Abstract
Background
The Bacillus cereus sensu lato group currently includes seven species (B. cereus, B. anthracis, B. mycoides, B. pseudomycoides, B. thuringiensis, B. weihenstephanensis and B. cytotoxicus) that recent phylogenetic and phylogenomic analyses suggest are likely a single species, despite their varied phenotypes. Although horizontal gene transfer and insertion-deletion events are clearly important for promoting divergence among these genomes, recent studies have demonstrated that a major basis for phenotypic diversity in these organisms may be differential regulation of the highly similar gene content shared by these organisms. To explore this hypothesis, we used an in silico approach to evaluate the relationship of pathogenic potential and the divergence of the SigB-dependent general stress response within the B. cereus sensu lato group, since SigB has been demonstrated to support pathogenesis in Bacillus, Listeria and Staphylococcus species.
Results
During the divergence of these organisms from a common “SigB-less” ancestor, the placement of SigB promoters at varied locations in the B. cereus sensu lato genomes predict alternative structures for the SigB regulon in different organisms. Predicted promoter changes suggesting differential transcriptional control of a common gene pool predominate over evidence of indels or horizontal gene transfer for explaining SigB regulon divergence.
Conclusions
Four lineages of the SigB regulon have arisen that encompass different gene contents and suggest different strategies for supporting pathogenesis. This is consistent with the hypothesis that divergence within the B. cereus sensu lato group rests in part on alternative strategies for regulation of a common gene pool.
Keywords: Microbial evolution, Generalized stress response, SigB sigma factor
Background
The genus Bacillus is a heterogeneous group of Gram-positive heterotrophic aerobic or facultative anaerobic bacilli that form environmentally resistant, metabolically inert spores [1]. These soil-borne organisms are ubiquitous, and occupy surprisingly diverse environments [2,3]. Within this large genus, the B. cereus sensu lato group consists of seven species B. anthracis (Ba), B. cereus (Bc), B. mycoides, B. pseudomycoides, B. thuringiensis (Bt), B. weihenstephanensis (Bw) and B. cytotoxicus, based on classical microbial taxonomy [4,5]. More recent molecular phylogenies and comparative genome sequence analysis indicate that these organisms should more accurately be viewed as a single species [6,7] despite their phenotypic diversity. Indeed, the various species of the Bc sensu lato group are phylogenetically interspersed among one another in several phylogenies [8-10]. Although the population has a clonal character, there do not appear to be clonal lineages that are species-specific, with the exception of the Ba lineage [11]. Some Bc sensu lato organisms are thermophiles [12], while Bw is a psychrophile [4]. Nevertheless, most Bc sensu lato isolates are mesophiles, found in a breadth of locales including the soil, on plant surfaces and the mammalian gastrointestinal microflora [13]. Some Bc sensu lato members appear to be nonpathogenic, while others cause diverse disease ranging from food poisoning (intoxication without colonization) to gastroenteritis [13], endophthalmitis [14], tissue abscesses [15,16], and aggressively invasive systemic disease, including anthrax [3]. Bt strains can cause disease in insects [17,18] and possibly nematodes [19-21], while some Bc strains are part of the normal insect gut flora [13,22].
Thus, these organisms appear to have arisen from a common ancestor to display impressive phenotypic diversity while nevertheless occupying a close phylogenetic space. What mechanisms produced this dichotomy? Horizontal gene transfer (HGT), mobile genetic elements, and the routine processes of insertion/deletion (indel) formation have typically been invoked to explain the diversity in these organisms [23,24]. Clearly, the presence of mobile genetic elements such as the virulence plasmids pXO1 and pXO2 in the Ba lineage, and the Cry toxin plasmids in various Bt strains, are essential for the signature phenotypes of these organisms [3,25]. On the other hand, it is unclear how much of the phenotypic diversity in these organisms can be explained by these mechanisms. As a whole, Bc sensu lato organisms have an extremely high degree of chromosomal synteny [26], and whole genome comparisons between these organisms reveal a highly similar gene content [26,27]. Han et al.[27] suggested “that differential regulation [of gene content] modulates virulence rather than simple acquisition of virulence factor genes”, a conclusion confirmed by other studies [28]. This proposition is consistent with observations that the most evolutionarily flexible portions of the bacterial genome are regulatory sequences and transcriptional networks [29-31].
We decided to explore the divergence within the Bc sensu lato group by examining the divergence of the SigB regulons in these organisms. The sigB locus encodes an alternative sigma factor with orthologs confined to the Bacillus, Staphylococcus and Listeria genera [32]. In each of these, the SigB protein is responsible for transcriptionally activating the generalized stress response when induced by a variety of stressors, including heat, osmolarity, organic solvents, low pH or cell-wall active antibiotics [33-35]. In these organisms, SigB appears to control virulence-related functions including biofilm formation and invasion [36,37]. Some data that suggest that components of the SigB-mediated stress response vary between strains of Listeria and of S. aureus in a lineage-dependent manner [38-40]. This suggests that part of the evolutionary differentiation that occurred within these genera included divergence of the structure of the SigB-controlled regulons.
The divergence of the SigB regulon in the Bc sensu lato group appears to be similar. Lapidus et al.[4] observed that B. cytotoxicus [formerly Bc biovar cytotoxis[41]] is likely the most similar of the Bc sensu lato organisms to the nearest common ancestor of the group, based on comparative genome analysis and 16S rRNA phylogeny. Our whole genome single-gene phylogeny [7] supports the placement of B. cytotoxicus at the base of the phylogeny of the Bc sensu lato group. Important in the present context, the B. cytotoxicus genome lacks the entire SigB operon [4], including the SigB gene and the primary regulatory loci that control SigB activity, RsbV (anti-sigB antagonist) and RsbW (anti-sigB factor). Consequently, the B. cytotoxicus genome does not encode either the sigma factor or associated SigB regulatory genes, and cannot mount a SigB-activated stress response. Lapidus et al. suggested that an organism similar to B. cytotoxicus was likely the ancestor of the remainder of the Bc sensu lato lineage, which arose after receiving the SigB operon during an HGT event. Consistent with this, we were unable to find convincing evidence of SigB binding sites in the B. cytotoxicus genome that correspond to SigB promoters in other members of the Bc sensu lato group (see below). An alternative explanation, that B. cytotoxicus had lost the SigB operon and other genomic information during streamlining of a larger genome [42,43], is less likely. In that instance, we expect that detectable remnants of the SigB regulon (e.g., SigB promoters, pseudogenes) would remain in the B. cytotoxicus genome. Thus, the introduction of an intact SigB operon into a B. cytotoxicus-like ancestor, likely by HGT as suggested by Lapidus et al., appears to have set the stage for the emergence of the SigB-controlled stress response regulon in the entire Bc sensu lato group. From this perspective, the B. cytotoxicus genome therefore is a convenient lens through which to view the appearance and divergence of the SigB regulon in the remainder of the Bc sensu lato group. Exploiting experimental data derived from Bc strain ATCC14579 [44,45], we used an in silico approach to predict and compare the SigB regulons of the completed genomes of 20 members of the Bc sensu lato group. Not surprisingly, protein coding sequence indels play an important role in the divergence of these regulons. However, changes in promoter sequence between members of the Bc sensu lato group that ‘re-purpose’ conserved genes into/out of the SigB regulon appear to be more common than indel formation for remodeling the structure of this regulon during divergence. Four different lineages of the SigB regulon appear to have arisen during this process. One lineage appears to carry the core SigB regulon that arose after the emergence of these organisms from a B. cytotoxicus-like ancestor. This lineage appears to have given rise to three additional groups that each appropriated different genes from a common gene pool into the SigB regulon, suggesting different strategies for the support of pathogenesis by the SigB-mediated generalized stress response.
Results and discussion
SigB binding site model building and regulon predictions
We began with nine 150bp DNA sequences encompassing the SigB-dependent promoters identified by Van Schaik et al.[44], expanded to 166 sequences by phylogenetic footprinting [46]. Redundant sequences were removed from this collection to yield a final training set of 130 sequences. The training set was then used in parallel to develop separate HMM and PWM models for the SigB-dependent promoters in the Bc sensu lato group. Our models are virtually identical to previously derived models for this DNA binding site in Bc ATCC14579 [44] and B. subtilis[47] (Figure 1). Each model was used to scan all genomes for potential SigB binding sites, and this information was coupled with transcriptome-derived transcriptional unit (TU) predictions to arrive at a predicted regulon structure for each organism.
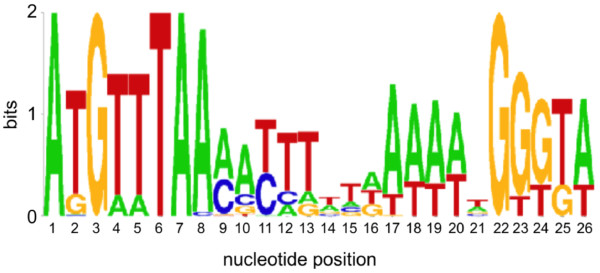
Figure 1.
Seqlogo describing the SigB binding site model derived in this study. This model is virtually identical to the Bc ATCC14579 SigB binding site derived by van Schaik et al.[44], except that the A at position 8 has a stronger signal, and the spacer region between the −35 and −10 sequences is shorter by 1bp. Since the Bc ATCC14579 binding site was derived from 10 sequences, while our SigB binding site model relied on 130 sequences, our model may be somewhat more robust, although the differences appear negligible. A model of the binding site for the orthologous B. subtilis SigB protein also is very similar, lacking only the strong A at position 7; the B. subtilis SigB binding site was extracted from a training set of 63 sequences [47].
