Abstract
Structures of human cytochrome P450 2B6 and rabbit cytochrome P450 2B4 in complex with two molecules of the calcium channel blocker amlodipine have been determined by X-ray crystallography. The presence of two drug molecules suggests clear substrate access channels in each P450. According to a previously established nomenclature, amlodipine molecules were trapped in access pathway 2f in P450 2B6 and in pathway 2a or 2f in P450 2B4. These pathways overlap for part of the length and then diverge as they extend toward the protein surface. A previously described solvent channel was also found in each enzyme. The results indicate that key residues located on the surface and at the entrance of the substrate access channels in each of these P450s may play a crucial role in guiding substrate entry. In addition, the region of P450 2B6 and 2B4 involving helices B’, F, F’, G’ and part of helix G is substantially more open in the amlodipine complexes compared with the corresponding 4-(4-chlorophenyl)imidazole complexes. The increased active site volume observed results from the major retraction of helices F, F’ and B’ and the β4 sheet region located close to the binding cavity to accommodate amlodipine. These structures demonstrate novel insight into distinct conformational states not observed with previous P450 2B structures and provide clear evidence of the substrate access channels in two drug metabolizing P450s. In addition, the structures exhibit the versatility that can be exploited in silico studies with other P450 2B6 ligands as large as raloxifene and itraconazole.
Cytochrome P450 (P450) enzymes are a superfamily of monooxygenases involved in the metabolism of a vast array of xenobiotics (1). In addition to their critical role in drug clearance, these heme-containing enzymes are involved in the biosynthesis of steroids and prostaglandins, and can also oxidize a wide variety of endogenous fatty acids (2). X-ray crystal structures and biochemical analysis of cytochromes P450 have led to a wealth of information regarding their substrate specificity and conformational flexibility.
The anchoring of microsomal P450s into the membrane occurs primarily via an N-terminal transmembrane segment, although the enzymes retain membrane-binding properties even upon removal of this domain (3). Our laboratory has used N-terminally truncated and engineered constructs of P450 2B enzymes with internal mutations to achieve the increased solubility, purity and stability required for biophysical and structural studies (4). Various techniques such as site-directed mutagenesis, isothermal titration calorimetry, and deuterium exchange mass spectrometry have been employed previously to characterize the members of the 2B subfamily, primarily rat P450 2B1 and rabbit P450 2B4 (5-7). Additionally, as a result of its superior solubility compared with other 2B enzymes, P450 2B4 has been an excellent model for X-ray crystallography. To date, a total of twelve structures of P450 2B4 have been determined. There are six structures that represent closed conformations of the enzyme in complex with small imidazole inhibitors, the antiplatelet drugs ticlopidine and clopidogrel, and the covalently bound mechanism-based inactivator tert-butylphenylacetylene (8, 9). There are also two distinct ligand free structures, one open and one closed (7, 10), as well as one expanded (11) and three intermediate (9, 12) conformations with various inhibitors.
Of the 57 human P450 enzymes identified, P450 2B6 contributes extensively to the metabolism of pharmaceuticals that include bupropion, efavirenz, propofol, selegiline and artemisinin (13). Moreover, human P450 2B6 is known for its polymorphic nature, with Q172H and K262R representing the most common amino acid substitutions. Years of research efforts on expression, purification, and crystallization have recently led to the structure determination of a human P450 2B6 genetic variant in complex with the nonpharmaceutical ligands 4-(4-chlorophenyl)imidazole (4CPI), 4-benzylpyridine (4BP), and 4-(4-nitrobenzyl)pyridine (4NBP), yielding details on rearrangement of active site residues to accommodate small inhibitors (14, 15). However, information on the conformational behavior of P450 2B4 and 2B6 in the presence of bulkier ligands and/or clinical drugs has been lacking. Interestingly, recent structural analysis of bacterial P450cam revealed distinct conformational states of the protein in the presence or absence of ligand and demonstrated the role of certain secondary structural elements in the conformational changes (16).
The substrate access or exit channels that connect the protein surface to the deeply buried active site have been defined previously in several P450s using X-ray crystallography and molecular dynamics (MD) studies. The structure of P450 2C8 revealed channels on either side of helix B’ extending to the solvent from the buried active site (17), while the P450 2E1 structure described the channel location between the B-B’ loop, the β1 sheet system, and beneath the F’ and G’ helices (18). However, in the vitamin D 25-hydroxylase P450 2R1 structure, vitamin D3 enters the active site via a channel between the G and I helices, the B’ helix and the B-C loop (19), whereas helices B’, F and the β1 sheet region form such an access channel in the cholesterol hydroxylase P450 46A1 structure (20). In addition, the recent structural analysis of Cyp51 revealed substrate access channels near the F-G loop, A’ helix, and β4 loop (21). It is noteworthy that MD simulations on P450 2C9 and P450 3A4 demonstrated opening and closing of several putative substrate access and exit channels (22, 23). Knowledge of substrate access/egress channels was advanced substantially by recent structure determination of dual ligand complexes of P450 24A1, P450 101D2, P450 21A2 and P450 2A13 (24-27). The second ligand molecule was located at a distal site in a channel extending to the protein surface near the B-B’ loop, β1 sheet, and F-G loop in P450 24A1; B’, F/G helices and F-G loop in P450 101D2; between loops β1/β2, β3/β4, and helix F’ in P450 21A2; and close to the β4 sheet system, above K’-L loop in P450 2A13. Accordingly, the existing evidence suggests that the diversity of substrate access channels results from intrinsic differences in the P450s as well as structural differences among the substrates.
An intriguing feature of P450 2B enzymes is the ability to bind ligands of widely diverse size and shape. This may reflect not only the flexibility of the active site but also the dynamic nature of the access channel(s). To elucidate how substrates can gain access to the buried active site in human P450 2B6 and rabbit P450 2B4 and to determine the conformational flexibility in the presence of larger drugs, X-ray crystallography studies were initiated using P450 2B4a in complex with amlodipine (Supplementary Figure 1). Amlodipine is a calcium channel blocker drug used in the treatment of hypertension and coronary artery disease, and has been previously shown to inhibit human P450 2B6 with high affinity (28, 29). Subsequently, P450 2B6b was crystallized in the presence of amlodipine. The resulting structures reveal substrate access channels with two molecules of drug bound to both the rabbit and human P450 2B enzymes, and demonstrate the movement of the structural elements important for substrate access and binding.
MATERIALS AND METHODS
Materials
Amlodipine besylate was obtained from Sigma-Aldrich (St. Louis, MO). CHAPS was from Calbiochem (EMD Chemicals, San Diego, CA). CYMAL-5 (5-cyclohexyl-1-pentyl-ß-D-maltoside) was acquired from Anatrace (Maumee, OH). Nickelnitrilotriacetic (Ni2+-NTA) acid affinity resin was from Thermo Scientific (Rockford, IL). Macroprep CM cation exchange resin was received from Bio-Rad Laboratories (Hercules, CA). Amicon ultrafiltration devices were from Millipore (Billerica, MA). The pGro7 plasmid was from Takara Bio (Shiba, Japan). Escherichia coli JM109 and TOPP3 cells were from Stratagene (La Jolla, CA). Crystal Screen HR2-110 and Wizard II crystallization screen were from Hampton Research (Aliso Viejo, CA) and Emerald Biosciences (Seattle, WA) respectively. 3R-Hydroxy-7R,12Rbis(ethyloxy)cholane (234-chol) is a custom-made facial amphiphile (30). All protein figures were created using PyMOL (31). BKchem version 0.13.0, 2009, (http://bkchem.zirael.org/index.html) was used for drawing the chemical structure of amlodipine.
Expression and Purification of Rabbit P450 2B4 and Human P450 2B6
Cytochrome P450 2B4 was heterologously expressed as described previously (7). Briefly, an overnight Luria-Bertani broth culture of E. coli TOPP3 cells containing the cDNA for 2B4dH(H226Y) in the pKK2B4 plasmid was used to inoculate Terrific broth in the presence of tetracycline and ampicillin. Terrific broth cultures were grown until A600 reached approximately 0.7 at 37 °C. Isopropyl β-D-1-thiogalactopyranoside and δ-aminolevulinic acid were added to a final concentrations of 0.5 mM and 1 mM, respectively, and protein expression was induced for 72 h at 30 °C. Cells were harvested by centrifugation at 4,000 g and resuspended in 10% of the original culture volume in buffer containing 20 mM potassium phosphate (pH 7.4 at 4 °C), 20% (v/v) glycerol, 10 mM 2-mercaptoethanol (BME), and 0.5 mM phenylmethanesulfonyl fluoride (PMSF). The cell suspension was treated with lysozyme (0.3 mg/mL) for 2 h at 4 °C, and centrifuged at 8,000 g. The resulting spheroplasts were then resuspended in 5% of the original culture volume in buffer containing 500 mM potassium phosphate (pH 7.4 at 4 °C), 20% (v/v) glycerol, 10 mM BME, and 0.5 mM PMSF, and were sonicated on ice. The detergent CHAPS was added to a final concentration of 0.8% (w/v), and the sample was stirred for 90 min at 4 °C before subjecting it to ultracentrifugation for 45 min at 245,000 g in a Beckman Coulter Optima L-80 XP Ultracentrifuge using a Ti 50.2 rotor. The concentration of P450 in the supernatant was measured using the reduced CO difference spectra (32).
Histidine tagged P450 2B4 was purified using nickel-affinity chromatography in the presence of CHAPS. The protein bound Ni2+-NTA column was washed using buffer containing 100 mM potassium phosphate (pH 7.4 at 4 °C), 100 mM NaCl, 20% (v/v) glycerol, 10 mM BME, 0.5 mM PMSF, 0.5% CHAPS, and 5 mM histidine. The protein was eluted using the above buffer containing 50 mM histidine. The P450-containing fractions were pooled and quantitated as described above prior to diluting them in buffer with 5 mM potassium phosphate, pH 7.4 at 4 °C, 20% (v/v) glycerol, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.2 mM dithiothreitol (DTT), 0.5 mM PMSF and 0.5% (w/v) CHAPS. The protein was then loaded onto a Macroprep CM column that was washed using the buffer containing 50 mM potassium phosphate (pH 7.4 at 4 °C), 20 mM NaCl, 20% (v/v) glycerol, 1 mM EDTA and 0.2 mM DTT, and eluted with the above buffer containing 500 mM NaCl. Protein fractions containing protein of the highest quality as measured by the A417/A280 ratios were pooled, and the P450 concentration was measured using the reduced CO difference spectra (32).
