Abstract
Heterotrimeric G protein signaling is essential for normal hyphal growth in the filamentous fungus Neurospora crassa. We have previously demonstrated that the non-receptor guanine nucleotide exchange factor RIC8 acts upstream of the Gα proteins GNA-1 and GNA-3 to regulate hyphal extension. Here we demonstrate that regulation of hyphal extension results at least in part, from an important role in control of asexual spore (conidia) germination. Loss of GNA-3 leads to a drastic reduction in conidial germination, which is exacerbated in the absence of GNA-1. Mutation of RIC8 leads to a reduction in germination similar to that in the Δgna-1, Δgna-3 double mutant, suggesting that RIC8 regulates conidial germination through both GNA-1 and GNA-3. Support for a more significant role for GNA-3 is indicated by the observation that expression of a GTPase-deficient, constitutively active gna-3 allele in the Δric8 mutant leads to a significant increase in conidial germination. Localization of the three Gα proteins during conidial germination was probed through analysis of cells expressing fluorescently tagged proteins. Functional TagRFP fusions of each of the three Gα subunits were constructed through insertion of TagRFP in a conserved loop region of the Gα subunits. The results demonstrated that GNA-1 localizes to the plasma membrane and vacuoles, and also to septa throughout conidial germination. GNA-2 and GNA-3 localize to both the plasma membrane and vacuoles during early germination, but are then found in intracellular vacuoles later during hyphal outgrowth.
Introduction
Since the discovery of the first heterotrimeric G protein in filamentous fungi in the 1990's [1], G proteins have been found to play key roles in diverse fungal processes ranging from asexual and sexual development to pathogenicity of animal and phytopathogenic fungi (reviewed in Li et al, 2007). Most fungi possess three Gα subunits and a single Gβ and Gγ protein, therefore allowing for the assembly of three different heterotrimers. These three Gα subunits can act independently to regulate separate pathways, leading to differing phenotypes for single Gα mutants. For example, Neurospora crassa GNA-1 is required for normal vegetative growth, aerial hyphae formation and female fertility [2], whereas GNA-3 is required for normal production of asexual spores (conidia) and maturation of sexual spores (ascospores) [3]. In contrast, the N. crassa Δgna-2 mutant displays only a mild phenotype during growth on poor carbon sources [4]. However, loss of GNA-2 exacerbates phenotypes of the Δgna-1 and Δgna-3 mutants, indicating that GNA-2 shares overlapping functions with the other two Gα subunits [5], [6]. Indeed, all three G proteins are thought to act together to regulate certain processes, as mutants lacking GNA-1 and GNA-3 or all three Gα subunits are severely impaired in growth on solid medium, inappropriately conidiate in submerged liquid culture and do not produce female reproductive structures [6].
G protein coupled receptors (GPCRs), act as guanine nucleotide exchange factors (GEFs) for Gα subunits, facilitating exchange of GDP for GTP, thereby leading to activation and dissociation from the Gβγ dimer (reviewed in Li et al, 2007). However, recently a non-receptor GEF capable of activating Gα proteins, RIC8, has been identified in both animals and some fungi [7], [8], [9]. In N. crassa, loss of ric8 leads to a severe growth impairment phenotype similar to that in mutants lacking both gna-1 and gna-3 or all three Gα subunit genes [9]. Expression of GTPase-deficient gna-1 or gna-3 alleles rescued many of the defects of the Δric8 mutant during asexual growth on solid medium, and biochemical analyses showed that RIC8 can act as a GEF for both GNA-1 and GNA-3 in vitro, suggesting RIC8 acts upstream of both GNA-1 and GNA-3, particularly during regulation of asexual growth on solid medium [9]. Asexual hyphal growth is important for nutrient scavenging and for the organism to spread throughout the environment. In addition, it is important for encountering a mate of the opposite mating type, which allows the sexual cycle to proceed and produce the environmentally resistant sexual spores (ascospores).
Using a strain expressing a functional RIC8-GFP fusion, we have previously shown that RIC8 is a cytoplasmic protein [9]. Production of Gα-fluorescent protein fusions is problematic, as an N or C-terminal tag can interfere with normal functioning of the Gα protein. However, Dictyostelium discoideum Gα2 and mammalian Gαo have been successfully tagged by insertion of GFP in a fold where it does not interfere with Gαo function [10], [11].
In this study we further probe the role of RIC8, GNA-1 and GNA-3 in asexual hyphal growth and development. We analyze conidial morphology and determine conidial germination rates in ric8 and G protein subunit mutants and in Δric8 strains carrying GTPase-deficient alleles of gna-1 or gna-3. We produce strains expressing GNA-1, GNA-2 and GNA-3 proteins as internal TagRFP fusions. We present here the first use of this internal tagging method for localization of Gα proteins in filamentous fungi. Using these strains, we determine the localization pattern of Gα proteins in conidia and during conidial germination and test for colocalization of RIC8 with GNA-1 and GNA-3.
Materials and Methods
Strains and growth conditions
Neurospora strains used in this study are listed in Table 1. For vegetative growth analysis, strains were grown on Vogel's minimal medium (VM; [12]). To induce the formation of female structures (protoperithecia) required for sexual crossing strains were grown on synthetic crossing medium (SCM; [13]). Cultures were inoculated with conidia and grown as described previously [4], [14].
Table 1. Strains used in this study.
| Strain | Relevant genotype | Comments | Source |
| 74-OR23-IVA | Wild type, mat A | FGSC 2489a | FGSC |
| r81-5a | Δric8::hph+ mat a | Δric8 mutant | [9] |
| 3B10 | Δgna-1::hph+ mat a | Δgna-1 mutant | [38] |
| FGSC 12378 | Δgna-2::hph+ mat a | Δgna-2 mutant | FGSC |
| 31c2 | Δgna-3::hph+ mat A | Δgna-3 mutant | [3] |
| g1.3 | Δgna-1::hph+ Δgna-3::hph+ mat a | Δgna-1 Δgna-3 double mutant | [6] |
| noa | Δgna-1::hph+ Δgna-2::pyrG+Δgna-3::hph+ mat A | Δgna-1 Δgna-2 Δgna-3 triple mutant | [6] |
| R81* | Δric8::hph+ gna-1 Q204L ::his-3+ mat A | gna-1 Q204L in Δric8 background | [9] |
| R82* | Δric8::hph+ gna-2 Q205L ::his-3+ mat A | gna-2 Q205L in Δric8 background | [9] |
| R83* | Δric8::hph+ gna-3 Q208L ::his-3+ mat A | gna-3 Q208L in Δric8 background | [9] |
| 42-8-3 | Δgnb-1, mat A | Δgnb-1 mutant | [43] |
| 5-5-3 | Δgng-1, mat A | Δgng-1 mutant | [18] |
| Δ1his3 | Δgna-1::hph+, his-3, mat A | This Study | |
| Δ2his3 | Δgna-2::pyrG+, his-3, mat a | This Study | |
| 31 h | Δgna-3::hph+, his-3, mat A | This Study | |
| 2-1 | Δgna-1::hph+, gna-1-TagRFP::his-3+ mat A | Expresses GNA-1-TagRFP | This Study |
| 5-1 | Δgna-2::hph+, gna-2-TagRFP::his-3+ mat a | Expresses GNA-2-TagRFP | This Study |
| 12-1 | Δgna-3::hph+, gna-3-TagRFP::his-3+ mat A | Expresses GNA-3-TagRFP | This Study |
| R8GFP | Δric8::hph+, ric8-GFP::his-3+, mat a | Expresses RIC8-GFP | [9] |
FGSC, Fungal Genetics Stock Center, Kansas City, MO.
Gα-TagRFP strain construction
To observe the cellular localization of GNA-1, GNA-2 and GNA-3, TagRFP [15] was inserted into a conserved loop region within the Gα protein. This conserved loop region was found to be optimal for insertion of tags with conservation of Gα protein function in Dictyostelium discoideum Gα2 [11] and chinese hamster Gnao [10]; see Fig. 1). Primers were designed to prepare the Gα-TagRFP fusion constructs using yeast recombinational cloning, and are listed in Table 2. TagRFP was amplified from pAL3-Lifeact [15]; provided by Nick Read, University of Edinburgh) and the appropriate Gα N- and C-terminal fragments were amplified from cDNA clones. These fragments were inserted into pRS426 using yeast recombinational cloning [16]. The Gα-TagRFP fusion construct was then subcloned from pRS426 into pMF272 [17] as an EcoRI/XbaI fragment, resulting in replacement of sgfp from pMF272 with the Gα-TagRFP fusion and placing it under the control of the ccg-1 promoter. The fusion constructs were then transformed into his-3 Δgna-1, Δgna-2 or Δgna-3 gene replacement mutants (See Table 1). Transformants were then screened by Southern blotting to ensure correct integration of the construct (data not shown).
