingly, this part of the insert contains 2 putative modification DNA methylases and a putative endonuclease, possibly enabling a form of molecular vaccination as described by Kobayashi et al. [23]. During this process methylation protects the incoming element from host endonucleases and, following integration, will protect the host chromosome from endonucleases present on other mobile genetic elements. This sequence is followed by a complete prophage of approximately 39.5 kb (module B), which shows 92% sequence identity to a Thermoanaerobacter sp. prophage (Genbank accession no. CP002210). The next 4.5 kb stretch (module C) is 99% identical to part of the Enterococcus faecalis plasmid pEF418 containing, amongst others, a putative methyltransferase and a putative spectinomycin adenyltransferase (ant(9)Ia) [24]. It is also described to be part of a pathogenicity island in Streptococcus suis[25]. Finally, an insertion of approximately 4.5 kb (module D) with 90% sequence identity to the transferable pathogenicity island of Campylobacter fetus subsp fetus[26] is present within the sequence of Tn1806. This sequence contains, amongst others, putative tet(44) and ant(6)-Ib genes, which could respectively confer tetracycline and streptomycin resistance.
The G + C content of the entire insert (34%) was significantly higher than that of the entire genome (29%), clearly indicating that the insert was of foreign origin (see Additional file 1). In addition, within the insert the different modules could be distinguished by their G + C contents. The G + C content of module A, B, C, D and E was 31%, 41%, 35%, 28% and 31%, respectively.
The 100 kb insert is a transposon
Based on the bioinformatic comparison of the insert described above, the possible excision of 3 (independent) elements was predicted. Primers were designed (primers 14–20, see Table 3) to amplify the circular intermediates of the complete insert (primers 14 and 15), the putative Thermoanaerobacter sp. phage (module B, primers 15 and 16) and the C. fetus pathogenicity island (module D, primers 17 and 18) of the element. PCR confirmed only the excision and circularisation of the entire insert (results not shown). It is expected that the serine recombinase at the 3’ end of the element is responsible for excision (see Table 1). Sequencing of the circular intermediate was used to determine the precise ends of the element, showing the element is flanked by a TG dinucleotide; serine recombinases prefer a 2 bp crossover site identical in the target site and joint of the circular intermediate [27]. In silico extraction of this sequence from the genome confirms that the element is present in the homologous target site of CTn2 in strain 630 [7]. The precise size of the element is 106,711 bp and it runs from bp 418,525-525,236 (including the TG dinucleotide at both ends) in the M120 genomic sequence (GenBank accession no. FN665653). Upon our request, the transposon number Tn6164 was provided by the Transposon registry [28] (http://www.ucl.ac.uk/eastman/tn/index.php).
Table 3.
Oligonucleotides used in this study
| Name | Sequence | Purpose |
|---|---|---|
| 1 |
GAGATATGGTTATGAGATTAGG |
Presence/absence of insert |
| 2 |
CCCACCTTTATAGCATCATATAG |
Absence of insert |
| 3 |
CTAACCTATCAACTCAACCCC |
Presence of insert |
| 4 |
AGGATAAGACCGCAGCAGAA |
Presence 5’half of insert |
| 5 |
AAAAACGACGGTTTTCCTGTG |
Presence 5’half of insert |
| 6 |
GGGCAAATAGAAAGTCAAAACG |
Presence 3’half of insert |
| 7 |
AAGTGGTGTTTTCTTTGGAGGA |
Presence 3’half of insert |
| 8 |
CCACAGGGATACCTTTCTCGTGC |
Presence of tet(44) gene |
| 9 |
TTCCATATCCTCGGGTTTTTGCAT |
Presence of tet(44) gene |
| 10 |
CAGGTGTTGAAATAGATATTGAG |
Detect 3' end half insert |
| 11 |
CAGAAGTCGATCCTTTCTGGG |
Detect 3' end half insert |
| 12 |
GGTGGCTGAACTCGTTAATC |
Detect 3' end half insert |
| 13 |
CTCCACATGGCTCGAGTTG |
Detect 3' end half insert |
| 14 |
GAGGAATTTAACAGAACAGTATTT |
Excision studies |
| 15 |
TCTATCCTGCCTTCTCAACC |
Excision studies |
| 16 |
CGAATCGCTGAAATGACTGA |
Excision studies |
| 17 |
GCGAATGATTTCATGGAAGG |
Excision studies |
| 18 |
CGACTGCATTACCAGTTCCA |
Excision studies |
| Lok1 [13] |
AAAATATACTGCACATCTGTATAC |
Transconjugant screening |
| Lok3 [13] |
TTTACCAGAAAAAGTAGCTTTAA |
Transconjugant screening |
| 19 |
CAGCTGCAGTTTTTCCATGA |
Transconjugant screening |
| 20 |
GCAGCTAACGGTGATGACAA |
Transconjugant screening |
| Tn916 Fw [30] |
GACGGAAGATACTTATACA |
Transconjugant screening |
| Tn916 Rev [30] | GCCTTTGGATTCATTCCTGC | Transconjugant screening |
To test the conjugative transfer of the element, filter mating assays were performed, selecting for the possible tetracycline resistance by means of the tet(44) gene. However, M120 contains also a copy of tet(M) present on a conjugative transposon with 97% sequence identity to Tn916[16], which we have designated Tn6190. This element has inserted intragenically in the homologue of C. difficile strain 630 ORF CD2015. Tn6190 contains homologues to all Tn916 ORFs except orf12 which is involved in regulation of tet(M) through transcriptional attenuation [29].