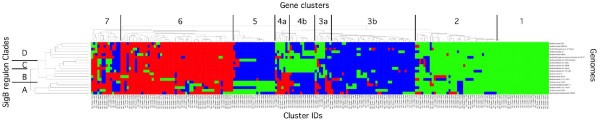
Horizontal gene transfer leading to insertion of novel genes, gene duplication and divergence, or gene deletion events, are thought to be the most common mechanisms responsible for remodeling bacterial genome function. Collectively, these events can be easily identified as the insertion or deletion (indel) of protein coding sequence, although the underlying mechanism responsible for the appearance of an indel is not so easily surmised. Surprisingly, preliminary manual comparison of the predicted SigB regulons of the Bc sensu lato group suggested that the appearance of indels was not the predominant mechanism of divergence of these SigB regulons. In the majority of instances, differences between SigB regulon structure appeared to arise from the placement of SigB-specific promoters in the 5’ region of TUs that were shared among these organisms. That is, SigB regulon divergence appeared to rely on a process of promoter ‘re-assignment’ of genes from a common gene pool into/out of the SigB regulon. This promoter re-assignment appears to predominate over instances of indels that changed the total coding capacity of a given SigB regulon. To get a better quantitative measure of this impression, we performed pairwise comparisons of the predicted SigB regulons of all Bc sensu lato genomes (Figure 2), by calculating a simple ratio of the number of predicted promoter changes to the number of observed indel events, for each pair of regulons. A promoter change occurs during the comparison of DNA sequence upstream of orthologous genes in which one gene contains a predicted binding site with a significant score while the second either does not contain a predicted binding site, or contains a predicted binding site that has a score that is not significant. The results confirmed that the predominant mechanism for divergence of these regulons relied on the assignment of new SigB promoters to genes from a common gene pool (Figure 2). We extended this comparison by constructing a heat map summarizing the structures of the predicted SigB regulons in the Bc sensu lato group (Figure 3; a larger version of this figure with legible annotation is presented as Additional file 1: Figure S1). This heat map is color-coded: green blocks indicate the presence of a gene predicted to be in a TU controlled by a SigB promoter, while blue blocks indicate the presence of an orthologous gene not controlled by SigB. Red blocks indicate that the ortholog in question is absent from the genome.
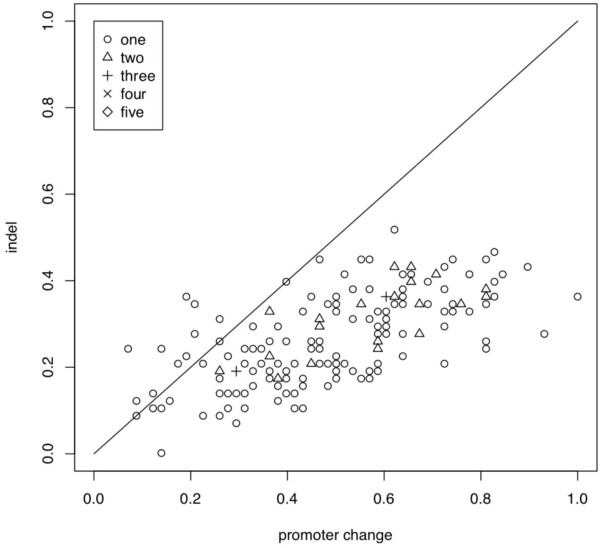
Figure 2.
Pairwise comparisons of the structure of the predicted SigB regulons of the Bc sensu lato group. Comparisons expressed as the ratio of promoter changes vs. indels for each pairwise comparison. If the number of promoter changes and indels for a given pairwise comparison were equivalent, then the value of this ratio would appear on the diagonal line across the center of the plot. Points above this line indicate a pairwise comparison where differences in the SigB regulons relied more commonly on indels rather than promoter changes. Conversely, points below this line denote a ratio arising from pairwise comparison where the differences between SigB regulons revealed a greater number of occurrences of reassignment of common genes into/out of the SigB regulon due to the appearance/disappearance of a predicted SigB promoter. The number of paired genomes yielding each ratio are plotted as one (○), two (), three (+), four (×) or five (◊) pairs.
Figure 3.
Heat map comparing the SigB regulons of 19 Bc sensu lato genomes. This heat map is color-coded as: Red, gene absent from the genome; Blue, gene present in the genome, but lacking a SigB promoter binding site 5’ to the beginning of the predicted TU; Green, gene present in the genome and in a TU predicted to be controlled by a SigB promoter. Cluster analysis (tree shown on top of heat map) segregated these genes into 7 clusters, and we operationally divided two of these clusters (3 and 4) into subdivisions (a and b). This was done to emphasize that clusters 3a, 4a and 4b appear to contribute to functional differences between these organisms, while cluster 4b does not (see text for explanation). Cluster analysis also indicated that the genomes included in this figure could be assigned to one of four clades, designated A-D, as noted at the one end of the heat map; at the other end, the identity of each genome used in these analyses is listed. This information is summarized in Table 2. The orthologous cluster ID tags for the genes that are predicted to be in these SigB regulons are displayed across the bottom of the heat map. The genome annotations and cluster ID tags are legible in Additional file 1: Figure S1, and are cataloged in Additional file 2: Table S1.
Cluster analysis of the genes predicted in the SigB regulons of these genomes grouped these genes into seven clusters; within these, we identified an additional three subclusters that appear to have functional significance (Figure 3, clusters 3a, 4a, and 4b). This cluster analysis defined a core SigB regulon (clusters 1 and 2) that included genes common to the SigB regulon in all Bc sensu lato organisms. Additions to this core regulon (clusters 3–7) appear to be clade-specific. It is important to note that the HMM and PWM scores for each of the predicted promoters in clusters 3–7 were statistically indistinguishable from those in clusters 1 and 2. That is, the predictions for the promoters for gene clusters 3–7 were as strongly supported as those in the core SigB regulon, including TUs for which supporting experimental evidence exists [44]. Note also that the bulk of gene cluster 3 (excepting cluster 3a), and clusters 6 and 7 provide relatively modest additions to the core SigB regulons of these organisms. In contrast to gene clusters 1, 2, 3a, 4a/b and 5, there does not appear to be any pattern to the additions in clusters 3b, 6 and 7. Instead, these gene clusters appear to represent noise in the data set. Since the genomes examined in this study were annotated by as many as eight different sequencing groups over a span of seven years (Table 1), the lack of standardization in these various annotation schemes appears to be the basis for much of the noise in these gene clusters (I. Toby and D. Dyer, manuscript in preparation).
Table 1.
Genomes used in this study
| |
|
|
|
|
Predicted Operons |
||
|---|---|---|---|---|---|---|---|
| Organism1 | Genbank Accession | Annotation date | Sequencing group3 | Locus tag | monocistronic | polycistronic | total |
| Bw KBAB4 |
NC_010184 |
05/2010 |
HAU |
KBAB |
2106 |
809 |
2915 |
| Bc B4264 |
NC_011725 |
12/2008 |
JCVI |
BCB |
2187 |
855 |
3042 |
| Bc ATCC14579 |
NC_004722 |
03/2003 |
INRAGM |
BC |
2163 |
840 |
3003 |
| Bt BMB171 |
NC_014171 |
05/2010 |
HAU |
BMB |
2135 |
837 |
2972 |
| Bc G9842 |
NC_011772 |
10/2008 |
JCVI |
BCG |
2271 |
847 |
3118 |
| Bc Q1 |
NC_011969 |
01/2009 |
MGCC |
BCQ |
2005 |
855 |
2860 |
| Bc AH187 |
NC_011658 |
12/2008 |
JCVI |
BCAH187 |
2199 |
858 |
3057 |
| Bc ATCC10987 |
NC_003909 |
09/2004 |
TIGR |
BCE |
2397 |
825 |
3222 |
| Ba Ames Ancestor |
NC_007530 |
03/2003 |
TIGR |
GBAA |
2146 |
841 |
2987 |
| Ba Ames |
NC_003997 |
03/2003 |
TIGR |
BA |
2081 |
834 |
2915 |
| Ba A0248 |
NC_012659 |
05/2009 |
LANL |
BAA |
1995 |
821 |
2816 |
| Ba Sterne2 |
NC_005945 |
01/2004 |
JGI |
BAS |
2245 |
938 |
3183 |
| Ba CDC 684 |
NC_012581 |
04/2009 |
LANL |
BAMEG |
2245 |
861 |
3106 |
| Bc E33L |
NC_006274 |
11/2004 |
JGI |
BCZK |
1930 |
842 |
2772 |
| Bc 03BB102 |
NC_012472 |
03/2009 |
LANL |
BCA |
2135 |
857 |
2992 |
| Bt Al-Hakam |
NC_008600 |
11/2006 |
JGI |
BALH |
1812 |
817 |
2629 |
| Bc CI |
NC_014335 |
07/2010 |
GGL |
BACI |
1963 |
862 |
2825 |
| Bc AH820 |
NC_011773 |
10/2008 |
JCVI |
BCAH820 |
2168 |
858 |
3026 |
| Bt konkukian 97-27 |
NC_005957 |
12/2004 |
JGI |
BT |
1907 |
844 |
2751 |
| Bcy NVH-391-98 | NC_009674 | 07/2007 | JGI | Bcer98 | 1687 | 589 | 2276 |
1 Ba, B. anthracis, Bc, B. cereus, Bt, B. thuringiensis, Bw, B. weihenstephanensis, Bcy, B. cytotoxicus.
2 Operon data experimentally determined by Passalacqua et al.[28].
3JGI: Joint Genome Institute.
LANL: Los Alamos National Labs.
JCVI: J. Craig Venter Institute.
TIGR: The Institute for Genome Research.
INRAGM: INRA Genetique Microbienne.
MGCC: Microbial Genome Center of Chinese Ministry of Public Health.
HAU: Huazhong Agricultural University.
GGL: Goettingen Genomics Laboratory.