Figure 1. Alignment of Gα proteins.
Amino acid alignment of N. crassa GNA-1, GNA-2, GNA-3 and chinese hamster GNAO1 (Genbank accession number ABA77543.1), showing position of the conserved loop into which TagRFP was inserted.
Table 2. Primers used in this study.
| Name | Anneals to | Sequence (5′–3′) |
| pRS426g1-F1 | pRS426/N′ gna-1 | ACGCCAGGGTTTTCCCAGTCACGACTCTAGAATGGGTTGCGGAATGAGTACAGAGGAG |
| g1TagRFP-R1 | N′ gna-1/TagRFP | GCATGTTCTCCTTAATCAGCTCGCTCATCTCTAGGGACTCCATGGCCTCGAGAATG |
| g1TagRFP-F1 | TagRFP/C′ gna-1 | CGACCTCCCTAGCAAACTGGGGCACAAGTTGCCACTCGCCGATCAGCGCGTCGAG |
| pRS426g1-R1 | C′ gna-1/pRS426 | GCGGATAACAATTTCACACAGGAAACAGCGAATTCTCAAATCAAACCGCAGAGACGCAGG |
| TagRFP-F | TagRFP | ATGAGCGAGCTGATTAAGGAG |
| TagRFP-R | TagRFP | CTTGTGCCCCAGTTTGCTAGG |
| pRS426g3-F1 | pRS426/N′ gna-3 | GTAACGCCAGGGTTTTCCCAGTCACGACTCTAGAATGGGCGCATGCATGAGCAAGAACG |
| g3TagRFP-R1 | N′ gna-3/TagRFP | GCATGTTCTCCTTAATCAGCTCGCTCATATCAAACTGGTGCATAGCATTCACAAC |
| g3TagRFP-F1 | TagRFP/C′ gna-3 | GCGACCTCCCTAGCAAACTGGGGCACAAGATCCAGCCAGCAGATCCGTCGCTACGG |
| pRS426g3-R1 | C′ gna-3/pRS426 | CGGATAACAATTTCACACAGGAAACAGCGAATTCTCATAGAATACCGGAGTCTTTAAGGG |
| pRS426g2-F1 | pRS426/N′ gna-2 | GTAACGCCAGGGTTTTCCCAGTCACGACTCTAGAATGTGTTTCGGGGGTCGTGGAAAGG |
| g2TagRFP-R1 | N′ gna-2/TagRFP | GCATGTTCTCCTTAATCAGCTCGCTCATGTTGAACTCATTCATCGCATCAAAGATC |
| g2TagRFP-F1 | TagRFP/C′ gna-2 | GCGACCTCCCTAGCAAACTGGGGCACAAGATCAAGCTGGAGGATGAAGATAATGAG |
| pRS426g2-R1 | C′ gna-2/pRS426 | GGATAACAATTTCACACAGGAAACAGCGAATTCCTACAGGATAAGTTGTTTCAGGTTTCG |
| TagRFP-F2 | TagRFP | TCTAGAATGAGCGAGCTGATTAAGGAG |
| TagRFP-R2 | TagRFP | GAATTCTCACTTGTGCCCCAGTTTGC |
Western blot analysis
Western blotting was used to detect Gα-TagRFP fusion proteins in whole-cell extracts prepared from macroconidia. Conidia from 6–7 day flask cultures were collected using sterile water, pelleted and stored at −80°C. After thawing on ice, the conidia were resuspended using 1 ml of extraction buffer (10 mM HEPES, pH 7.5, 0.5 mM EDTA, 0.5 mM PMSF, 1 mM DTT and 0.1% v/v of fungal protease inhibitor (Sigma-Aldrich, St. Louis, MO; Product #T8215) and then transferred to a mortar. The conidia were vigorously crushed using a mortar and pestle under liquid nitrogen. Roughly equal amounts of ground tissue were then transferred to 2 ml screw cap tubes, and topped off with additional extraction buffer if necessary. Samples were spun at 3,300 RPM (1,000×g) for 10 minutes at 4°C in a microcentrifuge and the supernatants (whole cell extracts) retained.
Protein concentration was determined using the Bradford Protein Assay (Bio-Rad, Hercules, CA). Samples containing 50 µg of whole-cell protein were subjected to SDS-PAGE using 10% gels and gels transferred to nitrocellulose membranes as described previously [18]. Western analysis was performed using a polyclonal RFP antibody (#R10367, Invitrogen, Carlsbad, CA) as the primary antibody at a dilution of 1∶2000. A horseradish peroxidase conjugate antibody (Bio-Rad) was used as the secondary antibody at a dilution of 1∶5000 and chemiluminescent detection was performed as described previously [18].
Microscopy
For analysis of conidial germination, 8×106 conidia were spread on 100 mm 10 ml VM agarose plates and incubated at 30°C for 0, 4, 6 or 8 h. Cells were then visualized using differential interference contrast (DIC) microscopy using an Olympus IX71 inverted microscope (Olympus America, Center Valley, PA) with a 60× oil immersion objective (NA = 1.42). Images were captured using a QIClickTM digital CCD camera (QImaging, Surrey, British Columbia, Canada) and analyzed using Metamorph software (Molecular Devices Corporation, Sunnyvale, CA). For analysis of conidial anastomosis tubes, conidia were spread on VM agarose, and imaged after 5–16 h at 30°C as detailed above.
For observation of Gα localization, VM agarose plate cultures were prepared as described above. Germinating conidia were analyzed using a Leica TCS SP5 II confocal microscope with a 63× oil objective (NA = 1.40; Leica Microsystems Inc., Buffalo Grove, IL). The Gα-RFP strains were visualized with the Hybrid Detection system (HyD) laser at an excitation of 543 nm, and emission of 565–665 nm.
To confirm vacuolar localization, conidia from the Gα-TagRFP strains were inoculated onto VM agarose plates and incubated as described above. An aliquot containing 30 µl of a 20 µg/ml solution of Oregon Green 488 carboxylic acid diacetate (Carboxy-DFFDA; catalog number O6151; Molecular Probes) was applied to a coverslip. An agarose block containing germinating conidia was inverted onto the coverslip and incubated for 5 minutes at room temperature in the dark. Images were obtained using the Leica TCS SP5 II confocal microscope described above. GFP images were obtained by excitation at 488 nm, with emission collected from 500–535 nm. RFP images were obtained with excitation at 543 nm and emission from 555–700 nm. Images were captured sequentially in order to prevent crosstalk among samples.
The vacuolar and plasma membrane localization of GNA-1-TagRFP was further explored by imaging a heterokaryon that expresses GNA-1-TagRFP and a GFP fusion of the Ca2+ ATPase, NCA-3 [19]. NCA-3-GFP is known to localize to vacuoles and the plasma membrane [19]. The two strains were co-inoculated onto a VM slant in order to produce a heterokaryon with conidia expressing both fluorescent proteins. Conidia were inoculated onto VM agarose plates as described above and imaged 6 h later using a 543 nm HyD laser and 488 nm laser on the Leica TCS SP5 II confocal microscope. Images were captured sequentially.
Possible co-localization of RIC8 and GNA-1 or GNA-3 was investigated through co-culturing of the RIC8-GFP strain with the GNA-1-TagRFP or GNA-3-TagRFP strains on a VM slant to produce a heterokaryon expressing two fluorescent proteins. Conidia from the heterokaryons were inoculated onto VM agarose plates as described above, followed by incubation at 30°C for 0 or 6 h for GNA-1-TagRFP/RIC8-GFP and 0 or 4 h for the GNA-3-TagRFP/RIC8-GFP fusion strain. Images were obtained by confocal microscopy, as described above.