During filter mating experiments with M120 as a donor strain and CD37 as a recipient, all putative transconjugants were identified as the recipient strain. In total 70 transconjugants were tested by PCR, using primers Lok1, Lok3 [13],[19,20], Tn916 Fw, and Tn916 Rev [30]. However, none contained Tn6164, all contained only Tn6190 (results not shown).
Tn6164 is sporadically present in PCR ribotype 078
Simultaneously with the publication of the M120 sequence, we obtained Illumina sequence reads of the C. difficile strain 31618, which was isolated from a diarrheic piglet from a pig farm in the Netherlands [16]. Comparative genomic analysis of 31618 to M120 revealed an almost complete overlap of the two genomes. However, reference assembly of the 31618 reads to M120 showed that Tn6164 was not present in 31618 (results not shown). This prompted us to investigate the prevalence of Tn6164 in PCR ribotype 078 strains. We designed a PCR to show presence (primers 1 and 3) or absence (primers 1 and 2) of Tn6164 in PCR ribotype 078 genomic DNA (see Figure 1 top panel). In addition, in view of the heterogeneous origin of Tn6164 and to investigate the presence of both the Thermoanaerobacter prophage and Streptococcus DNA (Modules B and E, respectively), we designed two more PCRs (primers 4–5 and 6–7). Finally, we designed a PCR to detect the presence of the tet(44) gene present on Tn6164 (Module D, primers 8 and 9). Besides the sequenced 31618 strain, 173 human PCR ribotype 078 strains and 58 porcine PCR ribotype 078 strains (from 27 pig farms) were tested for the presence of these elements.
A minority of the isolates tested did contain a DNA insert at the indicated location in the genome; 18 of the 231 isolates (7.8%) were positive in the 1–3 PCR (Table 2). Remarkably, all 18 strains were tetracycline resistant human isolates. None of the porcine strains contained an insert at the position tested. Strains positive in the 1–3 PCR were negative in the 1–2 PCR, and vice versa, showing complete complementarity of the two PCRs in PCR ribotype 078 strains.
Table 2.
Detection of specific regions of Tn6164 in PCR ribotype 078 strains
| Strain | PCR 1-2* | PCR1-3§ | PCR 4-5# | PCR 6-7 | PCR 8-9† | PCR 12-2‡ |
|---|---|---|---|---|---|---|
| 56/69 |
- |
+ |
+ |
- |
- |
+ |
| 26222 |
- |
+ |
+ |
- |
- |
+ |
| 26114 |
- |
+ |
+ |
- |
- |
+ |
| 26247 |
- |
+ |
+ |
- |
- |
+ |
| 26235 |
- |
+ |
+ |
- |
- |
+ |
| ES1203 |
- |
+ |
+ |
- |
- |
n.t. |
| 6065935 |
- |
+ |
+ |
- |
n.t. |
n.t. |
| 7047337 |
- |
+ |
+ |
- |
n.t. |
n.t. |
| 8088158 |
- |
+ |
+ |
- |
n.t. |
n.t. |
| 50/19 |
- |
+ |
+ |
+ |
+ |
- |
| GR0106 |
- |
+ |
+ |
+ |
+ |
n.t. |
| DE1210 |
- |
+ |
+ |
+ |
+ |
n.t. |
| BG1209 |
- |
+ |
+ |
+ |
+ |
n.t. |
| NO1311 |
- |
+ |
+ |
+ |
+ |
n.t. |
| NO1307 |
- |
+ |
+ |
+ |
+ |
n.t. |
| IE1102 |
- |
+ |
+ |
+ |
+ |
n.t. |
| GR0301 |
- |
+ |
+ |
+ |
+ |
n.t. |
| 10053737 | - | + | + | + | n.t. | n.t. |
*PCR only positive when no insert is present, §PCR only positive when insert is present #PCR detects Module B, ¶PCR detects module E, †PCR detects module D. ‡ PCR only positive in strains containing half of the element. Location of the oligonucleotides used is indicated in Figure 1. +, PCR positive; -, PCR negative; n.t., not tested.
Evidence for multiple insertions in Tn6164
All the strains that contained an insert (based on the 1–3 PCR) were further analyzed for the presence of Module B and E present in Tn6164, using primer pairs 4–5 and 6–7 (see Figure 1 top panel and Table 3). Only nine of 18 strains positive for PCR 1–3 were positive for PCRs 4–5 and 6–7, suggesting the presence of the complete element as described for M120. The other 9 strains were only positive for Module B (PCR 4–5), showing the existence of alternative (shorter) elements (see Table 2), as predicted by the bioinformatic analysis. The strains that were positive for Module E (PCR 6–7) were also positive for Module D (PCR 8–9, see Table 2). In contrast, strains containing Module B, but not Module E, thus containing only half the element, also lacked Module D. This indicates that the 3’end of half the element was situated upstream of Module D.