The core SigB regulon
Analysis of the SigB regulons in each genome suggested that these organisms could be assigned to four clades A through D (Table 2), based on the constituents of the SigB regulon predicted for each organism. Clade C consists exclusively of B. anthracis strains; this is not surprising, as these organisms are highly monomorphic. The size of this clade in comparison to Clades A, B and D is somewhat misleading, due to the overrepresentation of these genomes in the data set. Nevertheless, this cluster analysis approximates relationships that closely mirror other phylogenetic hypotheses for the Bc sensu lato group [8]; this is significant in that the relationships in Figure 3 are based on the structure of the SigB regulons (SigB promoters and SigB-controlled genes), rather than from DNA or protein sequence alone. The core SigB regulon (gene clusters 1 and 2) includes essentially all of the SigB-controlled genes found in Clade B organisms, with few exceptions. Comparison of this core regulon to the B. cytotoxicus genome (Table 3; 14 TUs) revealed that 20 of 47 (43%) genes in the core regulon are orthologous to B. cytotoxicus genes. Assuming that the B. cytotoxicus genome reflects the ancestral state prior to the introduction of SigB into this lineage, this suggests that the genes listed in Table 3 pre-dated the introduction of SigB into the Bc sensu lato lineage. These genes were subsequently recruited into the SigB core regulon by the assignment of a SigB promoter after the appearance of SigB. Orthologs of the remaining 23 genes of the SigB core regulon (total of 43 genes) were not found in B. cytotoxicus, and presumably were added later. At least 20 of the core regulon genes encode hypothetical proteins, presumably arising in the Bc sensu lato group by HGT. Most of the remainder of the genes in this group are members of large gene families (e.g., sensor kinases, permease subunits) that likely became part of the SigB regulon by promoter re-assignment, possibly following a gene duplication event. However, the timing of the introduction of the SigB operon into the Bc sensu lato group and the appearance of these genes is uncertain, as is when these genes became incorporated into the SigB regulon.
Table 2.
SigB regulon clades
| SigB regulon Clade | Organism | Source or location of isolation |
|---|---|---|
|
Clade A |
B. weihenstephanensis KBAB4 |
Soil |
| |
B. cereus B4264 |
Bloodstream isolate from pneumonia patient |
| |
B. cereus ATCC 14579 |
Dairy product |
| |
B. thuringiensis BMB171 |
Soil |
| |
B. cereus G9842 |
Stool sample from food poisoning outbreak |
|
Clade B |
B. cereus Q1 |
Deep oil reservoir |
| |
B. cereus AH187 |
Dairy product |
| |
B. cereus ATCC 10987 |
Cheese spoilage |
|
Clade C |
B. anthracis Ames Ancestor |
Bovine carcass |
| |
B. anthracis Ames |
Bovine carcass |
| |
B. anthracis A0248 |
Human disease |
| |
B. anthracis Sterne |
Vaccine strain |
| |
B. anthracis CDC684 |
NA |
|
Clade D |
B. cereus E33L |
Zebra carcass |
| |
B. cereus 03BB102 |
Human blood isolate |
| |
B. thuringiensis Al-Hakam |
Iraq bioweapons facility |
| |
B. cereus Cl |
Chimpanzee carcass |
| |
B. cereus AH820 |
Human periodontitis |
| |
B. thuringiensis konkukian 97-27 |
Human tissue necrosis |
| No regulon evident | B. cytotoxicus NVH-391-98 | Food poisoning outbreak |
Table 3.
B. cytotoxicus genes that pre-date the Bc sensu lato core SigB regulon
| Cluster ID | TU | Representative locus tag | Annotation |
|---|---|---|---|
| bID_Cluster_3759 |
1 |
Bcer98_0620 |
thiamine/molybdopterin biosynthesis ThiF/MoeB-like protein |
| bID_Cluster_5471 |
1 |
Bcer98_0621 |
phosphomethylpyrimidine kinase |
| bID_Cluster_1960 |
2 |
Bcer98_2653 |
PhoH family protein |
| bID_Cluster_3960 |
3 |
Bcer98_3311 |
N-6 DNA methylase |
| bID_Cluster_9165 |
3 |
Bcer98_3312 |
redoxin domain-containing protein |
| bID_Cluster_10657 |
3 |
Bcer98_3313 |
hypothetical protein |
| bID_Cluster_10524 |
4 |
Bcer98_3648 |
nuclear protein SET |
| bID_Cluster_8180 |
5 |
Bcer98_3694 |
ATP-dependent Clp protease proteolytic subunit |
| bID_Cluster_2287 |
6 |
Bcer98_3853 |
transcription termination factor Rho |
| bID_Cluster_10550 |
7 |
Bcer98_0367 |
methyl-accepting chemotaxis sensory transducer |
| bID_Cluster_4143 |
7 |
Bcer98_0430 |
NAD-dependent epimerase/ dehydratase |
| bID_Cluster_1966 |
8 |
Bcer98_0498 |
citrate carrier protein |
| bID_Cluster_2625 |
8 |
Bcer98_0499 |
malate dehydrogenase, putative |
| bID_Cluster_3344 |
9 |
Bcer98_0651 |
hypothetical protein |
| bID_Cluster_2017 |
10 |
Bcer98_1017 |
hypothetical protein |
| bID_Cluster_6230 |
11 |
Bcer98_1200 |
two component transcriptional regulator, ResD |
| bID_Cluster_857 |
11 |
Bcer98_1201 |
multi-sensor signal transduction histidine kinase, ResE |
| bID_Cluster_9721 |
12 |
Bcer98_3007 |
ferric uptake regulator family protein |
| bID_Cluster_3378 |
13 |
Bcer98_3102 |
hypothetical protein |
| bID_Cluster_4858 | 14 | Bcer98_4017 | MscS mechanosensitive ion channel |
All of the SigB-controlled genes originally described by Van Schaik et al.[44] are found in this core regulon. We identified 27 additional genes that are predicted to be in the core SigB regulon (Figure 3 and Additional file 2: Table S1). Many of the additional genes encode regulatory proteins, whose altered transcription may be difficult to detect by microarray analysis, and whose regulatory control may be difficult to ascertain in the absence of experimental data. Another large group of genes added to the core regulon are predicted to encode hypothetical proteins. As a consequence, it is difficult in most instances to predict how these additional genes contribute to the function of the core SigB stress response.
The discrepancy between our predictions and the data of Van Schaik et al. is not surprising; similar difficulties in cataloguing the complete SigB-dependent response have been encountered in other systems. For instance, as noted by Nannapaneni et al.[48], three independent B. subtilis studies identified over 100 SigB-dependent target genes, but only 67 genes were commonly identified in all studies. After extensive testing in a variety of experimental conditions including ethanol and butanol shock, osmotic and oxidative stress, low-temperature growth and heat shock, Nannapaneni et al. demonstrated 166 genes that appeared to be part of the B. subtilis SigB-controlled response; 19 of these also were controlled by secondary regulators. Earlier discrepancies were likely due to differences in growth conditions, microarray platforms and the experimental setups employed. Similarly, the experiments of Van Schaik et al. likely identified a subset of the total SigB-dependent genes in Bc ATCC14579, as these experiments employed a short heat shock [44] or overexpression of the SigB protein [45], and were not as extensive as those of Nannapaneni et al. Secondary regulators interacting with the SigB regulon also are a likely complication. In S. aureus, SpoVG fine-tunes SigB-dependent regulation by antagonizing SigB [49]. In L. monocytogenes, SigB regulatory control is intertwined with that of HrcA [50], CtsR [51] and PrfA [52], complicating the analysis of SigB-mediated effects. In addition to the secondary regulators found in gene clusters 1 and 2 (Figure 3), additional regulatory proteins are included in clade-specific SigB regulons (see below), and may modulate the control of gene expression by SigB, confounding the results of in vitro experiments. Thus, gene clusters 1 and 2 in Figure 3 likely are a more comprehensive representation of the total SigB core regulon in the Bc sensu lato group than the early studies of Van Schaik et al..
Additions to the core SigB regulon
Clade A and Clades C/D organisms appear to have evolved different pathogenic potentials, and this likely is supported by the gene sets added to the core SigB regulon. Clade A organisms have added a group of 20 genes (gene cluster 5, in 4 TUs; Table 4) to the core regulon. The mechanism of addition of this gene cluster to the core SigB regulon almost exclusively rests on the assignment of SigB promoters to TUs found in the common gene pool. This can most easily be seen in Figure 3 (gene cluster 5) as an almost exclusive assignment of either green blocks (genes predicted to be driven by SigB promoters) or blue blocks (orthologous genes present, but lacking a SigB promoter). Only occasionally does one observe an orthologous gene that is absent from a given genome (denoted by a red block in this region of the heat map). Thus, HGT/indels appear to play only a minor role in the assembly of this gene cluster. Clade A includes one member (Bc B4264; Table 1) that was isolated from the bloodstream of a pneumonia patient. Although a second Clade A strain, Bc ATCC14579, was originally isolated from a dairy product (Table 1), this organism has been shown to cause experimental endophthalmitis following intravitreal injection [53]. This intravitreal injection model mimics the course of disease associated with the entry of the organism into the interior of the eye following traumatic injury. However, this infection model likely by-passes the requirement for direct invasion strategies such as those employed by Clade C/D organisms during anthrax or similarly aggressive disease. The remainder of Clade A includes soil-borne organisms and one organism associated with food-borne intoxication. Thus, Clade A organisms may be pathogenic opportunists, in contrast to Clades C/D organisms, which appear to be frank pathogens (Table 1).
Table 4.