Statistical analysis
Germination rates of strains relative to wild type were determined using Student's two-sided t test, with values paired by day of analysis [20]. Multiple comparisons within time points were corrected using the false discovery rate (FDR) approach of [21]. Differences in arthroconidiation and germination rates between strains were determined using Student's unpaired two-sided t test. Germination rates were first normalized for daily differences in wild type germination rates. All statistics were performed in R [22]; code is available on request. Details of the statistical analyses are presented in Tables 3, 4, 5, 6.
Table 3. Statistical analysis of arthroconidiation.
| Strain 1 | Strain 2 | t a | df b | p c | sigd |
| Δric8 | Wild type | −8.02 | 2.18 | 0.024 | * |
| Δgna-1Δgna-3 | ” | −11.02 | 2.18 | 0.018 | * |
| Δgna-1Δgna-2Δgna-3 | ” | −20.41 | 2.81 | 0.002 | ** |
| Δric8gna1 * | Δric8 | −0.19 | 3.51 | 0.991 | ns |
| Δric8gna2 * | ” | −0.28 | 3.92 | 0.991 | ns |
| Δric8gna3 * | ” | 0.01 | 3.90 | 0.991 | ns |
Student's t statistic,
degrees of freedom,
probability value, and d significance range, where ns is not significant;
, 0.05≥p>0.01;
, 0.01≥p>0.001. Statistically significant results are bolded.
Table 4. Statistical analysis of germination rates relative to wild type.
| Strain 1 | Strain 2 | 0 h | 4 h | 6 h | 8 h | ||||||||||||
| t a | df b | p c | sigd | t | df | p | sig | t | df | p | sig | t | df | p | sig | ||
| Δgna-1 | Wild type | 1.00 | 4 | 0.772 | ns | 1.94 | 4 | 0.140 | ns | 3.77 | 4 | 0.027 | * | 2.41 | 4 | 0.090 | ns |
| Δgna-2 | ” | 0.02 | 4 | 0.982 | ns | 1.92 | 4 | 0.140 | ns | −0.52 | 4 | 0.627 | ns | −1.41 | 4 | 0.254 | ns |
| Δgna-3 | ” | 0.15 | 4 | 0.982 | ns | 5.59 | 4 | 0.008 | ** | 7.91 | 4 | 0.003 | ** | 5.63 | 4 | 0.009 | ** |
| Δric8 | ” | −1.10 | 5 | 0.772 | ns | 7.63 | 5 | 0.002 | ** | 21.86 | 5 | 0.000 | *** | 26.88 | 5 | 0.000 | *** |
| Δgna-1Δgna-3 | ” | 1.73 | 3 | 0.772 | ns | 19.80 | 3 | 0.001 | *** | 49.08 | 3 | 0.000 | *** | 23.66 | 3 | 0.001 | *** |
| Δgna-1Δgna-2Δgna-3 | ” | −0.06 | 3 | 0.982 | ns | 22.73 | 3 | 0.001 | *** | 21.47 | 3 | 0.001 | *** | 36.33 | 3 | 0.000 | *** |
| Δric8gna1 * | ” | 1.73 | 3 | 0.772 | ns | 21.36 | 3 | 0.001 | *** | 24.97 | 3 | 0.001 | *** | 9.71 | 3 | 0.005 | ** |
| Δric8gna2 * | ” | 1.00 | 2 | 0.772 | ns | 16.80 | 2 | 0.006 | ** | 14.89 | 2 | 0.007 | ** | 13.34 | 2 | 0.009 | ** |
| Δric8gna3 * | ” | −0.47 | 3 | 0.922 | ns | 10.23 | 3 | 0.004 | ** | 7.90 | 3 | 0.007 | ** | 12.48 | 3 | 0.003 | ** |
| Δgnb1 | ” | −0.84 | 2 | 0.772 | ns | 1.65 | 2 | 0.240 | ns | 1.32 | 2 | 0.349 | ns | 0.51 | 2 | 0.660 | ns |
| Δgng1 | ” | −1.40 | 2 | 0.772 | ns | 5.08 | 2 | 0.050 | * | 1.95 | 2 | 0.233 | ns | 3.90 | 2 | 0.083 | ns |
Student's t statistic,
degrees of freedom,
probability value, and d significance range, where ns is not significant;
, 0.05≥p>0.01;
, 0.01≥p>0.001;
, p≤0.001. Statistically significant results are bolded.
Table 5. Statistical analysis of germination rates between strains other than wild type.
| Strain 1 | Strain 2 | 0 h | 4 h | 6 h | 8 h | ||||||||||||
| t a | df b | p c | sigd | t | df | p | sig | t | df | p | sig | t | df | p | sig | ||
| Δric8 | Δric8gna1 * | −2.05 | 4.9 | 0.097 | ns | −1.47 | 6.7 | 0.188 | ns | 0.39 | 7.9 | 0.710 | ns | 2.30 | 4.3 | 0.078 | ns |
| Δric8 | Δric8gna2 * | −1.38 | 3.0 | 0.260 | ns | −1.42 | 6.9 | 0.198 | ns | −0.41 | 3.8 | 0.705 | ns | −0.07 | 3.0 | 0.946 | ns |
| Δric8 | Δric8gna3 * | 0.21 | 3.3 | 0.847 | ns | 3.60 | 6.3 | 0.011 | * | 13.06 | 8.0 | 0.000 | *** | 14.34 | 8.0 | 0.000 | *** |
| Δric8 | Δgna-3 | −0.67 | 6.4 | 0.527 | ns | 0.86 | 8.5 | 0.411 | ns | 1.79 | 5.6 | 0.128 | ns | 3.18 | 4.9 | 0.025 | * |
| Δric8 | Δgna-1Δgna-3 | −2.05 | 4.9 | 0.097 | ns | −1.77 | 7.0 | 0.119 | ns | −1.81 | 7.0 | 0.114 | ns | −1.22 | 6.6 | 0.263 | ns |
| Δgna-3 | Δgna-1Δgna-3 | −1.12 | 6.9 | 0.301 | ns | −2.66 | 5.4 | 0.042 | * | −2.80 | 4.4 | 0.044 | * | −3.70 | 5.2 | 0.013 | * |
| Δgna-1Δgna-3 | Δgna-1Δgna-2Δgna-3 | 1.04 | 5.4 | 0.343 | ns | 0.07 | 5.9 | 0.946 | ns | 0.17 | 4.1 | 0.877 | ns | −0.64 | 5.3 | 0.551 | ns |
Student's t statistic,
degrees of freedom,
probability value, and d significance range, where ns is not significant;
, 0.05≥p>0.01;
, 0.01≥p>0.001;
, p≤0.001. Statistically significant results are bolded.
Table 6. Statistical analysis of germination rates between wild type and Gα-TagRFP expressing strains.
| Strain 1 | Strain 2 | 0 h | 4 h | 6 h | 8 h | ||||||||||||
| t a | df b | p c | sigd | t | df | p | sig | t | df | p | sig | t | df | p | sig | ||
| Δgna-1/GNA-1-TagRFP | Wild type | −0.95 | 2 | 0.515 | ns | −1.48 | 2 | 0.651 | ns | −0.24 | 2 | 0.833 | ns | −3.60 | 2 | 0.158 | ns |
| Δgna-2/GNA-2-TagRFP | Wild type | 0.82 | 2 | 0.515 | ns | 0.93 | 2 | 0.651 | ns | 3.29 | 2 | 0.122 | ns | 0.46 | 2 | 0.692 | ns |
| Δgna-3/GNA-3-TagRFP | Wild type | 0.78 | 2 | 0.515 | ns | −0.53 | 2 | 0.651 | ns | 3.38 | 2 | 0.122 | ns | 2.83 | 2 | 0.158 | ns |
Student's t statistic,
degrees of freedom,
probability value, and d significance range, where ns is not significant.
Results
Loss of ric8 or both gna-1 and gna-3 leads to overproduction of arthroconidia
We have previously demonstrated that Δgna-1 and Δric8 single mutants, Δgna-1 Δgna-3 double mutants, and the triple Gα mutant have smaller colony sizes than wild type [2], [6], [9]. The defects of the latter three mutants are similar and much more severe than those of the Δgna-1 strain [6], [9]. Smaller colony size could result from slow germination, reduced hyphal extension or both phenomena. In order to determine whether germination defects contribute to the overall reduction in colony size, we investigated early events during germination of conidia in Δric8 mutants and in strains lacking G protein subunit genes.