Of the isolates that were only positive for the PCR 4–5, the exact 3’end of the insert was determined by sequencing the PCR product obtained with primers 12 and 2 (see Figure 1 bottom panel), which yielded a 350 bp product. The border of the 3’end was between the 3’ end of Module C and the 5’end of Module E. A similar sequence was found at the homologous site when the full element was present, but also at the 3’ end of the full element, the 5’ end of the element, the joint of the circular intermediate and the predicted target site as based on the 630 sequence (see Table 4). This indicates that Tn6164 was created by two elements integrating in the same target site (next to each other) and fusing, with a second copy of the target site still present between the two original elements within Tn6164.
Table 4.
Sequences of the joints between the genome and Tn 6164 and the joint of the circular form
| CGCATTGCG-AGACTATAG |
3’ends of half insert |
| CGCATTGCG-AGACTATAG |
3’ends of full insert |
| CTCA-TGTGGAGTGCGTGG |
5’end of full insert |
| GCCA-TGTGGAGACTATAG |
middle section of full element |
| CACA-TGCGTTGTCTTGTG |
Joint of circular intermediate Tn6164 |
| CACATTGTG-AGACTGTAG | CTn2 target site in strain 630 |
Absence of Tn6164 sequences in other PCR ribotypes
Since PCR ribotype 126 has been shown to be very closely related to PCR ribotype 078, with an almost indistinguishable PCR ribotype banding pattern, we also tested a small collection of PCR ribotype 126 strains with the 1–2 and 1–3 PCRs. In none of the 10 PCR ribotype 126 strains tested could we demonstrate the presence of an insert at the site in which Tn6164 was inserted in M120 (results not shown).
In addition, a collection of 66 other PCR ribotypes was tested as well. This collection consisted of the 25 most frequently found PCR ribotypes in Europe, supplemented with the Leeds-Leiden collection [31]. None of the other PCR ribotypes, was positive for PCR 1–3, 4–5 or 6–7.
No antibiotic resistance phenotype linked to presence of Tn6164
Since several putative antibiotic resistance genes were found to be present on the element (see Figure 1 and Table 1), strains containing full Tn6164, only half of the element, or no element at all were tested for antibiotics resistance. Resistance to tetracycline, spectinomycin and streptomycin was tested using several methods (see materials and methods). Surprisingly, no correlation was found between the presence of tet(44), ant(6)Ib or ant(9)Ia and resistance to tetracycline, spectinomycin or streptomycin (see Table 5).
Table 5.
Antibiotic sensitivity of PCR ribotype 078 strains with.doc
| Genes present (transposon) | Strain | MIC Tet (μg/ml) | MIC Spec (μg/ml) | Strep |
|---|---|---|---|---|
| |
56/69 |
24 |
> 750 |
N.D. |
| |
26222 |
16 |
N.D. |
R |
|
ant(9)Ia (Tn6164) |
26114 |
32 |
N.D. |
R |
|
tet(M) (Tn6190) |
26247 |
16 |
> 750 |
R |
| |
26235 |
48 |
N.D. |
N.D. |
| |
06065935 |
8 |
N.D. |
R |
| |
50/19 |
48 |
>750 |
S |
| |
GR0106 |
12 |
>750 |
R |
|
ant(9)Ia (Tn6164) |
DE1210 |
8 |
>750 |
R |
|
ant(6) (Tn6164) |
BG1209 |
8 |
>750 |
R |
|
tet(44) (Tn6164) |
NO1311 |
12 |
>750 |
R |
|
tet(M) (Tn6190) |
NO1307 |
8 |
>750 |
R |
| |
IE1102 |
12 |
>750 |
R |
| |
GR0301 |
8 |
>750 |
R |
| |
10053737 |
N.D |
N.D |
R |
|
tet(M) (Tn6190) |
45/22 |
8 |
>750 |
N.D. |
| |
29/74 |
<8 |
>750 |
N.D. |
| |
31618 |
N.D. |
<250 |
N.D. |
| None |
07053152 |
<8 |
N.D. |
R |
| R20291(027) | N.D. | <250 | N.D. |
R, resistant (no halo around diffusion disk); S, sensitive (15 mm halo).
Strains containing full Tn6164 are all genetically related
Since we could not find many isolates containing Tn6164, we reasoned that the element could be relatively recently acquired and that the isolates thus might be genetically closely related. Therefore, we applied MLVA [3,16] on all the isolates containing Tn6164, or only half of it, supplemented with a number of isolates without the element, to investigate the genetic relatedness of the strains. In Figure 2, a minimal spanning tree of all the isolates containing an element is shown, with control strains. Based on the MLVA, all the isolates containing full Tn6164 (n = 9) are genetically related (STRD < 10) and four of them are in one clonal complex. Six isolates containing half of the element are also in this genetically related cluster, whereas the other three isolates containing half the element are not (STRD > 10).
Figure 2.