Gene clusters in Clades A, C and D that augment the SigB core regulon
| Cluster | Cluster ID | TU | Locus tag | Annotation |
|---|---|---|---|---|
|
Clades C and D | ||||
|
3a |
bID_Cluster_13222 |
1 |
BT9727_2119 |
spore germination protein PF |
| |
bID_Cluster_527 |
2 |
BT9727_2420 |
FtsI ortholog |
| |
bID_Cluster_845 |
2 |
BT9727_2419 |
sensor histidine kinase |
| |
bID_Cluster_2071 |
3 |
BT9727_3346 |
C4-dicarboxylate transporter DctA |
| |
bID_Cluster_2862 |
4 |
BT9727_3495 |
Hypothetical protein |
|
4a |
bID_Cluster_8996 |
1 |
GBAA_0583 |
acetyltransferase |
| |
bID_Cluster_1542 |
1 |
GBAA_0584 |
sensor histidine kinase |
| |
bID_Cluster_6986 |
1 |
GBAA_0585 |
DNA-binding response regulator |
| |
bID_Cluster_4984 |
2 |
GBAA_1077 |
Hypothetical protein |
| |
bID_Cluster_1988 |
3 |
GBAA_5500 |
phosphoglycerate transporter family protein |
| |
bID_Cluster_3664 |
3 |
GBAA_5501 |
putative lipoprotein |
| |
bID_Cluster_5866 |
3 |
GBAA_5502 |
Hypothetical protein |
| |
bID_Cluster_1603 |
3 |
GBAA_5503 |
sensor histidine kinase |
| |
bID_Cluster_6528 |
3 |
GBAA_5504 |
DNA-binding response regulator |
| |
bID_Cluster_3959 |
3 |
GBAA_5505 |
UDP-glucose 4-epimerase |
| |
bID_Cluster_4585 |
3 |
GBAA_5506 |
membrane-bound transcriptional regulator LytR |
| |
bID_Cluster_1186 |
4 |
GBAA_5678 |
ABC transporter ATP-binding protein |
|
4b |
bID_Cluster_10476 |
1 |
GBAA_1939 |
Hypothetical protein |
| |
bID_Cluster_5559 |
2 |
GBAA_2162 |
Hypothetical protein |
| |
bID_Cluster_11158 |
3 |
GBAA_2384 |
Hypothetical protein |
| |
bID_Cluster_13854 |
4 |
GBAA_2523 |
HTH DNA-binding protein |
| |
bID_Cluster_618 |
5 |
GBAA_3291 |
Methyl-accepting chemotaxis protein |
| |
bID_Cluster_3242 |
6 |
GBAA_3338 |
S-layer protein |
| |
bID_Cluster_2077 |
7 |
GBAA_5674 |
Hypothetical protein |
|
Clade A | ||||
|
5 |
bID_Cluster_6809 |
1 |
BMB171_C1598 |
two-component response regulator, LuxR family |
| |
bID_Cluster_3224 |
1 |
BMB171_C1599 |
sensory transduction protein kinase |
| |
bID_Cluster_4364 |
1 |
BMB171_C1600 |
ABC transporter ATP-binding protein |
| |
bID_Cluster_3195 |
1 |
BMB171_C1601 |
ABC transporter permease |
| |
bID_Cluster_2768 |
1 |
BMB171_C1602 |
ABC transporter permease |
| |
bID_Cluster_2336 |
1 |
BMB171_C1603 |
cardiolipin synthetase |
| |
bID_Cluster_5196 |
2 |
BMB171_C2914 |
Hypothetical protein |
| |
bID_Cluster_3683 |
3 |
BMB171_C3448 |
RecA recombinase |
| |
bID_Cluster_2435 |
3 |
BMB171_C3449 |
Competence damage-inducible protein A |
| |
bID_Cluster_8205 |
3 |
BMB171_C3450 |
Phosphatidylglycerophosphate synthase |
| |
bID_Cluster_4493 |
3 |
BMB171_C3451 |
Hypothetical protein |
| |
bID_Cluster_5830 |
3 |
BMB171_C3452 |
Hypothetical protein, ACT-binding domain |
| |
bID_Cluster_12831 |
3 |
BMB171_C3453 |
Hypothetical protein |
| |
bID_Cluster_6635 |
3 |
BMB171_C3454 |
3-ketoacyl-(acyl-carrier-protein) reductase |
| |
bID_Cluster_2214 |
3 |
BMB171_C3455 |
Predicted Zn-dependent peptidases |
| |
bID_Cluster_2266 |
3 |
BMB171_C3456 |
Predicted Zn-dependent peptidases |
| |
bID_Cluster_3669 |
3 |
BMB171_C3457 |
ABC-type transporter, permease component |
| |
bID_Cluster_3507 |
3 |
BMB171_C3458 |
ABC-type transporter, permease component |
| |
bID_Cluster_1276 |
3 |
BMB171_C3459 |
ABC-type transporter, ATPase component |
| bID_Cluster_9779 | 4 | BMB171_C3659 | Hypothetical protein | |
Although Clades C/D genomes encode cluster 5 genes, they are not functionally included in the SigB generalized stress response. Instead, Clades C/D have augmented the core SigB regulon with 24 genes from three different clusters (3a, 4a/b, in 15 TUs; Table 4). Similar to gene cluster 5 in Clade A, gene clusters 3a and 4a have been added to the core SigB regulon primarily by SigB promoter assignment from a common gene pool. By contrast, gene cluster 4b was added to the SigB regulon in Clades C/D by a mixture of HGT/indels and promoter assignment (Figure 3); most of these genes are not present in Clades A and B (red blocks in Figure 3, cluster 4b).
Clade A and Cluster 5
Cluster 5 includes a two-component regulator not orthologous to those included in Clusters 3a/4a. This suggests that Cluster 5 adds to the core regulon a regulatory cascade that extends further into the Bc sensu lato transcriptome, and specific to Clade A. This extension of the core SigB regulon would allow the stress response to be coordinated with the environmental signal(s) to which this secondary regulator independently responds. Gene cluster 5 also appears to coordinate the enhanced synthesis of cardiolipin (CL), stimulated by at least three genes in this cluster, including CL synthase (TU #1), and phosphatidylglycerophosphate synthase and 3-ketoacyl-(acyl-carrier-protein) reductase (TU #2). Increased CL synthesis would change the composition of the cell membrane, increasing the hydrophobicity and viscosity of this membrane [54], which could have varied phenotypic effects. Increased CL levels stimulate protein translocation across the cell membrane in B. subtilis[55] and E. coli[56], and are important for high osmolarity survival in B. subtilis[57] and S. aureus[58]. High CL concentrations decrease cell envelope permeability, affecting nonspecific antibiotic resistance in E. coli[59], resistance to organic solvents [60], daptomycin resistance in S. aureus[61] and Enterococcus faecalis[62], and resistance to antimicrobial peptides such as enterococcal AS-48 [63] and platelet microbicidal peptide tPMP-1 [64]. CL-rich domains in B. subtilis are preferentially distributed at the medial septa and the poles during exponential growth, and the polar septal membrane and the engulfment and forespore membranes during sporulation [65]. B. subtilis spore membranes have a significantly higher CL content than membranes of exponentially growing cells, and CL appears to be essential to the proper functioning of germinant receptors [65].
Due to the extensive divergence within ABC transporter gene family, functional predictions may be difficult to substantiate by sequence similarity alone. Nevertheless, the ABC transporter genes included in TU#1 and TU#3 appear to be most similar to the ABC A superfamily [66], typically responsible for export of hydrophobic compounds. Prokaryotic members of this large gene family include the Streptomyces peucetius[67] DrrAB exporter for hydrophobic compounds such as daunomycin and doxorubicin, and the B. licheniformis BcrABC bacitracin-resistance proteins [68]. Consequently, the ABC transporters in Cluster 5 may be efflux systems either responsible for CL export or working in concert with enhanced CL production to increase resistance to deleterious hydrophobic compounds or antimicrobial peptides.
Cluster 5 TU #3 also includes the genes encoding RecA and CinA. In S. pneumoniae, the CinA ortholog appears to direct RecA to the membrane and enhance competency [69], although in B. subtilis, CinA appears to be a nucleoid-associated protein [70]. However, the most obvious consequence of enhanced RecA expression is an increased likelihood of activation of the SOS stress response by cleavage of LexA. In B. subtilis, at least 33 genes in 18 TUs appear to be directly under RecA/LexA control [71]. If the Bc sensu lato SOS response is similar, this also would extend the SigB generalized stress response into a region of the Clade A transcriptome to make these organisms more apt to express, among others, components of the excision, recombinational and error-prone repair pathways for dealing with DNA damage. Consequently, the collective inclusion of the genes in cluster 5 in the SigB-mediated stress response may fine-tune the stress response for competition with other microorganisms in a mixed microbial milieu. Cluster 5 genes also may confer some advantage during pathogenesis, for example by increasing resistance to the bactericidal effects of neutrophil-induced DNA damage or antimicrobial peptides.
Clades C/D and Clusters 3a, 4a and 4b
These clusters include the genes for three signal transduction histidine kinases, two of which are in TUs with a (probable) cognate response regulator (Table 4). Two additional transcriptional regulators and a methyl-accepting chemotaxis protein also are included in these gene clusters. This suggests that an entirely different regulatory effector gene cascade extends the core SigB regulon in Clade C/D organisms, presumably coordinating the SigB generalized stress response with different environmental signals than that of Clade A organisms. Note that the two Cluster 4b regulatory proteins are unique to the genomes of invasive Clade C/D organisms, suggesting that these genes likely control functions unique to the invasive phenotype. By contrast, the SigB-responsive regulators in Clusters 3a/4a are part of the common gene pool shared with all genomes, although not included in the SigB regulon in those other organisms. This suggests that regulators found in Clusters 3a/4a are important for supervising aspects of the common metabolism shared with other Bc sensu lato organisms, but their inclusion in the generalized stress response is uniquely important for Clades C/D organisms.
Together, these gene clusters allow SigB to coordinate the expression of functions devoted to cell wall and spore structure and biosynthesis. Cluster 3a encodes an FtsI ortholog, also known as PBP-2B [72], which appears to be recruited late to the division septum in B. subtilis[73]. One of the cluster 4a regulatory proteins is the ortholog of the Bacillus LytR protein, a member of the LytR-CpsA-Psr gene family found in most members of the Firmicutes [74]. In B. subtilis, LytR lies in an operon with and controls expression of genes encoding the major autolytic amidase LytC, and a modifier gene LytB [75]. In Streptococcus pneumoniae, LytR is essential for normal septum formation [76], while the S. mutans LytR controls autolytic activity [77]. The Staphylococcus aureus LytR ortholog MsrR plays a role in cell envelope maintenance, cell separation and virulence, and appears to be connected to the sarA attenuation and virulence regulatory network [78,79]. Thus, inclusion of LytR in the Clades C/D SigB regulons suggests that controlling cell wall-specific functions is important for supporting invasive disease.
Cluster 4b includes an S-layer protein with three SLH domains, previously designated as BslI by Kern and Schneewind [80]. While no function has yet been ascribed to BslI, the genome of Ba Sterne encodes a suite of at least 24 S-layer proteins. Eight other S-layer proteins are involved in peptidoglycan metabolism, with an additional seven important for virulence [80], suggesting that the function of the cell-surface S-layer protein BslI is similar.