Heterologous expression of P450 2B6 was carried out in E. coli JM109 cells containing the pKK2B6dH (Y226H/K262R) plasmid. In addition, GroEL/ES chaperones (pGro7 plasmid) were coexpressed with the pKK2B6 plasmid as described previously (15, 33). The purification protocol was exactly the same as that described above for P450 2B4 except that 150 μM of amlodipine from a stock solution in DMSO was included in all the buffers used throughout the purification of P450 2B6. Crystals of P450 2B6-amlodipine complex were not obtained without such inclusion of amlodipine during the purification procedure.
Crystallization and Data Collection
Two mL of each pooled sample of P450 2B4 and P450 2B6 from the ion-exchange column were diluted to 18 μM in 50 mM potassium phosphate (pH 7.4 at 4 °C), 500 mM NaCl, 500 mM sucrose, 1 mM EDTA and 0.2 mM DTT. Amlodipine was added to a final concentration of 180 μM. The P450 2B4 and P450 2B6 were concentrated to 550 μM and 280 μM, respectively, by centrifugation using 50 kD molecular weight cutoff Amicon Ultrafiltration devices. The protein aliquots were again diluted to 18 μM with the above buffer containing amlodipine at a concentration of 180 μM. This process was repeated twice before each aliquot was concentrated to a final concentration of 550 μM and 280 μM, respectively. The concentrated protein samples were supplemented with 4.8 mM CYMAL-5 and 0.077% (w/v) 234-chol (15) before crystallization. Crystal screening for P450 2B4 and P450 2B6 was performed by the sitting drop vapor diffusion method. Crystals of P450 2B4 were obtained from the Wizard II screen (Emerald Biosciences) at 18 °C after 7 days of incubating the protein in a 1:1 ratio with the precipitant containing 1 M K/Na tartrate, 0.1 M Tris pH 7 and 0.2 M Li2SO4.
Crystals of P450 2B6 grew over a period of one month after incubating the protein at 18 °C in a 1:1 ratio with the precipitant containing 0.2 M sodium acetate trihydrate, 0.1M Tris HCl pH 8.5 and 30% w/v PEG4000 using Crystal Screen HR2-110 from Hampton Research. Crystals of P450 2B4 and P450 2B6 were transferred to mother liquor containing 20% (v/v) sucrose as cryoprotectant before being flash frozen in liquid nitrogen. Crystallographic data were collected on each P450 crystal remotely at Stanford Synchrotron Radiation Lightsource (SSRL) beamline 11-134 (2B4) and 7-134 (2B6) using 1° oscillations over 240 frames and 20 s exposures using Marmosaic 325 CCD detector for P450 2B4 crystal and Quantum 315 CCD detector for P450 2B6 crystal at 100 K. Crystals of P450 2B4 diffracted to 2.25 Å, while the crystals of human P450 2B6 diffracted to 2.8 Å. Data were integrated using iMOSFLM (35) and scaled via SCALA in CCP4i (36).
Structure Determination and Refinement
To determine the structure of P450 2B4, coordinates of the closed ligand-free P450 2B4 structure (PDB entry 3MVR) were used as a search model in the molecular replacement program Phaser (37) in CCP4i. The space group was determined to be P3121 and Matthews coefficient determination suggested the presence of one molecule in the asymmetric unit with 67.8% solvent content. The output model from Phaser was submitted to rigid body and restrained refinement in REFMAC (38), and COOT (39) was used to further build the model using Fo-Fc and 2Fo-Fc electron density maps contoured at 3-sigma and 1-sigma, respectively. The library description for amlodipine was created using the PRODRG server (40). Iterative model building and refinement was continued until the R-factor and R-free stopped improving. The overall geometry of the structure analyzed by MOLPROBITY (41) ranked in the 98th percentile among structures of comparable resolution, with no Ramachandran outliers or bad bond lengths or angles. The final model of the crystal structure contains protein residues 28 to 492, with the terminal histidine residue being a part of the larger C-terminal His-tag. There were 206 water molecules found in the model, and a single CYMAL-5 detergent molecule was observed in the hydrophobic pocket near residues Phe202 and Phe296. In addition, during refinement, two alternate conformations of the amlodipine chlorophenyl ring were observed in the molecule ligated to heme. Residue 136, located within the C-D loop region, which was previously shown to interact with cytochrome P450 reductase (42), was disordered in the final model. This was probably due to high mobility and is consistent with several other P450 2B4 structures. Moreover, density for residue 48 was missing, as was part of density for the side chains of R49 and R73, which thus were not modeled. Coordinates and structure factors were deposited in the Protein Data Bank (PDB entry 3TMZ). The refinement statistics for the above structure are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics.
| Cytochrome P450 | 2B4 | 2B6 |
| Ligand | Amlodipine | |
| Crystal Space group | P3121 | P 212121 |
| Crystal unit cell parameters | ||
| a | 92.9 Å | 58.0 Å |
| b | 92.9 Å | 78.3 Å |
| c | 152.7 Å | 247.3 Å |
| α = β | 90 ° | 90 ° |
| γ | 120 ° | 90 ° |
| Molecules per asymmetric unit | 1 | 2 |
| Data Collection statistics a | ||
| Beamline | SSRL 11-1 | SSRL 7-1 |
| Wavelength (Å) | 0.98 | 0.98 |
| Resolution range (Å) | 38.93 - 2.25 | 82.45 - 2.80 |
| Completeness (%) | 96.4 (65.0) | 90.4 (82.5) |
| Redundancy | 6.2 (3.3) | 4.3 (3.2) |
| Rmerge(%) | 4.3 (49.8) | 13.1 (60.3) |
| I/σ | 13.6 (1.5) | 4.7 (1.3) |
| No. of unique reflections | 36,346 | 25,912 |
| Refinement statistics | ||
| R-factor | 19.1 % | 24.2 % |
| R-free | 25.1 % | 29.4 % |
| RMS deviations | ||
| Bond lengths (Å) | 0.023 | 0.012 |
| Bond angles (°) | 2.085 | 1.430 |
| No. of atoms. Average B-values Å2 are in brackets | ||
| Protein | 3694 [48.3] | 7379 [50.8] |
| Heme | 43 [33.4] | 86 [38.2] |
| Proximal Amlodipine Distal Amlodipine |
28 [47.06] 28 [80.2] |
28 [48.9] 28 [67] |
| Waters | 203 [51.2] | 78 [36.34] |
| Detergent | 34 [86.3] | 0 |
| Ramachandran Plot | ||
| Preferred (%) | 96.7 | 94.7 |
| Allowed (%) | 3.3 | 5.3 |
Parenthesis indicate highest resolution shell
The structure of P450 2B6 was determined using the automated molecular replacement pipeline Balbes (43). The space group was found to be P212121, containing 52.3% solvent, assuming two monomers per asymmetric unit. A solution was determined using the P450 2B6-4CPI structure (PDB entry 3IBD) from the PDB database as the search model. The output model was subjected to a rigid body refinement and a restrained refinement in REFMAC using the tight non-crystallographic symmetry options. The water molecules were added manually and the iterative process of model building and refinement yielded the final R-factor and R-free of 0.24 and 0.29, respectively. MOLPROBITY (41) analysis, which scored the structure at 91st percentile among structures of comparable resolution with no Ramachandran outliers, is presented in Table 1, along with overall geometry and final refinement statistics of the structure. The final model of P450 2B6 consisted of residues 28 to 492 in chain A and chain B as observed with P450 2B4, and 78 water molecules. Density for residues 28, 280 and 281 in chain B was poorly defined so they could not be modeled. The atomic coordinates and structure factors were deposited in the Protein Data Bank (PDB entry 3UA5).
Determination of Substrate Access Channels in P450 2B4 and P450 2B6 and Active Site Cavity Volume Calculations
The substrate access channels in P450 2B4 and P450 2B6 were analyzed using CAVER software (44). The water, CYMAL-5, and amlodipine molecules were deleted from the coordinates of P450 2B4 and P450 2B6 chain A, and the region above the active site heme iron was chosen as the starting point of access channel calculation using a 4 Å probe in CAVER. PyMOL (31) was used for visualizing the substrate access channels. Active site cavity volumes of the P450 2B-amlodipine complexes were calculated using Voidoo (45), and probe occupied volumes were determined with a probe radius of 1.40 Å. Because of the connection between the active site and the substrate access channel 2f leading to the protein exterior (vide infra), water molecules were added at the location of the second molecule of amlodipine to limit the active site to the region surrounding the first amlodipine.
Ligand Docking
Docking of amlodipine, raloxifene, and itraconazole into the amlodipine-bound complexes of 2B4 (PDB ID: 3TMZ) and 2B6 (PDB ID: 3UA5) was done by using AutoDock Vina version 1.1.1 (46). Residues not modeled into the experimental electron density were added using COOT, and amlodipine and CYMAL-5 molecules were removed from the file. Experiments were performed with a rigid receptor molecule and Gasteiger charges were utilized for the small molecule. Heme charges were modified using a separate script to provide previously reported values (47). The docking experiment included 20 events with a box 70Å × 70Å × 70Å centered on the heme iron.
Enzyme Inhibition Studies
Rates of the O-dealkylation of 7-ethoxy-4-(trifluoromethyl) coumarin (7-EFC) and 7-methoxy-4-(trifluoromethyl)coumarin (7-MFC) to the metabolite 7-hydroxy-4-(trifluoromethyl)coumarin for P450 2B4 and P450 2B6 respectively, were measured using a fluorometric assay. A reconstituted enzyme system contained P450 2B4 or P450 2B6, rat cytochrome P450 reductase (CPR) and rat cytochrome b5 (48) at a molar ratio of 1:4:2. The reactions were performed in a 100 μl volume containing 50 mM HEPES, pH 7.4, 15 mM MgCl2, 10 pmol P450, 40 pmol CPR, 20 pmol cytochrome b5, 50 μM 7-EFC for P450 2B4 or 7-MFC for P450 2B6, and 0-1 mM of amlodipine. Reactions were initiated by adding NADPH after 5 min of preincubation at 37 °C. Trichloroacetic acid (20% v/v) was added to quench the reaction after 5 min of incubation. In addition, a single reaction was terminated without the addition of substrate in a control experiment. An F-2000 Fluorescence Spectrophotometer (Hitachi, Tokyo, Japan) with λex at 410 nm and λem at 500 nm was used to measure the fluorescence, and the IC50 values were determined using the Michaelis Menten equation with the scientific package Igor Pro 6.1 (Wavemetrics, Inc., Lake Oswego, OR). IC50 values of 2 μM and 1.6 μM were determined for inhibition by amlodipine of P450 2B4 and P450 2B6, respectively. These values were similar to the previously reported values using wild-type P450 2B6 (29).