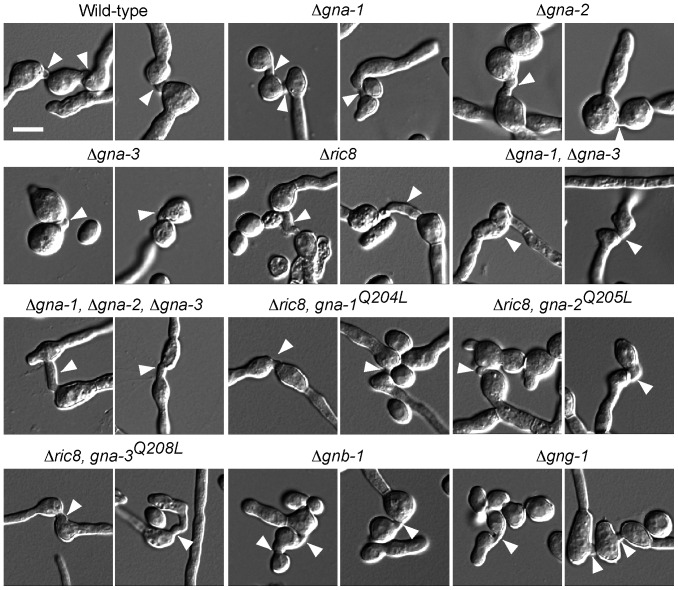
We began with an analysis of conidial morphology. All five N. crassa G protein subunit mutants (Δgna-1, Δgna-2, Δgna-3, Δgnb-1 and Δgng-1) produce normal-looking macroconidia (Fig. 2; Fig. 3). However, Δric8, the Δgna-1 Δgna-3 double mutant and the triple Gα mutant all display an increased proportion of arthroconidia (wild type 6.2%±0.8%; Δric8 36.0%±3.6%; Δgna-1 Δgna-3 47.1%±3.6%; and Δgna-1 Δgna-2 Δgna-3 43.8%±1.7%) (Fig. 2, Table 3). Arthroconidia are formed by fragmentation of hyphae [23]. Introduction of GTPase-deficient alleles for any of the three Gα genes did not lead to a significant reduction in the proportion of arthroconidia in the Δric8 background (Δric8, gna-1 Q204L 36.8%±2.5%; Δric8, gna-2 Q205L 37.6%±4.2%; Δric8, gna-3 Q208L 36.0%±4.3%) (Fig. 2, Table 3). Therefore, inhibition of arthroconidiation depends on RIC8, GNA-1 and GNA-3, and constitutive activation of a Gα subunit is not sufficient to override the phenotype in Δric8 mutants.
Figure 2. G proteins and the RIC8 GEF are required for normal conidiation and conidial germination.
Conidia were harvested as described in the Materials and Methods. An aliquot containing 8×106 conidia was spread on a 100 mm 10 ml VM solid medium plate and spore germination monitored at 30°C over time. DIC (differential interference contrast) micrograph images were obtained using an Olympus IX71 microscope with a QIClick digital CCD camera and analyzed using Metamorph software. Scale bar = 5 µm. Strains are wild type (74-OR23-IVA), Δgna-1 (3B10), Δgna-2 (FGSC 12378), Δgna-3 (31c2), Δric8 (r81-5a), Δgna-1, Δgna-3 (g1.3), Δgna-1, Δgna-2, Δgna-3 (noa), Δric8, gna-1 Q204L (R81*), Δric8, gna-2 Q205L (R82*), Δric8, gna-3 Q208L (R83*). Arrowheads indicate arthroconidia.
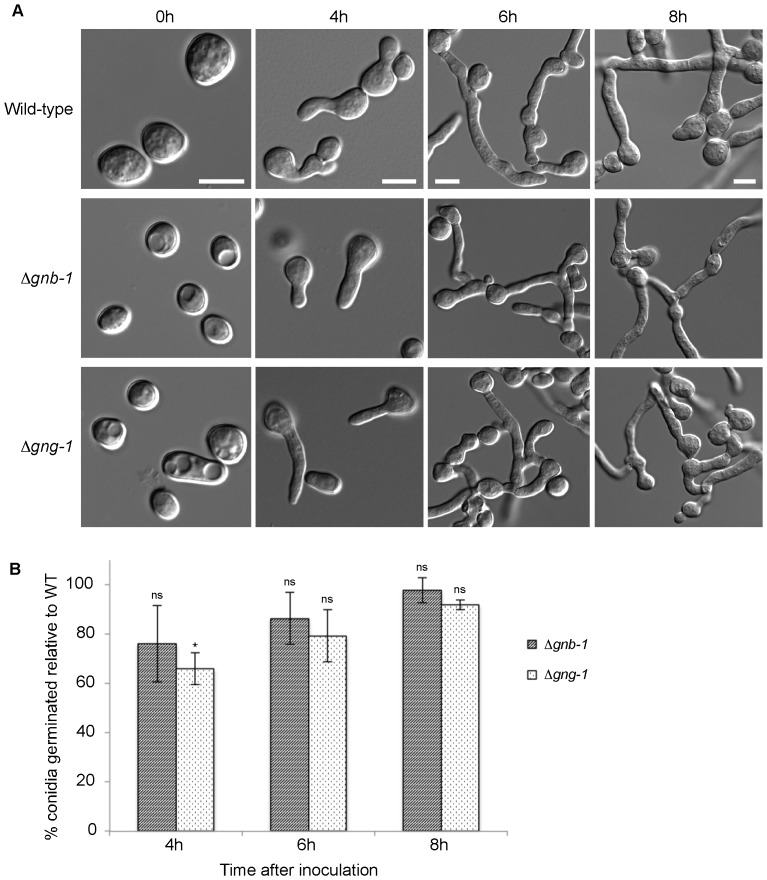
Figure 3. Germination of Δgnb-1 and Δgng-1 mutants.
A. Conidia of wild-type (74-OR23-IVA), Δgnb-1 (42-8-3) and Δgng-1 (5-5-3) strains were harvested as described in the Materials and Methods. An aliquot containing 8×106 conidia was spread on VM solid medium and germination monitored at 30°C over time. DIC images were obtained using an Olympus IX71 microscope with a QIClick™ digital CCD camera and analyzed using Metamorph software. Bar = 5 µm. B. Proportion of germinated conidia of N. crassa wild-type, Δgnb-1 and Δgng-1 strains at various times after inoculation onto VM medium. Error bars are ±SE for a minimum of three independent experiments (n = minimum of 100 cells for all strains). Strains are the same as in A.
In N. crassa, arthroconidiation has been proposed to be the default conidiation pathway in mutants with defects in macroconidiation [24]. Arthroconidia are the major cell type used to disseminate many animal fungal pathogens, such as the valley fever fungus Coccidioides immitis and dermatophytes in the genera Mycosporum, Trichophyton and Epidermophyton [25], [26], [27]. To date, little is known about the regulation of arthroconidiation in these fungi, but it seems clear that RIC8, GNA-1 and GNA-3 impact this process in N. crassa.
G protein signaling is required for conidial germination, but is not essential for conidial anastomosis tube formation
We next investigated the ability of macroconidia from the various strains to form conidial anastomosis tubes (CATs) and to germinate on solid medium. CATs are small tube-like structures produced by conidia early during colony initiation (rev. in [28]). The results demonstrate that Δric8 and all G protein subunit single and double mutants produce CATs from macroconidia (Fig. 4).
Figure 4. G protein mutants produce conidial anastomosis tubes (CATs).
Conidia were used to inoculate 100 mm 10 ml solid VM agarose plates as described in Fig. 2. Plates were incubated at 30°C for 5–16 h depending on the germination rate of the mutant strain. Images were captured and analyzed and strains are the same as in Fig. 2. Arrowheads indicate positions of CATs. Bar = 5 µm.