Cluster 4a also encodes UDP-glucose 4-epimerase, which catalyzes the conversion of UDP-glucose to UDP-galactose [81]. Transcriptionally linked to this gene by SigB, Cluster 3a includes a member of the GNAT-transacetylase superfamily [82]. Some members of this superfamily use phosphoglucosamine as a substrate for transacetylation [82]. Collectively, these enzymes could collaborate to promote a burst in the synthesis of galactose-N-acetylglucosamine (GalNAc) during the stress response; this disaccharide is an important component of secondary cell wall polysaccharides found in Ba and some virulent Bc strains [83]. Additionally, GalNAc is an important linker molecule for tethering oligosaccharides to the collagen-like exosporium glycoprotein BclA [81], an immunodominant Ba spore antigen [84].
Cluster 3a encodes the gene for spore germination protein GerPF. In Ba spores, impairment of GerPF expression causes a germination defect in response to nutrient germinants [85]. This suggests that the SigB stress response in Clades C/D organisms has been adjusted to ensure that a sufficient amount of GerPF is made for proper spore germination potential. Targeting expression of GerPF via SigB does not appear necessary for Clades A/B organisms; possibly GerPF is more important for the proper functioning of spores produced by invasive pathogens. For instance, this protein could be important for proper germination of spores within macrophages, an event that is associated with the ability of Ba strains to cause disease [86]. Note also that some cluster 4a genes, while present in Clade D organisms, are not included in the SigB regulon (red blocks in the heat map in the cluster 4a region). This may suggest that the generalized stress response in Clade D microbes is somewhat less capable of augmenting the pathogenic potential of these organisms, in contrast to Clade C organisms, which include only virulent Ba.
Conclusions
Evolution of the SigB regulon and pathogenesis of the Bc sensu lato group
This sample size is admittedly small, and so the generalizations leading to these hypotheses require further exploration. Nevertheless, the hypotheses drawn from this heat map and cluster analyses are provocative. The simplest interpretation of the accumulated data (Figure 4) suggests that a Clade B-like organism arose first from a B. cytotoxicus-like progenitor after receiving the SigB operon during an HGT event. Following this, a nascent core SigB regulon was assembled by the addition of SigB promoters to TUs from the B. cytotoxicus-like progenitor. Other genes were added to the core regulon by a combination of HGT and gene duplication/divergence, both events accompanied by SigB promoter assignment. Clade A and Clades C/D organisms subsequently diverged from this common Clade B-like organism by the addition of gene clusters 5 and 3a/4a/b, respectively, to the SigB core regulon. These additions occurred primarily by the assignment of SigB promoters to genes from a common gene pool, while the appearance of new genes, presumably by HGT or gene duplication, appears to be a much less common event. The TUs augmenting the SigB regulon of Clade A organisms may enhance the basal pathogenic potential of this clade, but may also make these organisms more capable of competing in the soil or other microbial consortia. Clades C/D organisms are invasive pathogens; we hypothesize that the augmentation to the SigB core regulon in these organisms is important for supporting this phenotype. Note also that some of the Clade D organisms have been referred to as ‘anthrax-like’ bacteria [27,87], despite the tight clonality of the Ba phylogeny that excludes these organisms. The close similarity of the SigB regulon of Clade C and D organisms suggests a functional relationship between these two groups that may be important for their similar pathogenic potential.
Figure 4.
Proposed pathway for the divergence of the SigB regulons within the Bc sensu lato group.
Methods
Genome sequences used in this study
The completed genome sequences of 20 members of the B. cereus sensu lato group used in these studies are listed in Table 1. In this study, we made the simplifying assumption to focus exclusively on whole genome sequences and excluded the extensive collection of draft sequences available for the Bc sensu lato group. This was due to our intent to compare the frequency of indels to promoter differences in the SigB regulons of these organisms. Lack of a particular protein coding sequence in a draft genome could be due either to deletion of this coding sequence from the genome, or a lack of sampling of this region during the sequencing for the draft assembly. A related concern was that many draft sequences likely contain regions of misassembly or low sequence quality that could complicate the analyses that we intended, since searching for specific promoter sequences in the genome would require high quality data at all points in the sequence.
Sigma factor B comparisons
To confirm the identity of the annotated SigB orthologs in each genome, all sigma factor proteins in this genome set were identified using blastp, with the Bc ATCC14579 SigB (locus tag BC1004) as the query sequence. These sequences were compared with a maximum likelihood algorithm using PhyML [Seaview [88]] with 100 bootstrap iterations and then analyzed manually to confirm the identity of all BC1004 orthologs. We further examined these proteins to confirm that the DNA binding sites encoded in each protein were the same, and were distinct from those of other sigma factors. The DNA binding sites of all sigma factor proteins encoded in the Bc ATCC14579 genome were compared using Sigma70_r2.hmm and Sigma70_r4.hmm (hidden Markov models of sigma factor region 2 and region 4, respectively), obtained from the Pfam website [89]. The utility hmmalign [hmmer-3.0b3 [90]] was used to create alignments of region 2 and region 4 of all Bc ATCC14579 primary alternative sigmas, which were manually inspected. A similar comparison was made with all SigB sequences using the utility hmmalign along with Sigma70_r2.hmm and Sigma70_r2.hmm. These comparisons confirmed that all SigB orthologs encoded in these genomes had a conserved DNA binding domain, and therefore are expected to recognize a conserved DNA binding site.
SigB binding site model building and validation
We began model building by using experimentally-derived SigB binding sites derived from Bc ATCC14579 [44,45]. This data set was expanded for training purposes using phylogenetic footprinting [91]. To do this, we began with the 5’ gene of each Bc ATCC14579 operon regulated by SigB, including BC0863, BC0998, BC1001, BC1002, BC1005, BC1009, BC1010, BC3132, and BC4641. Blastp [92] was used with default parameters, except an e-value cut-off of 1e-5, to identify similar sequences in the remaining 18 Bc sensu lato genomes. We did not use the B. cytotoxicus genome in these model-building experiments, as this organism lacks a SigB operon [4]. Orthologs to the Bc ATCC14579 proteins in each organism were identified by constructing gene phylogenies for each gene family using Maximum Likelihood analysis (Seaview) and 100 bootstrap iterations. Tblastn searches were performed to ensure that any missing protein sequences in this data set were due to deletion events in the genome in question, rather than being missed during annotation. Then, the 150bp DNA sequence 5’ to each ortholog were collected and analyzed using the MEME default parameters [93] to identify the SigB binding motif 5’ to each gene. These sequences defined the initial training set used for model building. Redundancy in this training was reduced using the purge function from the MEME package. We built two complementary models of the SigB binding site to be used in parallel for all subsequent analyses. A hidden Markov model (HMM) of the training sequences was created using hmmer-1.8.5 [94], while a position weight matrix (PWM) was derived using MEME. Leave-one-out cross validation [95] was used to determine an appropriate empirical p-value cut-off of .00273 for the HMM and of 4.61e-7 for the PWM. Background scores for empirical p-value calculations for the HMM analysis were determined by searching the SigB HMM against the randomly shuffled Bc ATCC14579 genome [shuffleseq from EMBOSS-6.3.1 [96]]. Empirical p-values for the SigB HMM alignments were determined by comparing the alignment score to the background scores using a Perl script and R-2.13.0.
Orthologous Gene Clustering, Operon and Regulon predictions
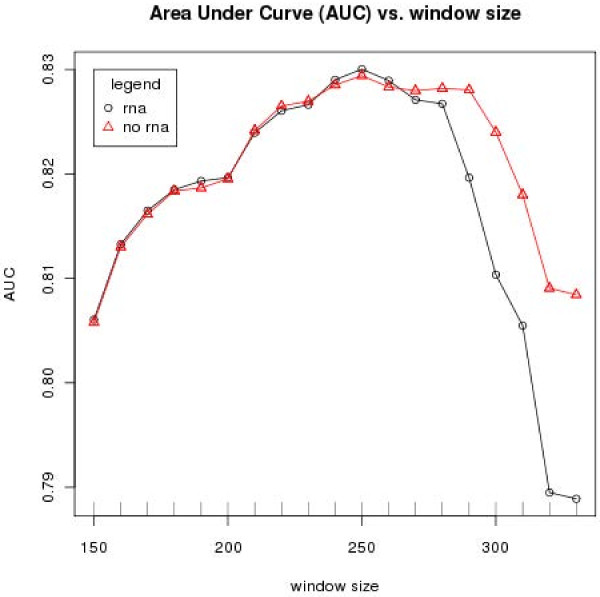
Initially, the Ba str. Sterne transcriptome structure was extracted from the supplementary information provided by Passalacqua et al.[28]. For comparing the predicted transcriptomes from all genomes, we clustered all annotated protein-coding sequences using CD-hit [97] to identify orthologous gene clusters. This resulted in a total of 16,835 gene clusters, with cluster sizes ranging from 1 to 19 genes; each cluster was assigned a unique identifier (bID_Cluster_number; see Additional file 3: Table S2). All predicted SigB-regulated genes were combined into a separate file and also clustered with CD-hit. On both data sets, we used the CD-hit default parameters except requiring an 85% sequence identity and an 80% sequence length difference cut-off. Operon predictions were performed using operonMBP [98]. Receiver operator characteristic analysis [99] was applied to the Ba str. Sterne transcriptome data to determine the appropriate cut-off to maximize true positive and minimize false positive transcription unit (TU) predictions (Figure 5). The appropriate window size was determined to be 250, and the appropriate cut-off for pairing two consecutive genes as co-operonic was 14. All TU predictions were then stored in a MySQL database (see Additional file 4: Table S3 for a summary of the TU predictions for each genome). To predict SigB regulon structure in each organism, the SigB HMM and PWM were used to independently scan across all 20 Bacillus genomes, and TUs 3’ to a significant binding site were extracted. We defined significant SigB binding sites as DNA regions that aligned to both the SigB HMM and PWM models, with p-values equal to or below at least one of their respective p-value cut-offs. Further, we allowed a 5’ untranslated region of up to 4.2 kb, based on the observations of Nicholas et al. in B. subtilis[100]. This information was then added to the MySQL database.