RESULTS
Structural Analysis of P450 2B4 and P450 2B6 Complexes with Amlodipine
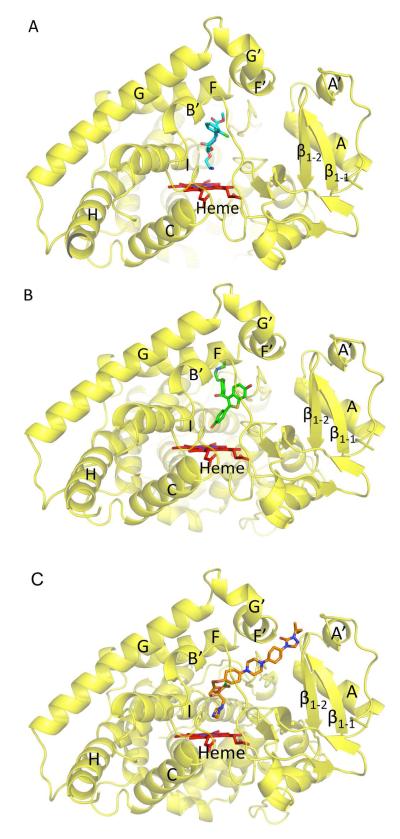
Crystal structures of P450 2B4 and P450 2B6 were solved in the presence of amlodipine at 2.25 Å and 2.8 Å resolution, respectively. The structures described here displayed the global P450 fold and similar overall conformation to each other. Unbiased electron density for the drug amlodipine (Figures 1A, B and Supplemental figure 2) was clearly observed bound to heme in P450 2B4 and P450 2B6 via the amine nitrogen atom. In addition, unbiased electron density for another molecule of amlodipine near the active site was located close to the first molecule in both enzymes. The second molecule of amlodipine in P450 2B6 was found in a different orientation than in P450 2B4. Attempts to model second amlodipine molecule observed in P450 2B4 in the orientation similar to the second amlodipine in P450 2B6 resulted in worsening of the B-values. The two P450 2B6 molecules present in the asymmetric unit are identical to each other with a root mean square deviation (rmsd) of 0.1 Å for the Cα atoms between chains A and B. The complete structures of P450 2B4 and P450 2B6 are shown in Figures 1C and 1D, respectively.
Figure 1.
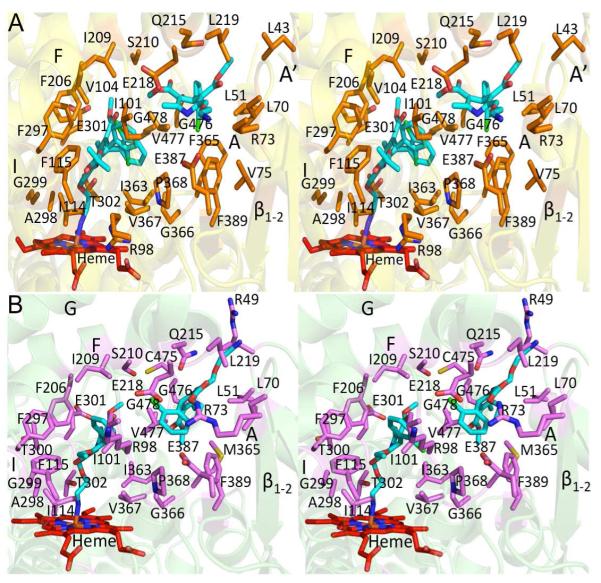
Unbiased electron density map (green mesh) of the ligand and structures of P450 2B4 and P450 2B6 in complex with amlodipine. Heme is shown in red and amlodipine is shown in cyan. A, an unbiased Fo-Fc omit map calculated prior to adding the ligand in the P450 2B4-amlodipine complex contoured at 3σ shows the presence of electron density corresponding to amlodipine above the heme. Density corresponding to an alternate conformation of the chlorophenyl ring was also observed. In addition, an unbiased Fo-Fc omit map before the inclusion of second amlodipine molecule is also shown. B, an unbiased Fo-Fc omit map calculated prior to inclusion of amlodipine in the P450 2B6-complex contoured at 3σ shows the electron density above heme. Such unbiased electron density corresponding to the presence of another molecule of amlodipine is also shown from chain B. The density for this second molecule of amlodipine in chain A was partially disordered in P450 2B6. C, a ribbon diagram of P450 2B4-amlodipine structure is shown with labeled α-helices (orange) and β-sheets and loops (yellow). D, a ribbon diagram of the P450 2B6-amlodipine structure is shown (chain B) with labeled α-helices (magenta) and β-sheets and loops (green).
Figures 2A and 2B show the residues located within a 5 Å radius of either amlodipine molecule in P450 2B4 or P450 2B6. The residues found within such distance from the heme-bound amlodipine molecule in the active site are located mainly on the B/C loop and helices F and I in each of these proteins. The orientation of amlodipine and the majority of the active site residues are identical in both enzymes (Supplemental Table 1A), except for residues T300 and C475 in P450 2B4 and R98 and V104 in P450 2B6, which move out of the 5 Å radius in the other structure. C475 in P450 2B6 and V104 in P450 2B4 have been shown previously to interact with ligands in the active site in other 2B structures (49, 50). The orientation of the F206 and F297 side chains, which were shown recently to rearrange in order to accommodate various ligands within the active site of human P450 2B6 (15), were found in the same orientations observed in the respective P450 2B4 and 2B6 4CPI complexes.
Figure 2.
Stereo view of the active site of P450 2B4 and P450 2B6 structures showing residues located within a 5 Å radius of either molecule of bound amlodipine. Heme and amlodipine are shown in red and cyan respectively. A, the active site residues of P450 2B4 are depicted as orange sticks. B, the active site residues of P450 2B6 are depicted as magenta sticks.
The second molecule of amlodipine was sandwiched between helices F, F’ and A’, and A in both P450 2B4 and P450 2B6, suggesting a secondary binding site for the ligand. Additionally, as listed in Supplemental Table 1B, 15 residues in P450 2B4 (Figure 2A) and 17 residues in P450 2B6 (Figure 2B) are within a 5 Å radius of this amlodipine. In P450 2B4, the chlorophenyl moiety and the ethyl ester of this secondary amlodipine face helices A and A’ respectively, while the amine nitrogen orients toward the solvent region near the β1-2 sheet. However, in P450 2B6, the chlorophenyl group of the secondary amlodipine molecule is facing the heme-bound amlodipine, with its amine group extending toward the solvent region between helices A’ and F’ and the ethyl ester located near the β1-2 sheet.
Substrate Access Channels Revealed in Mammalian P450 2B4 and P450 2B6
The different orientations of the second amlodipine molecule are not explained readily by differences in amino acid sequences between the rabbit and the human P450 2B enzymes. Because of the location of this molecule extending to the protein surface in both enzymes, we hypothesized that the drug was trapped in a substrate access channel. Similar channels have been identified previously in several P450 crystal structures using CAVER software (44). Recent computational studies of P450 2C9 (51, 52) explicitly described such channels using a well-defined nomenclature (53). Using CAVER a total of three channels in P450 2B4 and two channels in P450 2B6 were identified, each running from the active site to the bulk solvent. In both enzymes, the second amlodipine molecule is poised to enter the active site via an entrance pathway that lies between helices A’, A, and F’. Inside each enzyme, the entrance path for amlodipine is surrounded by helices B-B’, K-K’ and the β4 loop system, which guide the substrate into the active site. Residues L43, M46, R48, K/R49, F/V212, V/L216, and L219 flank this substrate access channel.
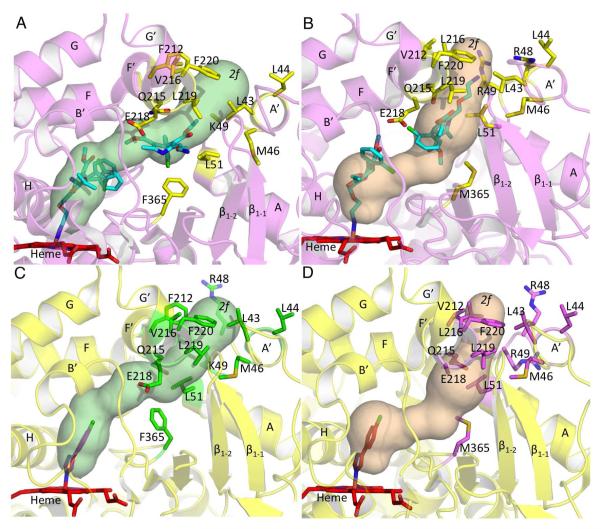
When compared with the corresponding residues of P450 2C9, the access channel observed in P450 2B4 and P450 2B6 closely resembles channel 2f based on the previously defined nomenclature (Figure 3A, 3B and Table 2). Channel 2f, which was mostly closed during P450 2C9 simulations, clearly opens up in P450 2B6 and P450 2B4 for amlodipine access. In P450 2B6, the primary amine of the second amlodipine molecule faces the entrance of the 2f channel. However, in P450 2B4, the ethyl ester group of this drug molecule is located near this entrance. When compared with the respective P450 2B-4CPI complexes, the patch of hydrophobic residues near the entrance of the channel, (F/V212, V/L216, L219, F220 on the F’ helix, and L43, L44 on the A’ helix) is pushed out significantly, opening the region by more than 6 Å to allow the bulky amlodipine to enter (Figure 3C and 3D, respectively). In addition, residues L51, Q215, and E218, which protrude into the channel in the 4CPI complexes, move out in the amlodipine complexes. Interestingly, F365 in P450 2B4 flips by more than 60° compared with the 4CPI complex to make space for the chlorophenyl ring of the second amlodipine molecule. This movement also contributes to opening of the 2f channel in P450 2B4. Such a marked movement of residue M365 is not observed in P450 2B6, mainly due to an alternate conformation of amlodipine. However, the sulfur side chain of methionine does move out slightly to accommodate the methyl ester group of amlodipine.