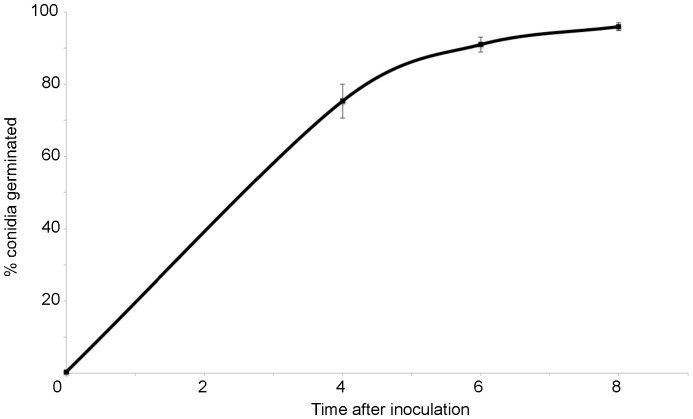
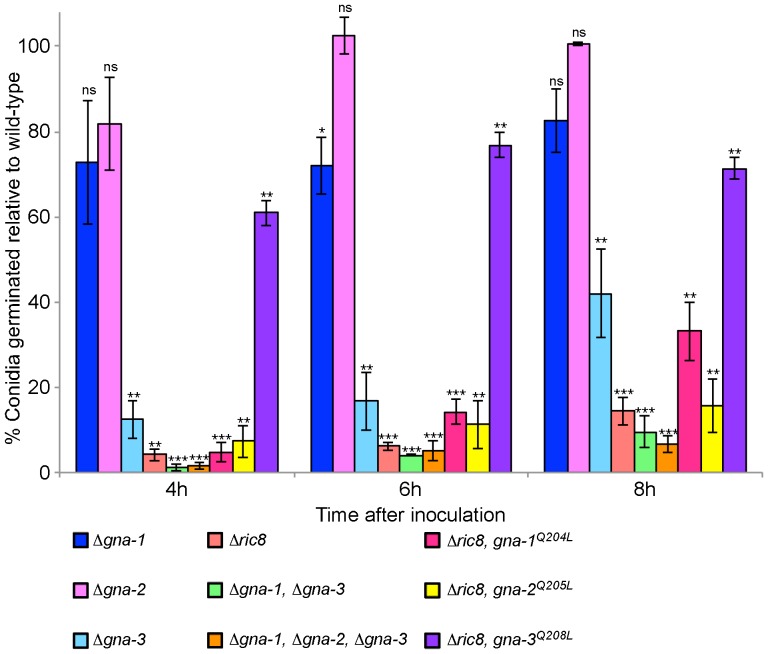
We next analyzed conidial germination (both macroconidia and arthroconidia) in the various strains at 4, 6 and 8 h after plating on solid medium. In wild-type, greater than 70% of the conidia have germinated by 4 hr after inoculation, with nearly 100% germination after 8 h (Fig. 5; Table 7). In contrast, germination is significantly reduced in Δric8, Δgna-3, Δgna-1 Δgna-3 and the triple Gα mutants relative to wild type at all time points (Fig. 2, 6). Germination of the Δgna-1 mutant also appears to be slower than wild-type, but this is only statistically significant at 6 h. However, the observation that germination of the Δgna-1 Δgna-3 mutant is significantly lower than the Δgna-3 mutant supports a compensatory role for GNA-1 in conidial germination (Fig. 2, 6). Loss of gna-2 had no effect on conidial germination, and did not exacerbate the germination defect of the Δgna-1 Δgna-3 mutant (Fig. 2). Taken together, the results indicate that RIC8, GNA-3, and to a lesser extent GNA-1, play important roles during conidial germination.
Figure 5. Germination rate of wild-type N. crassa.
Values are taken from Table 7. Error bars are ±SE for twelve independent experiments (n = minimum of 60 cells).
Table 7. Germination rate of wild-type N. crassa.
| 0 h | 4 h | 6 h | 8 h | |
| Average percentage of conidia germinated | 0.4 | 75.3 | 91.0 | 96.0 |
| Standard error | 0.1 | 4.7 | 2.1 | 1.1 |
Figure 6. Quantitation of conidial germination rates in G protein and ric8 mutants.
The proportion of germinated conidia of wild-type, Gα mutant and Δric8 strains was determined at various times after inoculation onto solid medium. Error bars are ±SE for a minimum of three independent experiments (n = minimum of 60 cells for all strains). Strains and conditions are the same as in Fig. 2.
Subsequently, we tested whether constitutive activation of any of the three Gα subunits could rescue the conidial germination defect of Δric8 strains. Introduction of the GTPase-deficient gna-1 or gna-2 alleles did not significantly improve germination, (Fig. 2, 6; Table 4, 5). Strikingly, constitutive activation of gna-3 in the Δric8 background resulted in a significant increase in conidial germination, approaching that of the wild-type strain (Fig. 6; Table 4, 5). The ability of the GTPase-deficient gna-3 allele to bypass the Δric8 germination defect supports RIC8 acting through GNA-3 to regulate conidial germination in N. crassa. We also probed a possible role for the Gβ and Gγ subunits during conidial germination (Fig. 3, Table 4). Similar to the results exploring conidial morphology, germination is largely unaffected in Δgnb-1 and Δgng-1 mutants, aside from a slight delay in Δgng-1 4 h after inoculation.
The three Gα proteins localize to the plasma membrane and vacuoles during conidial germination
Having demonstrated that Gα proteins are important for conidial germination, we next investigated the subcellular localization of these proteins during this process. For these experiments, we took advantage of previous studies in Dictyostelium and mammalian cells demonstrating that insertion of a fluorescent tag in a loop region of the Gα subunit does not disrupt the interaction with the Gβγ dimer or interfere with Gα function (Fig. 1; [10], [11].
We expressed all three Gα-TagRFP fusions under control of the highly expressed ccg-1 promoter from the his-3 locus in the corresponding Gα mutant strain (see Materials and Methods). Expression of GNA-1-TagRFP complemented the conidial germination (Table 6) and female fertility defects and partially complemented the growth rate phenotype of the Δgna-1 mutant (data not shown). Expression of GNA-3-TagRFP in the Δgna-3 mutant restored conidial germination back to wild-type levels at all time points investigated (Table 6).
We confirmed that the three TagRFP constructs are expressed as full-length fusion proteins in conidia using Western analysis with an RFP antiserum (Fig. 7). TagRFP is 27 kD and each Gα is approximately 41 kD. Correspondingly, each of the three Gα fusion proteins migrated close to their predicted molecular mass of 68 kD. Interestingly, although all three genes are expressed under control of the same promoter from a common genomic site, levels of GNA-1 are much higher than those of GNA-2 and GNA-3. This suggests some post-transcriptional regulation of Gα protein levels, with GNA-1 accumulating to higher levels in N. crassa. In addition, potential degradation products were observed for all three proteins (Fig. 7; asterisks), suggesting at least some of the variation in protein levels may be due to differences in protein turnover.
Figure 7. Western blot detection of Gα-TagRFP fusion proteins.
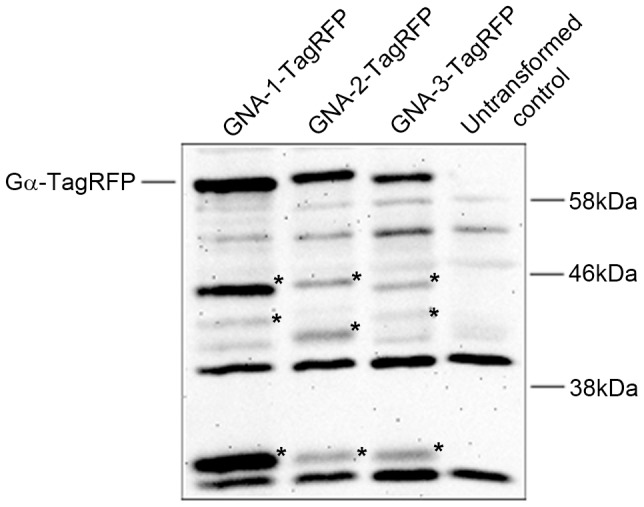
Samples containing 50 µg of protein from conidial extracts were subjected to western blot analysis with a RFP primary antiserum as described in the Materials and Methods. Strains are Δgna-1, gna-1-TagRFP (2.1), Δgna-2, gna-2-TagRFP (5.1), Δgna-3, gna-3-TagRFP (12.1) and wild type (untransformed control; 74-OR23-IVA). TagRFP is 27 kD, while the predicted size of the three TagRFP fusion proteins is 68 kD. Potential degradation products for each RFP fusion are noted with asterisks.