Figure 5.
Receiver-operator characteristic analysis of the predicted Bc sensu lato transcriptome. Plot of the area under the curve (auc) versus the window size parameter of operonMBP for two different datasets. One data set contained RNA annotations while the RNA annotations were not in the second annotation.
Finally, the regulon predictions were tabulated in a table of genomes vs. orthologous gene clusters and manually edited for accuracy. Scores of 0, 1, or 2 were assigned to the following states: 0, a gene not present in an orthologous cluster for a specific organism; 1, gene present but not predicted to be regulated by SigB in an orthologous cluster for a specific organism; and 2, gene present and predicted to be regulated by SigB in an orthologous cluster for a specific organism. Then, two comparisons were performed. We first used a pairwise comparison to assess the contribution of SigB promoters and indels to the structure of each regulon, and the changes between genomes. A promoter change occurs during the comparison of DNA sequence upstream of orthologous genes in which one gene contains a predicted binding site with a significant score while the second either does not contain a predicted binding site, or contains a predicted binding site that has a score that is not significant. And, we defined indels simply as the presence or lack of a protein coding sequence at a specific location; other more complex alterations, such as deletion of untranslated regions, SNPs or inversions, were not tabulated. All pairwise regulon comparisons were made in which the number of changes from 2 to 1, or 2 to 0 were counted. These numbers were normalized to the largest value counted and then plotted as indels vs. promoter changes (Figure 2). Secondly, the regulon scores were placed in a table that was converted to a phylip file and analyzed using PHYLIP with Maximum Parsimony and 100 bootstrap iterations to create a SigB regulon phylogeny. This table was also analyzed in R to create a heatmap (Figure 3 and Additional file 1: Figure S1) using heatmap.2 from the library package gplots. Cluster analysis for this heatmap was performed using the default hclust function in R, which specifies a complete-linkage clustering method similar to neighbor-joining.
Abbreviations
Ba: Bacillus anthracis; Bc: Bacillus cereus; Bw: Bacillus weihenstephanensis; HGT: Horizontal gene transfer; TU: Transcriptional unit; HMM: Hidden Markov Model; PWM: Position Weight Matrix; CL: Cardiolipin; PBP-2B: Penicillin binding protein 2B; SLH: S-layer homology.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
ES and DD designed the studies described in this manuscript. ES performed the initial analyses and ES and DD analyzed the data and co-wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Heat map of predicted SigB regulons of 19 Bc sensu lato genomes. This heat map is color-coded: green blocks indicate the presence of a gene that is predicted to be in a TU controlled by a SigB promoter, while blue blocks indicate the presence of an orthologous gene that is not controlled by SigB. Red blocks indicate that the ortholog in question is absent from the genome.
Table S1. Cluster IDs found in each Gene Cluster identified in Figure 3 and Additional file 1: Figure S1. Cluster IDs generated by CD-hit analysis.
Table S2. Cluster IDs for all genes in this study.
Table S3. Promoters and TU predictions from this study, for each genome.
Contributor Information
Edgar Scott, II, Email: edgar-scott@ouhsc.edu.
David W Dyer, Email: david-dyer@ouhsc.edu.
Acknowledgments
The authors wish to thank Lydgia Jackson, Inimary Toby, Michelle Callegan and Jimmy Ballard for suggestions during the preparation of this manuscript. These studies were supported by Grant # P20RR016478 from the National Institute of General Medical Sciences (NIGMS), a component of the National Institutes of Health (NIH).
References
- Rooney AP, Price NP, Ehrhardt C, Swezey JL, Bannan JD. Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. inaquosorum subsp. nov. Int J Syst Evol Microbiol. 2009;59(Pt 10):2429–2436. doi: 10.1099/ijs.0.009126-0. [DOI] [PubMed] [Google Scholar]
- Hadjifrangiskou M, Chen Y, Koehler TM. The alternative sigma factor sigmaH is required for toxin gene expression by Bacillus anthracis. J Bacteriol. 2007;189(5):1874–1883. doi: 10.1128/JB.01333-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler TM. Bacillus anthracis physiology and genetics. Mol Aspects Med. 2009;30(6):386–396. doi: 10.1016/j.mam.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus A, Goltsman E, Auger S, Galleron N, Segurens B, Dossat C, Land ML, Broussolle V, Brillard J, Guinebretiere MH. et al. Extending the Bacillus cereus group genomics to putative food-borne pathogens of different toxicity. Chem Biol Interact. 2008;171(2):236–249. doi: 10.1016/j.cbi.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Soufiane B, Cote JC. Discrimination among Bacillus thuringiensis H serotypes, serovars and strains based on 16S rRNA, gyrB and aroE gene sequence analyses. Antonie Van Leeuwenhoek. 2009;95(1):33–45. doi: 10.1007/s10482-008-9285-4. [DOI] [PubMed] [Google Scholar]
- Helgason E, Okstad OA, Caugant DA, Johansen HA, Fouet A, Mock M, Hegna I, Kolsto AB. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis–one species on the basis of genetic evidence. Appl Environ Microbiol. 2000;66(6):2627–2630. doi: 10.1128/AEM.66.6.2627-2630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TR, Scott EJ II, Dyer DW. Whole-genome phylogenies of the family Bacillaceae and expansion of the sigma factor gene family in the Bacillus cereus species-group. BMC Genomics. 2011;12:430. doi: 10.1186/1471-2164-12-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourasse NJ, Helgason E, Klevan A, Sylvestre P, Moya M, Haustant M, Okstad OA, Fouet A, Mock M, Kolsto AB. Extended and global phylogenetic view of the Bacillus cereus group population by combination of MLST, AFLP, and MLEE genotyping data. Food Microbiol. 2011;28(2):236–244. doi: 10.1016/j.fm.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Tourasse NJ, Helgason E, Okstad OA, Hegna IK, Kolsto AB. The Bacillus cereus group: novel aspects of population structure and genome dynamics. J Appl Microbiol. 2006;101(3):579–593. doi: 10.1111/j.1365-2672.2006.03087.x. [DOI] [PubMed] [Google Scholar]
- Tourasse NJ, Kolsto AB. SuperCAT: a supertree database for combined and integrative multilocus sequence typing analysis of the Bacillus cereus group of bacteria (including B. cereus, B. anthracis and B. thuringiensis) Nucleic Acids Res. 2008;36(Database issue):D461–D468. doi: 10.1093/nar/gkm877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmaster AR, Novak RT, Marston CK, Gee JE, Helsel L, Pruckler JM, Wilkins PP. Genetic diversity of clinical isolates of Bacillus cereus using multilocus sequence typing. BMC Microbiol. 2008;8:191. doi: 10.1186/1471-2180-8-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger S, Galleron N, Bidnenko E, Ehrlich SD, Lapidus A, Sorokin A. The genetically remote pathogenic strain NVH391-98 of the Bacillus cereus group is representative of a cluster of thermophilic strains. Appl Environ Microbiol. 2008;74(4):1276–1280. doi: 10.1128/AEM.02242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenfors Arnesen LP, Fagerlund A, Granum PE. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol Rev. 2008;32(4):579–606. doi: 10.1111/j.1574-6976.2008.00112.x. [DOI] [PubMed] [Google Scholar]
- Moyer AL, Ramadan RT, Novosad BD, Astley R, Callegan MC. Bacillus cereus-induced permeability of the blood-ocular barrier during experimental endophthalmitis. Invest Ophthalmol Vis Sci. 2009;50(8):3783–3793. doi: 10.1167/iovs.08-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latsios G, Petrogiannopoulos C, Hartzoulakis G, Kondili L, Bethimouti K, Zaharof A. Liver abscess due to Bacillus cereus: a case report. Clin Microbiol Infect. 2003;9(12):1234–1237. doi: 10.1111/j.1469-0691.2003.00795.x. [DOI] [PubMed] [Google Scholar]
- Psiachou-Leonard E, Sidi V, Tsivitanidou M, Gompakis N, Koliouskas D, Roilides E. Brain abscesses resulting from Bacillus cereus and an Aspergillus-like mold. J Pediatr Hematol Oncol. 2002;24(7):569–571. doi: 10.1097/00043426-200210000-00016. [DOI] [PubMed] [Google Scholar]
- Steggles JR, Wang J, Ellar DJ. Discovery of Bacillus thuringiensis virulence genes using signature-tagged mutagenesis in an insect model of septicaemia. Curr Microbiol. 2006;53(4):303–310. doi: 10.1007/s00284-006-0037-2. [DOI] [PubMed] [Google Scholar]
- Fedhila S, Nel P, Lereclus D. The InhA2 metalloprotease of Bacillus thuringiensis strain 407 is required for pathogenicity in insects infected via the oral route. J Bacteriol. 2002;184(12):3296–3304. doi: 10.1128/JB.184.12.3296-3304.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei JZ, Hale K, Carta L, Platzer E, Wong C, Fang SC, Aroian RV. Bacillus thuringiensis crystal proteins that target nematodes. Proc Natl Acad Sci USA. 2003;100(5):2760–2765. doi: 10.1073/pnas.0538072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello M, Bungiro RD, Harrison LM, Bischof LJ, Griffitts JS, Barrows BD, Aroian RV. A purified Bacillus thuringiensis crystal protein with therapeutic activity against the hookworm parasite Ancylostoma ceylanicum. Proc Natl Acad Sci USA. 2006;103(41):15154–15159. doi: 10.1073/pnas.0607002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae R, Riebesell M, Dinkelacker I, Wang Q, Herrmann M, Weller AM, Dieterich C, Sommer RJ. Isolation of naturally associated bacteria of necromenic Pristionchus nematodes and fitness consequences. J Exp Biol. 2008;211(Pt 12):1927–1936. doi: 10.1242/jeb.014944. [DOI] [PubMed] [Google Scholar]