Figure 3.
Substrate access channel 2f shown in P450 2B4 and P450 2B6 as computed by CAVER, and the residues located at the channel entrance. Heme is shown in red and amlodipine and 4CPI molecules are shown in cyan and orange, respectively. Tunnels are labeled according to the previously defined nomenclature (53). In addition, residue 365 located further in the substrate access channel is also shown to represent the marked movement of the side chain. A, the 2f channel observed in the P450 2B4-amlodipine complex (magenta) and the residues (yellow sticks) located at the entrance of the channel. B, the 2f substrate access channel found to open in human P450 2B6 amlodipine complex and the residues (yellow sticks) located at the channel entrance. C and D, corresponding residues in the P450 2B4-4CPI (green sticks) and P450 2B6-4CPI (magenta sticks) complexes, respectively that protrude into channel 2f of the P450 2B4- (yellow) and P450 2B6-amlodipine complex (magenta).
TABLE 2.
Residues and secondary structural elements located at the entrance channels in P450 2B4 and P450 2B6 are shown along with the corresponding residues in P450 2C9 proposed previously.
| Channel | Residues Lining the access channels in corresponding P450s |
Secondary Structure |
||
|---|---|---|---|---|
| P4502B4 | P450 2B6 | P450 2C9 | ||
| Solvent | S207, E301, R308, V477 |
S207, E301, R308, V477 |
C206, E300, R307, F476 |
E, F, I helices, β4 sheet |
| 2f | P38, L43, M46, R48, F212, V216, L219, G222 |
P38, L43, M46, R48, V212, L216, L219, G222 |
P37, I42, I45, I47, I215, N218, P211, P221 |
F’, G’ helices, A’, A helices |
| 2a | L70, R73, A102, S221, G222 |
L70, R73, A102, S221, G222 |
K69, K72, P101, S220, P221 |
F’, G’ helices, β1 sheets |
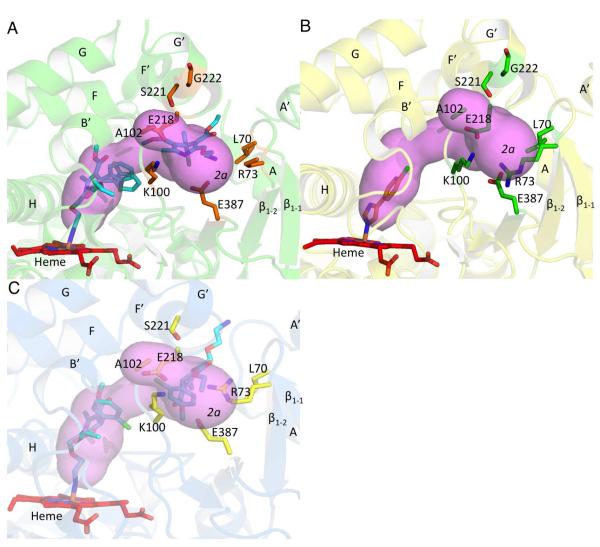
Further analysis revealed a second substrate access channel in P450 2B4 between helices B’ and G’ near the β1 sheet system (Figure 4A). Comparison of the corresponding residues lining the channel entrance with P450 2C9 indicates that this channel closely resembles channel 2a according to the nomenclature (Table 2). Additionally, as seen from comparison of figures 3a and 4a, channels 2a and 2f start at the same location and then diverge as they extend toward the protein surface. In the P450 2B4-4CPI complex, channel 2a is blocked by E218 on the F’ helix and R73 on the β1-2 sheet, which protrude into the channel (Figure 4B). The movement of the F’ helix as a result of amlodipine binding displaces E218 in both P450 2B complexes, which contributes to branching of channel 2f and 2a in P450 2B4 and P450 2B6. However, channel 2a is blocked in the P450 2B6-amlodipine structure (Figure 4C) because of the orientation of R73. In P450 2B4, the density for the guanidino group of R73 is missing, but based on the side chain carbons that are observed, the group must point away from this channel towards the solvent. This side chain orientation opens up the branched out channel 2a in P450 2B4. The presumed high mobility of R73 likely allows it to avoid any clash with the primary amine of amlodipine in P450 2B4. Residues lining the entrance of this tunnel are R73, K100, A102, E387, on the β1 sheet system and S221 on the G’ helix.
Figure 4.
Substrate access channel 2a as computed by CAVER. A, access channel 2a observed in the P450 2B4-amlodipine complex is shown and residues located at the entrance near the protein surface are shown in orange sticks. The primary amine of amlodipine (cyan sticks) is seen to extend near the entrance of the 2a channel. B, corresponding residues in the P450 2B4-4CPI complex that need to move are shown in green sticks near the 2a channel of the amlodipine complex. C, access channel 2a is not observed in P450 2B6 mainly as a result of R73 (yellow sticks), which projects into the channel entrance. The primary amine tail of amlodipine (cyan sticks) is now seen to extend at the 2f channel entrance.
The solvent channel, which has been described, previously in several MD simulation and structural studies (24, 51, 54-57), was located between helices F and I and the β4 loop region near helix E. A recent study of P450 2C9 (51) predicted that the solvent channel is used for metabolite egress due to the polar residues lining the channel. However, this channel may also be involved in substrate entry. The residues lining the entrance of this channel in P450 2B4 and P450 2B6 are shown in Table 2 along with the corresponding residues previously suggested in P450 2C9. In the P450 2B4-4CPI complex, the side chain of residue F203 protrudes into the solvent channel, while in the amlodipine complex, F203 flips ~90° outward to open this channel, thus creating a large access pathway for possible substrate or metabolite to entrance or egress (Supplemental Figure 3). In P450 2B6, residue 203 is a tyrosine and is observed to move ~30° compared with the 4CPI complex (Supplemental Figure 4). Again, this creates significantly more room for larger ligands. Additionally, residue E474 in P450 2B6 flips away from the channel by about 180°, allowing C475 to be closer to the access path than in the 4CPI complex. Such displacement of E474 in P450 2B4 by about 90° is also observed compared with the respective P450 2B4-4CPI complex, creating an even larger space for the solvent channel. The roles of E474 and C475 have been investigated recently in P450 2B4 and P450 2B6 respectively, using site-directed mutagenesis. The P450 2B4 E474 mutants exhibited a significant change in binding affinity with several ligands, while P450 2B6 C475S showed altered mechanism-based inactivation by 2-oxo clopidogrel (50, 58).
Conformational and Active Site Adaptations of P450 2B4 and P450 2B6 upon Amlodipine Binding
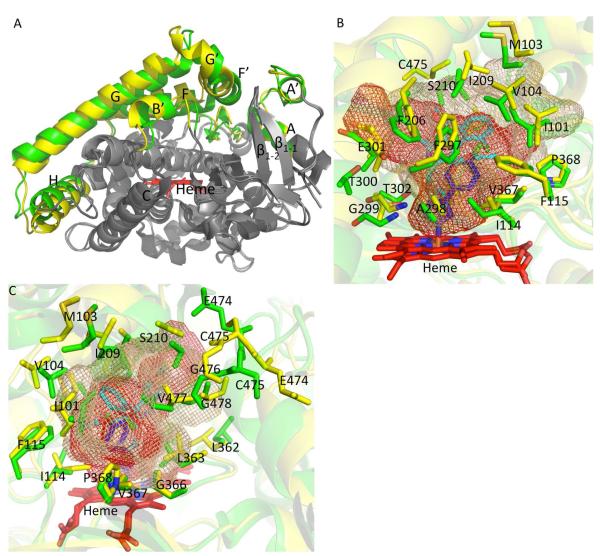
An overlay of the structure of the P450 2B4-amlodipine complex onto the open ligand free structure of the protein (10) and the closed conformation of the 4CPI complex (49) is shown in Figure 5A. The closed 4CPI-bound conformation of P450 2B4 is essentially identical to the closed ligand free structure (rmsd of 0.49 Å) as well as the complexes with the drugs ticlopidine or clopidogrel (rmsd of 0.37 Å and 0.32 Å, respectively). Relative to these closed P450 2B4 structures, the P450 2B4-amlodipine complex exhibits a substantial displacement of helices F, F’, G’, and G, which coordinate as a unit, and of helices A’, A, and B’. Additionally, the β1-1 and β1-2 sheets were also displaced considerably. The average rmsd between the structure of the amlodipine complex and the closed 2B4 structures was 0.66 Å, with the largest differences resulting from the movement of the secondary structural elements discussed above. A comparison with the open ligand free 2B4 structure reveals changes in the orientations of helices I and C and G-H and H-I loop, in addition to similar differences seen in the closed conformation structures. In the active site, eight additional residues R98, F206, I209, S210, P368, and three glycines 299, 366, and 478 compared with the 4CPI complex of P450 2B4 circumscribe the bulkier amlodipine molecule. Interestingly, each of these additional contact residues has been shown to contact ligands in the active site in at least one or more of the previously solved ligand bound structures of P450 2B4 (59).
Figure 5.
Overlay of P450 2B4 structures. A, superimposed structures of the P450 2B4 amlodipine complex (yellow), P450 2B4 open ligand free (1PO5) (pink), and 4CPI (1SUO) bound (green) structures with secondary structural elements that are displaced colored as described above. Ligand molecules were removed for better clarity. B and C (orthogonal views of the same figure), overlay of residues in sticks lining the active site of P450 2B4-amlodipine (yellow) and P450 2B4-4CPI (green). The cavity volume of P450 2B4-amlodipine (green mesh) is increased significantly upon movement of active site residues. The cavity volume of P450 2B4-4CPI complex is shown in red mesh. Amlodipine and 4-CPI molecules are shown in cyan and blue, respectively.