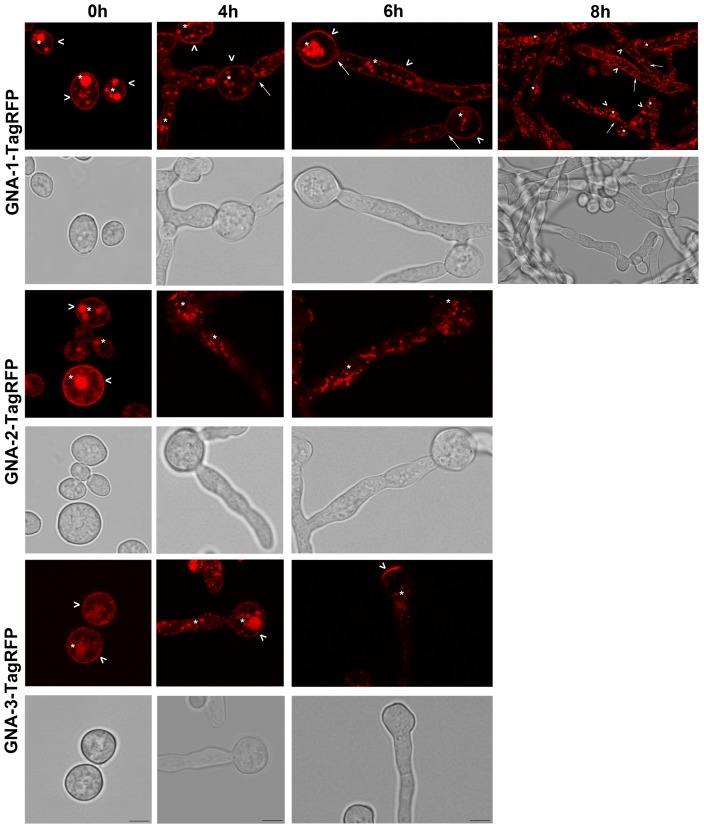
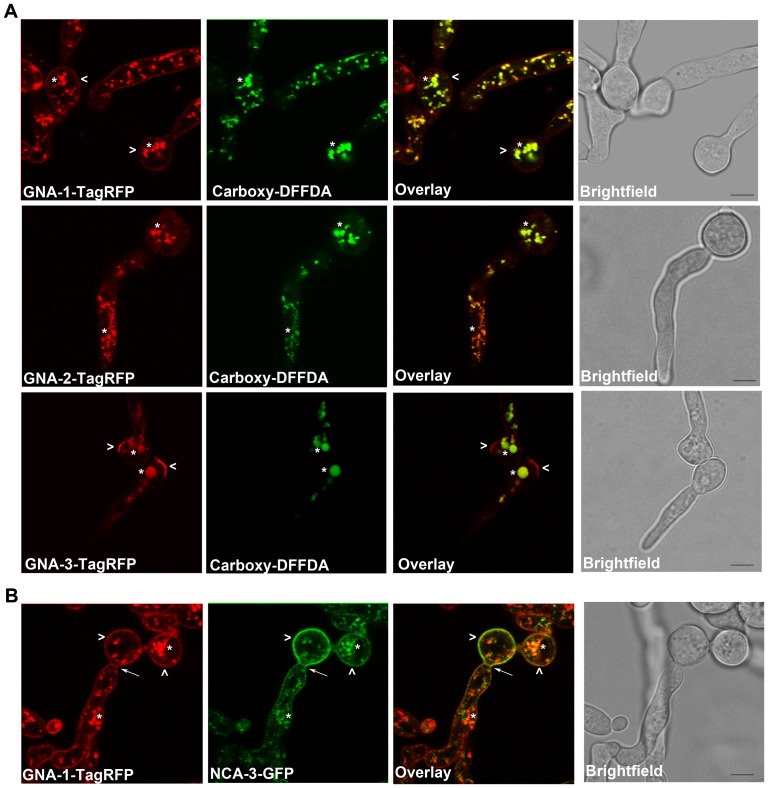
We examined the strains expressing TagRFP fusion proteins during conidial germination using confocal fluorescent microscopy (Fig. 8). Conidia were plated on solid medium and examined at four time points (0, 4, 6 and 8 h). At 0 h, all three Gα proteins localized to the plasma membrane and vacuoles (Fig. 8, 9A). The plasma-membrane localization is consistent with results previously observed during cell fractionation studies in our laboratory [2], [5], [6]; data not shown). The vacuolar localization of GNA-1-TagRFP, GNA-2-TagRFP, and GNA-3-TagRFP was validated by the overlapping signal observed between TagRFP and the carboxy-DFFDA vacuolar dye during confocal microscopy (Fig. 9A). In addition, a strain expressing a GFP-tagged protein, NCA-3, a Ca2+ ATPase known to localize to vacuoles and the plasma membrane, was combined with the GNA-1-RFP strain to produce a heterokaryon [19]. Many conidia and germinating hyphae from these heterokaryons exhibit green and red fluorescence, indicative of the presence of both NCA-3-GFP and GNA-1-TagRFP. Confocal microscopy of conidia from this heterokaryon confirmed that GNA-1 localizes to the plasma membrane and vacuoles (Fig. 9B). It is unclear whether the vacuolar fluorescence represents a functional localization or a site of protein turnover.
Figure 8. Localization of Gα proteins in germinating conidia.
Conidia from strains expressing GNA-1-TagRFP, GNA-2-TagRFP, GNA-3-TagRFP and untransformed controls were inoculated on solid medium and analyzed after 0, 4, 6 and 8 h of growth. Images were captured by bright field and the 543 nm HyD laser using the Leica TCS SP5 II inverted confocal microscope. The arrowhead, asterisk and solid arrow correspond to plasma membrane, vacuole and septa localization, respectively. Panels are only shown for time points in which fluorescence can be detected above background. All panels are 4× zoom, with the exception of GNA-1 at 8 h, which is 2×. Scale bar = 5 µm.
Figure 9. All three Gα proteins localize to vacuoles.
A. Conidia expressing the corresponding Tag-RFP Gα were inoculated onto 100 mm solid VM at a concentration of 8×106 conidia per plate. Plates were incubated at 30°C for 4 h (GNA-2 and GNA-3) or 6 h (GNA-1). The vacuolar dye Carboxy-DFFDA was applied at a concentration of 20 µg/ml. Images were captured using the Leica TCS SP5 II inverted confocal microscope. The RFP and GFP panels were merged to create the overlay. B. Conidia expressing both GNA-1-TagRFP and NCA-3-GFP were inoculated onto 100 mm solid VM at a concentration of 8×106 conidia per plate. Plates were incubated at 30°C for 6 h. Images were captured using the Leica TCS SP5 II inverted confocal microscope. The RFP and GFP panels were merged to create the overlay. All panels are 4× zoom. Bar = 5 µm.
At 4 h of conidial germination, GNA-1 continued to display plasma membrane and vacuolar localization, and could also be observed on the first septum separating the conidium and the developing hypha (Fig. 8). In contrast, GNA-2 and GNA-3 were predominantly found in vacuoles, with less apparent plasma membrane localization (Fig. 8). At 6 h, GNA-1 was present in the plasma membrane, vacuoles and septa. (Fig. 8). GNA-2 exhibited a similar localization as observed at 4 h. Interestingly, in addition to vacuolar localization, GNA-3 was also found in distinct patches on the plasma membrane of the original conidium (Fig. 8). These patches were observed in 50% or more of the cells being sampled. At 6 h, the plasma membrane patches were even more easily observed in GNA-3-TagRFP strains, as the fluorescence in vacuoles and other regions of the plasma membrane began to weaken (Fig. 8). At 8 h, GNA-1-TagRFP fluorescence was still relatively strong in plasma membrane, vacuoles and septa (Fig. 8). In contrast, GNA-2- and GNA-3-TagRFP fluorescence was too dim to analyze in 8 h germlings (data not shown).
We have previously demonstrated that RIC8 exhibits cytosolic localization in conidia and mature hyphae using fluorescent microscopy with an inverted compound microscope [9]. In this study, we utilized the more discriminating method of confocal fluorescence microscopy to explore possible co-localization of GNA-1-TagRFP and/or GNA-3-TagRFP with RIC8-GFP in cellular compartments during conidial germination. For these experiments, we cultured strains carrying the RIC8-GFP and GNA-1-TagRFP constructs together to produce heterokaryons expressing both fluorescent proteins. Similar to previous results, RIC8-GFP is cytosolic and excluded from vacant areas that appear to be vacuoles (Fig. 10). In contrast, GNA-1-TagRFP is located in the plasma membrane and vacuoles (Fig. 10) and the merged image did not reveal co-localization with RIC8-GFP in conidia or young germlings (Fig. 10). Similarly, co-localization was also not observed between GNA-3-TagRFP and RIC8-GFP (Fig. 10). In addition, there was no evidence for co-localization between the GNA-3 fluorescent patches and RIC8 (Fig. 10).