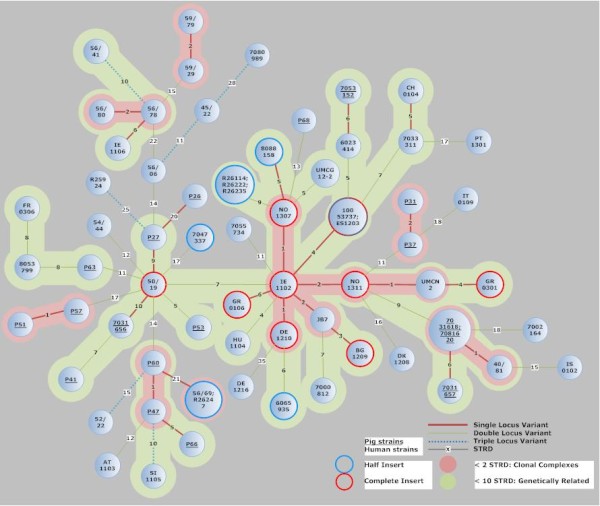
Minimum spanning tree of all the PCR ribotype 078 isolates that contained an insert (50 or 100 kb), supplemented with strains not containing the element. Each circle represents either one unique isolate or more isolates that have identical MLVA types. Red circles indicate strains with full Tn6164 and blue circles indicate strains with half the element. The numbers between the circles represent the summed tandem-repeat differences (STRD) between MLVA types. Underlined numbers represent porcine strains and normal numbers represent human isolates. Thick red lines represent single-locus variants; thin green lines represent double-locus variants and dotted blue lines represent triple locus variants between MLVA types. Clonal clusters are defined by an STRD of <2 (pink area), and genetically related clusters are defined by an STRD of <10 (green area)
Suggestive link between the 100 kb insert and increased virulence
To investigate a possible increased virulence of strains containing the element, clinical parameters of patients with a C. difficile infection due to a strain that contained Tn6164 were compared to parameters of patients that suffered from a strain that did not contain the full element. Patients with Tn6164 resembled patients without the element concerning demographic characteristics. Clinical characteristics were only known for patients from the ECDIS study [32] and patients registered in the CDRL (n = 84). Patients with and without the element suffered from severe diarrhea in similar proportions. Mortality due to CDI was more common in patients infected with C. difficile::Tn6164 (29% vs 3%). This suggests that Tn6164 might convert PCR ribotype 078 strains to a more virulent strain. However, since the number of patients infected with a Tn6164-positive strain, and for which the clinical data was available, was very low (n = 7), no multivariate analysis could be performed, which means that a bias cannot be ruled out. Further research is needed to confirm a possible link between increased virulence and the presence of Tn6164.
Discussion
PCR ribotype 078 has recently emerged as a hypervirulent C. difficile strain [2,3]. Previously published MLVA studies have shown that all PCR ribotype 078 strains are closely related [3], irrespective of human or porcine origin [16], fostering the notion that PCR ribotype 078 infection could be a zoonosis. Recently, the full genome sequence of a C. difficile PCR ribotype 078 strain was published [5]. This M120 strain was shown to contain a unique insert of approximately 100 kilobases. In this paper we show that this insert is a transposable element, Tn6164. It is not representative for all PCR ribotype 078 strains. On the contrary, we found that the majority of the PCR ribotype 078 strains do not contain the element. Moreover, some strains contain only half of the element. So, three different kinds of PCR ribotype 078 can now be distinguished: Those with a full length element, those with half the element, and those with no element at all. Tn6164 was exclusively found in tetracycline resistant PCR ribotype 078 strains, isolated from humans. We tested a collection of other PCR ribotypes, of which none contained the element. Since we only tested 1 strain per PCR ribotype, we cannot rule out the possibility that Tn6164 is present in other PCR ribotypes. We covered the whole genomic spectrum of C. difficile since we tested multiple samples of each genetic clade previously identified [10,33-35]. In addition, Tn6164 has not been found in any other C. difficile genome that has been published so far than M120.
Although Tn6164 contained a tet(44) gene, we could not demonstrate increased tetracycline resistance of strains containing the element. Previously, it has been shown that this gene, present on a homologues resistance island, is active in C. fetus[26]. In C. difficile, the copresence of the tet(44) gene on Tn6164 and the tet(M) gene on the Tn6190 in one bacterium does not result in an increased resistant phenotype. Also the spectinomycin and streptomycin resistance genes did not result in a phenotype, despite the presence of two potential aminoglycoside resistance genes (ant(9)Ia) and ant(6)) on Tn6164 (see Figure 1 and Table 1). We do not know if the resistance genes are expressed in M120. However, since we show the presence of the circular intermediate transposon DNA, some activity of transposon related genes is expected.
Since we have only found Tn6164 in strains also containing Tn6190, it is possible that Tn6164 transfer is dependent on Tn6190. Further research is needed to investigate the possibility of Tn6190-dependent transfer of Tn6164. In addition, remarkably, Tn6164 (the whole or half the element) was significantly (p = 0.01) more found in strains isolated from humans than in strains isolated from pigs. Although the same strains circulate in humans and pigs [16], and also Tn6190 circulates in pig strains [16], we did not find any porcine strain that contained the element. We have no explanation for this difference.
None of the transconjugants tested showed the presence of Tn6164, but all contained Tn6190. These results indicate that Tn6164 has a (much) lower transfer frequency than Tn6190. Nevertheless, a complete set of proteins, required for transfer, is present on Tn6164. Loss of Tn6190 or introduction of another selection marker in Tn6164[11] could prove to be a strategy to further study the capability of conjugative transfer of this element.
Tn6164 has integrated intergenically between homologs of the 630 ORFs CD0406 and CD0437, a tRNA methyltransferase and a hypothetical protein respectively. In strain 630, this target site is occupied by the conjugative transposon CTn2[7,11]. There is no significant homology between Tn6164 and CTn2. The empty target site is present in many sequenced strains of C. difficile. However, no other mobile genetic elements have been reported to integrate at this site.