When compared to the 4CPI complex, the binding of the much larger amlodipine results in significant alteration of the size and topology of the active site (Figure 5B and 5C), yielding an active site cavity (605 Å3) that is much larger than previously observed in P450 2B4 structures such as the 4CPI complex (253 Å3) (8). This difference results from the location and orientation of the residues that line the heme-binding pocket region. Cα displacement by ~2 Å in the amlodipine relative to the 4CPI complex was observed for S475, G476 and V477 located near the β4 sheet region in P450 2B4. In addition the backbone of residues I101 and V104 and I209 and, S210 on the B’ and F helix respectively, were retracted considerably by about 1.5 Å compared with the 4CPI complex. Moreover, the cluster of phenylalanine residues F115, F206, F297 and F365 exhibited marked side chain adjustments to accommodate amlodipine, and E301 and F365 rotate out in the amlodipine complex, further contributing to the enlarged binding cavity.
While structural studies have revealed a range of P450 2B4 conformations, human P450 2B6 has only been crystallized in the presence of the small inhibitors 4CPI, 4BP, and 4NBP, resulting in similar closed protein structures. The conformational changes observed in the P450 2B6-amlodipine complex when compared with the above closed conformation structures solved previously were more pronounced (average rmsd of 0.88 Å) than in P450 2B4. Binding of amlodipine to P450 2B6 is accompanied by conformational shifts of the G-H loop and helices A’, A, B’, F, F’, G’, G, and H (Figure 6A). Moreover, there were marked differences between the amlodipine and 4CPI complexes of P450 2B6 near the C-terminal coil (the β3-3 sheet, helix L’, the β4-1, β4-2, and β3-2 region), as compared with the corresponding P450 2B4 structures (Supplemental Figure 5). The β4-1 and β4-2 sheets observed in the 4CPI complexes of the respective P450s are now seen as a continuous loop extending to the C-terminal end close to the β3-2 sheet in the amlodipine complexes of P450 2B4 and P450 2B6.
Figure 6.
Overlay of human P450 2B6 structures. A, P450 2B6 complexed with amlodipine and 4CPI (3IBD) superimposed onto each other with differences in secondary structural elements colored in yellow and green, respectively. Amlodipine and 4CPI were deleted for a clear view. B and C, (orthogonal views of the same figure) overlay of cavity volume and residues in sticks lining the active site of P450 2B6-amlodipine (yellow) and 4CPI complexes (green). The cavity volume of the amlodipine complex and the 4CPI complex of P450 2B6 is shown in brown and red mesh, respectively.
As shown in Figure 6B and 6C, the active site of the P450 2B6-amlodipine complex extends to residues F115, S210, G299, T300, E301, L362, G366, P368, and G478, which were not found to be a part of the P450 2B6-4CPI binding pocket. The calculated volume of the P450 2B6 active site cavity is 755 Å3 compared with the P450 2B6-4CPI complex of 582 Å3 previously observed (8). Residues I101, M103 and V104 on the B’ helix and I209 and S210 on the F helix in the P450 2B6 amlodipine complex structure are now shifted out even more compared with the 4CPI complex or the P450 2B4-amlodipine complex, and this contribute to the significant increase in the cavity dimensions compared with the other structures. Additionally, movement of residues 474-477 with the Cα of E474 and C475 displaced by as much as ~3 Å, expands the volume of the cavity to accommodate amlodipine within the active site. Differences in the cavity volume between the two P450 2B6 complexes also reflect the movement of phenylalanine side chains as observed with P450 2B4.
Interestingly, the part of the cavity bounded by residues T300, E301, and T302 on the I-helix is actually reduced in volume in the amlodipine structure compared with the 4CPI complex. Specifically, the side chain of E301 now protrudes into the cavity observed in the 4CPI complex. Moreover, as observed previously in the 4BP complex (15), the side chain of L362 swings in by 90° towards the active site and directs T302 in close proximity to the amlodipine molecule.
Differences in Specific Amlodipine Interactions between P450 2B4 and P450 2B6
An overlay of the P450 2B4 and P450 2B6 amlodipine bound structures (Supplemental Figure 6) resulted in an rmsd of 0.66 Å and revealed considerable differences in the C-D loop, H-I loop, C-terminal loop and the β1-1 and β1-2 sheets, which bulge out more toward the solvent in the P450 2B4 structure (Supplemental Figure 6 – inset). The only polar residue interaction evident between the proteins and amlodipine within the hydrophobic active site is with T302. This side chain in the P450 2B4-amlodipine complex makes a hydrogen bond with the pyridine nitrogen of amlodipine, while the polar side chain of T302 interacts with the ethoxy oxygen of amlodipine within the active site of P450 2B6 (Supplemental Figure 7A and 7B). Moreover, the distinct orientation of the second amlodipine in the two structures revealed interesting differences in polar interactions with this molecule. In P450 2B4, hydrogen bonding occurs between the primary amine of this amlodipine molecule, which extends into the 2a substrate access channel, and the carboxylate of E387 on the β1-3 sheet (Supplemental Figure 7A). Interestingly, the alternate conformation of this molecule located in the 2f substrate access channel in P450 2B6 structure now allows the polar side chain of Q215 to hydrogen bond with the pyridine nitrogen and ethoxy oxygen of the amlodipine (Supplemental Figure 7B). However, Q215 and the pyridine nitrogen and ethoxy oxygen of amlodipine contact a water molecule in the P450 2B4 structure.
Ligand Docking
Computer aided docking was used to explore the utility of the amlodipine-bound complexes of P450 2B4 and P450 2B6 in predicting protein-ligand interactions of larger drugs. First, the methodology was validated by docking amlodipine, which inhibited P450 2B4 and 2B6 with IC50 values of 2 μM and 1.6 μM respectively. Subsequently, two larger compounds, the selective estrogen receptor modulator raloxifene (MW = 473.5 g/mol) and the antifungal agent itraconazole (MW = 705.5 g/mol) were studied. Both have been shown previously to be among the most potent inhibitors of P450 2B6 (29). As presented in more detail below, amlodipine runs produced the largest variations in binding site, and raloxifene runs produced binding events predominantly in the active site or the 2f substrate access channel. While itraconazole runs largely produced poses with the imidazole end of the molecule located in the active site, roughly equal numbers of poses placed the imidazole or the p-chloro of the dichlorophenyl group closest to the heme iron. Representative poses of P450 2B6 docking experiments are shown in Figure 7.
Figure 7.
Representative pose showing amlodipine (cyan sticks), raloxifene (green sticks) and itraconazole (orange sticks) docking using the P450 2B6-amlodipine structure shown in yellow ribbon. Heme is shown in red sticks. A, Docking of amlodipine shows the amine nitrogen orienting towards the heme iron in the highest affinity pose consistent with the x-ray crystal structure. (B) Raloxifene docking with the hydroxyphenyl oriented toward the heme as the lowest-energy and highest affinity pose located within the active site in P450 2B6. C, depiction of itraconazole with the imidazole nitrogen facing the heme in the most populous and highest affinity poses.
Amlodipine
As expected, this molecule adopted multiple poses in the active site and substrate access channels of both P450 2B4 and P450 2B6; the energy distributions were roughly equal for rigid poses (P450 2B4: –7.4 to –5.4 kcal mol−1; P450 2B6: –7.5 to –5.2 kcal mol−1). Docking utilizing P450 2B4 as a receptor produced a variety of poses of a single molecule of amlodipine. These were distributed: 1) throughout the active site, 2) partially located in the solvent access channel, 3) in the 2a and 2f substrate access channels as in the structure, 4) only in the 2f substrate access channel, or 5) in various locations across the enzyme’s external surface. With P450 2B6 as the receptor, most poses were: 1) in the active site, 2) solely in the 2f channel, or 3) at the interface of the 2a and 2f substrate access channels. The three lowest affinity poses were located between the C- and H-helices on the periphery of the enzyme. In each experiment, the most populated poses that include the one with highest affinity for P450 2B4 and P450 2B6 had the terminal amine nitrogen oriented toward the heme iron in the active site (Figure 7A).
Raloxifene
For both P450 2B4 and P450 2B6, all poses were found in the active site, substrate access channel 2f, or at the interface of these two regions; docked complexes resulted in similar energy distributions for the two enzymes in the rigid docking events (P450 2B4: –10.1 to –9.4 kcal mol−1; P450 2B6: –10.8 to –9.5 kcal mol−1). For P450 2B4, raloxifene showed approximately equal preference for the active site or the substrate access channel interface involving mainly channel 2f. In contrast, raloxifene was found in the active site of P450 2B6 in the 18 events with lowest energy, the junction of substrate access channels 2a and 2f in one event and at the interface of the active site and the substrate access channels in one event. The majority of the clusters within the active site had either the hydroxyphenyl group or the thiophene group closer to the heme. Moreover, the thiophene group was facing the heme iron in the highest affinity pose with P450 2B4, whereas P450 2B6 had the hydroxyphenyl group towards heme in the highest affinity orientation (Figure 7B).
Itraconazole
This relatively linear molecule adopted multiple poses in the active sites and access channels of both P450 2B4 and P450 2B6, and the energy distributions were comparable for both enzymes (P450 2B4: −10.9 to −8.5 kcal mol−1; P450 2B6: −11.5 to −8.5 kcal mol−1). Events with P450 2B4 as the receptor produced 17 poses with part of the ligand in the active site and the remainder protruding into the 2f substrate access channel or exiting the solvent access channel; three poses bridged the 2a and 2f substrate access channels and did not enter the active site. Similarly, P450 2B6 produced 17 poses, 16 of which entered the active site and one of the access channels, while one pose was on the distal face of the protein. With each of the above proteins, the highest affinity pose of itraconazole in the active site had the imidazole nitrogen facing the heme iron as shown in Figure 7C.
Spectral Binding Titrations
To confirm the orientation of the first amlodipine molecule, spectral binding titrations were performed with both P450 2B4 and P450 2B6. As shown in Supplemental Figure 8, amlodipine induced a type II spectrum in P450 2B4 indicative of an iron-nitrogen coordinate bond. The spectral dissociation constant (Ks) from three independent experiments was 2.06 +/− 0.37 μM for P450 2B4, which is very similar to the IC50. The maximal % conversion to the type II complex was 23 +/− 3 %. P450 2B6 produced a similar type II spectral change and binding affinity upon amlodipine addition as P450 2B4 (data not shown).