- Gunawan S, Tufts DM, Bextine BR. Molecular identification of hemolymph-associated symbiotic bacteria in red imported fire ant larvae. Curr Microbiol. 2008;57(6):575–579. doi: 10.1007/s00284-008-9245-2. [DOI] [PubMed] [Google Scholar]
- Cardazzo B, Negrisolo E, Carraro L, Alberghini L, Patarnello T, Giaccone V. Multiple-locus sequence typing and analysis of toxin genes in Bacillus cereus food-borne isolates. Appl Environ Microbiol. 2008;74(3):850–860. doi: 10.1128/AEM.01495-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko KS, Kim JW, Kim JM, Kim W, Chung SI, Kim IJ, Kook YH. Population structure of the Bacillus cereus group as determined by sequence analysis of six housekeeping genes and the plcR Gene. Infect Immun. 2004;72(9):5253–5261. doi: 10.1128/IAI.72.9.5253-5261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson A. Sporulation and delta-endotoxin synthesis by Bacillus thuringiensis. Cell Mol Life Sci. 2002;59(3):417–425. doi: 10.1007/s00018-002-8434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, Altherr MR, Han CS, Ravel J. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol Rev. 2005;29(2):303–329. doi: 10.1016/j.femsre.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Han CS, Xie G, Challacombe JF, Altherr MR, Bhotika SS, Brown N, Bruce D, Campbell CS, Campbell ML, Chen J. et al. Pathogenomic sequence analysis of Bacillus cereus and Bacillus thuringiensis isolates closely related to Bacillus anthracis. J Bacteriol. 2006;188(9):3382–3390. doi: 10.1128/JB.188.9.3382-3390.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passalacqua KD, Varadarajan A, Byrd B, Bergman NH. Comparative transcriptional profiling of Bacillus cereus sensu lato strains during growth in CO2-bicarbonate and aerobic atmospheres. PLoS One. 2009;4(3):e4904. doi: 10.1371/journal.pone.0004904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen MA, Bork P. Measuring genome evolution. Proc Natl Acad Sci USA. 1998;95(11):5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozada-Chavez I, Angarica VE, Collado-Vides J, Contreras-Moreira B. The role of DNA-binding specificity in the evolution of bacterial regulatory networks. J Mol Biol. 2008;379(3):627–643. doi: 10.1016/j.jmb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozada-Chavez I, Janga SC, Collado-Vides J. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 2006;34(12):3434–3445. doi: 10.1093/nar/gkl423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittenhuber G. A phylogenomic study of the general stress response sigma factor sigmaB of Bacillus subtilis and its regulatory proteins. J Mol Microbiol Biotechnol. 2002;4(4):427–452. [PubMed] [Google Scholar]
- Becker LA, Cetin MS, Hutkins RW, Benson AK. Identification of the gene encoding the alternative sigma factor sigmaB from Listeria monocytogenes and its role in osmotolerance. J Bacteriol. 1998;180(17):4547–4554. doi: 10.1128/jb.180.17.4547-4554.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker LA, Evans SN, Hutkins RW, Benson AK. Role of sigma(B) in adaptation of Listeria monocytogenes to growth at low temperature. J Bacteriol. 2000;182(24):7083–7087. doi: 10.1128/JB.182.24.7083-7087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley M, Hill C, Ross RP. Tolerance of Listeria monocytogenes to cell envelope-acting antimicrobial agents is dependent on SigB. Appl Environ Microbiol. 2006;72(3):2231–2234. doi: 10.1128/AEM.72.3.2231-2234.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak MJ, Mithoe SC, Boor KJ, Wiedmann M. Listeria monocytogenes sigma B regulates stress response and virulence functions. J Bacteriol. 2003;185(19):5722–5734. doi: 10.1128/JB.185.19.5722-5734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintens V, Massonet C, Merckx R, Vandecasteele S, Peetermans WE, Knobloch JK, Van Eldere J. The role of sigmaB in persistence of Staphylococcus epidermidis foreign body infection. Microbiology. 2008;154(Pt 9):2827–2836. doi: 10.1099/mic.0.2007/015768-0. [DOI] [PubMed] [Google Scholar]
- Oliver HF, Orsi RH, Wiedmann M, Boor KJ. Listeria monocytogenes {sigma}B has a small core regulon and a conserved role in virulence but makes differential contributions to stress tolerance across a diverse collection of strains. Appl Environ Microbiol. 2010;76(13):4216–4232. doi: 10.1128/AEM.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inose Y, Takeshita SL, Hidaka T, Higashide M, Maruyama A, Hayashi H, Morikawa K, Ohta T. Genetic characterization of the natural SigB variants found in clinical isolates of Staphylococcus aureus. J Gen Appl Microbiol. 2006;52(5):259–271. doi: 10.2323/jgam.52.259. [DOI] [PubMed] [Google Scholar]
- Singh VK, Schmidt JL, Jayaswal RK, Wilkinson BJ. Impact of sigB mutation on Staphylococcus aureus oxacillin and vancomycin resistance varies with parental background and method of assessment. Int J Antimicrob Agents. 2003;21(3):256–261. doi: 10.1016/S0924-8579(02)00359-X. [DOI] [PubMed] [Google Scholar]
- Guinebretiere MH, Auger S, Galleron N, Contzen M, De Sarrau B, De Buyser ML, Lamberet G, Fagerlund A, Granum PE, Lereclus D, Bacillus cytotoxicus sp. nov. is a new thermotolerant species of the Bacillus cereus group occasionally associated with food poisoning. Int J Syst Evol Microbiol. 2012. [Epub Feb. 17] [DOI] [PubMed]
- Novichkov PS, Wolf YI, Dubchak I, Koonin EV. Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. J Bacteriol. 2009;191(1):65–73. doi: 10.1128/JB.01237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambukkana A. M. leprae genome sequence. Trends Microbiol. 2001;9(4):157. doi: 10.1016/s0966-842x(01)02021-2. [DOI] [PubMed] [Google Scholar]
- van Schaik W, van der Voort M, Molenaar D, Moezelaar R, de Vos WM, Abee T. Identification of the sigmaB regulon of Bacillus cereus and conservation of sigmaB-regulated genes in low-GC-content gram-positive bacteria. J Bacteriol. 2007;189(12):4384–4390. doi: 10.1128/JB.00313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik W, Zwietering MH, de Vos WM, Abee T. Identification of sigmaB-dependent genes in Bacillus cereus by proteome and in vitro transcription analysis. J Bacteriol. 2004;186(13):4100–4109. doi: 10.1128/JB.186.13.4100-4109.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin V, Severinov K, Gelfand MS. Systematic prediction of control proteins and their DNA binding sites. Nucleic Acids Res. 2009;37(2):441–451. doi: 10.1093/nar/gkn931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierro N, Makita Y, de Hoon M, Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008;36(Database issue):D93–96. doi: 10.1093/nar/gkm910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannapaneni P, Hertwig F, Depke M, Hecker M, Mader U, Volker U, Steil L, van Hijum SA. Defining the structure of the general stress regulon of Bacillus subtilis using targeted microarray analysis and random forest classification. Microbiology. 2012;158(Pt 3):696–707. doi: 10.1099/mic.0.055434-0. [DOI] [PubMed] [Google Scholar]
- Schulthess B, Bloes DA, Francois P, Girard M, Schrenzel J, Bischoff M, Berger-Bachi B. The sigmaB-dependent yabJ-spoVG operon is involved in the regulation of extracellular nuclease, lipase, and protease expression in Staphylococcus aureus. J Bacteriol. 2011;193(18):4954–4962. doi: 10.1128/JB.05362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Oliver HF, Raengpradub S, Palmer ME, Orsi RH, Wiedmann M, Boor KJ. Transcriptomic and phenotypic analyses suggest a network between the transcriptional regulators HrcA and sigmaB in Listeria monocytogenes. Appl Environ Microbiol. 2007;73(24):7981–7991. doi: 10.1128/AEM.01281-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Raengpradub S, Schwab U, Loss C, Orsi RH, Wiedmann M, Boor KJ. Phenotypic and transcriptomic analyses demonstrate interactions between the transcriptional regulators CtsR and Sigma B in Listeria monocytogenes. Appl Environ Microbiol. 2007;73(24):7967–7980. doi: 10.1128/AEM.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGann P, Wiedmann M, Boor KJ. The alternative sigma factor sigma B and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins. Appl Environ Microbiol. 2007;73(9):2919–2930. doi: 10.1128/AEM.02664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegan MC, Kane ST, Cochran DC, Gilmore MS, Gominet M, Lereclus D. Relationship of plcR-regulated factors to Bacillus endophthalmitis virulence. Infect Immun. 2003;71(6):3116–3124. doi: 10.1128/IAI.71.6.3116-3124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CS, Heras H, Garda H, Ruzal S, Sanchez-Rivas C, Rivas E. Biochemical and biophysical studies of Bacillus subtilis envelopes under hyperosmotic stress. Int J Food Microbiol. 2000;55(1–3):137–142. doi: 10.1016/s0168-1605(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Kontinen VP, Helander IM, Tokuda H. The secG deletion mutation of Escherichia coli is suppressed by expression of a novel regulatory gene of Bacillus subtilis. FEBS Lett. 1996;389(3):281–284. doi: 10.1016/0014-5793(96)00602-3. [DOI] [PubMed] [Google Scholar]
- Mikhaleva NI, Golovastov VV, Zolov SN, Bogdanov MV, Dowhan W, Nesmeyanova MA. Depletion of phosphatidylethanolamine affects secretion of Escherichia coli alkaline phosphatase and its transcriptional expression. FEBS Lett. 2001;493(2–3):85–90. doi: 10.1016/s0014-5793(01)02288-8. [DOI] [PubMed] [Google Scholar]
- Lopez CS, Alice AF, Heras H, Rivas EA, Sanchez-Rivas C. Role of anionic phospholipids in the adaptation of Bacillus subtilis to high salinity. Microbiology. 2006;152(Pt 3):605–616. doi: 10.1099/mic.0.28345-0. [DOI] [PubMed] [Google Scholar]