Figure 10. Localization of GNA-1-TagRFP and GNA-3-TagRFP with RIC8-GFP in germinating conidia.
Conidia from a fused strain expressing GNA-1-TagRFP or GNA-3-TagRFP and RIC-8-GFP were harvested as described in the Materials and Methods. An aliquot containing 8×106 conidia was spread on a 100 mm VM solid medium plate. Images were captured using a Leica TCS SP5 II inverted confocal microscope. Conidia were imaged immediately after inoculation on the solid medium plate. Conidia were allowed to germinate for 6 h or 4 h at 30°C before imaging. The first two panels were merged to create the third panel. All panels are 4× zoom. Scale bar = 5 µm.
Discussion
G proteins regulate nearly every facet of growth and development in fungi. During asexual development, in particular, they have been found to regulate both the timing and level of conidiation (rev. in [29], and in recent years have also been shown to regulate conidial germination [30], [31], [32], [33], [34]. Here, we report evidence showing that in N. crassa G protein signaling regulates both conidial development and germination, and this is regulated through the non-receptor GEF RIC8.
We observed that loss of any single G protein subunit has no effect on conidial morphology in N. crassa. However, loss of both gna-1 and gna-3 Gα genes leads to a dramatic increase in arthroconidia formation, which is also observed in the Δric8 mutant. Arthroconidia are formed by the fragmentation of vegetative hyphae [23] and their production is proposed to be a default pathway in mutants defective in macroconidiation [24]. Expression of constitutively active (GTPase deficient) GNA-1 or GNA-3 in the Δric8 mutant background does not lead to reduced arthroconidiation, suggesting RIC8 controls arthroconidia formation through Gα-dependent and independent mechanisms. While the exact mechanism by which RIC8 and the Gα proteins negatively regulate arthroconidiation is unclear, it may involve the MAK-1 MAP kinase signaling pathway, as deletion of any of the three kinases of this pathway leads to an overproduction of arthroconidia [35]. Additionally, loss of rgb-1, homologous to the B subunit of type 2A Ser/Thr phosphatases, also leads to production of large amounts of arthroconidia, perhaps through a negative effect on the MAK-1 pathway [24]. Interestingly, in spite of the requirement for G protein signaling genes during conidial development and germination, Δric8 and all G protein subunit single and double mutants were found to produce conidial anastomosis tubes from macroconidia. However, it remains unclear whether the conidia from these mutants that do not germinate are also unable to produce conidial anastomosis tubes.
Conidial germination was found to be significantly reduced in the Δgna-3 mutant, indicating GNA-3 regulates conidial germination in N. crassa, as has been reported for homologs in Penicillium marneffei [34], Aspergillus nidulans [30] and Botrytis cinerea [31]. The germination defect is exacerbated in the Δgna-1 Δgna-3 double mutant, revealing a role for GNA-1 in the absence of GNA-3. GNA-1 homologs have been shown to regulate conidial germination in other fungi [32], [33], [36]. Germination is reduced in the Δgna-1 mutant relative to wild type, but this is only statistically significant 6 h after inoculation, consistent with GNA-1 playing a more minor role in N. crassa. The observation that GNA-3 is more important during germination in N. crassa than GNA-1 is interesting, given the Δgna-1 mutant displays a greater reduction in hyphal extension rate than the Δgna-3 mutant [2], [3]. This suggests that GNA-1 is more important for overall extension of hyphae rather than for early events during germination, while the converse is true for GNA-3. Germination of the Δgna-2 mutant is no different than wild type and germination is similar in Δgna-1 Δgna-2 Δgna-3 triple and Δgna-1 Δgna-3 double mutants, implying that GNA-2 is not required for conidial germination in N. crassa.
Our results demonstrate that the non-receptor GEF RIC8 regulates conidial germination primarily through the GNA-3 Gα subunit, with minor contribution from GNA-1. This observation is consistent with results from GTP binding assays demonstrating that RIC8 exhibits greater GEF activity towards GNA-3 than GNA-1 [9]. Interestingly, in M. oryzae loss of MoRIC8 does not affect conidial germination [7]. Instead, MoRIC8 is required for normal asexual growth, conidiation, appressorium formation and pathogenicity [7].
It is likely that RIC8 and the Gα proteins regulate conidial germination in N. crassa through modulation of cAMP levels [3], [9], [37], [38]. The N. crassa Δric8 mutant has decreased adenylyl cyclase protein and the Δric8 mutation can be suppressed by a mutation in the protein kinase A regulatory subunit [9]. A mutation in the regulatory subunit leads to hyperactivation of the PKA catalytic subunit, thereby bypassing the need for wild-type levels of adenylyl cyclase. The Gα subunit GNA-3 is required to maintain wild-type levels of adenylyl cyclase, further supporting a role for G proteins in cAMP-dependent conidial germination [3]. In B. cinerea and A. nidulans cAMP plays a key role in conidial germination regulated by GNA-3 homologs [31], [39].
In this study we pioneered the use of an internal Gα tagging method in filamentous fungi based on a successful approach used in Dictyostelium and mammalian cells, inserting TagRFP into a conserved loop region in the Gα subunits [10], [11]. Using this approach we were able to localize GNA-1, GNA-2 and GNA-3 during conidial germination, revealing that all three Gα proteins localize to the plasma membrane in ungerminated conidia and young germlings and that GNA-1 can be detected in septa in older hyphae. All three Gα subunits were also detected in vacuoles, however, it is unclear if this is a functional localization, as proteins are often targeted for degradation by the vacuole [40]. Perhaps the most intriguing finding was the observation of patches of GNA-3-TagRFP on the plasma membrane at later time points during germination of conidia. This change in localization during the transition from conidium to germling may be related to the requirement for GNA-3 during conidial germination; further analysis is necessary to explore this hypothesis.
Using purified proteins in in vitro assays, we have previously demonstrated that RIC8 can bind to and act as a GEF for GNA-1 and GNA-3 [9]. However, confocal microscopy of strains expressing GNA-1-TagRFP or GNA-3-TagRFP and RIC8-GFP did not reveal any evidence for co-localization of either Gα protein with RIC8 in conidia or hyphae, suggesting the interaction may be transient. Studies in Drosophila neuroblasts have demonstrated cytoplasmic localization of Ric-8, while Gαi was found in the apical cortex [41]. Ric-8 was also observed in ‘spot-like’ structures close to the apical cortex that partially colocalized with Gαi, indicating that their interaction may take place on the cytoplasmic face of the plasma membrane or in the cytoplasm [41].
Interestingly, the GNA-1-TagRFP signal was stronger than that of GNA-2 and GNA-3, and detectable over a longer period, suggesting this protein fusion may be more stable. This conclusion is also supported by the results from western analysis using RFP antibodies. In yeast, the Ste2p GPCR is ubiquitinated and targeted to the vacuole for degradation [42]. Analysis of N. crassa GNA-1, GNA-2 and GNA-3 using UbPred (http://www.ubpred.org/) predicts that GNA-1 and GNA-2 contain potential ubiquitination target sites, which may explain the vacuolar localization of these proteins. Future studies will explore whether this vacuolar localization is functional, and if so investigate the importance of G protein signaling in the vacuole.
Acknowledgments
We are indebted to David Carter for advice on confocal microscopy. We acknowledge Amanda Ortiz and Yoselyn Sanchez for technical assistance and Patrick Schacht for help with image analysis. We thank Amruta Garud and Arit Ghosh for critical reading of the manuscript. We acknowledge Alex Lichius and Nick Read for the gift of the TagRFP plasmid and for information on its use. We thank Doug Ivey, Ann Kays and Hyojeong Kim for strain construction.