It was impossible to phenotypically distinguish strains containing Tn6164 from strains without the element. Although we have no transcriptional data available of the genes that are located on Tn6164 it is clear that it could provide an advantage under certain circumstances. In this respect it is interesting to note that the patients suffering from an element-containing strain are suggested to undergo a more severe illness than patients with a strain not containing Tn6164. However, because of the low number of strains containing the insert no multivariate analysis could be carried out. Therefore, we cannot rule out that these data are biased. Further research is needed to confirm this observation.
Isolates containing the full element originated from all over Europe, including Ireland, England, Norway, Germany, Bulgaria, Greece and the Netherlands, whereas isolates containing half the element were only found in the United Kingdom, Spain and the Netherlands. MLVA showed genetic relatedness between most of the strains, although no epidemiologic link between the strains from different countries could be found. It has recently been shown that PCR ribotype 078 strains show a lot less heterogeneity in MLVA than for instance PCR ribotype 027 or PCR ribotype 017 [36-38]. This could indicate a higher level of relatedness, or it could mean that the mechanism behind the MLVA variability is different in PCR ribotype 078 strains than in other PCR ribotypes [16].
Altogether, we show the presence of a 100 kb transposon in some C. difficile PCR ribotype 078 strains. Although we could not show any evolutionary benefits of the transposon, it could very well serve as a reservoir of antibiotic resistance [26], for commensal bacteria in the human gut.
Conclusions
Tn6164 is a novel transposon of approximately 100 kb, found sporadically in Clostridium difficile PCR ribotype 078 strains, isolated from humans. Tn6164 has a modular composition and is the product of multiple insertions of separate elements from various origins, as evidenced by the existence of strains containing only half the element. Strains containing Tn6164 were all genetically related. We were not able to find a readily distinguishable phenotype for strains containing the element, although several potential antibiotic resistance genes were present on Tn6164. Tn6164 may act as a source of antibiotic resistance genes in the human gut. Further research is needed to investigate if Tn6164 plays a role in the virulence of PCR ribotype 078 Clostridium difficile strains.
Methods
Bacterial Isolates and culture conditions
PCR ribotype 078 C. difficile strain 31618 was obtained from a pig farm in the eastern part of the Netherlands where neonatal diarrhea was present. Culturing of the feces yielded C. difficile, as determined by an in-house PCR for the presence of the gluD gene encoding the glutamate dehydrogenase specific for C. difficile[39]. PCR ribotype was determined as previously described [40].
The other PCR ribotype 078 strains used in this study were obtained from a previously described PCR ribotype 078 strain collection [16], consisting of strains isolated from humans and pigs, supplemented with human PCR ribotype 078 strains from the ECDIS (European Clostridium difficile Infection Survey) study in 2010 [32]. In addition, recently isolated PCR ribotype 078 strains from Dutch diarrheic piglets (2007–2010) and human (2006–2010) strains collected by the Dutch C. difficile Reference Laboratory (CDRL) were used. The 58 Pig strains were collected on 27 pig farms in the Netherlands.
PCR ribotype 126 strains used in this study originate from the ECDIS study, isolated in 2010, from several countries in Europe [32]. PCR ribotype reference strains (n = 68) were obtained from the CDRL.
The nontoxinogenic strain CD37 [41,42] was used as a recipient in filter mating experiments as this has previously been shown to be a good recipient for mobile genetic elements from other C. difficile strains [11].
C. difficile strain M120 was kindly provided by Dr. Trevor Lawley (Sanger Institute). Standard culturing of C. difficile isolates was carried out on blood agar plates at 37°C and anaerobic conditions.
DNA Sequencing, reference assembly and annotation
DNA was isolated from one colony of the 31618 strain by standard techniques [43]. The isolate was sequenced using the Illumina platform (Solexa) at the Leiden Genome Technology Center (LGTC) at the LUMC, using the manufacturers’ protocols. Single end reads were generated and submitted to the NCBI sequence read archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRX030155. A reference assembly of the reads was carried out against strain C. difficile PCR ribotype 078 strain M120 (GenBank accession no. FN665653), using CLC genomics workbench (CLCbio, Aarhus, Denmark). Number of reads used was 5267302, of which 2968638 reads could be mapped to the M120 genome sequence. The unique 100 kb insert present in M120 was readily identified with the CLC genomics workbench. The ORFs present in the insert were identified by CLC genomics workbench and annotation was carried out manually, using BLAST and SMART. ORFs identified as “protein of unknown function” were further analyzed by profile-profile searches through HHpred (http://toolkit. tuebingen.mpg.de/hhpred).
Bioinformatic comparison of the mixed origin of Tn6164
The genome of strain M120 was compared to the genomes of C. difficile 630 (Genbank accession no. AM180355), Thermoanaerobacter sp. (GenBank accession no. CP002210), S. pneumonia (Genbank accession no. CP002121) and C. fetus (Genbank accession no. FN594949) using the Artemis Comparison Tool [44].
Circularization of the transposon
In order to investigate if the putative element could excise itself from the genome, PCR analysis was performed to amplify the joint region of a circular molecule using primers at the ends of the element, facing outward (primers 14 and 15 in Table 3). PCR amplifications were carried out using the NEB Taq Polymerase kit (New England Biolabs, Herts, UK) according to the manufacturer’s instructions with 10 mM dNTPs (NEB). The primers that were used are listed in Table 3 (Sigma-Genosys, UK).