- Kanemasa Y, Takatsu T, Sasai K, Kojima I, Hayashi H. The salt-resistance mechanism of Staphylococcus aureus examined by salt-sensitive mutants. Acta Med Okayama. 1976;30(4):271–276. [PubMed] [Google Scholar]
- Ivanisevic R, Milic M, Ajdic D, Rakonjac J, Savic DJ. Nucleotide sequence, mutational analysis, transcriptional start site, and product analysis of nov, the gene which affects Escherichia coli K-12 resistance to the gyrase inhibitor novobiocin. J Bacteriol. 1995;177(7):1766–1771. doi: 10.1128/jb.177.7.1766-1771.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DP, Beard JP. Altered phospholipid composition in mutants of Escherichia coli sensitive or resistant to organic solvents. J Gen Microbiol. 1979;113(2):267–274. doi: 10.1099/00221287-113-2-267. [DOI] [PubMed] [Google Scholar]
- Peleg AY, Miyakis S, Ward DV, Earl AM, Rubio A, Cameron DR, Pillai S, Moellering RC Jr, Eliopoulos GM. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One. 2012;7(1):e28316. doi: 10.1371/journal.pone.0028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Daniel A, Hardy C, Silverman J, Gilmore MS. Genetic basis for daptomycin resistance in enterococci. Antimicrob Agents Chemother. 2011;55(7):3345–3356. doi: 10.1128/AAC.00207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez A, Valdivia E, Martinez M, Maqueda M. Effect of peptide AS-48 on Enterococcus faecalis subsp. liquefaciens S-47. Antimicrob Agents Chemother. 1989;33(5):641–645. doi: 10.1128/AAC.33.5.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay K, Whitmire W, Xiong YQ, Molden J, Jones T, Peschel A, Staubitz P, Adler-Moore J, McNamara PJ, Proctor RA. et al. In vitro susceptibility of Staphylococcus aureus to thrombin-induced platelet microbicidal protein-1 (tPMP-1) is influenced by cell membrane phospholipid composition and asymmetry. Microbiology. 2007;153(Pt 4):1187–1197. doi: 10.1099/mic.0.2006/003111-0. [DOI] [PubMed] [Google Scholar]
- Kawai F, Shoda M, Harashima R, Sadaie Y, Hara H, Matsumoto K. Cardiolipin domains in Bacillus subtilis marburg membranes. J Bacteriol. 2004;186(5):1475–1483. doi: 10.1128/JB.186.5.1475-1483.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski WE, Piehler A, Wenzel JJ. ABC A-subfamily transporters: structure, function and disease. Biochim Biophys Acta. 2006;1762(5):510–524. doi: 10.1016/j.bbadis.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Malla S, Niraula NP, Liou K, Sohng JK. Self-resistance mechanism in Streptomyces peucetius: overexpression of drrA, drrB and drrC for doxorubicin enhancement. Microbiol Res. 2010;165(4):259–267. doi: 10.1016/j.micres.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Podlesek Z, Comino A, Herzog-Velikonja B, Zgur-Bertok D, Komel R, Grabnar M. Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol Microbiol. 1995;16(5):969–976. doi: 10.1111/j.1365-2958.1995.tb02322.x. [DOI] [PubMed] [Google Scholar]
- Masure HR, Pearce BJ, Shio H, Spellerberg B. Membrane targeting of RecA during genetic transformation. Mol Microbiol. 1998;27(4):845–852. doi: 10.1046/j.1365-2958.1998.00732.x. [DOI] [PubMed] [Google Scholar]
- Kaimer C, Graumann PL. Bacillus subtilis CinA is a stationary phase-induced protein that localizes to the nucleoid and plays a minor role in competent cells. Arch Microbiol. 2010;192(7):549–557. doi: 10.1007/s00203-010-0583-7. [DOI] [PubMed] [Google Scholar]
- Au N, Kuester-Schoeck E, Mandava V, Bothwell LE, Canny SP, Chachu K, Colavito SA, Fuller SN, Groban ES, Hensley LA. et al. Genetic composition of the Bacillus subtilis SOS system. J Bacteriol. 2005;187(22):7655–7666. doi: 10.1128/JB.187.22.7655-7666.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel RA, Harry EJ, Errington J. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol Microbiol. 2000;35(2):299–311. doi: 10.1046/j.1365-2958.2000.01724.x. [DOI] [PubMed] [Google Scholar]
- Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2003;67(1):52–65. doi: 10.1128/MMBR.67.1.52-65.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubscher J, Luthy L, Berger-Bachi B, Stutzmann Meier P. Phylogenetic distribution and membrane topology of the LytR-CpsA-Psr protein family. BMC Genomics. 2008;9:617. doi: 10.1186/1471-2164-9-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V, Margot P, Soldo B, Karamata D. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-L-alanine amidase and its modifier. J Gen Microbiol. 1992;138(9):1949–1961. doi: 10.1099/00221287-138-9-1949. [DOI] [PubMed] [Google Scholar]
- Johnsborg O, Havarstein LS. Pneumococcal LytR, a protein from the LytR-CpsA-Psr family, is essential for normal septum formation in Streptococcus pneumoniae. J Bacteriol. 2009;191(18):5859–5864. doi: 10.1128/JB.00724-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatfield CH, Koo H, Quivey RG Jr. The putative autolysin regulator LytR in Streptococcus mutans plays a role in cell division and is growth-phase regulated. Microbiology. 2005;151(Pt 2):625–631. doi: 10.1099/mic.0.27604-0. [DOI] [PubMed] [Google Scholar]
- Hubscher J, McCallum N, Sifri CD, Majcherczyk PA, Entenza JM, Heusser R, Berger-Bachi B, Stutzmann Meier P. MsrR contributes to cell surface characteristics and virulence in Staphylococcus aureus. FEMS Microbiol Lett. 2009;295(2):251–260. doi: 10.1111/j.1574-6968.2009.01603.x. [DOI] [PubMed] [Google Scholar]
- Rossi J, Bischoff M, Wada A, Berger-Bachi B. MsrR, a putative cell envelope-associated element involved in Staphylococcus aureus sarA attenuation. Antimicrob Agents Chemother. 2003;47(8):2558–2564. doi: 10.1128/AAC.47.8.2558-2564.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern J, Schneewind O. BslA, the S-layer adhesin of B. anthracis, is a virulence factor for anthrax pathogenesis. Mol Microbiol. 2010;75(2):324–332. doi: 10.1111/j.1365-2958.2009.06958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Chesnokova ON, Turnbough CL Jr, Pritchard DG. Identification of the UDP-N-acetylglucosamine 4-epimerase involved in exosporium protein glycosylation in Bacillus anthracis. J Bacteriol. 2009;191(22):7094–7101. doi: 10.1128/JB.01050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetting MW, LP SC, Yu M, Hegd SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433(1):212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Forsberg LS, Choudhury B, Leoff C, Marston CK, Hoffmaster AR, Saile E, Quinn CP, Kannenberg EL, Carlson RW. Secondary cell wall polysaccharides from Bacillus cereus strains G9241, 03BB87 and 03BB102 causing fatal pneumonia share similar glycosyl structures with the polysaccharides from Bacillus anthracis. Glycobiology. 2011;21(7):934–948. doi: 10.1093/glycob/cwr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen C, Chen P, Kearney JF, Turnbough CL Jr. Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J Bacteriol. 2003;185(6):1903–1910. doi: 10.1128/JB.185.6.1903-1910.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr KA, Janes BK, Hanna PC. Role of the gerP operon in germination and outgrowth of Bacillus anthracis spores. PLoS One. 2010;5(2):e9128. doi: 10.1371/journal.pone.0009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel AE, Kuo SR, Dostal D, Watson L, Duesbery NS, Cheng CP, Cheng HJ, Leppla SH. Pathophysiology of anthrax. Front Biosci. 2009;14:4516–4524. doi: 10.2741/3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee SR, Brzuszkiewicz EB, Nattermann H, Bruggemann H, Dupke S, Wollherr A, Franz T, Pauli G, Appel B, Liebl W. et al. The genome of a Bacillus isolate causing anthrax in chimpanzees combines chromosomal properties of B. cereus with B. anthracis virulence plasmids. PLoS One. 2010;5(7):e10986. doi: 10.1371/journal.pone.0010986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 2009;537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A. Prediction of regulatory elements. Methods Mol Biol. 2008;453:233–244. doi: 10.1007/978-1-60327-429-6_11. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34(Web Server issue):W369–373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR, Mitchison G, Durbin R. Maximum discrimination hidden Markov models of sequence consensus. J Comput Biol. 1995;2(1):9–23. doi: 10.1089/cmb.1995.2.9. [DOI] [PubMed] [Google Scholar]
- Picard R, Cook D. Cross-validation of Regression Models. J Am Stat Assoc. 1984;79(387):575–583. doi: 10.1080/01621459.1984.10478083. [DOI] [Google Scholar]
- Letondal C. A Web interface generator for molecular biology programs in Unix. Bioinformatics. 2001;17(1):73–82. doi: 10.1093/bioinformatics/17.1.73. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Li G, Che D, Xu Y. A universal operon predictor for prokaryotic genomes. J Bioinform Comput Biol. 2009;7(1):19–38. doi: 10.1142/S0219720009003984. [DOI] [PubMed] [Google Scholar]
- Bergman NH, Passalacqua KD, Hanna PC, Qin ZS. Operon prediction for sequenced bacterial genomes without experimental information. Appl Environ Microbiol. 2007;73(3):846–854. doi: 10.1128/AEM.01686-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S. et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012;335(6072):1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Heat map of predicted SigB regulons of 19 Bc sensu lato genomes. This heat map is color-coded: green blocks indicate the presence of a gene that is predicted to be in a TU controlled by a SigB promoter, while blue blocks indicate the presence of an orthologous gene that is not controlled by SigB. Red blocks indicate that the ortholog in question is absent from the genome.
Table S1. Cluster IDs found in each Gene Cluster identified in Figure 3 and Additional file 1: Figure S1. Cluster IDs generated by CD-hit analysis.
Table S2. Cluster IDs for all genes in this study.
Table S3. Promoters and TU predictions from this study, for each genome.