Funding Statement
This study was funded by National Institutes of Health grant GM086565 to KAB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Turner GE, Borkovich KA (1993) Identification of a G protein α subunit from Neurospora crassa that is a member of the Gi family. J Biol Chem 268: 14805–14811. [PubMed] [Google Scholar]
- 2. Ivey FD, Hodge PN, Turner GE, Borkovich KA (1996) The Gαi homologue gna-1 controls multiple differentiation pathways in Neurospora crassa . Mol Biol Cell 7: 1283–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kays AM, Rowley PS, Baasiri RA, Borkovich KA (2000) Regulation of conidiation and adenylyl cyclase levels by the Gα protein GNA-3 in Neurospora crassa . Mol Cell Biol 20: 7693–7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li L, Borkovich KA (2006) GPR-4 is a predicted G-protein-coupled receptor required for carbon source-dependent asexual growth and development in Neurospora crassa . Eukaryot cell 5: 1287–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baasiri RA, Lu X, Rowley PS, Turner GE, Borkovich KA (1997) Overlapping functions for two G protein α subunits in Neurospora crassa . Genetics 147: 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kays AM, Borkovich KA (2004) Severe impairment of growth and differentiation in a Neurospora crassa mutant lacking all heterotrimeric Gα proteins. Genetics 166: 1229–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Y, Yan X, Wang H, Liang S, Ma WB, et al. (2010) MoRic8 Is a novel component of G-protein signaling during plant infection by the rice blast fungus Magnaporthe oryzae . MPMI 23: 317–331. [DOI] [PubMed] [Google Scholar]
- 8. Wilkie TM, Kinch L (2005) New roles for Gα and RGS proteins: Communication continues despite pulling sisters apart. Curr Biol 15: R843–854. [DOI] [PubMed] [Google Scholar]
- 9. Wright SJ, Inchausti R, Eaton CJ, Krystofova S, Borkovich KA (2011) RIC8 is a guanine-nucleotide exchange factor for Gα subunits that regulates growth and development in Neurospora crassa . Genetics 189: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Azpiazu I, Gautam N (2004) A fluorescence resonance energy transfer-based sensor indicates that receptor access to a G protein is unrestricted in a living mammalian cell. J Biol Chem 279: 27709–27718. [DOI] [PubMed] [Google Scholar]
- 11. Janetopoulos C, Jin T, Devreotes P (2001) Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science 291: 2408–2411. [DOI] [PubMed] [Google Scholar]
- 12. Vogel HJ (1964) Distribution of lysine pathways among fungi - Evolutionary implications. Am Nat 98: 435–446. [Google Scholar]
- 13. Westergaard M, Mitchell HK (1947) Neurospora-V - a synthetic medium favoring sexual reproduction. Am J Bot 34: 573–577. [Google Scholar]
- 14. Krystofova S, Borkovich KA (2006) The predicted G-protein-coupled receptor GPR-1 is required for female sexual development in the multicellular fungus Neurospora crassa . Eukaryot cell 5: 1503–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berepiki A, Lichius A, Shoji JY, Tilsner J, Read ND (2010) F-actin dynamics in Neurospora crassa . Eukaryot Cell 9: 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, et al. (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Freitag M, Hickey PC, Raju NB, Selker EU, Read ND (2004) GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa . Fungal Genet Biol 41: 897–910. [DOI] [PubMed] [Google Scholar]
- 18. Krystofova S, Borkovich KA (2005) The heterotrimeric G-protein subunits GNG-1 and GNB-1 form a Gβγ dimer required for normal female fertility, asexual development, and Gα protein levels in Neurospora crassa . Eukaryot Cell 4: 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bowman BJ, Draskovic M, Freitag M, Bowman EJ (2009) Structure and distribution of organelles and cellular location of calcium transporters in Neurospora crassa . Eukaryot Cell 8: 1845–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Student (1908) The probable error of a mean. Biometrika 6: 1–25. [Google Scholar]
- 21. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300. [Google Scholar]
- 22.R Development Core Team (2012) R: A Language and Environment for Statistical Computing.
- 23.Griffin DH (1994) Fungal Physiology. New York: Wiley-Liss.
- 24. Yatzkan E, Yarden O (1999) The B regulatory subunit of protein phosphatase 2A is required for completion of macroconidiation and other developmental processes in Neurospora crassa . Mol Microbiol 31: 197–209. [DOI] [PubMed] [Google Scholar]
- 25. Chermette R, Ferreiro L, Guillot J (2008) Dermatophytoses in animals. Mycopathologia 166: 385–405. [DOI] [PubMed] [Google Scholar]
- 26. DiCaudo DJ (2006) Coccidioidomycosis: a review and update. J Am Acad Dermatol 55: 929–942. [DOI] [PubMed] [Google Scholar]
- 27. Nweze EI (2010) Dermatophytosis in Western Africa: a review. Pak J Biol Sci 13: 649–656. [PubMed] [Google Scholar]
- 28. Read ND, Lichius A, Shoji JY, Goryachev AB (2009) Self-signalling and self-fusion in filamentous fungi. Curr Opin Microbiol 12: 608–615. [DOI] [PubMed] [Google Scholar]
- 29. Li L, Wright SJ, Krystofova S, Park G, Borkovich KA (2007) Heterotrimeric G protein signaling in filamentous fungi. Annu Rev Microbiol 61: 423–452. [DOI] [PubMed] [Google Scholar]
- 30. Chang MH, Chae KS, Han DM, Jahng KY (2004) The GanB Gα-protein negatively regulates asexual sporulation and plays a positive role in conidial germination in Aspergillus nidulans . Genetics 167: 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doehlemann G, Berndt P, Hahn M (2006) Different signalling pathways involving a Gα protein, cAMP and a MAP kinase control germination of Botrytis cinerea conidia. Mol Microbiol 59: 821–835. [DOI] [PubMed] [Google Scholar]
- 32. García-Rico RO, Chavez R, Fierro F, Martin JF (2009) Effect of a heterotrimeric G protein α subunit on conidia germination, stress response, and roquefortine C production in Penicillium roqueforti . Int Microbiol 12: 123–129. [PubMed] [Google Scholar]
- 33. García-Rico RO, Martin JF, Fierro F (2011) Heterotrimeric Gα protein Pga1 from Penicillium chrysogenum triggers germination in response to carbon sources and affects negatively resistance to different stress conditions. Fungal Genet Biol 48: 641–649. [DOI] [PubMed] [Google Scholar]
- 34. Zuber S, Hynes MJ, Andrianopoulos A (2003) The G-protein α-subunit GasC plays a major role in germination in the dimorphic fungus Penicillium marneffei . Genetics 164: 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park G, Pan S, Borkovich KA (2008) Mitogen-activated protein kinase cascade required for regulation of development and secondary metabolism in Neurospora crassa . Eukaryot cell 7: 2113–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Truesdell GM, Yang Z, Dickman MB (2000) A Gα subunit gene from the phytopathogenic fungus Colletotrichum trifolii is required for conidial germination. Physiol Mol Plant Path 56: 131–140. [Google Scholar]
- 37. Ivey FD, Kays AM, Borkovich KA (2002) Shared and independent roles for a Gαi protein and adenylyl cyclase in regulating development and stress responses in Neurospora crassa . Eukaryot cell 1: 634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ivey FD, Yang Q, Borkovich KA (1999) Positive regulation of adenylyl cyclase activity by a Gαi homolog in Neurospora crassa . Fungal Genet Biol 26: 48–61. [DOI] [PubMed] [Google Scholar]
- 39. Lafon A, Seo JA, Han KH, Yu JH, d'Enfert C (2005) The heterotrimeric G-protein GanB(α)-SfaD(β)-GpgA(γ) is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans . Genetics 171: 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klionsky DJ, Herman PK, Emr SD (1990) The fungal vacuole: composition, function, and biogenesis. Microbiol Rev 54: 266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang H, Ng KH, Qian H, Siderovski DP, Chia W, et al. (2005) Ric-8 controls Drosophila neural progenitor asymmetric division by regulating heterotrimeric G proteins. Nature cell biology 7: 1091–1098. [DOI] [PubMed] [Google Scholar]
- 42. Hicke L, Riezman H (1996) Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84: 277–287. [DOI] [PubMed] [Google Scholar]
- 43. Yang Q, Poole SI, Borkovich KA (2002) A G-protein β subunit required for sexual and vegetative development and maintenance of normal Gα protein levels in Neurospora crassa . Eukaryot cell 1: 378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]