Filter-matings assays
Filter-matings were carried out as described previously [45]. C. difficile strains M120 and CD37 were cultured on Brain heart infusion (BHI) (Oxoid Ltd.) agar supplemented with 5% Horse blood (E&O laboratories). C. difficile strain CD37 was used as recipient. Transconjugants were selected for on BHI plates supplemented with 25 μg/ml rifampicin (Sigma Aldrich) and 10 μg/ml tetracycline (Sigma Aldrich). Transconjugants were examined using PCR with primer pair Lok1/Lok3 to confirm identity of the recipient strain and primer pairs Tn6164 accessory region Fw + Rev and Tn916 Fw + Rev to confirm the transfer of Tn6164 or Tn6190.
Inverse PCR
C. difficile genomic DNA was digested with PstI or EcoRI. After purification, the genomic DNA fragments were self-ligated to create circular DNAs. Subsequently, the DNA was precipitated and dissolved in H2O. PCR was carried out on the DNA, using primers 4-rev and 5-rev or 14 and 15 (annealing at 58°C, 35 cycles). PCR products were visualized by gel electrophoresis and sequences were determined through direct sequencing on the purified PCR amplicons or through cloning into pCR2.1/TOPO (Invitrogen) and subsequent sequencing with the plasmid-located primers T7 and M13 reverse.
Antibiotic resistance
The MIC for tetracycline was determined using E tests (BioMérieux, Boxtel, the Netherlands) on blood plates under anaerobic conditions at 37°C. Breakpoint for tetracycline was 8 μg/ml. Spectinomycin resistance was determined by an agar dilution method of C. difficile colonies on BHI agar plates, supplemented with increasing amounts of spectinomycin. Streptomycin resistance was tested by disk diffusion method, using Sensi-Neotabs (Rosco, Denmark) (Streptomycin 500 ug disks) on blood plates under anaerobic conditions at 37°C.
Oligonucleotides
Oligonucleotides used in this study are shown in Table 3.
PCR
PCRs were carried out using Gotaq polymerase (Promega, Leiden, the Netherlands). Reactions contained 0.4 mM dNTPs, 0.4 uM oligonucleotides. Annealing temperature of the PCR was set at 50°C and PCRs were standardized at 30 cycles.
Statistical analyses
Patients samples with the full 100 kb insert were compared to patients samples with a part of the insert or no insert. The Chi-square test and t-test were used to calculate the p-value. Analyses were performed using the SPSS for Windows software package, version 17.0.
MLVA
Sixty eight strains were subjected to MLVA, of which 39 were previously characterized [16]. MLVA and construction of the minimal spanning tree based on the MLVA results were carried out as described previously [16].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JC designed the study, carried out PCRs, antibiotic resistance assays, analyzed the data and wrote the paper; DB carried out sequencing and analyzed the data; MB carried out the circularization and filter mating experiments and wrote the paper; CH managed the strain collections and carried out MLVA; MH carried out statistical analysis and wrote the paper; AM carried out filter mating experiments and wrote the paper; LL gathered pig samples; EK designed the study and wrote the paper; HL designed the study, analyzed data and wrote the paper. All authors read and approved the final manuscripts.
Supplementary Material
Circular representation of the genome of C. difficile strain M120.The two concentric circles represent the genome (outer circle) and the G + C content (inner circle; window size 10,000; Step size 200). Green represents values higher than average (29%), purple below average. In between the two circles, the presence of the two transposable elements is indicated in red (Tn6164) and blue (Tn6190). Figure was created using DNA plotter [46].
Contributor Information
Jeroen Corver, Email: j.corver@lumc.nl.
Dennis Bakker, Email: d.bakker@lumc.nl.
Michael S M Brouwer, Email: m.brouwer@ucl.ac.uk.
Céline Harmanus, Email: c.harmanus@lumc.nl.
Marjolein P Hensgens, Email: m.p.m.hensgens@lumc.nl.
Adam P Roberts, Email: adam.roberts@ucl.ac.uk.
Len J A Lipman, Email: l.j.a.lipman@uu.nl.
Ed J Kuijper, Email: e.j.kuijper@lumc.nl.
Hans C van Leeuwen, Email: h.c.van_leeuwen@lumc.nl.
Acknowledgements
This study was supported by HYPERDIFF-The Physiological Basis of Hypervirulence in Clostridium difficile: a Prerequisite for Effective Infection Control (Health-F3-2008-223585), and by ZonMW (NWO; the Netherlands Organization for Scientific Research) grant “Reduction of community health risks of animal-associated Clostridium difficile” (project number 50-50800-98-075).
APR is supported by the Medical Research Council (grant no. G0601176).