DISCUSSION
The first structure of human P450 2B6 in complex with a drug and the P450 2B4 complex determined by X-ray crystallography in this study show the existence of substrate access channels in these enzymes. The detailed information obtained advances significantly our mechanistic understanding of how P450s recognize and bind larger drugs and the conformational changes that a P450 undergoes in the process. Previous in silico studies of several mammalian and bacterial P450s, including P450 2C9, P450 3A4, P450 101A1 and P450 2A6 (23, 51, 60, 61) have used MD simulations to predict the presence of a substrate entrance pathway that connects the active site and the surface of the protein. The channels 2f and 2a in P450 2C9 have been suggested based on MD simulations to point toward the hydrophobic core of the membrane and to be involved in substrate access, whereas the solvent channel is hypothesized to be involved in metabolite egress. However, substrate access channel 2f was found mostly closed in the simulations performed for P450 2C9 and was thought to facilitate wider opening of the protein by merging with channel 2a (51). A similar MD study using the P450 2C9 substrate ibuprofen also suggested that the 2f and 2a channels could be important for substrate binding, whereas the solvent channel is involved in metabolite release (52). Our X-ray structures indicate that both amlodipine molecules could have entered the active site via channel 2f in human P450 2B6 and rabbit P450 2B4 as well as via the additional channel 2a in P450 2B4. However, the possibility remains that the first amlodipine molecule enters the protein via several available channels, with the more distal amlodipine molecule representing a secondary binding site after the binding of first amlodipine to the heme. The precise role of the solvent channel lined with several hydrophilic residues remains unknown; nevertheless, it is accessible for ligands to enter or exit from protein exterior. Furthermore, results from computer-aided docking corroborated structural studies and the use of these channels for substrate access not only to fit amlodipine into the active site pocket, but also other 2B ligands as large as raloxifene and itraconazole.
Multiple binding of ligands has been observed previously with P450 3A4-ketoconazole and P450 2C8-9-cis-retinoic acid complexes, but the second ligand molecule was confined to the active site and packed on top of the first molecule near the heme (17, 62). Recent structural analysis of P450 2A13 found two NNK molecules in a closed conformation of the protein (24). From a structural overlay with the current 2B4-amlodipine complex, (Supplementary Figure 9) it appears that the second NNK molecule in P450 2A13 is located right at the interface of the solvent channel and the 2a channel. However, neither the 2a or 2f channels were observed in the dual ligand complex of P450 2A13 as a result of the closing of helices F, F’, G’, G and the β4 sheet system, which retracted as much as 4 Å compared to the current 2B structures bound to amlodipine. Clearly, the presence of a solvent channel, which was also observed in the recent P450 2D6 structure (57), suggests a possible route for ligand entry into these enzymes that is located near the β4 sheets and the F helix. Interestingly, the second molecule of 17α-hydroxyprogesterone in the P450 21A2 (25) complex is in a perfect overlay with the second molecule of amlodipine in the 2a channel of P450 2B4, further illustrating the use of different substrate access routes in various cytochromes P450.
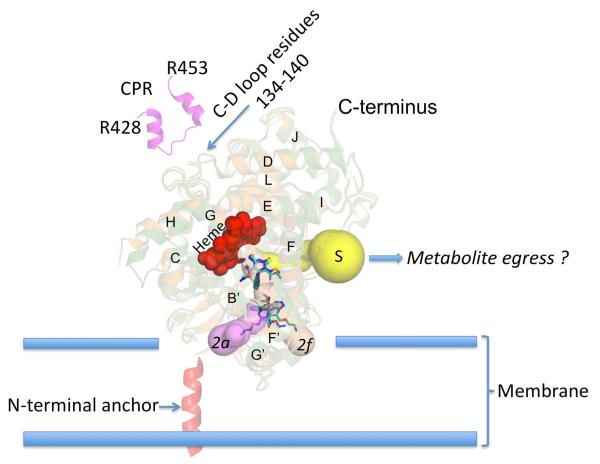
Interestingly, a CYMAL-5 molecule has been shown to be located at the interface of channel 2f and the surface of the protein in several P450 2B4 and P450 2B6 structures solved previously (Supplemental Figure 10) (14, 15, 63). In addition, the P450 2B4-ticlopidine complex shows a different detergent molecule (232-chol) located at the 2f channel entrance close to helix F’ where CYMAL-5 has been observed. This is indicative of a putative surface recognition site in P450 2B4 and P450 2B6 prior to ligand entry into the protein. Positioning P450 2B4 and P450 2B6 as described previously with P450 2C9 (52), shows that channels 2a and 2f are located within the membrane (Figure 8), and the solvent channel is close to the membrane-water interface. The residues that interact with CPR in the cytosol are also shown. This representation mimics the mechanism previously proposed (23, 51), where the substrate molecule, amlodipine in this case, enters access channel 2f (or 2a for P450 2B4), and the polar metabolite is then released into the cytosol via the solvent channel. Moreover, membrane insertion predictions for P450s 2B4 and 2B6 were made using the Orientations of Proteins in Membranes server (64). The location of channel 2f pointing towards the membrane, further suggests that this is the major route for substrate access into P450 2B4 and P450 2B6. Furthermore, a CYMAL-5 molecule could also be seen pointing from the membrane into the channel 2f upon similar analysis of the 2B4-ticlopidine complex.
Figure 8.
Cartoon of substrate access and solvent channels in P450 2B4 and P450 2B6 based on the current study with the enzymes anchored to the membrane via N-terminal residues (adopted from (52). Amlodipine molecules (blue and cyan) are shown to enter the protein via substrate access channels 2a or 2f rabbit P450 2B4 and channel 2f in human P450 2B6. The possible exit route for metabolites via the solvent channel (S) is also shown. Residues of CPR (428 to 453 in magenta) shown previously (42) to bind at the CD loop region (residues 134 to 140) are located at the proximal side of the protein predicted to lie within the cytosol.
In addition to P450 2C9, channel 2a has been suggested via MD simulation to be one of the channels used for substrate access in P450 3A4 (56) and proposed as the main ligand exit channel in some bacterial P450s (60). The opening of channel 2a in the P450 2B4-amlodipine complex is caused by the shift of the R73 side chain. In a protein alignment, R73 of P450 2B4 aligns to K72 of P450 2C9 and R47 of P450 BM3, both of which are proposed to interact with the substrate and play an important role in recognition at the substrate access channel entrance (65, 66). The side chain of K72 in P450 2C9 makes a similar turn at the β1-1 and β1-2 sheets (67), closing channel 2a, similar to what is observed in P450 2B6 or the closed conformations of P450 2B4. In addition, recent structural analysis of rat mitochondrial P450 24A1 in the presence of multiple detergent molecules showed that the membrane directed substrate access channel 2a is the major access route (27). Interestingly, F104 in P450 24A1, which was predicted to be one of the important residues to line the above membrane-accessible substrate access channel, aligns with R73 in P450 2B4.
The availability of the new P450 2B4-amlodipine complex enables us to extract considerably more detail from two previously solved structures containing two or more molecules of imidazole inhibitors (11, 12). An overlay of the structures of the amlodipine and bifonazole complexes revealed that the position of a second bifonazole molecule coincides with the entrance of the 2f channel observed in the current structure (Supplemental Figure 11). This structural evidence is supported by a recent computational analysis that suggested that channel 2f is used for bifonazole access in P450 2B4 (68). Moreover, superimposing the P450 2B4 amlodipine complex and the triple ligand occupancy structure of P450 2B4 with 1-biphenyl-4-methyl-1H–imidazole(1PBI) (12) positions one molecule of 1PBI at the entrance of channel 2a (Supplemental Figure 12). Interestingly, the side chain of R73 bulges out towards the solvent in the 1PBI complex, further supporting our hypothesis of a putative substrate recognition residue involved in channel opening. In addition, the open ligand free structure of P450 2B4 demonstrated a wide opening of channel 2a to allow another protein molecule into the active site (69). Thus, although the markedly open conformations observed due to dimerization of these previous P450 2B4 structures precluded the precise determination of substrate access channels, the results are very consistent with the channels that are now apparent from the P450 2B4 amlodipine complex.
Recent analysis of P450cam structures (16), revealed several conformational states that contained significant differences in locations of helices B’, F, and G and the F-G loop and to some extent helices C, H and I, which are close to the substrate access channel. Furthermore, the amlodipine complexes of P450 2B4 and P450 2B6 display conformational changes largely similar to those in P450cam. This is in sharp contrast to earlier studies using imidazoles or ligand-free P450 2B4, which showed much more wide-open conformations. In that context, it is noteworthy that recent MD simulations on several P450s indicate substrate induced conformational changes that enable substrate access without large opening of the protein (70).
Analysis of adaptive changes within the hydrophobic active sites of P450 2B4 and P450 2B6 reveal the polar interaction of T302 with the inner amlodipine and distinct polar interactions of E387 and Q215 with the outer amlodipine in P450 2B4 and P450 2B6, respectively. In addition, residue E218 on the F’ helix located at the intersection of substrate access channels 2a and 2f in P450 2B4 is retracted substantially in both enzymes, now allowing amlodipine access (Supplemental Figure 13). Gating of this entrance by movement of E218 allows hydrogen bonding between E387 or Q215 with amlodipine. Moreover, E218 aligns structurally with E222 in P450 2D6, which has been proposed recently as the “bait” for guiding initial substrate binding or entry and proper orientation for metabolism (57). Interestingly, the E222A mutant in P450 2D6 has exhibited significantly altered ligand binding (71), and the corresponding polar residue in P450 3A4 and P450 2C9 is critical for the productive orientation of the ligand (72, 73).
To conclude, the crystal structures of drug metabolizing enzymes presented here reveal two amlodipine molecules bound and show several possible substrate access channels in human P450 2B6 and rabbit P450 2B4. These structures also provide valuable insight into how active site topology is altered to accommodate large and/or multiple drug molecules. In addition to residues E218 and E387 or Q215 described above, the residues located at the respective substrate access channel entrance in human P450 2B6 and rabbit P450 2B4 could be crucial in substrate recognition, and efforts are underway to explore the functional role of these residues. Finally, these new structures of P450 2B6 and 2B4 in a conformation with an enlarged active site are amenable for the docking of larger drug molecules as demonstrated here, and will serve as important in silico models for designing novel drugs. Additional crystal structures of these enzymes with non-nitrogenous ligands will help explain how enzymes from different species recognize and bind the same drug.