References
- Pepin J, Valiquette L, Cossette B. Mortality attributable to nosocomial Clostridium difficile-associated disease during an epidemic caused by a hypervirulent strain in Quebec. CMAJ. 2005;173:1037–1042. doi: 10.1503/cmaj.050978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorhuis A, Debast SB, van Leengoed LA, Harmanus C, Notermans DW, Bergwerff AA. et al. Clostridium difficile PCR ribotype 078: an emerging strain in humans and in pigs? J Clin Microbiol. 2008;46:1157. doi: 10.1128/JCM.01536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorhuis A, Bakker D, Corver J, Debast SB, Harmanus C, Notermans DW. et al. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction Type 078. Clin Infect Dis. 2008;47:1162–1170. doi: 10.1086/592257. [DOI] [PubMed] [Google Scholar]
- Debast SB, van Leengoed LA, Goorhuis A, Harmanus C, Kuijper EJ, Bergwerff AA. Clostridium difficile PCR ribotype 078 toxinoType V found in diarrhoeal pigs identical to isolates from affected humans. Environ Microbiol. 2009;11:505–511. doi: 10.1111/j.1462-2920.2008.01790.x. [DOI] [PubMed] [Google Scholar]
- He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ. et al. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci USA. 2010;107:7527–7532. doi: 10.1073/pnas.0914322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C. et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R. et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- Forgetta V, Oughton MT, Marquis P, Brukner I, Blanchette R, Haub K. et al. Fourteen-Genome Comparison Identifies DNA Markers for Severe-Disease-Associated Strains of Clostridium difficile. J Clin Microbiol. 2011;49:2230–2238. doi: 10.1128/JCM.00391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden GL, Davis IJ, Wright VJ, Sebaihia M, Kuijper EJ, Minton NP. Array comparative hybridisation reveals a high degree of similarity between UK and European clinical isolates of hypervirulent Clostridium difficile. BMC Genomics. 2010;11:389. doi: 10.1186/1471-2164-11-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler RA, Gerding DN, Songer JG, Drudy D, Brazier JS, Trinh HT. et al. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J Bacteriol. 2006;188:7297–7305. doi: 10.1128/JB.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer MSM, Warburton PJ, Roberts AP, Mullany P, Allan E. Genetic Organisation, Mobility and Predicted Functions of Genes on Integrated, Mobile Genetic Elements in Sequenced Strains of Clostridium difficile. PLoS One. 2011;6:e23014. doi: 10.1371/journal.pone.0023014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan KS, Wee BY, Song KP. Evidence for holin function of tcdE gene in the pathogenicity of Clostridium difficile. J Med Microbiol. 2001;50:613–619. doi: 10.1099/0022-1317-50-7-613. [DOI] [PubMed] [Google Scholar]
- Braun V, Hundsberger T, Leukel P, Sauerborn M, von Eichel-Streiber C. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene. 1996;181:29–38. doi: 10.1016/S0378-1119(96)00398-8. [DOI] [PubMed] [Google Scholar]
- Govind R, Vediyappan G, Rolfe RD, Dupuy B, Fralick JA. Bacteriophage-mediated toxin gene regulation in Clostridium difficile. J Virol. 2009;83:12037–12045. doi: 10.1128/JVI.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulovic O, Meessen-Pinard M, Fortier LC. Prophage-Stimulated Toxin Production in Clostridium difficile NAP1/027 Lysogens. J Bacteriol. 2011;193:2726–2734. doi: 10.1128/JB.00787-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker D, Corver J, Harmanus C, Goorhuis A, Keessen EC, Fawley WN. et al. Relatedness of human and animal Clostridium difficile PCR ribotype 078 isolates determined on the basis of multilocus variable-number tandem-repeat analysis and tetracycline resistance. J Clin Microbiol. 2010;48:3744–3749. doi: 10.1128/JCM.01171-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams V, Lyras D, Farrow KA, Rood JI. The clostridial mobilisable transposons. Cell Mol Life Sci. 2002;59:2033–2043. doi: 10.1007/s000180200003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AP, Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 2009;17:251–258. doi: 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Brouwer MSM, Roberts AP, Mullany P, Allan E. In silico analysis of sequenced strains of Clostridium difficile reveals a related set of conjugative transposons carrying a variety of accessory genes. Mobile Genetic Elements. 2012;2 doi: 10.4161/mge.19297. http://dx.doi.org/10.4161/mge.2.1.19297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullany P, Wilks M, Lamb I, Clayton C, Wren B, Tabaqchali S. Genetic analysis of a tetracycline resistance element from Clostridium difficile and its conjugal transfer to and from Bacillus subtilis. J Gen Microbiol. 1990;136:1343–1349. doi: 10.1099/00221287-136-7-1343. [DOI] [PubMed] [Google Scholar]
- Wang H, Roberts AP, Lyras D, Rood JI, Wilks M, Mullany P. Characterization of the ends and target sites of the novel conjugative transposon Tn5397 from Clostridium difficile: excision and circularization is mediated by the large resolvase, TndX. J Bacteriol. 2000;182:3775–3783. doi: 10.1128/JB.182.13.3775-3783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli R, Del GM, Iannelli F, Pantosti A. New genetic element carrying the erythromycin resistance determinant erm(TR) in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2008;52:619–625. doi: 10.1128/AAC.01081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3") (9) Mol Gen Genet. 1985;200:33–39. doi: 10.1007/BF00383309. [DOI] [PubMed] [Google Scholar]
- Chen C, Tang J, Dong W, Wang C, Feng Y, Wang J. et al. A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS One. 2007;2:e315. doi: 10.1371/journal.pone.0000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abril C, Brodard I, Perreten V. Two novel antibiotic resistance genes, tet(44) and ant(6)-Ib, are located within a transferable pathogenicity island in Campylobacter fetus subsp. fetus. Antimicrob Agents Chemother. 2010;54:3052–3055. doi: 10.1128/AAC.00304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MC, Thorpe HM. Diversity in the serine recombinases. Mol Microbiol. 2002;44:299–307. doi: 10.1046/j.1365-2958.2002.02891.x. [DOI] [PubMed] [Google Scholar]
- Roberts AP, Chandler M, Courvalin P, Guedon G, Mullany P, Pembroke T. et al. Revised nomenclature for transposable genetic elements. Plasmid. 2008;60:167–173. doi: 10.1016/j.plasmid.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YA, He P, Clewell DB. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob Agents Chemother. 1992;36:769–778. doi: 10.1128/AAC.36.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciric L, Mullany P, Roberts AP. Antibiotic and antiseptic resistance genes are linked on a novel mobile genetic element: Tn6087. J Antimicrob Chemother. 2011;66:2235–2239. doi: 10.1093/jac/dkr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knetsch CW, Hensgens MPM, Harmanus C, van der Bijl MW, Savelkoul PH, Kuijper EJ. et al. Genetic markers for Clostridium difficile lineages linked to hypervirulence. Microbiology. 2011;157:3113–3123. doi: 10.1099/mic.0.051953-0. [DOI] [PubMed] [Google Scholar]
- Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, Rupnik M. et al. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377:63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R. et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. 2010;48:770–778. doi: 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler RA, Dawson LF, Valiente E, Cairns MD, Martin MJ, Donahue EH. et al. Macro and Micro Diversity of Clostridium difficile Isolates from Diverse Sources and Geographical Locations. PLoS One. 2012;7:e31559. doi: 10.1371/journal.pone.0031559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle KE, Griffiths D, Didelot X, Evans J, Vaughan A, Kachrimanidou M. et al. Clinical Clostridium difficile: clonality and pathogenicity locus diversity. PLoS One. 2011;6:e19993. doi: 10.1371/journal.pone.0019993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawley WN, Freeman J, Smith C, Harmanus C, van den Berg RJ, Kuijper EJ. et al. Use of highly discriminatory fingerprinting to analyze clusters of Clostridium difficile infection cases due to epidemic Type 027 strains. J Clin Microbiol. 2008;46:954–960. doi: 10.1128/JCM.01764-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg RJ, Schaap I, Templeton KE, Klaassen CH, Kuijper EJ. Typing and subtyping of Clostridium difficile isolates by using multiple-locus variable-number tandem-repeat analysis. J Clin Microbiol. 2007;45:1024–1028. doi: 10.1128/JCM.02023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorhuis A, Legaria MC, van den Berg RJ, Harmanus C, Klaassen CH, Brazier JS. et al. Application of multiple-locus variable-number tandem-repeat analysis to determine clonal spread of toxin A-negative Clostridium difficile in a general hospital in Buenos Aires, Argentina. Clin Microbiol Infect. 2009;15:1080–1086. doi: 10.1111/j.1469-0691.2009.02759.x. [DOI] [PubMed] [Google Scholar]
- Paltansing S, van den Berg RJ, Guseinova RA, Visser CE, van der Vorm ER, Kuijper EJ. Characteristics and incidence of Clostridium difficile-associated disease, The Netherlands, 2005. Clin Microbiol Infect. 2007;13:1058–1064. doi: 10.1111/j.1469-0691.2007.01793.x. [DOI] [PubMed] [Google Scholar]
- Bidet P, Barbut F, Lalande V, Burghoffer B, Petit JC. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett. 1999;175:261–266. doi: 10.1111/j.1574-6968.1999.tb13629.x. [DOI] [PubMed] [Google Scholar]
- Hachler H, Kayser FH, Berger-Bachi B. Homology of a transferable tetracycline resistance determinant of Clostridium difficile with Streptococcus (Enterococcus) faecalis transposon Tn916. Antimicrob Agents Chemother. 1987;31:1033–1038. doi: 10.1128/AAC.31.7.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer MS, Allan E, Mullany P, Roberts AP. Draft Genome Sequence of the Nontoxigenic Clostridium difficile Strain CD37. J Bacteriol. 2012;194:2125–2126. doi: 10.1128/JB.00122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg RJ, Claas EC, Oyib DH, Klaassen CH, Dijkshoorn L, Brazier JS. et al. Characterization of toxin A-negative, toxin B-positive Clostridium difficile isolates from outbreaks in different countries by amplified fragment length polymorphism and PCR ribotyping. J Clin Microbiol. 2004;42:1035–1041. doi: 10.1128/JCM.42.3.1035-1041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG. et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain HA, Roberts AP, Mullany P. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Deltaerm) and demonstration that the conjugative transposon Tn916DeltaE enters the genome of this strain at multiple sites. J Med Microbiol. 2005;54:137–141. doi: 10.1099/jmm.0.45790-0. [DOI] [PubMed] [Google Scholar]
- Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Circular representation of the genome of C. difficile strain M120.The two concentric circles represent the genome (outer circle) and the G + C content (inner circle; window size 10,000; Step size 200). Green represents values higher than average (29%), purple below average. In between the two circles, the presence of the two transposable elements is indicated in red (Tn6164) and blue (Tn6190). Figure was created using DNA plotter [46].