Supplementary Material
Acknowledgments
We are grateful to Dr. Sean Gay for critical reading of the manuscript. We thank Dr. Dmitri Davydov for assistance with SpectraLab software and Dr. Deepak Dalvie for helpful discussions. We also thank the staff at the Stanford Synchrotron Radiation Lightsource, operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences for assistance with data collection. The Stanford Synchrotron Radiation Lightsource is supported by the National Institute of Health, the National Center for Research Resources, the Biomedical Technology Program, and the United States Department of Energy of Biological and Environmental Research.
Abbreviations
- P450
cytochrome P450
- DTT
dithiothreitol
- BME
2-mercaptoethanol
- PMSF
phenylmethanesulfonyl fluoride
- CYMAL-5
5-cyclohexyl-1-pentyl-β-D-maltoside
- EDTA
ethylenediaminetetraacetic acid
- 4CPI
4-(4-chlorophenyl)imidazole
- 1PBI
1-biphenyl-4-methyl-1H–imidazole
- Ni2+-NTA
nickel-nitrilotriacetic acid
- 232-chol
3α,7α,12α-tris[(β-D-maltopyranosyl)ethyloxy]cholane
- SSRL
Stanford Synchrotron Radiation Lightsource
- BL
Beamline
- rmsd
root mean square deviation
- PDB
Protein Data Bank
- 234-chol
3R-hydroxy-7R,12Rbis({[2(trimethylamino)ethyl]phosphoryl}ethyloxy)cholane
- MD
Molecular Dynamics
- 7-EFC
7-ethoxy-4-(trifluoromethyl) coumarin
- 7-MFC
7-methoxy-4-(trifluoromethyl)coumarin
- CPR
Cytochrome P450 Reductase
- 4BP
4-benzylpyridine
- 4NBP
4-(4-nitrobenzyl)pyridine
- NADPH
reduced nicotine adenine dinucleotide phosphate
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- DMSO
dimethyl sulfoxide
Footnotes
This research was supported by NIH grants ES003619 to James R. Halpert and GM098538 to Qinghai Zhang.
Atomic coordinates and structure factors for the P450 2B4-amlodipine and 2B6-amlodipine complexes are deposited in the Protein Data Bank with accession number 3TMZ and 3UA5, respectively.
The initial P450 2B4 X-ray crystal structure was solved using a truncated and modified protein containing the wild-type His226. Because of the formation of a dimer involving coordination of His226 of each monomer with the heme iron of the other monomer, subsequent biochemical and crystallography work utilized the mutant H226Y. In this manuscript, P450 2B4 will refer to P450 2B4dH(H226Y) unless otherwise stated.
P450 2B6 stands for an N-terminally truncated and modified and C-terminally His-tagged form of the cytochrome P450 2B6 genetic variant K262R with an internal mutation Y226H.
Supporting Information Available Table showing the active site residues located within 5 Å of amlodipine molecules in P450 2B4 and P450 2B6. A diagram showing the amlodipine structure. Figure showing an orthogonal stereo-view of amlodipine electron density in P450 2B4 and P450 2B6 complex. Figures showing the residues lining the solvent channel in P450 2B4 and P450 2B6, and an overlay with the corresponding 4-CPI complexes. Overlay of P450 2B6 and P450 2B4 structures showing differences near the C-terminal region. Overlay of P450 2B4 and P450 2B6 amlodipine complexes. Structures of P450 2B4 and P450 2B6 showing interactions with amlodipine. A difference spectra of P450 2B4 representative of a type II change upon binding amlodipine. Overlay of P450 2B4-amlodipine complex with P450 2A13-NNK structure. Overlay of various P450 2B4 and P450 2B6 complexes showing the location of Cymal-5 molecule at the 2f substrate access channel entrance. Figures showing an overlay of P450 2B4-amlodipine with P450 2B4-bifonazole and P450-1PBI complexes. Overlay of P450 2B4 and P450 2B6 amlodipine and 4CPI complexes showing the location of residue E218. This material is available via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Ortiz de Montellano PR. Cytochrome P450: Structure, Mechanism and Biochemistry. 3rd ed Kluwer Academic/Plenum Publishers; New York: 2005. [Google Scholar]
- 2.Johnson EF, Stout CD. Structural diversity of human xenobioticmetabolizing cytochrome P450 monooxygenases. Biochem Biophys Res Commun. 2005;338:331–336. doi: 10.1016/j.bbrc.2005.08.190. [DOI] [PubMed] [Google Scholar]
- 3.Black SD, Martin ST, Smith CA. Membrane topology of liver microsomal cytochrome P450 2B4 determined via monoclonal antibodies directed to the halt-transfer signal. Biochemistry. 1994;33:6945–6951. doi: 10.1021/bi00188a025. [DOI] [PubMed] [Google Scholar]
- 4.Scott EE, Spatzenegger M, Halpert JR. A truncation of 2B subfamily cytochromes P450 yields increased expression levels, increased solubility, and decreased aggregation while retaining function. Arch Biochem Biophys. 2001;395:57–68. doi: 10.1006/abbi.2001.2574. [DOI] [PubMed] [Google Scholar]
- 5.Scott EE, Liu H, Qun He Y, Li W, Halpert JR. Mutagenesis and molecular dynamics suggest structural and functional roles for residues in the N-terminal portion of the cytochrome P450 2B1 id helix. Arch Biochem Biophys. 2004;423:266–276. doi: 10.1016/j.abb.2003.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y, Halpert JR. Structure-function analysis of cytochromes P450 2B. Biochim Biophys Acta. 2007;1770:402–412. doi: 10.1016/j.bbagen.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Wilderman PR, Shah MB, Liu T, Li S, Hsu S, Roberts AG, Goodlett DR, Zhang Q, Woods VL, Jr., Stout CD, Halpert JR. Plasticity of cytochrome P450 2B4 as investigated by hydrogen-deuterium exchange mass spectrometry and X-ray crystallography. J Biol Chem. 2010;285:38602–38611. doi: 10.1074/jbc.M110.180646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gay SC, Roberts AG, Halpert JR. Structural features of cytochromes P450 and ligands that affect drug metabolism as revealed by X-ray crystallography and NMR. Future Med Chem. 2010;2:1451–1468. doi: 10.4155/fmc.10.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gay SC, Zhang H, Wilderman PR, Roberts AG, Liu T, Li S, Lin HL, Zhang Q, Woods VL, Jr., Stout CD, Hollenberg PF, Halpert JR. Structural analysis of mammalian cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene: insight into partial enzymatic activity. Biochemistry. 2011;50:4903–4911. doi: 10.1021/bi200482g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. An open conformation of mammalian cytochrome P450 2B4 at 1.65A resolution. Proc Natl Acad Sci U S A. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, White MA, Muralidhara BK, Sun L, Halpert JR, Stout CD. Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction. J Biol Chem. 2006;281:5973–5981. doi: 10.1074/jbc.M511464200. [DOI] [PubMed] [Google Scholar]
- 12.Gay SC, Sun L, Maekawa K, Halpert JR, Stout CD. Crystal structures of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole: ligand-induced structural response through alpha-helical repositioning. Biochemistry. 2009;48:4762–4771. doi: 10.1021/bi9003765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanger UM, Klein K, Saussele T, Blievernicht J, Hofmann MH, Schwab M. Polymorphic CYP2B6: molecular mechanisms and emerging clinical significance. Pharmacogenomics. 2007;8:743–759. doi: 10.2217/14622416.8.7.743. [DOI] [PubMed] [Google Scholar]
- 14.Gay SC, Shah MB, Talakad JC, Maekawa K, Roberts AG, Wilderman PR, Sun L, Yang JY, Huelga SC, Hong WX, Zhang Q, Stout CD, Halpert JR. Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.05A resolution. Mol Pharmacol. 2010;77:529–538. doi: 10.1124/mol.109.062570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah MB, Pascual J, Zhang Q, Stout CD, Halpert JR. Structures of cytochrome P450 2B6 bound to 4-benzylpyridine and 4-(4-nitrobenzyl)pyridine: insight into inhibitor binding and rearrangement of active site side chains. Mol Pharmacol. 2011;80:1047–1055. doi: 10.1124/mol.111.074427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YT, Glazer EC, Wilson RF, Stout CD, Goodin DB. Three clusters of conformational states in p450cam reveal a multistep pathway for closing of the substrate access channel. Biochemistry. 2011;50:693–703. doi: 10.1021/bi101726d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoch GA, Yano JK, Sansen S, Dansette PM, Stout CD, Johnson EF. Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid. J Biol Chem. 2008;283:17227–17237. doi: 10.1074/jbc.M802180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porubsky PR, Meneely KM, Scott EE. Structures of human cytochrome P-450 2E1. Insights into the binding of inhibitors and both small molecular weight and fatty acid substrates. J Biol Chem. 2008;283:33698–33707. doi: 10.1074/jbc.M805999200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strushkevich N, Usanov SA, Plotnikov AN, Jones G, Park HW. Structural analysis of CYP2R1 in complex with vitamin D3. J Mol Biol. 2008;380:95–106. doi: 10.1016/j.jmb.2008.03.065. [DOI] [PubMed] [Google Scholar]
- 20.Mast N, Charvet C, Pikuleva IA, Stout CD. Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain. J Biol Chem. 2010;285:31783–31795. doi: 10.1074/jbc.M110.143313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strushkevich N, Usanov SA, Park HW. Structural basis of human CYP51 inhibition by antifungal azoles. J Mol Biol. 2010;397:1067–1078. doi: 10.1016/j.jmb.2010.01.075. [DOI] [PubMed] [Google Scholar]
- 22.Cojocaru V, Balali-Mood K, Sansom MS, Wade RC. Structure and dynamics of the membrane-bound cytochrome P450 2C9. PLoS Comput Biol. 2011;7:e1002152. doi: 10.1371/journal.pcbi.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fishelovitch D, Shaik S, Wolfson HJ, Nussinov R. Theoretical characterization of substrate access/exit channels in the human cytochrome P450 3A4 enzyme: involvement of phenylalanine residues in the gating mechanism. J Phys Chem B. 2009;113:13018–13025. doi: 10.1021/jp810386z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devore NM, Scott EE. Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) binding and access channel in human cytochrome P450 2A6 and 2A13 enzymes. J Biol Chem. 2012 doi: 10.1074/jbc.M112.372813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao B, Lei L, Kagawa N, Sundaramoorthy M, Banerjee S, Nagy LD, Guengerich FP, Waterman MR. Three-dimensional structure of steroid 21-hydroxylase (cytochrome P450 21A2) with two substrates reveals locations of disease-associated variants. J Biol Chem. 2012;287:10613–10622. doi: 10.1074/jbc.M111.323501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Bell SG, Wang H, Zhou W, Bartlam M, Wong LL, Rao Z. The structure of CYP101D2 unveils a potential path for substrate entry into the active site. Biochem J. 2011;433:85–93. doi: 10.1042/BJ20101017. [DOI] [PubMed] [Google Scholar]
- 27.Annalora AJ, Goodin DB, Hong WX, Zhang Q, Johnson EF, Stout CD. Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism. J Mol Biol. 2010;396:441–451. doi: 10.1016/j.jmb.2009.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murdoch D, Heel RC. Amlodipine. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic use in cardiovascular disease. Drugs. 1991;41:478–505. doi: 10.2165/00003495-199141030-00009. [DOI] [PubMed] [Google Scholar]
- 29.Walsky RL, Astuccio AV, Obach RS. Evaluation of 227 drugs for in vitro inhibition of cytochrome P450 2B6. J Clin Pharmacol. 2006;46:1426–1438. doi: 10.1177/0091270006293753. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Q, Ma X, Ward A, Hong WX, Jaakola VP, Stevens RC, Finn MG, Chang G. Designing facial amphiphiles for the stabilization of integral membrane proteins. Angew Chem Int Ed Engl. 2007;46:7023–7025. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]
- 31.DeLano WL. In: The PyMOL Molecular Graphics System. MacPyMOL, editor. DeLano Scientific; Palo Alto, CA: 2002. [Google Scholar]
- 32.Omura T, Sato R. The Carbon Monoxide-Binding Pigment of Liver Microsomes. I. Evidence for Its Hemoprotein Nature. J Biol Chem. 1964;239:23705–2378. [PubMed] [Google Scholar]
- 33.Mitsuda M, Iwasaki M. Improvement in the expression of CYP2B6 by co-expression with molecular chaperones GroES/EL in Escherichia coli. Protein Expr Purif. 2006;46:401–405. doi: 10.1016/j.pep.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 34.Soltis SM, Cohen AE, Deacon A, Eriksson T, Gonzalez A, McPhillips S, Chui H, Dunten P, Hollenbeck M, Mathews I, Miller M, Moorhead P, Phizackerley RP, Smith C, Song J, van dem Bedem H, Ellis P, Kuhn P, McPhillips T, Sauter N, Sharp K, Tsyba I, Wolf G. New Paradigm for Macromolecular Crystallography Experiments at SSRL: Automated Crystal Screening and Remote Data Collection. Acta Crystallographica Section D. 2008;64:1210–1221. doi: 10.1107/S0907444908030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leslie AGW. Integration of macromolecular diffraction data. Acta Crystallogr. Sect. D-Biol. Crystallogr. 1999;55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 36.Bailey S. The CCP4 Suite - Programs for Protein Crystallography. Acta Crystallogr. Sect. D-Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 37.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D-Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 41.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bumpus NN, Hollenberg PF. Cross-linking of human cytochrome P450 2B6 to NADPH-cytochrome P450 reductase: Identification of a potential site of interaction. J Inorg Biochem. 2010;104:485–488. doi: 10.1016/j.jinorgbio.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petrek M, Otyepka M, Banas P, Kosinova P, Koca J, Damborsky J. CAVER: a new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinformatics. 2006;7:316. doi: 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 46.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helms V, Wade RC. Thermodynamics of water mediating protein-ligand interactions in cytochrome P450cam: a molecular dynamics study. Biophys J. 1995;69:810–824. doi: 10.1016/S0006-3495(95)79955-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harlow GR, Halpert JR. Alanine-scanning mutagenesis of a putative substrate recognition site in human cytochrome P450 3A4. Role of residues 210 and 211 in flavonoid activation and substrate specificity. J Biol Chem. 1997;272:5396–5402. doi: 10.1074/jbc.272.9.5396. [DOI] [PubMed] [Google Scholar]
- 49.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-A resolution: insight into the range of P450 conformations and the coordination of redox partner binding. J Biol Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H, Amunugama H, Ney S, Cooper N, Hollenberg PF. Mechanism-based inactivation of human cytochrome P450 2B6 by clopidogrel: involvement of both covalent modification of cysteinyl residue 475 and loss of heme. Mol Pharmacol. 2011;80:839–847. doi: 10.1124/mol.111.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cojocaru V, Balali-Mood K, Sansom MS, Wade RC. Structure and dynamics of the membrane-bound cytochrome P450 2C9. PLoS Comput Biol. 2011;7:e1002152. doi: 10.1371/journal.pcbi.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berka K, Hendrychova T, Anzenbacher P, Otyepka M. Membrane Position of Ibuprofen Agrees with Suggested Access Path Entrance to Cytochrome P450 2C9 Active Site. J Phys Chem A. 2011;115:11248–11255. doi: 10.1021/jp204488j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cojocaru V, Winn PJ, Wade RC. The ins and outs of cytochrome P450s. Biochim Biophys Acta. 2007;1770:390–401. doi: 10.1016/j.bbagen.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 54.Rydberg P, Rod TH, Olsen L, Ryde U. Dynamics of water molecules in the active-site cavity of human cytochromes P450. J Phys Chem B. 2007;111:5445–5457. doi: 10.1021/jp070390c. [DOI] [PubMed] [Google Scholar]
- 55.Otyepka M, Berka K, Anzenbacher P. Is there a relationship between the substrate preferences and structural flexibility of cytochromes P450? Curr Drug Metab. 2012;13:177–189. doi: 10.2174/138920012798918372. [DOI] [PubMed] [Google Scholar]
- 56.Skopalik J, Anzenbacher P, Otyepka M. Flexibility of human cytochromes P450: molecular dynamics reveals differences between CYPs 3A4, 2C9, and 2A6, which correlate with their substrate preferences. J Phys Chem B. 2008;112:8165–8173. doi: 10.1021/jp800311c. [DOI] [PubMed] [Google Scholar]
- 57.Wang A, Savas U, Hsu MH, Stout CD, Johnson EF. Crystal structure of human cytochrome P450 2D6 with prinomastat bound. J Biol Chem. 2012;287:10834–10843. doi: 10.1074/jbc.M111.307918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilderman PR, Gay SC, Jang HH, Zhang Q, Stout CD, Halpert JR. Investigation by site-directed mutagenesis of the role of cytochrome P450 2B4 non-active site residues in protein-ligand interactions based on crystal structures of the ligand-bound enzyme. FEBS J. 2012;279:1607–1620. doi: 10.1111/j.1742-4658.2011.08411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilderman PR, Halpert JR. Plasticity of CYP2B Enzymes: Structural and Solution Biophysical Methods. Curr Drug Metab. 2012;13:167–176. doi: 10.2174/138920012798918417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winn PJ, Ludemann SK, Gauges R, Lounnas V, Wade RC. Comparison of the dynamics of substrate access channels in three cytochrome P450s reveals different opening mechanisms and a novel functional role for a buried arginine. Proc Natl Acad Sci U S A. 2002;99:5361–5366. doi: 10.1073/pnas.082522999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li W, Shen J, Liu G, Tang Y, Hoshino T. Exploring coumarin egress channels in human cytochrome P450 2A6 by random acceleration and steered molecular dynamics simulations. Proteins. 2011;79:271–281. doi: 10.1002/prot.22880. [DOI] [PubMed] [Google Scholar]
- 62.Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci U S A. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gay SC, Roberts AG, Maekawa K, Talakad JC, Hong WX, Zhang Q, Stout CD, Halpert JR. Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel. Biochemistry. 2010;49:8709–8720. doi: 10.1021/bi100914z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 65.Lewis DF, Dickins M, Weaver RJ, Eddershaw PJ, Goldfarb PS, Tarbit MH. Molecular modelling of human CYP2C subfamily enzymes CYP2C9 and CYP2C19: rationalization of substrate specificity and site-directed mutagenesis experiments in the CYP2C subfamily. Xenobiotica. 1998;28:235–268. doi: 10.1080/004982598239542. [DOI] [PubMed] [Google Scholar]
- 66.Haines DC, Tomchick DR, Machius M, Peterson JA. Pivotal role of water in the mechanism of P450BM-3. Biochemistry. 2001;40:13456–13465. doi: 10.1021/bi011197q. [DOI] [PubMed] [Google Scholar]
- 67.Ravichandran KG, Boddupalli SS, Hasermann CA, Peterson JA, Deisenhofer J. Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450’s. Science. 1993;261:731–736. doi: 10.1126/science.8342039. [DOI] [PubMed] [Google Scholar]
- 68.Otyepka M, Berka K, Anzenbacher P. Is there a relationship between the substrate preferences and structural flexibility of cytochromes P450? Curr Drug Metab. 2012;13:130–142. doi: 10.2174/138920012798918372. [DOI] [PubMed] [Google Scholar]
- 69.Wade RC, Winn PJ, Schlichting I, Sudarko A survey of active site access channels in cytochromes P450. J Inorg Biochem. 2004;98:1175–1182. doi: 10.1016/j.jinorgbio.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 70.Hendrychova T, Berka K, Navratilova V, Anzenbacher P, Otyepka M. Dynamics and hydration of the active sites of mammalian cytochromes P450 probed by molecular dynamics simulations. Curr Drug Metab. 2012;13:177–189. doi: 10.2174/138920012798918408. [DOI] [PubMed] [Google Scholar]
- 71.Masuda K, Tamagake K, Okuda Y, Torigoe F, Tsuzuki D, Isobe T, Hichiya H, Hanioka N, Yamamoto S, Narimatsu S. Change in enantioselectivity in bufuralol 1″-hydroxylation by the substitution of phenylalanine-120 by alanine in cytochrome P450 2D6. Chirality. 2005;17:37–43. doi: 10.1002/chir.20092. [DOI] [PubMed] [Google Scholar]
- 72.Sevrioukova IF, Poulos TL. Structural and Mechanistic Insights into the Interaction of Cytochrome P4503A4 with Bromoergocryptine, a Type I Ligand. J Biol Chem. 2012;287:3510–3517. doi: 10.1074/jbc.M111.317081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Williams PA, Cosme J, Ward A, Angove HC, Matak Vinkovic D, Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–468. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.